Significance
Chemical fixation has been widely used in the last decades, allowing science to unveil the complexity of biological specimens. However, the lack of control over the fixation process can lead to cell structure alterations. While cryo-fixation can offer alternatives, the easy and unexpensive chemical fixation is often a “necessary evil.” To avoid data misinterpretation, it is paramount to recognize possible changes induced by such a treatment, especially when fields related to cancer investigation and therapy are involved. The combination of scanning electron-assisted dielectric microscopy and Raman imaging allows an easy detection of PFA-induced artifacts, emphasizing the differences between living and fixed cells. The ultimate goal is to provide a practical tool ensuring safe data analysis for analogous specimens.
Keywords: PFA-fixation, MNT-1, melanosomes, SE-ADM, Raman imaging
Abstract
Aldehydes fixation was accidentally discovered in the early 20th century and soon became a widely adopted practice in the histological field, due to an excellent staining enhancement in tissues imaging. However, the fixation process itself entails cell proteins denaturation and crosslinking. The possible presence of artifacts, that depends on the specific system under observation, must therefore be considered to avoid data misinterpretation. This contribution takes advantage of scanning electron assisted-dielectric microscopy (SE-ADM) and Raman 2D imaging to reveal the possible presence and the nature of artifacts in unstained, and paraformldehyde, PFA, fixed MNT-1 cells. The high resolution of the innovative SE-ADM technique allowed the identification of globular protein clusters in the cell cytoplasm, formed after protein denaturation and crosslinking. Concurrently, SE-ADM images showed a preferential melanosome adsorption on the cluster’s outer surface. The micron-sized aggregates were discernible in Raman 2D images, as the melanosomes signal, extracted through 2D principal component analysis, unequivocally mapped their location and distribution within the cells, appearing randomly distributed in the cytoplasm. Protein clusters were not observed in living MNT-1 cells. In this case, mature melanosomes accumulate preferentially at the cell periphery and are more closely packed than in fixed cells. Our results show that, although PFA does not affect the melanin structure, it disrupts melanosome distribution within the cells. Proteins secondary structure, conversely, is partially lost, as shown by the Raman signals related to α-helix, β-sheets, and specific amino acids that significantly decrease after the PFA treatment.
Chemical fixation of culture cells has been known for decades as a method to preserve the cell structure in its most fine details, to allow observation through optical or electron microscopes (1–3). Aldehydes-based fixatives such as paraformaldehyde (PFA) and glutaraldehyde (GA) are among the most common reagents employed in chemical fixation. Aldehydes react with -NH2 or -NH- groups in proteins to form methylene bridge adducts, eventually forming an insoluble network which traps other cell components. However, the fixation process is not instantaneous: a redistribution of proteins can occur as long as the cell components are not completely immobilized, resulting in clusters not representative of live-cell conditions (4, 5).
The presence of abnormal protein clusters at the cell membrane of fixed cells has been proven through confocal laser microscopy (5–7) and atomic force microscopy (AFM) (8–11). However, the former requires fluorescent labeling of the proteins and allows observing only large objects (>0.3 µm), while the latter provides information about the morphology of very restricted areas of the cell membrane, losing track of organization inside the cell and at a wider spatial range.
The proteins state and their organization in unlabeled living cells can be investigated through the innovative scanning-electron assisted dielectric microscopy (SE-ADM) technique, recently developed by our group (12–15). SE-ADM measurements are performed in a conventional scanning electron microscope (SEM). However, the sample holder is customized: Liquid samples are sandwiched and sealed between two silicon nitride (SiN) films, with one of them further coated by a tungsten (W) film. The W film will absorb the electron beam once the sample-holder is placed in the SEM chamber, generating a local potential change, which propagates differently in different media. In other words, SE-ADM contrast is obtained by harnessing differences in the relative permittivity between water and the biological specimen (16). As a result, high-contrast and high-resolution imaging is achieved. SE-ADM allowed the investigation of the melanosome morphology in living unstained MNT-1 cells, revealing a toroidal conformation (17).
Melanosome shape and morphology have been thoroughly investigated through electron tomography (ET) and four growth stages were identified, with melanin contained only in the most mature melanosomes (stages III, IV) (18, 19). Even if extremely informative, ET techniques require cryofixation, an expensive method involving several steps that are difficult to control. The quality of freezing (vitrification is needed) and solvent substitution can affect proteins conformation (20, 21). Nonetheless, Cryo-EM stands as an invaluable tool for the structural analysis of proteins and cells, allowing structure determination with atomic, 3D resolutions (22, 23). On the other hand, PFA-fixation is largely used in the investigation of proteins location within the cells, through immunolabeling (5). The affordability and accessibility of chemical fixation methods are points of strength to be reckoned: Artifacts can be identified through advanced fluorescence microscopy imaging or labeling techniques (24, 25), while customized protocols and reagents can limit inner cells order disruption (26, 27).
PFA-fixation has been largely used to investigate melanocyte structure, as well (28–30).
The study of melanosome location and colocalization with specific proteins are of paramount importance to elucidate the melanin formation process and the characteristic of pigmentation-related diseases, from congenital disorders to malignant melanoma (31). Any disruption of the cells local order, induced by PFA, would lead to incorrect information.
In this work we identified specific patterns, that underline PFA-induced disruption of MNT-1 cells local order. SE-ADM imaging and confocal raman microscopy (CRM) were combined to identify protein aggregation and its effect on the melanosomes distribution within the cells. The comparison between living and PFA-fixed cells showed that PFA does not affect the melanin polymer itself but alters protein conformation and structure, compromising the localization of melanosomes within the cells. Thus, the position of melanosomes in PFA-fixed MNT-1 cells (PFA cells) should be interpreted with caution. The effects of PFA on melanosomes’ localization are easily identifiable through the combined SE-ADM and CRM imaging, making an easily accessible tool for the detection of artifacts in PFA-treated cells. The information gained from SE-ADM can also be used as a prescreening tool for Cryo-EM imaging, while the two imaging methods can be directly compared to investigate differences in live/fixed biological specimens.
