Abstract
Genome-wide in silico analysis identified thousands of natural sense–antisense transcript (SAT) pairs in the mouse transcriptome. We investigated their expression using strand-specific oligo-microarray that distinguishes expression of sense and antisense RNA from 1947 SAT pairs. The majority of the predicted SATs are expressed at various steady-state levels in various tissues, and cluster analysis of the array data demonstrated that the ratio of sense and antisense expression for some of the SATs fluctuated markedly among these tissues, while the rest was unchanged. Surprisingly, further analyses indicated that vast amounts of multiple-sized transcripts are expressed from the SAT loci, which tended to be poly(A) negative, and nuclear localized. The tendency that the SATs are often not polyadenylated is conserved, even in the randomly chosen SAT genes in the plant Arabidopsis thaliana. Such common characteristics imply general roles of the SATs in regulation of gene expression.
Recently, increasing numbers of natural antisense transcripts have been identified in a variety of eukaryotic organisms using large-scale transcriptome analysis. The sense–antisense transcript (SAT) pair is a pair of transcripts produced from the same locus on the chromosome, but from the DNA strands opposite each other. These include 2481 SAT pairs in mice (The FANTOM Consortium and The Riken Genome Exploration Research Group Phase I & II Team 2002; Kiyosawa et al. 2003), 1027 SATs in Drosophila melanogaster (Misra et al. 2002), 2667 SATs in humans (Yelin et al. 2003), and 601 SATs in rice (Osato et al. 2003). Whole-genome array expression analysis has also revealed ∼7600 SATs (∼30% of all annotated genes) in Arabidopsis thaliana (Yamada et al. 2003). These vast numbers prompt the notion that SATs may have regulatory roles in gene expression and/or RNA processing via the formation of double-stranded RNA (dsRNA) (Herbert 2004). On the other hand, small noncoding RNAs (ncRNA) have also drawn attention as regulators of gene expression. microRNAs (miRNA) are small (∼22 nucleotides), processed antisense RNAs, which bind to sense transcripts, thereby blocking further translation of the sense strand RNA (Nelson et al. 2003). Small interfering RNAs (siRNA) are of similar size to miRNA, but are generated from longer dsRNA, a product of the sense and antisense RNA duplex (Nelson et al. 2003). siRNA is responsible for mRNA degradation of matched sequences and is also implicated in gene silencing at the transcriptional level (Vance and Vaucheret 2001; Hall et al. 2002; Volpe et al. 2002). Another type of small dsRNAs (20–21 nucleotides) is implicated in gene regulation via transcription-factor binding in a mechanism distinct from that of miRNA/siRNAs (Kuwabara et al. 2004). The relationships between thousands of natural antisense transcripts identified within the genome, and these small RNAs are yet to be determined. In mammals, a small cohort of longer antisense RNAs has been located within imprinted loci. One of these, Air, was proven via gene-targeting to be involved in the maintenance of the local imprint status (Sleutels et al. 2002).
Although we now have massive amounts of data gleaned from genome/transcriptome analyses, the experimental evidence defining the extent of most SAT expression remains very limited; in part, because the majority of the described data have been obtained using in silico analysis exclusively. Here, we report actual transcriptional analyses of sense and antisense genes and show that these transcripts tend to be both poly(A) negative and nuclear localized.
Results
Expression analysis of SATs by Oligo DNA microarray
To determine whether SATs are actually transcribed, we performed expression analysis of available mouse SATs by using custom-made oligo DNA (60-mer) chips that distinguish the expression of sense versus antisense transcripts. From 2481 pairs of SATs identified in mouse transcriptome (The FANTOM Consortium and The RIKEN Genome Exploration Research Group Phase I & II Team 2002), we chose those pairs that consisted of either FANTOM2 full-length cDNA or RefSeq sequences (2097 pairs of SATs). The final DNA sequences on the microarray for which unique 60-mer sequences were successfully chosen were 1947 SAT pairs for the microarray probes. These SAT sequences can be classified as “coding” or “noncoding” genes based on the gene annotation by the FANTOM Consortium (2002). These distinctions of each gene are shown in Supplemental Table 1. Of 1947 sense–antisense pairs, 943, 828, and 173 were coding/coding, coding/noncoding, and noncoding/noncoding pairs, respectively. The coding status of three genes could not be determined. Conventionally, sense transcripts often represent coding genes. In this study, however, we define the “sense” transcripts as those that map to the (+) strand of the published mouse genome assembly, whereas “antisense” transcripts are on the (–) strand, because we found both of the SAT pair genes corresponded to coding or noncoding in many cases. In addition, the coding status of each gene is still tentative in a strict sense. In our definition, sense does not necessarily indicate the protein-coding status of the transcript, nor does antisense imply ncRNA.
