Abstract
In Bacillus sphaericus and other Bacillus spp., d-amino acid transaminase has been considered solely responsible for biosynthesis of d-glutamate, an essential component of cell wall peptidoglycan, in contrast to the glutamate racemase employed by many other bacteria. We report here the cloning of the dat gene encoding d-amino acid transaminase and the glr gene encoding a glutamate racemase from B. sphaericus ATCC 10208. The glr gene encodes a 28.8-kDa protein with 40 to 50% sequence identity to the glutamate racemases of Lactobacillus, Pediococcus, and Staphylococcus species. The dat gene encodes a 31.4-kDa peptide with 67% primary sequence homology to the d-amino acid transaminase of the thermophilic Bacillus sp. strain YM1.
Bacteria normally synthesize d-glutamate and d-alanine as essential components of cell wall peptidoglycan (24). In addition, certain species such as the bacilli synthesize a number of other d-amino acids, including d-phenylalanine, d-asparagine, and d-ornithine, as intermediates in the production of secondary metabolites such as peptide-based antibiotics (2, 13, 36). d-Alanine biosynthesis from l-alanine by alanine racemase appears to be ubiquitous in bacteria (1, 34), but two distinct enzymatic routes have been identified for bacterial d-glutamate biosynthesis. In organisms such as Escherichia coli, Lactobacillus spp., and Pediococcus spp., specific racemases are responsible for the direct biosynthesis of d-glutamate and d-alanine from their l-amino acids (6, 9, 20). Conversely, Bacillus spp., such as Bacillus sphaericus and Bacillus licheniformis, possess a d-amino acid transaminase capable of synthesizing d-glutamate among a broad range of d-amino acids from keto acid precursors, using d-alanine as the amino donor (13, 35). The d-amino acid transaminases of B. sphaericus ATCC 10208 and Bacillus sp. strain YM1 have been particularly well studied (18, 27, 31, 35). The bacilli have generally been considered to synthesize only d-alanine through the action of a racemase, with d-glutamate biosynthesis attributed to the d-amino acid transaminase using α-ketoglutarate and d-alanine as substrates (5, 20), although a strain of Bacillus pumilus which lacks d-amino acid transaminase does possess glutamate racemase activity (14). Only one organism, Staphylococcus haemolyticus, has been shown to possess both d-amino acid transaminase and glutamate racemase activities (22).
We have cloned and characterized genes encoding two distinct d-amino acid biosynthetic enzymes from B. sphaericus ATCC 10208 by using a complementation screen with E. coli WM335 (15). This mutant, dependent on exogenously supplied d-glutamate has been used previously to isolate genes encoding d-amino acid transaminase and glutamate racemase (14, 22, 32). Enzymatic assays indicate that the genes isolated in this work encode the d-amino acid transaminase purified from this strain earlier and a previously undetected glutamate racemase. The presence of the two enzymes in B. sphaericus suggests that the bacilli, like Staphylococcus spp., possess two biosynthetic routes to d-glutamate.
Cloning of d-glutamate biosynthetic genes from B. sphaericus.