Results
SE-ADM high-resolution imaging of unstained samples revealed new details of protein clusters formed after PFA-fixation of MNT-1 cells. The comparison of living and PFA cells SE-ADM images (Fig. 1) shows clear differences. In PFA cells, proteins form a thick, crosslinked network in the cell’s cytoplasm (see the area in the yellow square, Fig. 1D) that is not observed in living cells (Fig. 1A). Higher magnifications showed that the cytoplasm contains globular aggregates of denatured, crosslinked proteins. Melanosomes, i.e., the 3 to 600 nm black particles in Fig. 1 E and F, stick to protein’s surface, partially covering them (see the areas circled in yellow). No aggregates were found in living cells (Fig. 1 B and C). The circular patterns of melanosomes could be easily identified also in the Melanin component map of PFA cells: They are circled in Fig. 1G; the map was extracted through principal component analysis, PCA, of CRM images (see following section). Fig. 1H shows a scheme highlighting the inner cells melanosome arrangement in living and PFA cells, influenced by the presence of protein clusters in the latter case.
Fig. 1.
SE-ADM imaging of living (A–C) and PFA (D–F) MNT-1 cells. Globular protein clusters are circled in yellow in (E and F). (G) Melanin component map in PFA cells, extracted from PCA of CRM images: melanosomes are arranged in circular patterns, as well; (H) 2D scheme of proteins and melanosomes distribution in live and PFA cells; (I) Melanosomes abundance (number of melanosomes per µm2) in live and PFA cells. Scale bar in (A and D) is 20 µm. Scale bar in (B, C, E, and F) is 2 µm. Scale bar in (G) is 5 µm.
Besides, in PFA cells, the presence of aggregates leads to an increase of the average distance between melanosomes. In other words, the number of melanosomes/µm2 is significantly higher (P < 0.01) in living cells. From SE-ADM images, we counted 1.5 ± 0.3 and 0.8 ± 0.1 melanosomes/µm2 for living and PFA cells, respectively (Fig. 1I). In the following sections, PFA-induced alteration of melanosome distribution, melanin, and protein structures will be assessed.
Melanosome Distribution.
The melanosome’s location was obtained from CRM imaging, and in particular from the analysis of 2D maps obtained in different areas of living MNT-1 cell cultures. Raman spectra were acquired at low laser intensity and acquisition time, to avoid melanin particles disruption and the consequent increase of background fluorescence. Melanin signal could be easily identified among those registered inside cells, being the most intense. More specifically, the melanin polymer features two characteristic Raman bands, centered at approximately 1,380 and 1,584 cm−1 (17, 32, 33). These bands can be compared to the Raman signals of graphite, a carbonaceous layered material that shares with melanin characteristic structural features (34, 35). As Raman spectra of cells are extremely complex, the most relevant signals, mainly related to melanin, were initially isolated by implementing PCA on Raman 2D maps. Additional details about PCA are reported in Materials and Methods.
The most representative components extracted through this method feature the characteristic melanin peaks (SI Appendix, Fig. S1), for both living and PFA cells. The most intense peak is centered around 1,584 to 1,592 cm−1; a second peak is approximately centered at 1,380 cm−1. Fig. 2 shows the optical images (Fig. 2 A, D, G, and J) and the PCA-extracted melanin component (i.e., the most representative component extracted from each dataset, Fig. 2 B, E, H, and K) in living and PFA cells.
Fig. 2.
Living (A–F) and PFA (G–L) cells optical images and CRM mapping. (A, D, G, and J) Optical microcopy images, in areas where Raman mapping was performed; (B, E, H, and K) Melanin component, extracted through PCA; (C, F, I, and L) Normalized map of intensity, showing I1584/I1331 ratio in PCA reconstructed spectra. The right panel shows the magnifications of I1584/I1331 maps areas. Scale bar in (A, D, G, and J) is 20 µm. Scale bar in (B, E, H, and K) is 5 µm. Scale bar in (B and C), (E and F), (H and I), and (K and L) are the same.
The original Raman spectra showed that the 1,380 cm−1 band is characterized by sharper peaks, centered at approximately at 1,331 to 1,335, 1,351, and 1,404 to 1,412 cm−1. They can be related to melanin indole C–N, C–N–C vibrations, and pyrrole ring stretching, respectively (33).
The I1584/I1331 ratio (Fig. 2 C, F, I, and L), calculated from the PCA reconstructed signals (SI Appendix, Fig. S1), was used to trace areas where melanin is more closely packed, i.e., to localize mature melanosomes.
These two specific bands were chosen since they are easily identifiable, being 1,584 cm−1 one of the most intense and recurrent melanin signals, while 1,331 cm−1 is characteristic of the indole ring vibration (33, 36) and lower polymer packing (35). Fig. 2 suggests that, in living cells, mature melanosomes are preferentially located at the cell membrane (see also SI Appendix, Fig. S2, in which the 1,584 cm−1 signal is mapped in three samples of live and PFA cells).
However, PFA cells melanosomes show a different distribution. In this case, mature melanosomes, characterized by a high I1584/I1331 ratio (red spots in Fig. 2 I and L), are localized both in the internal areas of the cytoplasm and at the membrane. In I1584/I1331 maps, some circular arrangements of melanosomes, signaling protein clusters, can be identified (see magnifications of Fig. 2 I and L, Right panel in Fig. 2) and compared to those in Fig. 1. Such patterns were not found in living cells (see magnifications of Fig. 2 C and F, Right panel in Fig. 2). These observations are reproducible for different cell cultures and in different culture areas, ruling out possible artifact in the experimental data obtained with SE-ADM images from living and PFA cells.
Melanin Structure.