We found that most of the sense and antisense genes are indeed expressed at various levels in various cells and tissues (Fig. 1) (all of the processed signal data are found in Supplemental Table S1). Figure 1, A and B, show the expression of the SATs in ES cells (Tada et al. 2001), whereas Figure 1, C and D, show the same in fibroblast cells. In Figure 1, A and C, the dots represent expression levels of the coding/coding SAT pairs, while in Figure 1, B and D, the dots indicate expression signals of the noncoding/coding SATs. In light of quantitative PCR-validation experiments, a signal intensity value of 100 was defined as the threshold for calling a gene “expressed.” According to this criterion, 89.6% of the coding and 81.7% of the noncoding genes of the SATs were expressed in ES cells, whereas 94.6% of the coding and 91.3% of the noncoding genes of the SATs were expressed in fibroblast. The expression level varied, ranging from 100 to 100,000 (processed signals). The ratio of the expression level of the coding to that of the noncoding genes also varied (Fig. 1, A vs. B, C vs. D). We extended this type of analysis to samples from brain, heart, and testis (data not shown) and found that the ratio of coding and noncoding gene expression in each SAT pair fluctuated markedly among these cells and tissues.
Figure 1.
Overall expression of sense and antisense genes as determined by using an oligo DNA microarray. The combined expression of one SAT pair is represented by a dot. The expression in ES cells is presented in A and B, and that in fibroblast cells is presented in C and D. The pairs consisting of coding and noncoding genes are shown in A and C (x-axis, noncoding gene; y-axis, coding gene), whereas the pairs consisting of both coding genes are shown in B and D.
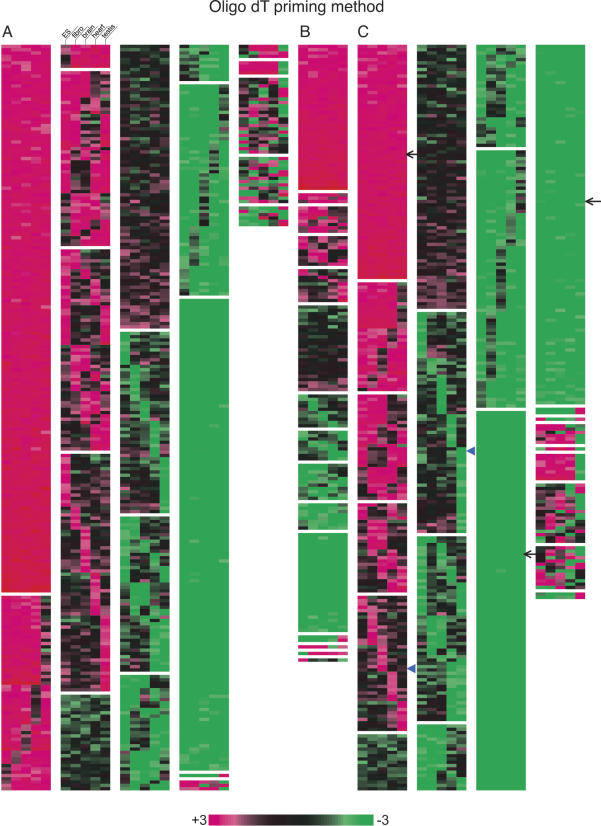
We next performed clustering analysis of the microarray data, in which the SATs pairs were grouped according to the log ratio of the expression levels for the sense and the antisense pairs (when both of the pairs consist of coding genes or noncoding genes only), or the coding and noncoding gene pairs (Fig. 2A–F). If the expression level of the noncoding gene is threefold more than that of the coding gene (in noncoding/coding pairs) (Fig. 2C), or when the expression of the sense gene is more than threefold compared with antisense gene (in coding/coding or noncoding/noncoding pairs) (Fig. 2A,B), the ratio is shown in red. In the reversed situation, it is shown in green. As shown in these expression heat maps, there are SAT pairs whose expression ratio is relatively unchanged among the five tissues/cell samples tested, whereas some of the SAT pairs exhibit tissue-specific expression patterns. For example, as shown in Figure 2C, ∼8.5% of the coding/noncoding SAT correspond to a monotonously red block (indicated by arrow), while ∼26% of the SATs belong to green block, suggesting that the expression ratio of these SAT pairs are fixed among the five different samples, and that there are SAT pairs in which expression of noncoding genes are higher than that of coding genes. On the other hand, the expression ratio of the rest of the SAT pairs fluctuated to a various extent. Some of the SAT pairs showed expression-ratio differences specific to each tissue sample (arrowhead in Fig. 2C), implying that noncoding RNA may be involved in tissue-specific regulation of the expression of their coding partners. Such trends are pertinent for coding/coding or noncoding/noncoding SAT pairs; expression ratios of 30%∼40% of the SAT pairs were relatively unchanged, while the rest fluctuated significantly among the tissues/cells (Fig. 2A,B).