Liquid cultures were grown with aeration at 37°C in Lennox broth (LB) (Gibco/BRL, Gaithersburg, Md.). Plate cultures were grown at 37°C on LB agar or M9 (23) minimal salts–1.5% agar supplemented with 0.2% glucose. E. coli WM335 cultures were supplemented with d-glutamate and thymine, both at 50 μg/ml. Where appropriate, chloramphenicol (10 μg/ml) and ampicillin (200 μg/ml) were added. pBR322 DNA was obtained from New England Biolabs (Beverly, Mass.). Partially MboI-digested chromosomal DNA (16) of B. sphaericus ATCC 10208 was size fractionated on a 0.8% agarose gel, and 2- to 10-kb fragments were ligated to BamHI- and BglII-cleaved pIF306. Plasmid pIF306 is a pBR322 (3) derivative containing a unique BglII site inserted between the vector BamHI and SphI sites. Genes inserted between the BamHI and BglII sites are transcribed from a strong constitutive promoter (21) derived from the pheA (10) promoter of E. coli K-12, located immediately upstream between the unique HindIII and BamHI sites of the vector. The plasmid library was used to transform the restriction-deficient strain XL1 Blue (Stratagene), and approximately 20,000 colonies were obtained. Plasmid DNA prepared from pooled isolates was then introduced into E. coli WM335 and plated on LB-thymine medium in the absence of supplemental d-glutamic acid. Approximately 50 clones which were able to complement the d-glutamic acid deficiency of the host were identified. Restriction analysis of these clones identified two classes of clones represented by the individual plasmid isolates pIF1001 and pIF1002. Each of these plasmids was used to retransform fresh cells of WM335 which were cultured and assayed for d-amino acid transaminase and glutamate racemase activity.
Characterization of the B. sphaericus glr gene.
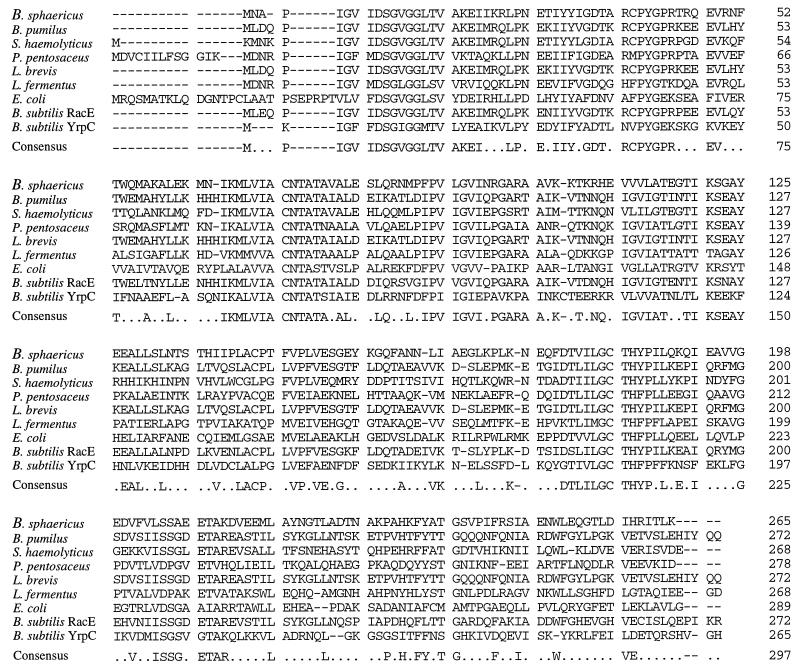
Plasmid DNA isolation, restriction analysis, and ligation were performed by standard methodology as previously described (16). Plasmid pIF1001 carried a 2.34-kb MboI fragment of B. sphaericus chromosome. Sequence analysis of this fragment (25) revealed a single large open reading frame of 684 bp encoding a 28,876-Da protein (GenBank accession no. U26733). Strong primary sequence identity was observed between this protein sequence and the glutamate racemases of B. pumilus (50%), S. haemolyticus (50%), Pediococcus pentosaceus (43%), Lactobacillus fermentus (37%), Lactobacillus brevis (40%), and to a lesser extent E. coli (26%). Strong sequence identity (50 and 30%, respectively) was also observed between B. sphaericus glutamate racemase and the putative products of the racE (GenBank accession no. Z75208) and yrpC (GenBank accession no. U93875) genes identified in the B. subtilis genomic sequence. The sequence alignment is shown in Fig. 1. The peptide also displayed a low level of primary sequence identity (21%) to the aspartate racemases of Streptococcus thermophilus and Desulfurococcus sp., but no significant identity was observed with alanine racemase of B. subtilis or Bacillus stearothermophilus or with B. subtilis amino acid racemase (GenBank accession no. Z94043). Identification of the ATG start codon of the protein was strongly supported by the sequence homology between the protein’s N terminus and the other known glutamate racemase sequences, the presence of a consensus Bacillus ribosome binding site GAGG 7 bp upstream of the ATG codon, and the absence of upstream ATG, TTG, and GTG start codons capable of initiating the same open reading frame. Downstream of the coding sequence, there is a region of strong dyad symmetry typical of a bacterial transcription terminator.