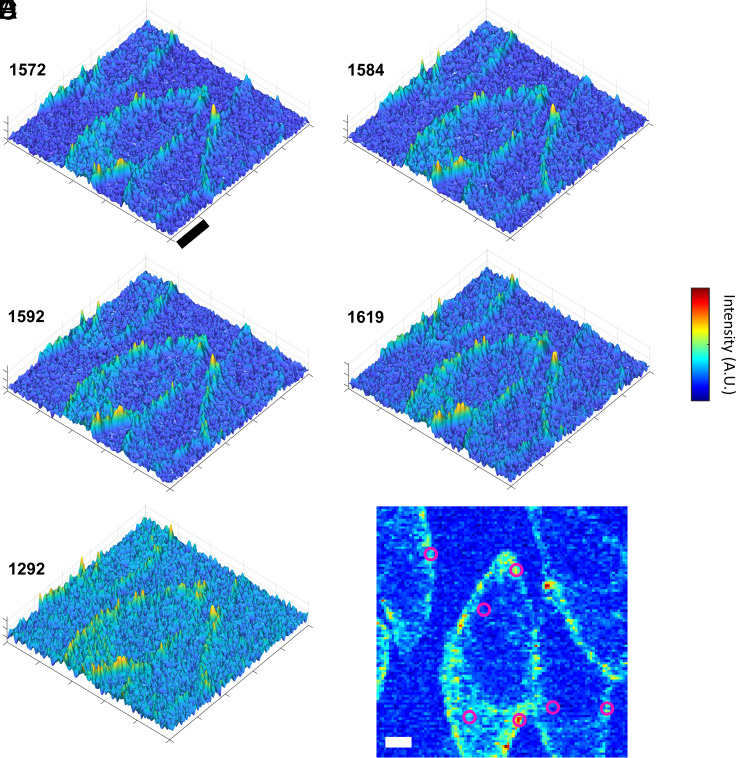
The above SE-ADM and CRM imaging results show that the distribution of melanosomes is altered by fixation. To further support this point, Raman spectra were analyzed more in detail. As the PCA-reconstructed signal is only a rough approximation of the original signal, the original datasets were further analyzed. More specifically, we focused our attention on the 1,570 to 1,620 cm−1 range (most intense melanin peaks). Specific signals can be associated with different “states” of the melanin polymer; therefore, they can ultimately provide information on melanosomes’ degree of maturity in specific cell areas. As already pointed out by PCA, melanosomes are characterized by two main signals, associated with the melanin: 1,584 cm−1 and 1,592 cm−1. Both signals have been largely reported in the literature as the main Raman signals of melanin, from synthetic and natural sources (17, 34, 37–40). However, other two recurring signals were also identified at 1,572 and 1,619 cm−1, probably indicating defects or holes in a dense polymer matrix (38, 41, 42). In addition to the previous absorption, a signal at 1,292 cm−1, related to the precursor 3-4 dihydroxyphenilalanine (DOPA), was detected (38). Fig. 3 shows 3D maps of the above-mentioned signals (1,572, 1,584, 1,592, and 1,619 cm−1 for melanin, 1,292 cm−1 for DOPA) in a live cell culture. In general, signals are more intense at the cell membrane.
Fig. 3.
3D CRM maps of specific melanin-related signals in living cells. (A–D) Melanin polymer signals; (E) DOPA (precursor) signal; black scale bar: 10 µm. (F) Points of sampling for signals comparison. White scale bar: 5 µm.
Melanin signals in PFA cells were sorted by using the same approach (Fig. 4). SI Appendix, Table S1 reports the average relative intensity of melanin-related signal in specific points of several sample maps (see Figs. 3F and 4F as examples), calculated as a ratio to the main 1,584 cm−1 peak. About 75 spots were probed in 13 samples, while spots at the cell membrane and in the cytoplasm were considered separately. Data in SI Appendix, Table S1 show that 1,584 and 1,592 cm−1 are the most intense signals in all probed areas. This is expected, as they are generic melanin signals. In living cells, 1,572 and 1,619 cm−1 signals show the same trend with higher intensity at the membrane. Moreover, 1,572 cm−1 signal is significantly higher (P < 0.05) in PFA than live cells cytoplasm (SI Appendix, Table S1) suggesting that the concentration of highly packed, defected melanin in the cytoplasm increases after fixation.
Fig. 4.
3D CRM maps of specific melanin-related signals in PFA cells. (A–D) Melanin polymer signals; (E) DOPA (precursor) signal; black scale bar: 10 µm. (F) Points of sampling for signals comparison. White scale bar: 5 µm.
To explore this finding, the signals were further analyzed. Specifically, the 1,572 to 1,619 and 1,584 to 1,592 cm−1 pairs were considered separately. The paired signals were normalized to their total intensity, ITOT = I1572 + I1584 + I1592 + I1619 cm−1 (SI Appendix, Table S2). As the 1,572 to 1,619 cm−1 pair indicates closely-PaCKed melanin its normalized intensity will be hereinafter referred to as IPCK (IPCK = I1572+1619/ITOT). On the other hand, the intensity of the Generic Melanin signals 1,584 to 1,592 cm−1 will be indicated as IGM (IGM = I1584+1592/ITOT).
Fig. 5 shows IPCK and IGM trends in living and PFA cells. Cells were probed in the cytoplasm and at the membrane (see Figs. 3F and 4F as examples of probing spots). In living cells, IPCK is significantly higher at the membrane than in the cytoplasm (Fig. 5A, P < 0.01); conversely, IGM is significantly higher in the cytoplasm (Fig. 5C, P < 0.05). Such difference was not found in PFA cells (Fig. 5 B and D), highlighting that highly packed, defected melanin can be found both at the membrane and in the cytoplasm of PFA cells, or preferentially accumulating at the membrane of living cells. This suggests that IPCK signals the presence of mature melanosomes.
Fig. 5.
Relative intensity of (A–E) melanin closely packed chains (ICP) and generic (IMel) signals in living and PFA cells and (F and G) Relative abundance of DOPA precursor (1,292 cm−1) and mature melanin (1,619 cm−1) in the cytoplasm of live cells. Error bars show the SDs. Signals marked by stars are significantly different: *(P < 0.05), **(P < 0.01).
The local abundance of the precursor DOPA (IDOPA = I1292/I1292+1619) with respect to the mature polymer (IMature Mel = I1619/I1292+1619) was evaluated in the cytoplasm of living cells. More specifically, high Raman signal (HS) and low Raman signal (LS) areas were considered separately (Fig. 5 F and G). As a matter of fact, IDOPA is higher than IMature Mel (P < 0.01) in LS spots, while the opposite occurs in HS areas.
Proteins Structure Alteration.
Since we have shown that protein denaturation occurs due to PFA fixation, protein Raman signals are expected to change.