Figure 2.
Clustering analysis of the expression balance between the sense and antisense genes, or the coding and noncoding genes in ES cells, fibroblast cells, brain, heart, and testis. A, B, C was based on the microarray data obtained with the oligo dT priming method, whereas the random priming method was used for the data in D, E, and F. (A,D) Clustering with the data of the pairs consisting of the coding genes only. When the sense gene expression is threefold more than the antisense gene expression, the color is shown in red. In the reversed situation, the color is shown in green. (B,E) Clustering with the data of the pairs consisting of the noncoding genes only. The coloring is the same as found in A. (C,F) Clustering with the data of the pairs consisting of the coding and noncoding genes. When the noncoding gene expression is threefold more than the coding gene expression, the color is shown in red. Note that this figure is intended to show the ratio between sense and antisense, or noncoding and coding expression. The absolute expression values for each gene are found in Supplemental Table 1.
Northern hybridization analysis of selected SATs
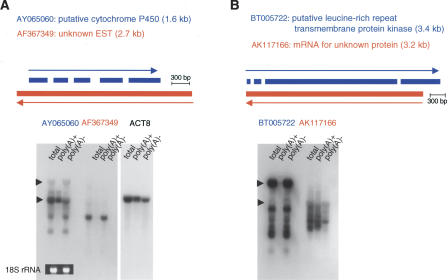
We next performed Northern analysis of six randomly chosen pairs of SATs. To distinguish the sense and antisense transcripts, we transcribed single-stranded riboprobes and used them in the analysis. The results from two SAT pairs are shown in Figures 3 and 4. The first pair comprises 6330439J10 (FANTOM2 clone ID, 3-oxoacid CoA transferase) and A230019L24 (unknown EST). A230019L24 lacks significant ORFs and appears to represent ncRNA. Clone 6330439J10 is 2099-bp long, which corresponds to the lower band on both the Northern blot loaded with total RNA and that with poly(A)+ RNA (Fig. 3D). The microarray data were in agreement with the signal intensities in the Northern blots (Fig. 3B). Very faint bands of ∼1.8 kb and 3 kb corresponding to A230019L24 were detected on the poly(A)+ RNA blot. However, the total RNA blot for A230019L24 showed an extremely large band (>10 kb), as well as a strong hybridization smear between 18S and 28S rRNA (Fig. 3E). This hybridization smear was not due to repetitive sequences in the probe, as the same probe gave a single band on a genomic Southern blot (data not shown).
Figure 3.
Microarray and Northern hybridization analysis of representative SAT pair. (A) Mapping patterns of sense (coding, 6330439J10) and antisense (noncoding, A230019L24) genes in the genome. The positions of exons are indicated as filled columns. The directions of transcription are indicated with arrows. In this figure, information associated with sense genes is shown in blue and that for antisense genes in red. (B) Microarray signal intensities obtained with samples labeled by oligo dT priming. (C) Microarray signal intensities obtained with samples labeled by random-nanomer priming. (D,E) Northern hybridization of sense (6330439J10) and antisense (A230019L24) genes. In each lane, 20 μg of total RNA or 1 μg of mRNA was loaded. In the Northern blot figures, the positions of 18S and 28S ribosomal RNA are indicated by arrowheads at the left edges of the blots.
Figure 4.
Microarray and Northern hybridization analysis of another SAT pair. (A–E) A similar analysis to that of Figs. 3A–E is presented for another pair of sense (noncoding, G430028I15) and antisense (coding, D930007L12) genes.