FIG. 1.
Primary sequence homology between glutamate racemase of B. sphaericus, glutamate racemases from other bacteria, and the putative gene products of the B. subtilis racE and yrpC genes. The consensus sequence shows the amino acids that are conserved in four or more sequences. Sequence identities (percentages) between B. sphaericus glutamate racemase and the individual enzymes are as follows: B. pumilis, 50%; S. haemolyticus, 50%; P. pentosaceus, 43%; L. brevis, 40%; L. fermentus, 37%; E. coli, 26%; B. subtilis RacE, 50%; and B. subtilis YrpC, 30%.
Glutamate racemase activity of pIF1001.
Cultures of strains to be assayed were inoculated from single colonies into 10 ml of LB and grown with shaking at 37°C. These were used to inoculate 50-ml LB cultures which were grown for 8 h in a 1-liter shake flask from an initial optical density at 600 nm of 0.05. Cells for glutamate racemase assay were washed, resuspended in 50 mM Tris HCl buffer (pH 8.0), and lysed in a French pressure cell at 1,000 lb/in2. Supernatant fluid was recovered for the assay following centrifugation at 14,000 × g for 30 min at 4°C. Glutamate racemase activity was assayed by an l-glutamate dehydrogenase-coupled assay as described previously (22). Assay results were corrected for nonspecific reduction of NADP, which was determined from control assays carried out in the absence of d-glutamate. One unit is defined as 1 μmol of NADP reduced per min at 37°C. The results are summarized in Table 1. Measurements of glutamate racemase activity in pIF1001-bearing cells were consistently three to four times higher than those of the control cells (WM335 carrying pIF306), while the activity of WM335 carrying pIF1002 was not higher than that of the control. The protein encoded on pIF1001 was concluded to be a glutamate racemase of B. sphaericus ATCC 10208, and the gene was designated glr. The glr gene does not appear to be part of an operon, as there are no significant open reading frames upstream or downstream of the gene. Moreover, glr is oriented on pIF1001 such that expression is convergent with the upstream pheA-derived promoter region. This suggests that the native glr promoter is present on the fragment cloned in pIF1001, although we have not yet identified the transcription start site in B. sphaericus. Analysis of the DNA sequence upstream of the coding region did not reveal sequences with significant homology to the consensus Bacillus ςa −35 (TTGACA) and −10 (TATAAT) regions reported for several B. sphaericus genes expressed during vegetative growth (4, 33). The codon usage of glr is generally consistent with that of moderately expressed E. coli genes (7).
TABLE 1.
d-Amino acid transaminase and glutamate racemase activity of dat and glr clonesa
| Strain | d-Amino acid trans- aminase sp act (Ub/ mg of protein) | Glutamate race- mase sp act (Uc/ mg of protein) | Complemen- tation of WM335d |
|---|---|---|---|
| WM335/pIF306 | 0.0080 | 0.007 | − |
| WM335/pIF1001 (glr) | 0.0065 | 0.025 | + |
| WM335/pIF1002 (dat) | 7.09 | 0.005 | + |
| WM335/pIF372 (dat) | 0.3 | NDe | + |
Results represent averages of three assays corrected for endogenous rates.
One unit of activity defined as 1 mmol of pyruvate (NAD) produced per min at 37°C.
One unit of activity defined as 1 mmol of NADPH produced per min at 37°C.
Complementation being normal overnight growth on LB agar. Symbols: −, no growth; +, growth.
ND, not determined.
Characterization of the B. sphaericus dat gene.