Raman spectra of MNT1 cells are extremely complex, displaying hundreds of signals; several protein signals overlap with those of melanin, and a complete separation of each contribution is not trivial. Nevertheless, recurrent peaks, not related to melanin but colocalized with melanosomes, were detected in the spectra. Several signals were identified. Peaks at 1,212 + 1,572, 1,212 + 1,552, and 1,212 + 1,615 cm−1 can be associated with His, Trp, and Tyr, respectively (43–48). Then, 1,572 and 1,616 cm−1 signals are superimposed to melanin signals (see previous sections). Generic protein signals at 1,550, 1,265, and 1,456 cm−1 can be related to Amide II, Amide III, and C–H stretching (both proteins and lipids), respectively (49, 50). Then, 1,265 and 1,552 cm−1 signal maps are shown in SI Appendix, Fig. S3.
Proteins-melanosomes colocalization is expected for different proteins, the most important being Pmel17 and TYRP1 (51, 52). According to the literature, Raman signals referable to Pmel17 were found at 1,552 (Trp), 1,248, and 1,655 to 1,665 cm−1 (β-sheet conformation) (49, 53). TYRP1 is characterized by a transmembrane α-helix domain (54), with signals at approximately 1,270 cm−1 (55) and in the 1,650 to 1,655 cm−1 range (56). Moreover, zinc domains (54) may give rise to a peak around 420 cm−1 (57). Intensity maps of 1,248, 1,270, and 420 cm−1 peaks are shown in SI Appendix, Fig. S4.
Fig. 6 A–F show the intensity maps, probed in the whole cells, of the signals related to His–Trp–Tyr, proteins–lipids backbone, and α-helix—β-sheet conformation of Pmel17 and TYRP (1,212, 1,456, and 1,655 cm−1, respectively).
Fig. 6.
(A–F) 3D Raman maps of specific protein signals (ratio to 1,584 cm−1 signal) in living and PFA cells: (A and B) His–Trp–Tyr related signal; (C and D) Generic protein/lipid signal; (E and F) β-sheet–α-helix related signal. (G and H) Comparison of His–Trp–Tyr and β-sheet–α-helix signals in live and PFA cells. Error bars show the SDs. **(P < 0.01). (Scale bar: 10 µm.)
Signals normalized to the 1,584 cm−1 melanin peak were compared, and those significantly different are shown in the bar plots (Fig. 6 G and H; IHis-Trp-Tyr = I1212/I1584, Iα-helix-β-sheet = I1655/I1584).
PFA-fixation leads to a general decrease of signal intensity: both IHis-Trp-Tyr and Iα-helix-β-sheet are lower in PFA cells (Fig. 6 G and H). As expected 1,456 cm−1 signal, simply related to proteins and lipids concentration, does not vary significantly after fixation (SI Appendix, Table S2).
Discussion
In this work, melanosome distribution and MNT-1 cells protein structure alterations, following PFA-fixation, were investigated. SE-ADM and confocal Raman imaging revealed the formation of micron-sized, globular aggregates, due to crosslinking of denatured proteins. The adsorption of melanosomes on the surface of such aggregates caused a disruption in the melanosome distribution within the cell. As a matter of fact, the formation of protein clusters after chemical fixation has already been observed through confocal laser microscopy (5–7) and AFM (8–11). However, while confocal microscopy required protein labeling, AFM allowed only the investigation of the external morphology of the cell membrane, leaving proteins organization in the cytoplasm unexplored. The SE-ADM method allows the direct observation of unstained samples in both fixed and live conditions. The comparison of SE-ADM and CRM images allowed the identification of clusters also in the PCA-extracted melanin components of fixed cells. Such aggregates were not detected in living MNT-1 cells. Therefore, Raman 2D imaging can be used as an easy diagnostic tool, to get a prompt response about inner cells alterations due to chemical fixation in melanocytes. PFA alteration to melanosomes distributions, melanin, and proteins structures were investigated sequentially.
Melanosomes Distribution and Melanin Raman Signals.
Melanosomes go through four different growth stages (18, 19, 58, 59). Melanin is contained only in the latter stages: in type III melanosomes, it forms a layered structure, while in type IV, it is arranged in a closely packed, dense matrix (18, 60–62). Mature melanosomes are transported, trough the cytoskeleton, to the cell membrane, accumulating in the cell peripheral areas (63). As melanin can be easily identified through SE-ADM and CRM, we focused our attention on type III–IV melanosomes.
Melanin Raman signal was initially identified and isolated by implementing PCA on Raman maps: two bands, centered at 1,584 to 1,592 cm−1 and about 1,380 cm−1, emerged. The 1,584 cm−1 signal can be related to the C=C in-plane vibrations of the aromatic structures constituting the polymer. This absorption is referred to as G band in the literature (from Graphite, that shares some structural features with melanin). The 1,380 cm−1 is referred to as D band and can be linked to loosely packed melanin chains, i.e., to younger melanosomes (35). In general, the lower the polymer packing, the higher the D band intensity (41). Therefore, the IG/ID ratio was used to gain information about the degree of melanin local packing inside melanosomes, i.e., the melanosomes’ growth stage. The D band was further analyzed in the original spectra that report sharper peaks centered at approximately 1,331 to 1,335, 1,351, and 1,404 to 1,412 cm−1. They can be related to melanin indole C–N, C–N–C vibrations and pyrrole ring stretching (33). The I1584/I1331 signals ratio (Fig. 2 C, F, I, and L) in living and PFA cells CRM maps indicated the areas where closely packed melanin was localized, i.e., the mature melanosomes. Mature melanosomes preferentially accumulate at the membrane in living cells. This is due to the active transport, through microtubule motors, from the perinuclear to peripheral areas (51, 61, 64). In PFA cells, after protein denaturation and crosslinking, active transport no longer occurred. As a result, melanosomes are evenly distributed in the cytoplasm and at the membrane.
A deeper analysis of melanin G band (Raman signals in the 1,572 to 1,619 cm−1 range) showed that melanin itself is not affected by PFA, as the signals characterizing mature melanosomes in live and PFA cells are the same. More specifically, two signal pairs were considered: IPCK and IGM, indicating closely packed melanin and a generic melanin signal, respectively. In living cells, IPCK is more intense at the membrane than in the cytoplasm. No difference in IPCK intensity was detected when the same areas were probed in PFA cells. As a matter of fact, IPCK can be related to an increased number of structural defects (38, 41, 42), which occur when melanin is highly packed and concentrated, i.e., in mature melanosomes. IPCK intensity does not significantly vary at the membrane of live and PFA cells and in PFA cells cytoplasm. As mentioned above, the active transport in living cells drives mature melanosomes toward the membrane; therefore, the inner areas of the cytoplasm are expected to be richer in younger melanosomes and melanin precursors. The analysis of IDOPA and IMature Mel signals in the cytoplasm (Fig. 5 F and G) supported this hypothesis, as higher concentrations of DOPA were not colocalized with the most mature melanosomes.