In the second SAT pair (G430028I15, unknown EST; D930007L12, Metaxin1), G430028I15 appears to encode ncRNA. It is intriguing that the ncRNA probe again gave a strong hybridization smear on the total RNA blot, but not on the poly(A)+ blot (Fig. 4D). The size of the Metaxin1 mRNA is 1.8 kb (Bornstein et al. 1995), which corresponds to a band in testis poly(A)+ RNA, whereas the probe identified a larger band in the heart (Fig. 4E). The total RNA blot showed multiple bands of different sizes (Fig. 4E). Some of them were quite large and may correspond to Metaxin1 transcripts that are contiguous with those of neighboring genes (Glucocerebrosidase and Thrombospondin 3), as previously suggested (Bornstein et al. 1995). Again, the observed hybridization patterns were not caused by repetitive sequences in the probe, according to the results of genomic Southern analysis.
To ascertain whether these banding patterns are not due to nonspecific cross hybridization to the RNA probes used, we designed 30-mer oligo DNA probes complementary to either 6330439J10/A230019L24 or G430028I15 (the probe positions indicated in Figs. 3A and 4A) and performed Northern analysis again (Fig. 5A,B). The hybridization patterns with 30-mer probes were very similar to those obtained with the full-length RNA probes, confirming that both kinds of probes specifically detected multiple-sized transcripts in the total RNA fraction. The multiple bands thus detected likely are not degradation products produced during the RNA preparation, since other probes such as 6330439J10 detected bands with distinct sizes on the same blot (Fig. 3D). These results suggest that multiple-sized transcripts can be generated from a single SAT locus. As shown in the lower part of Figure 5A, a small band of ∼60–100 bases was detected with A230019L24-30-mer probe. A similarly sized band was also detected with another 30-mer probe only when the probe position was chosen from where the sense and antisense exons overlap (Fig. 5C, probe positions #3 and #4).
Figure 5.
(A,B) Northern hybridization with oligo DNA (30-mer) as a probe. Total RNA (10 μg) from adult mouse brain was loaded in each lane. The 30-mer oligo DNA probes used for 6330439J10 and A230019L24 were exactly complementary to each other. (C) Northern hybridization with a series of 30-mer probes chosen from introns and exons. The mapping pattern of the sense and antisense genes (6330439J10/A230019L24) and expanded map at exons 1 of both genes were diagramed at top. The numbers and arrows in red indicate approximate positions of 30-mer probes selected. Each lane contained 10 μg of total RNA from adult mouse brain. The positions of 18S and 28S ribosomal RNA are indicated by arrowheads at the left edges of the blots. (D,E) Northern hybridization of total (20 μg), nuclear (4 μg), cytoplasmic (16 μg), poly(A)+ (0.5 μg), and poly(A)– (19.5 μg) RNA samples from fibroblast cells with A230019L24 and G430028I15 probes. Each lane contained equivalent amounts of RNA on a per-cell basis. The hybridization to U1 RNA is shown at the bottom of the blot as a control for each fraction of RNA. (F) Overlapping mapping pattern of G430028I15 and D930007L12 (Metaxin1), and the results from 5′ RACE analysis of G430028I15 gene with fibroblast cells are summarized at the top of the figure. The positions of the initial and nested primers for the 5′ RACE analysis are indicated as black arrows above the G430028I15 gene position. The GC boxes identified in the intron of Metaxin1 genes are denoted “G.” The square-boxed G is the one to which Sp1 actually bound (Collins et al. 1998). The transcriptional start sites identified by 5′ RACE analysis are indicated with asterisks and bent arrows. The genomic region from –20 kb to +40 kb of the G430028I15 gene and Northern hybridization with the probes from the –20-, +20-, and +40-kb regions are presented at the bottom of the figure. Total (20 μg), nuclear (4 μg), and cytoplasmic (16 μg) RNA from fibroblast cells were used in each lane. The regions of the probes used for Northern hybridization are shown in the genome map, along with the nearby genes (mouse genome browser at http://genome.ucsc.edu/, October 2003 version).
The SAT gene probes often detected many bands or strong hybridization smears in total RNA, but not in poly(A)+ RNA. These hybridization patterns were not specific to ncRNA, but appeared to be associated with the SAT loci in general. Of the six SAT pairs analyzed (6 SATs; 2 probes per SAT = 12 probes total), we found the smear hybridization pattern with eight (66.7%) probes, five of which came from protein-coding genes (Table 1).