Plasmid pIF1002 contains a 2.5-kb insert, and sequence analysis revealed an open reading frame of 848 bp encoding a protein of 31,392 Da (GenBank accession no. U26732). The N-terminal coding sequence was subsequently determined by using additional primers and was found to agree entirely with that of the purified enzyme (data not shown). This confirmed that the gene cloned in pIF1002 encodes d-amino acid transaminase, and we have named it dat. The C-terminal amino acid sequence of the B. sphaericus ATCC 10208 d-amino acid transaminase has been previously reported to consist of the peptide sequence VI (FY)LAL-COOH (5). The VI(FY)LAL peptide does not occur anywhere in the peptide sequence, and the origin of this stretch of sequence is unknown. The coding sequence is preceded by a strong Bacillus ribosome binding site (AAAGGA) located 9 bp upstream of the ATG start codon. The gene is followed by a region of extensive dyad symmetry typical of bacterial transcription terminators.
d-Amino acid transaminase activity of pIF1002.
Cells for d-amino acid transaminase assay were prepared as described above for the glutamate racemase assay but were washed and resuspended in 1 ml of 50 mM potassium phosphate buffer (pH 8.5). Extracts of E. coli WM335 carrying pIF1002 were assayed for d-amino acid transaminase activity by the lactate dehydrogenase-coupled assay as described previously (11). Assay results were corrected for nonspecific oxidation of NADH, which was determined by assaying in the absence of d-alanine. One unit of activity is defined as that which produces 1 μmol of pyruvate (NAD) per min at 37°C. The results shown in Table 1 indicated levels of d-alanine-dependent NADH oxidation approximately 1,000-fold higher than those of control cultures carrying only the vector pIF306. Similar assays conducted with WM335 transformed with pIF1001 had shown no increase in d-alanine-dependent NADH oxidation over the levels in the controls. The specific activity of the purified B. sphaericus d-amino acid transaminase has been reported to be 116 to 160 U/mg, indicating that the WM335/pIF1002 cells express Dat to approximately 5% of releasable cell protein. The high level of dat expression may be due in part to the colinear orientation of the dat coding sequence with the constitutive pheA promoter located upstream on the vector. To determine if transcription occurred in the absence of a heterologous promoter, the dat gene and 400 bp of upstream untranslated sequence were isolated by PCR and subcloned onto the low-copy-number vector pLG338 (28) between the unique SalI and EcoRI sites. Oligonucleotides used to isolate the B. sphaericus dat gene by PCR had the sequences 5′-GATGTCGACGTTAATCCAAACGTTAGC-3′ and 5′-GACGAATTCTTTTAGGTAGCTCTTTTTAATC-3′. The resulting plasmid, pIF372, was able to confer wild-type growth upon WM335 in the absence of exogenous d-glutamate and showed d-amino acid transaminase activity of 0.3 U/mg. These results indicated that dat expression could originate from the 400-bp upstream sequence. However, neither this transcription start site nor that used in B. sphaericus has been identified.
Primary sequence conservation among known d-amino acid transaminase enzymes.
Genes encoding d-amino acid transaminases have previously been isolated from the thermophilic Bacillus sp. strain YM1 (30) and from S. haemolyticus (22). In addition, we have previously identified and characterized the dat gene of B. licheniformis (32) (GenBank accession no. U26947). The sequence identity between the enzymes encoded by these genes and the d-amino acid transaminase of B. sphaericus was examined as shown in Fig. 2. The B. sphaericus d-amino acid transaminase shows 67% identity to that of the thermophilic Bacillus sp. strain YM1, 48% identity to the d-amino acid transaminase of S. haemolyticus, and 42% identity to the B. licheniformis d-amino acid transaminase. The sequence also shows 42% identity to the putative d-amino acid transaminase encoded by the B. subtilis yheM gene (GenBank accession no. Y14082) identified by the sequencing of the genome. The crystal structure of the YM1 enzyme has been determined to 1.9-Å resolution, identifying a novel enzyme fold for a transaminase and enabling the assignment of residues believed to be important to the catalytic mechanism (29). Alignment of the primary sequence of B. sphaericus d-amino acid transaminase with those of the YM1 enzyme and the other d-amino acid transaminases not surprisingly shows complete conservation of K145 (important for substrate-cofactor proton transfer) and residues Y31, R50, E177, I204, and T205, which are all involved in cofactor ion pairing. There is also partial conservation of the serine residues S179 to S181, which contribute to cofactor positioning. Interestingly, the region from Ser240 to Ser243, which has been suggested to be important in substrate specificity through side chain discrimination, shows sequence variations in the known d-amino acid transaminase enzymes.