Proteins Structure.
Formaldehyde is known to react with proteins (65, 66) and lipids (67, 68), forming an insoluble network which traps other cell components. Tyrosine rings are among the most reactive moieties, followed by phenylalanine and tryptophan (66). Therefore, being tyrosine also one of the precursors of melanin, melanosome environment is expected to be significantly affected by PFA fixation. Moreover, protein denaturation implies the disruption of proteins’ secondary structures. Pmel17 and TYRP1, two of the most important proteins (51, 52) colocalized with melanosomes, are characterized by β-sheet (49, 53) and α-helix (54) domains.
Our data show that PFA fixation led to an intensity loss of all the above-mentioned protein signals. From the previous arguments, it could be argued that protein denaturation, PFA-crosslinking, and α-helix–β-sheet unfolding occurred. As a result, melanosomes that were being transported toward the cell membranes through the fibrils ended up being trapped in a network of denatured and crosslinked proteins, located in random areas of the cells; this alters melanosome distribution in PFA-treated cell cultures. On the other hand, Raman signals related to protein backbone remained unaltered: a lower chemical reactivity toward PFA is expected in this case.
PFA-fixation is still widely used to immobilize cell cultures, being a readily accessible, low-cost method. Our results, far from invalidating the method itself, suggest that in specific circumstances (such as the presence of solid-like, nanometric particles inside the cytoplasm) surface energies may drive the rearrangements of the cells’ inner structure. Such phenomenon must be considered to avoid data misinterpretation not only in the imaging of melanocytes but also in the investigation of solid-like nanocarriers or nanoparticles (e.g., for specific cell therapies) location within cells.
Furthermore, our approach is promising to investigate cell properties in live specimens in general: The high resolution of SE-ADM technique allows the investigation of biological mechanisms in unstained systems.
Conclusions
In this paper, we used the innovative SE-ADM technique to unveil the proteins state and their organization in unlabeled living cells, and Raman 2D imaging to confirm the possible presence and the nature of artifacts in unstained, and paraformaldehyde, PFA, fixed MNT-1 cells. To the best of our knowledge we demonstrated for the first time that the paraformaldehyde fixation practice, commonly used in the histological field, might produce significant cell structure alteration in MNT-1 cells, as chemical fixation disrupts melanosomes distribution within the cells without affecting the melanin structure. Proteins secondary structure is partially lost. The high-resolution SE-ADM images suggest that this technique would be a suitable low-cost and easily accessible tool for the prescreening of fixed biological specimens. Moreover, these findings are of paramount importance to elucidate the melanin formation process and the characteristic of pigmentation-related diseases, from congenital disorders to malignant melanoma, and in general in the characterization of all the biological systems where fixation is routinely employed.
Materials and Methods
MNT-1 Cell Cultures.
MNT-1 human melanoma cell line (61) was obtained from American Type Culture Collection (ATCC) (CRL-3450). MNT-1 cells were cultured in D-MEM (Thermo Fisher Scientific #11995065) containing 10 % AIM-V (Thermo Fisher Scientific #31035025), 10% MEM NEAA (Thermo Fisher Scientific #10370021), and 20% fetal bovine serum (Thermo Fisher Scientific), at 37 °C, under 5% CO2. Cells (4 × 104, 1.5 mL/dish) were seeded in dish holders or glass bottom dishes, 4 d before Raman or SE-ADM imaging.
SE-ADM.
The SE-ADM used in the current study is a handmade system. It was connected to a field-emission SEM (SU5000, Hitachi High-Tech Corp., Japan). A hand-made liquid sample holder was mounted into the SEM chamber, while the detector terminal was connected to a preamplifier under the holder (15, 16). The preamplifier electrical signal was fed into the SEM external input. SEM images (1,280 × 1,020 pixels) were acquired at 2,000 to 20,000× magnification. Scanning time was set to 40 s, working distance to 7 mm, with an EB acceleration voltage of 6 to 10 kV and a current of 1 to 10 pA. High-resolution SE-ADM images were processed from the LPF signal and scanning signal using the image-processing toolbox of MATLAB R2021a (Math Works Inc.). The original SE-ADM images were filtered using a 2D Gaussian filter (GF) with a kernel size of 11 × 11 pixels and a radius of 1.2 s. Background subtraction was achieved by subtracting SE-ADM images from the filtered images using a broad GF (400 × 400 pixels, 200σ). Melanosomes in SE-ADM images were counted through Fiji-Image J “Particle Analysis” tool. Seven images per sample were analyzed.
Melanosomes Extraction from MNT-1 Cells.
MNT-1 cells (5 ×106) were suspended in water (1 mL) and sonicated for 6 h. Then, they were centrifuged by a 5.0-µm Poly Vinylidene DiFluoride (PVDF) centrifugal filter (Ultrafree Centrifugal Filter UFC30SV00, Merk Millipore) at 12,000 rpm for 5 min. The pellet was harvested, suspended in water (5.0 µL), and observed through CRM and SE-ADM. For the extraction of melanosomes from fixed cells, cells were fixed by 4% PFA (FUJIFILM Wako Pure Chemical Co.) for 10 min and washed twice before the sonication. SE-ADM images, shown in SI Appendix, Fig. S5, prove the successful extraction of melanosomes from both living and PFA-fixed cells.
CRM.
MNT-1 cell cultures were prepared in a glass-bottomed dish (Matsunami glass Ltd.). The dish, containing also the culture medium, was sealed with a handmade glass lid and set in the field of the Raman microscope. Raman imaging was performed through a Raman Confocal microscope, equipped with a 532 nm Nd:YAG laser (alpha300R, WITech). Raman signal was acquired with a Peltier-cooled charge-coupled device detector (DV401-BV, Andor, UK) featuring 600 gratings/mm (UHTS 600, WITec). Raman spectra were acquired at very low laser power (0.2 mW) with a 4.5 cm−1 accuracy to reduce the measuring time, a critical parameter to avoid melanin disruption. For Raman mapping, 24 × 24, 30 × 30, 50 × 50 μm areas were probed, 120 × 120 pixel. 50× objective lens was used, with a 0.2 mW laser power. Raman 2D maps consist of 10,000 to 40,000 spectra. The probing area did not affect the accuracy of spot sampling as the size of the laser spot remained constant (diameter of approximately 400 nm).