Table 1.
Smearing Northern hybridization pattern in sense and antisense genes
| FANTOM2 clone ID | Protein coding | Smear on total RNA blot |
|---|---|---|
| 6330439J10 | Yes | No |
| A230019L24 | No | Yes |
| G430028I15 | No | Yes |
| D930007L12 | Yes | Yes |
| E430007F07 | Yes | No |
| 4930435G24 | Yes | Yes |
| 1300008P06 | Yes | Yes |
| 2810025E10 | No | Yes |
| 2410043H24 | Yes | No |
| C820017J03 | Yes | No |
| C130011I02 | Yes | Yes |
| 9130207N01 | Yes | Yes |
Because we obtained different results with total RNA and poly(A)+ RNA, we prepared poly(A)+ and poly(A)– RNA from fibroblasts and repeated the Northern analysis. The transcripts revealed by the A230019L24 and G430028I15 probes occurred in the poly(A)– fraction, indicating that they were not polyadenylated (Fig. 5D,E). We also found that these transcripts were localized predominantly in the nuclear fraction (Fig. 5D,E); in particular, G430028I15 transcripts occurred exclusively in the nucleus. We performed a similar Northern analysis with nuclear/cytoplasmic RNA fractions and the genes producing smears on the total RNA blots. Including A230019L24 and G430028I15 above, four genes produced almost nuclear-exclusive transcripts such as found in G430028I15 (Fig. 5E), and another four genes produced both nuclear and cytoplasmic transcripts similar to those found in A230019L24 (Fig. 5D) (data not shown). Thus, nuclear localization of the SATs may be an intrinsic nature of the SAT genes. It is generally believed that primary transcripts destined to be mRNAs move quickly to the cytoplasm after their synthesis (Jackson et al. 2000). However, our findings suggest that at least some SATs go through a processing pathway quite different from the typical mRNA maturation steps.
Estimations of expression levels of SATs by dot blots
As we detected strong hybridization signals for SATs on the Northern blots, we attempted to estimate the RNA expression levels of 6330439J10/A230019L24 and G430028I15/D930007L12 by quantitative dot blot analysis. 32P-labeled probes were hybridized to a serial concentration (50 ng, 100 ng, 250 ng, 500 ng, 1 μg, and 2.5 μg) of mouse brain RNA containing 40 pg/μg of in vitro-transcribed plant RNA as a spike control. Hybridization signal intensities changed proportionally to a serially diluted series of RNA samples for every probe used (Supplemental Fig. 1). With the dots of 1-μg RNA, we obtained the following hybridization intensities relative to that of β-actin: 6.1% (6330439J10), 10% (A230019L24), 104% (G430028I15), 20% (D930007L12). These intensity values approximately corresponded to 9, 14, 150, and 30 pg per 1 μg of mouse brain total RNA, respectively, based on the calibrations with a spike control.
Identification of start sites of SATs
As we found multiple transcripts of varying sizes for many of the SATs, we attempted to identify transcription start sites upstream of the G430028I15 gene using 5′ RACE. There are at least five start sites residing in the intron–exon region of Metaxin1 on the opposite strand of G430028I15 (Fig. 5F, top). We also performed Northern analysis with probes derived from regions –20 kb, +20 kb, and +40 kb from the G430028I15 gene (Fig. 5F, bottom). The probe at –20 kb gave a hybridization pattern almost identical to the one detected by the G430028I15 probe, suggesting the presence of transcription start site(s) further upstream. In contrast, the probes at +20 and +40 kb yielded different patterns. These results indicate that multiple transcription start sites are distributed along a relatively large genomic region, resulting in transcripts larger than predicted from the size of the cDNA.
Global comparison of microarray data: Difference between Oligo dT and random priming methods
Since the SATs are often localized in nucleus and not polyadenylated, we re-examined SAT expression by using the oligo microarray with a different cDNA priming method. The cDNA target was cyanine labeled with random nanomers this time, instead of the oligo dT primer used in the previous experiments. We reasoned that the results obtained with random nanomers would differ from the previous results if the majority of SATs are not polyadenylated. As expected, the random-priming method yielded increased signals only for the genes showing the hybridization smear on the Northern blots (Figs. 3B,C, 4B,C). In contrast, the signal intensity for the 6330439J10 probe was decreased greatly with the random-primed targets. This result is reasonable, because the 6330439J10 cDNA probe detected distinct bands on both poly(A)+ and total RNA blots (Fig. 3D).