FIG. 2.
Primary sequence homology between the d-amino acid transaminase, other known microbial d-amino acid transaminases, and the gene product of the B. subtilis yheM gene. The consensus sequence shows the amino acids that are conserved in three or more sequences. Individual percentages for sequence conservation with B. sphaericus d-amino acid transaminase are as follows: Bacillus sp. strain YM1, 67%; S. haemolyticus, 48%; B. licheniformis, 42%; and B. subtilis YheM, 42%. Asterisks indicate residues assigned active functions in the YM1 enzyme (29): substrate-cofactor proton transfer (K145), cofactor ion pairing (Y31, R50, E177, S179 to S181, I204, and T205), and substrate side chain recognition (S240 to S243).
Conclusions.
In this report we describe the isolation from B. sphaericus ATCC 10208 of the dat gene encoding the d-amino acid transaminase and a second gene we have designated glr, which encodes a hitherto unknown glutamate racemase. This is the first clear report of a Bacillus strain possessing both d-glutamate biosynthetic enzymes, although the genomic sequence of B. subtilis indicates that it may also possess both enzymes. The B. sphaericus enzymes display high primary sequence identity to other known d-amino acid transaminases and glutamate racemases.
The physiological significance of the utilization of two independent types of d-amino acid biosynthetic enzymes and the relative contribution and regulation of the two B. sphaericus enzymes in d-glutamate biosynthesis remain undetermined. Pucci et al. (22) have demonstrated that the d-amino acid transaminase and glutamate racemase of S. haemolyticus can each complement the d-glutamate auxotrophy of E. coli WM335 when these genes are present on low-copy-number plasmid vectors. Similarly, we have observed complementation of WM335 by both B. sphaericus genes on the low-copy-number plasmid vector pLG338, suggesting that either enzyme may be able to synthesize sufficient d-glutamate to sustain cell growth. Definitive experiments to disable each gene independently in the chromosome of B. sphaericus are required to determine the respective roles of the enzymes in d-glutamate biosynthesis. It is interesting to speculate that the glutamate racemase, by analogy to the alanine racemase, may be sufficient to provide the necessary d-glutamate for peptidoglycan synthesis, whereas the d-amino acid transaminase is required to provide a broader range of d-amino acids necessary for secondary metabolite biosynthesis. This would be consistent with the narrow substrate specificity typically displayed by glutamate and alanine racemases in contrast to the much broader substrate profile of the d-amino acid transaminases. Bacilli are known to produce a number of antibiotics, such as bacillomycins and bacitracins, which incorporate a variety of d-amino acids, such as d-phenylalanine, d-tyrosine, and d-asparagine (2, 13, 36).