Raman maps were analyzed through the WITec suite (version 5.0, Lab Co.). Cosmic rays removal, smoothing (Average, window: 4 points), and background subtraction were performed. MATLAB R2021a was used to plot signal maps and spectra. Single spectra were extracted from maps to compare the signal composition in specific cell areas. Data extracted from images with different areas did not differ significantly. Datasets were tested through the two-tailed t test, assuming different variances.
PCA.
The WITec suite also features 2D PCA analysis. PCA was run independently on each dataset, to preserve the inner variability of melanocytes samples. PCA was used to reduce data dimensionality, achieving an oversimplification of Raman spectra (in the “reconstructed” signal), useful to identify the most representative peaks, characteristic of melanin. Moreover, the S/N ratio improved, allowing the identification of melanosomes shape and distribution within the cells.
Details of PCA data processing are given below and exemplified by SI Appendix, Fig. S6.
SI Appendix, Fig. S6A shows 250 of the 22,500 Raman spectra randomly extracted from an original 2D Raman map (PFA-fixed cell sample in Fig. 2, third row), after cosmic rays removal, smoothing, and subtraction. In spite of spectra complexity, peaks around 1,380 and 1,580 cm−1 are evident, signaling the presence of melanin. The 2D map in the 1,560 to 1,630 cm−1 range, SI Appendix, Fig. S6B, shows low S/N ratio, preventing a clear identification of melanosomes within the cell. Principal components, extracted through PCA, are plotted in SI Appendix, Fig. S6C. Component 1 (Comp1, green solid line) is the most representative of melanin polymer signals. Such a profile is strictly comparable to Raman spectra acquired on purified melanosomes, extracted from living and PCA-fixed cells (SI Appendix, Fig. S6 D and E, respectively). Here, prominent peaks at about 1,331 and 1,584 cm−1 clearly emerge, among many others. Then, 3 to 5 PCA components were considered for each dataset, chosen in accordance to the Scree plot. PC maps associated with Comp1, 2, and 3 are shown in SI Appendix, Fig. S6 F–H, respectively. In Comp1 map, melanosome shapes and arrangement can be clearly distinguished. For illustrative purposes only, Comp1 map (i.e., the map related to the main principal component, ascribed to melanin) was referred to as “melanin component” in Fig. 2 and throughout the text.
SI Appendix, Fig. S6I shows 250/22,500 PCA reconstructed spectra, matching those reported in SI Appendix, Fig. S6A. Reconstructed spectra are an oversimplification of the original signals, but peaks associated with melanin can be clearly identified. The calculation of the I1584/I1331 ratio from reconstructed spectra allowed a first coarse, but fundamental step of data analysis. The 2D map of the reconstructed signal (1,560 to 1,630 cm−1 range, SI Appendix, Fig. S6J) shows a significantly higher S/N ratio than the original data map (SI Appendix, Fig. S6B), allowing the identification of melanosomes within the cell.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank Prof. Dr. S. Murakami and Dr. T. Iwayama (Osaka University) for their helpful advice regarding MNT-1 cells. We thank Ms. Miho Iida for her excellent technical assistance. Japan Science and Technology Agency CREST (JPMJCR19H2), Nikko Chemicals Co. Ltd. (Tokyo, Japan), and the Italian Consorzio Interuniversitario per lo Sviluppo dei Sistemi a Grande Interfase (Center for Colloid and Surface Science) are gratefully acknowledged for the financial support. The publication was made by a researcher (R.M.) with a research contract cofunded by the European Union—PON Research and Innovation 2014 to 2020 in accordance with Article 24, paragraph 3a of Law No. 240 of December 30, 2010, as amended and Ministerial Decree No. 1062 of August 10, 2021.
Author contributions
R.M., T. Okada, Taku Ogura, Toshihiko Ogura, and P.B. designed research; R.M., T. Okada, Toshihiko Ogura, and P.B. performed research; R.M. and Toshihiko Ogura contributed new reagents/analytical tools; R.M. and Toshihiko Ogura analyzed the data; Taku Ogura, Toshihiko Ogura, and P.B. acquired funding; and R.M., T. Okada, Taku Ogura, Toshihiko Ogura, and P.B. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission. E.O.P. is a guest editor invited by the Editorial Board.
Contributor Information
Toshihiko Ogura, Email: t-ogura@aist.go.jp.