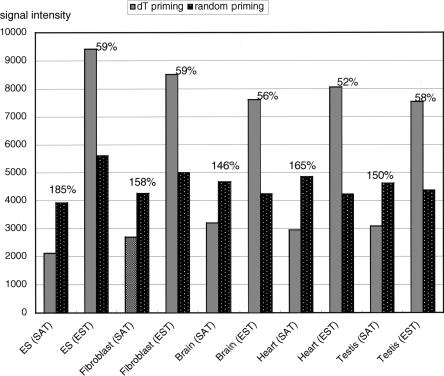
As described in the Methods section, our custom microarray contains 2697 probe sequences derived from ESTs that are unrelated to the SAT gene sequences. Homology search analysis showed that the EST sequences did not overlap with the SAT sequences used in this study (data not shown). Therefore, this EST set likely was derived from typical polyadenylated mRNA and could serve as a convenient control against the SATs. As expected, the sum of the signals detected by these EST probes was reduced markedly when random priming was used (Fig. 6). In contrast, the opposite trend was apparent for SATs; random-primed targets gave higher signals in all cases tested (Fig. 6). It is therefore likely that the SAT transcripts generally are enriched in the poly(A)– fraction.
Figure 6.
Average signal intensities on microarray chips after oligo dT priming were compared with those from random priming methods. In every cell type and tissue analyzed, the average signal intensity of SAT genes was higher than that of typical genes (ESTs) when the random priming method was used for sample labeling. The change in signal intensity after the change from oligo dT priming to random priming is shown at the top of the bars.
Cluster analysis of the expression ratio for the SAT pairs were again performed using the data obtained with random-primed targets (Fig. 2D,E,F). Global patterns of the heat maps were similar to those obtained with the oligo dT priming method; there were clusters with fixed ratios and clusters showing ratio fluctuations or tissue-specific ratio differences. However, members belonging to each cluster were quite different between the data obtained with two different priming methods. Side-by-side comparisons of cluster analysis data clearly showed that hybridization patterns for a given probe pair often disagreed when a different priming method was chosen (see Supplemental Fig. 2).
Some of the plant SATs are also present in poly(A)- fraction
To assess whether the SAT transcripts generally are enriched in the poly(A)– fraction in plants also, we conducted expression analyses of SATs identified by Seki and Shinozaki (pers. comm.; Seki et al. 2005) in the transcriptome of A. thaliana. We randomly chose four probe pairs from Arabidopsis SATs and hybridized them to total, poly(A)+, and poly(A)– RNA. Interestingly, seven of the eight transcripts were enriched in the poly(A)– fraction (data from two pairs shown in Fig. 7A,B), suggesting that SATs predominantly exist in the poly(A)–; fraction in both plants and animals. The greatest difference we found between mice and Arabidopsis was that strong, high molecular-weight hybridization smears were not apparent in the plant. This difference may reflect the difference in the genome size between mouse (2500 Mb) and A. thaliana (120 Mb) (The Arabidopsis Genome Intitiative 2000; Mouse Genome Sequencing Consortium 2002), a difference that may affect the mode of gene expression regulation that operates at the chromatin-domain level.
Figure 7.
(A,B) Northern hybridization analysis of A. thaliana sense and antisense gene pairs. The format of the figures is the same as for the mouse Northern analyses. In each lane, 20 μg of total RNA, 0.5 μg of poly(A)+ RNA, or 19.5 μg of poly(A)– RNA was loaded. The photograph of ethidium-bromide-stained 18S rRNA and hybridization to the Actin8 transcript are presented as controls.
Discussion
In this study, we present the first experimental evidence for the expression of SATs identified by genome-wide analyses in silico. The steady-state RNA levels of the SATs generally were high. In light of dot-blot hybridizations, we estimated the amounts of the noncoding transcripts, A230019L24 and G430028I15, to be ∼10% and ∼100% of the level of the β-actin RNA, respectively. More importantly, we disclosed several previously hidden characteristics of SAT transcripts; they generate multiple-sized transcripts that are not polyadenylated and tend to be nuclear localized. These traits suggest that SATs belong to a hitherto unknown category of transcripts. In this context, it is interesting to note that a smear hybridization pattern on the Northern blot with a noncoding gene probe was recently reported by Imamura et al. (2004). They identified an antisense RNA (Khps1a; antisense to the Sphk1 gene), which showed a hybridization smear ranging from 0.6 to 20 kb on Northern blots. The Sphk1–Khps1a pair was found to be included in our SAT pair collections.