The function of d-amino acid transaminase in the biosynthesis of d-glutamic acid (18) has been well documented, as has the ability of the enzyme to synthesize a wide range of d-amino acids from keto acid substrates (35). d-Amino acid transaminases from B. subtilis and B. licheniformis as well as the enzymes from Bacillus sp. strain YM1 and B. sphaericus have been shown to display diverse but distinct substrate specificities. The recent determination of the YM1 enzyme backbone fold and the identity of residues participating in the catalytic mechanism has provided a clearer understanding of the effects of the site-directed enzyme mutants created earlier (17, 19, 29). The isolation of the B. sphaericus dat gene now facilitates structure-function analyses between two well characterized and highly homologous Bacillus d-amino acid transaminases and potentiates three-dimensional molecular modeling studies. Molecular modeling of the B. sphaericus d-amino acid transaminase sequence on the YM1 enzyme backbone may permit mutagenesis studies to probe differences in the properties of the enzymes. The 67% exact sequence identity between the YM1 and B. sphaericus d-amino acid transaminases is considerably greater than the 43% identity used successfully to conduct earlier studies using E. coli l-amino acid transaminases in which molecular models of the aromatic transaminase encoded by the tyrB gene (8) were derived from the known three-dimensional structure of the aspartate transaminase (26) encoded by aspC. Site-directed mutagenesis studies of aspC and tyrB were then used to test predictions regarding the residues influencing the substrate specificity of those enzymes towards aromatic and dicarboxylic substrates (12).
Distinct differences in substrate preferences have been observed between the d-amino acid transaminases of Bacillus sp. strain YM1 and B. sphaericus. Amino acids, such as d-methionine, d-phenylalanine, and d-norleucine, which are good substrates for the B. sphaericus enzyme, are poor substrates for the YM1 enzyme (31). Many of the d-amino acid transaminase residues implicated in the active-site architecture of the YM1 d-amino acid transaminase are conserved in the B. sphaericus d-amino acid transaminase, but there are differences in the Ser240-to-Ser243 region proposed as part of a side chain binding pocket for d-amino acid substrate side chains. It will be interesting to explore this observation and to probe the differences in d-amino acid transaminase substrate specificities through mutagenesis selection procedures, site-directed mutagenesis, and gene shuffling experiments.
Acknowledgments
We thank Anna Kootstra for excellent technical assistance and Dave Ager and David Pantaleone for critical appraisal of this manuscript.
We are grateful to Monsanto Corporation for providing excellent research facilities for this work.
REFERENCES
- 1.Adams E. Amino acid racemases and epimerases. In: Boyer P D, editor. The enzymes. VI. New York, N.Y: Academic Press Inc.; 1972. pp. 479–507. [Google Scholar]
- 2.Besson F, Hourdou M L. Effect of amino acids on the biosynthesis of β-amino acids, constituents of bacillomycins F. J Antibiot. 1987;40:221–223. doi: 10.7164/antibiotics.40.221. [DOI] [PubMed] [Google Scholar]
- 3.Bolivar F, Rodriguez R L, Green P J, Betlach M C, Heyneker H L, Boyer H W, Crosa J H, Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2:95–113. [PubMed] [Google Scholar]
- 4.Bowditch R D, Baumann P, Yousten A A. Cloning and sequencing of the gene encoding a 125-kilodalton surface-layer protein from Bacillus sphaericus 2362 and of a related cryptic gene. J Bacteriol. 1989;171:4178–4188. doi: 10.1128/jb.171.8.4178-4188.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christen P, Metzler D E, et al., editors. Transaminases. New York, N.Y: John Wiley and Sons; 1985. pp. 463–467. [Google Scholar]