Piero Baglioni, Email: baglioni@csgi.unifi.it.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Sabatini D. D., Bensch K., Barrnett R. J., Cytochemistry and electron microscopy. The preservation of cellular ultrastructure and enzymatic activity by aldehyde fixation. J. Cell Biol. 17, 19–58 (1963). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hopwood D., Cell and tissue fixation, 1972–1982. Histochem. J. 17, 389–442 (1985). [DOI] [PubMed] [Google Scholar]
- 3.Ramos-Vara J. A., Principles and methods of immunohistochemistry. Methods Mol. Biol. Clifton NJ 1641, 115–128 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Brock R., Hamelers I. H. L., Jovin T. M., Comparison of fixation protocols for adherent cultured cells applied to a GFP fusion protein of the epidermal growth factor receptor. Cytometry 35, 353–362 (1999). [DOI] [PubMed] [Google Scholar]
- 5.Stanly T. A., et al. , Critical importance of appropriate fixation conditions for faithful imaging of receptor microclusters. Biol. Open 5, 1343–1350 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y., et al. , The effects of chemical fixation on the cellular nanostructure. Exp. Cell Res. 358, 253–259 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pereira P. M., et al. , Fix your membrane receptor imaging: Actin cytoskeleton and CD4 membrane organization disruption by chemical fixation. Front. Immunol. 10, 675 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ichikawa T., et al. , Chemical fixation creates nanoscale clusters on the cell surface by aggregating membrane proteins. Commun. Biol. 5, 1–9 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braet F. C., Rotsch C., Wisse E., Radmacher M., Comparison of fixed and living liver endothelial cells by atomic force microscopy. Appl. Phys. A 66, S575–S578 (1998). [Google Scholar]
- 10.Liu B. Y., Zhang G. M., Li X. L., Chen H., Effect of glutaraldehyde fixation on bacterial cells observed by atomic force microscopy. Scanning 34, 6–11 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Le Grimellec C., Giocondi M.-C., Pujol R., Lesniewska E., Tapping mode atomic force microscopy allows the in situ imaging of fragile membrane structures and of intact cells surface at high resolution. Single Mol. 1, 105–107 (2000). [Google Scholar]
- 12.Ogura T., A high contrast method of unstained biological samples under a thin carbon film by scanning electron microscopy. Biochem. Biophys. Res. Commun. 377, 79–84 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Ogura T., Direct observation of unstained biological specimens in water by the frequency transmission electric-field method using SEM. PLoS One 9, e92780 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ogura T., Non-destructive observation of intact bacteria and viruses in water by the highly sensitive frequency transmission electric-field method based on SEM. Biochem. Biophys. Res. Commun. 450, 1684–1689 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Ogura T., Nanoscale analysis of unstained biological specimens in water without radiation damage using high-resolution frequency transmission electric-field system based on FE-SEM. Biochem. Biophys. Res. Commun. 459, 521–528 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Okada T., Ogura T., Nanoscale imaging of untreated mammalian cells in a medium with low radiation damage using scanning electron-assisted dielectric microscopy. Sci. Rep. 6, 29169 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okada T., Iwayama T., Ogura T., Murakami S., Ogura T., Structural analysis of melanosomes in living mammalian cells using scanning electron-assisted dielectric microscopy with deep neural network. Comput. Struct. Biotechnol. J. 21, 506–518 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hurbain I., et al. , Electron tomography of early melanosomes: Implications for melanogenesis and the generation of fibrillar amyloid sheets. Proc. Natl. Acad. Sci. U.S.A. 105, 19726–19731 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bárcena M., Koster A. J., Electron tomography in life science. Semin. Cell Dev. Biol. 20, 920–930 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ameye L., Hermann R., DuBois P., Flammang P., Ultrastructure of the echinoderm cuticle after fast-freezing / freeze substitution and conventional chemical fixations. Microsc. Res. Tech. 48, 385–393 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Matsko N., Mueller M., Epoxy resin as fixative during freeze-substitution. J. Struct. Biol. 152, 92–103 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Topf M., et al. , Protein structure fitting and refinement guided by cryo-EM density. Structure 16, 295–307 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yip K. M., Fischer N., Paknia E., Chari A., Stark H., Atomic-resolution protein structure determination by cryo-EM. Nature 587, 157–161 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Schnell U., Dijk F., Sjollema K. A., Giepmans B. N. G., Immunolabeling artifacts and the need for live-cell imaging. Nat. Methods 9, 152–158 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Whelan D. R., Bell T. D. M., Image artifacts in single molecule localization microscopy: Why optimization of sample preparation protocols matters. Sci. Rep. 5, 7924 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Melan M. A., “Overview of cell fixatives and cell membrane permeants” in Immunocytochemical Methods and Protocols, Methods in Molecular Biology™, Javois L. C., Ed. (Humana Press, 1999), pp. 45–55. [DOI] [PubMed] [Google Scholar]
- 27.Magee A. I., Adler J., Parmryd I., Cold-induced coalescence of T-cell plasma membrane microdomains activates signalling pathways. J. Cell Sci. 118, 3141–3151 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Singh S. K., et al. , The silver locus product (Silv/gp100/Pmel17) as a new tool for the analysis of melanosome transfer in human melanocyte–keratinocyte co-culture. Exp. Dermatol. 17, 418–426 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Hou L., Panthier J. J., Arnheiter H., Signaling and transcriptional regulation in the neural crest-derived melanocyte lineage: Interactions between KIT and MITF. Development 127, 5379–5389 (2000). [DOI] [PubMed] [Google Scholar]
- 30.Valencia J. C., et al. , Sorting of Pmel17 to melanosomes through the plasma membrane by AP1 and AP2: Evidence for the polarized nature of melanocytes. J. Cell Sci. 119, 1080–1091 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamaguchi Y., Hearing V. J., Melanocytes and their diseases. Cold Spring Harb. Perspect. Med. 4, a017046 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yakimov B. P., et al. , Melanin distribution from the dermal–epidermal junction to the stratum corneum: Non-invasive in vivo assessment by fluorescence and Raman microspectroscopy. Sci. Rep. 10, 14374 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Centeno S. A., Shamir J., Surface enhanced Raman scattering (SERS) and FTIR characterization of the sepia melanin pigment used in works of art. J. Mol. Struct. 873, 149–159 (2008). [Google Scholar]