The processing pathways from primary transcript to mRNA or other classes of RNA are not yet fully understood. For example, ∼95% of the RNA synthesized by RNA polymerase II in mammalian cells remains in the nucleus; only a minority is processed to mRNA (Jackson et al. 2000). The predominant population of RNA appears to lack polyadenylation and seems to be a poor substrate for splicing (Salditt-Georgieff and Darnell Jr. 1982). The role of this category of RNA and identity of its transcripts have eluded understanding for more than two decades. Our present study indicates that SATs share several characteristics with this class of RNA; both are present in vast quantities in the nucleus, and neither is polyadenylated. The biological functions of this category of RNA, including those of SATs, remain to be elucidated. The evolutionary conservation of SATs and their characteristics in plants and animals strongly suggest their importance in the regulation of gene expression. The information we present likely will promote better understanding of a new class of RNA that has been puzzling researchers for a long time.
The regulatory roles played by the SAT genes may occur at the chromatin-domain level. Expression of imprinted genes is subjected to such domain-level regulation. Imprinted genes often are accompanied by a large antisense transcript, which is involved in the regional regulation of expression (e.g., Air–Igf2r, LIT1–KvLQT1) (Mitsuya et al. 1999; Sleutels et al. 2002). Nikaido et al. (2003) showed close relationships between SATs and imprinted genes. We also have discovered a number of novel SATs within imprinted loci (Kiyosawa and Abe 2002; Kiyosawa et al. 2003). Although the two SAT gene pairs we present in this study (6330439J10/A230019L24 and G430028I15/D930007L12) are not imprinted (data not shown, imprinting status of G430028I15 only could not be determined), complex transcription seems to be occurring at these loci; see, for example, the Metaxin1 locus. The intron sequence between exons 6 and 7 of Metaxin1 was reported to serve as a far-upstream enhancer element of Thrombospondin3 gene, which is downstream of the G430028I15 gene. There are four putative GC-boxes (Sp1-binding sites) in this region (marked as “G” in Fig. 5F), and two of these (square-boxed G) have been proven to bind to Sp1 (Collins et al. 1998). We now know that a long ncRNA is expressed from this region. Interestingly, deletion of this region markedly reduced the expression of the Thrombospondin3 gene, but not of the Metaxin1 gene (Collins et al. 1998). It is thus possible that the long ncRNA is involved in the regulation of Thrombospondin3 gene expression, although more detailed functional analysis of this ncRNA is needed. It is also possible that complex transcripts found at SAT loci merely reflect unusual chromatin status associated with the SAT region, generating transcripts with no apparent function. However, another example suggests that at least some of the antisense transcript has a functional role in epigenetic regulation. Sphk1 and Khps1a found in our SAT collections were expressed in a mutually exclusive manner in a single cell, and Khps1a transcript was localized in both cytoplasm and nucleus, implying that Khps1a RNA may act in the nucleus (Imamura et al. 2004). The forced expression of the Khps1a resulted in alterations of the methylation pattern of the CpG island at the Sphk1 locus (Imamura et al. 2004). Therefore, it is possible that antisense RNA controls gene expression at transcription level by changing methylation status of the sense gene.
Another interesting aspect of SATs is their relationship to RNA interference (RNAi). It is of great interest whether naturally occurring SATs can form dsRNA and become targets of RNAi machinery. Recently, dependence on natural RNAi via dsRNA formation in transposon silencing was demonstrated in Caenorhabditis elegans (Sijen and Plasterk 2003). In mice, when its expression was confined to nuclei, long dsRNA eventually produced siRNA and knocked down the expression of the corresponding protein-coding gene (Shinagawa and Ishii 2003). At least superficially, this situation resembles that produced with the SAT pairs, suggesting that some SATs may be involved in the repression of gene expression via natural RNAi. We actually detected small bands of <100 bases on the Northern blots with the probes only, from where the exons of the sense and antisense genes overlap, namely, where dsRNA is possibly formed (Fig. 5C).