- 6.Doublet P, Van Heijenoort J, Bohin J P, Mengin-Lecreulx D. The murI gene of Escherichia coli is an essential gene that encodes a glutamate racemase activity. J Bacteriol. 1993;175:2970–2979. doi: 10.1128/jb.175.10.2970-2979.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ernst J F. Codon usage and gene expression. Trends Biotechnol. 1988;6:196–199. [Google Scholar]
- 8.Fotheringham I G, Dacey S A, Taylor P P, Smith T J, Hunter M G, Finlay M E, Primrose S B, Parker D M, Edwards R M. The cloning and sequence analysis of the aspC and tyrB genes from Escherichia coli K12. Comparison of the primary structures of the aspartate aminotransferase and aromatic aminotransferase of E. coli with those of the pig aspartate aminotransferase isoenzymes. Biochem J. 1986;234:593–604. doi: 10.1042/bj2340593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallo K A, Knowles J R. Purification, cloning, and cofactor independence of glutamate racemase from Lactobacillus. Biochemistry. 1993;32:3981–3990. doi: 10.1021/bi00066a019. [DOI] [PubMed] [Google Scholar]
- 10.Hudson G S, Davidson B E. Nucleotide sequence and transcription of the phenylalanine and tyrosine operons of Escherichia coli K12. J Mol Biol. 1984;180:1023–1051. doi: 10.1016/0022-2836(84)90269-9. [DOI] [PubMed] [Google Scholar]
- 11.Jones W M, Soper T S, Ueno H, Manning J M. D-glutamate-D-amino acid transaminase from bacteria. Methods Enzymol. 1985;113:108–113. doi: 10.1016/s0076-6879(85)13024-7. [DOI] [PubMed] [Google Scholar]
- 12.Koehler E, Seville M, Jaeger J, Fotheringham I, Hunter M, Edwards M, Jansonius J N, Kirschner K. Significant improvement to the catalytic properties of aspartate aminotransferase: role of hydrophobic and charged residues in the substrate binding pocket. Biochemistry. 1994;33:90–97. doi: 10.1021/bi00167a012. [DOI] [PubMed] [Google Scholar]
- 13.Kuramitsu H K, Snoke J E. The biosynthesis of D-amino acids in Bacillus licheniformis. Biochim Biophys Acta. 1962;62:114–121. doi: 10.1016/0006-3002(62)90496-1. [DOI] [PubMed] [Google Scholar]
- 14.Liu L, Yoshimura T, Endo K, Esaki N, Soda K. Cloning and expression of the glutamate racemase gene of Bacillus pumilus. J Biochem. 1997;121:1155–1161. doi: 10.1093/oxfordjournals.jbchem.a021709. [DOI] [PubMed] [Google Scholar]
- 15.Lugtenberg E J J, Wijsman H J W, Van Zaane D. Properties of a d-glutamic acid-requiring mutant of Escherichia coli. J Bacteriol. 1973;114:499–506. doi: 10.1128/jb.114.2.499-506.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 17.Martinez del Pozo A, Merola M, Ueno H, Manning J M, Tanizawa K, Nishimura K, Asano S, Tanaka H, Soda K, et al. Activity and spectroscopic properties of bacterial D-amino acid transaminase after multiple site-directed mutagenesis of a single tryptophan residue. Biochemistry. 1989;28:510–516. doi: 10.1021/bi00428a015. [DOI] [PubMed] [Google Scholar]
- 18.Martinez del Pozo A, Merola M, Ueno H, Manning J M, Tanizawa K, Nishimura K, Soda K, Ringe D. Stereospecificity of reactions catalyzed by bacterial D-amino acid transaminase. J Biol Chem. 1989;264:17784–17789. [PubMed] [Google Scholar]
- 19.Merola M, Martinez del Pozo A, Ueno H, Recsei P, Di Donato A, Manning J M, Tanizawa K, Masu Y, Asano S, et al. Site-directed mutagenesis of the cysteinyl residues and the active-site serine residue of bacterial D-amino acid transaminase. Biochemistry. 1989;28:505–509. doi: 10.1021/bi00428a014. [DOI] [PubMed] [Google Scholar]
- 20.Nakajima N, Tanizawa K, Tanaka H, Soda K. Cloning and expression in Escherichia coli of the glutamate racemase gene from Pediococcus pentosaceus. Agric Biol Chem. 1986;50:2823–2830. [Google Scholar]