- 34.Huang Z., et al. , Raman spectroscopy of in vivo cutaneous melanin. J. Biomed. Opt. 9, 1198–1205 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Ferrari A., Robertson J., Reich S., Thomsen C., Raman spectroscopy of graphite. Philos. Trans. R. Soc. Lond. Ser. Math. Phys. Eng. Sci. 362, 2271–2288 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Perna G., Lasalvia M., Gallo C., Quartucci G., Capozzi V., Vibrational characterization of synthetic eumelanin by means of Raman and surface enhanced Raman scattering. Open Surf. Sci. J. 5, 1–8 (2013). [Google Scholar]
- 37.Roldán M. L., Centeno S. A., Rizzo A., An improved methodology for the characterization and identification of sepia in works of art by normal Raman and SERS, complemented by FTIR, Py-GC/MS, and XRF. J. Raman Spectrosc. 45, 1160–1171 (2014). [Google Scholar]
- 38.Mondal S., Thampi A., Puranik M., Kinetics of melanin polymerization during enzymatic and nonenzymatic oxidation. J. Phys. Chem. B 122, 2047–2063 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Krafft C., Kirsch M., Beleites C., Schackert G., Salzer R., Methodology for fiber-optic Raman mapping and FTIR imaging of metastases in mouse brains. Anal. Bioanal. Chem. 389, 1133–1142 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Gaspard D., Paris C., Loubry P., Luquet G., Raman investigation of the pigment families in recent and fossil brachiopod shells. Spectrochim. Acta. A. Mol. Biomol. Spectrosc. 208, 73–84 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Kudin K. N., et al. , Raman spectra of graphite oxide and functionalized graphene sheets. Nano Lett. 8, 36–41 (2008). [DOI] [PubMed] [Google Scholar]
- 42.Obraztsova E. D., et al. , Raman identification of onion-like carbon. Carbon 36, 821–826 (1998). [Google Scholar]
- 43.Leonhardt R. M., Vigneron N., Rahner C., den Eynde B. J. V., Cresswell P., Endoplasmic reticulum export, subcellular distribution, and fibril formation by Pmel17 require an intact N-terminal domain junction *. J. Biol. Chem. 285, 16166–16183 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu G., Zhu X., Fan Q., Wan X., Raman spectra of amino acids and their aqueous solutions. Spectrochim. Acta. A. Mol. Biomol. Spectrosc. 78, 1187–1195 (2011). [DOI] [PubMed] [Google Scholar]
- 45.Kuhar N., Sil S., Umapathy S., Potential of Raman spectroscopic techniques to study proteins. Spectrochim. Acta. A. Mol. Biomol. Spectrosc. 258, 119712 (2021). [DOI] [PubMed] [Google Scholar]
- 46.Dean D. N., J. C., Lee, pH-Dependent fibril maturation of a Pmel17 repeat domain isoform revealed by tryptophan fluorescence. Biochim. Biophys. Acta BBA - Proteins Proteomics 1867, 961–969 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li X., et al. , Different classification algorithms and serum surface enhanced Raman spectroscopy for noninvasive discrimination of gastric diseases. J. Raman Spectrosc. 47, 917–925 (2016). [Google Scholar]
- 48.Lee H.-I., Kim M.-S., Suh S.-W., Raman spectroscopy of L-Phenylalanine, L-Tyrosine, and their peptides adsorbed on silver surface. Bull. Korean Chem. Soc. 9, 218–223 (1988). [Google Scholar]
- 49.Talaikis M., Strazdaitė S., Žiaunys M., Niaura G., Far-Off Resonance: Multiwavelength Raman spectroscopy probing amide bands of Amyloid-β-(37–42) peptide. Molecules 25, 3556 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang Z., et al. , In vivo detection of epithelial neoplasia in the stomach using image-guided Raman endoscopy. Biosens. Bioelectron. 26, 383–389 (2010). [DOI] [PubMed] [Google Scholar]
- 51.Jiang M., et al. , Microtubule motor transport in the delivery of melanosomes to the actin-rich, apical domain of in the retinal pigment epithelium. J. Cell Sci. 133, jcs.242214 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berson J. F., Harper D. C., Tenza D., Raposo G., Marks M. S., Pmel17 initiates premelanosome morphogenesis within multivesicular bodies. Mol. Biol. Cell 12, 3451–3464 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Raposo G., Tenza D., Murphy D. M., Berson J. F., Marks M. S., Distinct protein sorting and localization to premelanosomes, melanosomes, and lysosomes in pigmented melanocytic cells. J. Cell Biol. 152, 809–824 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lai X., Wichers H. J., Soler-Lopez M., Dijkstra B. W., Structure of human tyrosinase related protein 1 reveals a binuclear zinc active site important for melanogenesis. Angew. Chem. Int. Ed. 56, 9812–9815 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feng X., et al. , Raman active components of skin cancer. Biomed. Opt. Express 8, 2835–2850 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kuhar N., Sil S., Verma T., Umapathy S., Challenges in application of Raman spectroscopy to biology and materials. RSC Adv. 8, 25888–25908 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tomas M., et al. , Comparative Raman study of four plant metallothionein isoforms: Insights into their Zn(II) clusters and protein conformations. J. Inorg. Biochem. 156, 55–63 (2016). [DOI] [PubMed] [Google Scholar]
- 58.Hurbain I., et al. , Melanosome distribution in keratinocytes in different skin types: Melanosome clusters are not degradative organelles. J. Invest. Dermatol. 138, 647–656 (2018). [DOI] [PubMed] [Google Scholar]
- 59.Daniele T., et al. , Mitochondria and melanosomes establish physical contacts modulated by Mfn2 and involved in organelle biogenesis. Curr. Biol. 24, 393–403 (2014). [DOI] [PubMed] [Google Scholar]
- 60.D’Alba L., Shawkey M. D., Melanosomes: Biogenesis, properties, and evolution of an ancient organelle. Physiol. Rev. 99, 1–19 (2019). [DOI] [PubMed] [Google Scholar]
- 61.Kushimoto T., et al. , A model for melanosome biogenesis based on the purification and analysis of early melanosomes. Proc. Natl. Acad. Sci. U.S.A. 98, 10698–10703 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hoashi T., et al. , Glycoprotein nonmetastatic melanoma protein b, a melanocytic cell marker, is a melanosome-specific and proteolytically released protein. FASEB J. 24, 1616–1629 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moreiras H., Seabra M. C., Barral D. C., Melanin transfer in the epidermis: The pursuit of skin pigmentation control mechanisms. Int. J. Mol. Sci. 22, 4466 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.DePina A. S., Langford G. M., Vesicle transport: The role of actin filaments and myosin motors. Microsc. Res. Tech. 47, 93–106 (1999). [DOI] [PubMed] [Google Scholar]
- 65.Kiernan J. A., Formaldehyde, Formalin, paraformaldehyde and glutaraldehyde: What they are and what they do. Microsc. Today 8, 8–13 (2000). [Google Scholar]
- 66.Thavarajah R., Mudimbaimannar V. K., Elizabeth J., Rao U. K., Ranganathan K., Chemical and physical basics of routine formaldehyde fixation. J. Oral. Maxillofac. Pathol. 16, 400–405 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jones D., Gresham G. A., Reaction of formaldehyde with unsaturated fatty acids during histological fixation. Nature 210, 1386–1388 (1966). [DOI] [PubMed] [Google Scholar]
- 68.Kim S.-O., Kim J., Okajima T., Cho N.-J., Mechanical properties of paraformaldehyde-treated individual cells investigated by atomic force microscopy and scanning ion conductance microscopy. Nano Converg. 4, 5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.