- 21.Nelms J, Edwards R M, Warwick J, Fotheringham I. Novel mutations in the pheA gene of Escherichia coli K-12 which result in highly feedback inhibition-resistant variants of chorismate mutase/prephenate dehydratase. Appl Environ Microbiol. 1992;58:2592–2598. doi: 10.1128/aem.58.8.2592-2598.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pucci M J, Thanassi J A, Ho H-T, Falk P J, Dougherty T J. Staphylococcus hemolyticus contains two d-glutamic acid biosynthetic activities, a glutamate racemase and a d-amino acid transaminase. J Bacteriol. 1995;177:336–342. doi: 10.1128/jb.177.2.336-342.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roberts R B, Abelson P H, Cowie D B, Bolton E T, Britten R J. Studies of biosynthesis in E. coli. Carnegie Inst. Washington Publication; 1955. p. 607. [Google Scholar]
- 24.Rogers H J, Perkins H R, Ward J B. Microbial cell walls and membranes. London, England: Chapman and Hall; 1980. [Google Scholar]
- 25.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seville M, Vincent M G, Hahn K. Modeling the three-dimensional structures of bacterial aminotransferases. Biochemistry. 1988;27:8344–8349. doi: 10.1021/bi00422a009. [DOI] [PubMed] [Google Scholar]
- 27.Soda K, Yonaha K, Misono H, Osugi M. Purification and crystallization of D-amino acid aminotransferase of Bacillus sphaericus. FEBS Lett. 1974;46:359–363. doi: 10.1016/0014-5793(74)80407-2. [DOI] [PubMed] [Google Scholar]
- 28.Stoker N G, Fairweather N F, Spratt B G. Versatile low-copy-number plasmid vectors for cloning in Escherichia coli. Gene. 1982;18:335–341. doi: 10.1016/0378-1119(82)90172-x. [DOI] [PubMed] [Google Scholar]
- 29.Sugio S, Petsko G A, Manning J M, Soda K, Ringe D. Crystal structure of a D-amino acid aminotransferase: how the protein controls stereoselectivity. Biochemistry. 1995;34:9661–9669. doi: 10.1021/bi00030a002. [DOI] [PubMed] [Google Scholar]
- 30.Tanizawa K, Asano S, Masu Y, Kuramitsu S, Kagamiyama H, Tanaka H, Soda K. The primary structure of thermostable D-amino acid aminotransferase from a thermophilic Bacillus species and its correlation with L-amino acid aminotransferases. J Biol Chem. 1989;264:2450–2454. [PubMed] [Google Scholar]
- 31.Tanizawa K, Masu Y, Asano S, Tanaka H, Soda K. Thermostable D-amino acid aminotransferase from a thermophilic Bacillus species. Purification, characterization, and active site sequence determination. J Biol Chem. 1989;264:2445–2449. [PubMed] [Google Scholar]
- 32.Taylor P P, Fotheringham I G. Nucleotide sequence of the Bacillus licheniformis ATCC 10716 dat gene and comparison of the predicted amino acid sequence with those of other bacterial species. Biochim Biophys Acta. 1997;1350:38–40. doi: 10.1016/s0167-4781(96)00204-7. [DOI] [PubMed] [Google Scholar]
- 33.Thanabalu T, Hindley J, Jackson-Yap J, Berry C. Cloning, sequencing, and expression of a gene encoding a 100-kilodalton mosquitocidal toxin from Bacillus sphaericus SSII-1. J Bacteriol. 1991;173:2776–2785. doi: 10.1128/jb.173.9.2776-2785.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wasserman S A, Walsh C T, Botstein D. Two alanine racemase genes in Salmonella typhimurium that differ in structure and function. J Bacteriol. 1983;153:1439–1450. doi: 10.1128/jb.153.3.1439-1450.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yonaha K, Misono H, Yamamoto T, Soda K. D-amino acid aminotransferase of Bacillus sphaericus. Enzymologic and spectrometric properties. J Biol Chem. 1975;250:6983–6989. [PubMed] [Google Scholar]
- 36.Zimmer T L. Gramicidin S synthetase. Methods Enzymol. 1975;43:567–579. doi: 10.1016/0076-6879(75)43119-6. [DOI] [PubMed] [Google Scholar]