Studies on poly(A)– RNAs has been limited, because conventional belief in molecular biology suggests that poly(A)+ mRNAs are major mediators in flows of genetic information. However, the information obtained from this study implies that some class of poly(A)– nuclear RNA may have important biological functions, and will promote a new field of research on the regulatory mechanisms mediated by the poly(A)– RNA.
Methods
DNA microarray experiment and analysis
The sequences of 60-mer DNA specific to the sense and antisense genes were chosen by K.K. DNAFORM (Japan), and Agilent Technologies manufactured custom oligo DNA microarray chips by using this information. RNA was labeled and hybridized using Fluorescent Direct Label Kits (oligo dT-primed labeling) (Agilent Technologies), according to the manufacturer's protocols. For labeling with random nanomers, we used the CyScribe First-Strand cDNA Labeling Kit (Amersham). The RNA samples used for microarray experiments were from mouse ES cells, SL10 cells (fibroblast cell line), brain, heart, and testis. The total RNA of brain, heart, and testis for array experiments was purchased from Ambion. The total RNA of ES and fibroblast cells was isolated using Trizol reagent (Invitrogen). The same total RNA samples were reciprocally labeled with Cy3 or Cy5, hybridized to the oligo DNA on the chip, and dye-normalized, and processed signals were obtained using Feature Extraction software (Agilent Technologies). The custom oligo DNA microarray chip contained 2097 pairs of the sense–antisense genes and 592 pairs of nonantisense bidirectional genes (total 5078 genes), along with an additional 3013 genes unrelated to natural antisense analysis. These 3013 genes include 2697 EST sequences mentioned in the section of “Global Comparison of Microarray Data” of the results. For the Feature Extraction software to produce the processed signals, the data for all genes on the chip were used. For further analysis, the average of Cy3-labeled and Cy5-labeled processed signals was used as the processed signal of a particular gene expression. The correlation coefficient between the Cy3- and Cy5-labeled processed signals from oligo dT-primed samples was 0.951 (ES cells), 0.944 (fibroblast cells), 0.982 (brain), 0.972 (heart), and 0.973 (testis). The correlation coefficient of random primed samples was 0.923 (ES cells), 0.970 (fibroblast cells), 0.985 (brain), 0.983 (heart), and 0.970 (testis). The total gross signal on the chip in each hybridization experiment was adjusted to that with the ES cell sample, so that the relative differences in gene expression among cell lines and tissues could be compared with one another. The processed signals in the Supplemental data (Table S1) were these averaged signals. The microarray data were analyzed using cluster analysis implemented in the statistical package R. A MIAME-compliant description of our array analysis is provided as an online Supplemental document. The raw data of array experiments were deposited to the Gene Expression Omnibus (GEO) at the National Center for Biotechnology Information (NCBI) under the accession number of GSE2185 (http://www.ncbi.nlm.nih.gov/geo/).
Northern hybridization
Total RNA was isolated from fibroblast cells; the brain, heart (male/female mixed), and testis of C57BL/6J mice; and 3-week-old whole-rosette A. thaliana plants by using Trizol reagent (Invitrogen). mRNA was further prepared using the Oligotex-MAG mRNA Purification Kit (Takara). The fractions that did not bind to oligo dT beads were recovered, precipitated, and used as poly(A)– RNA samples. Nuclear and cytoplasmic RNA were isolated using the PARIS Protein and RNA Isolation System (Ambion). RNA was denatured with glyoxal prior to size fractionation by 1% agarose gel electrophoresis, blotted onto nylon membranes, hybridized to 32P-labeled probes, washed, and exposed to X-ray film by standard methods. The labeled probes were single-stranded RNAs generated by in vitro transcription, so that the strand specificity enabled us to distinguish the sense and antisense transcripts. Mouse full-length cDNA clones were used for the in vitro transcription: FANTOM clone ID G430028I15 (unknown EST; GenBank accession no. AK089957), D930007L12 (Metaxin1, AK086135), 1300008P06 (Ankyrin repeat structure-containing protein, AK004948), 2810025E10 (unknown EST, AK012809). As for dot-blot hybridization, the sequences excluding repeated elements were used for probes. The GenBank accession numbers of the A. thaliana full-length cDNA clones used for the Northern hybridization were AY065060, AF367349, BT006722, and AK117166.
To obtain the probes for the –20 kb, +20 kb, and +40 kb regions of the Metaxin1 locus, DNA fragments were cloned into the pGEM-T Easy vector (Promega) after being amplified by PCR using C57BL/6J mouse genomic DNA and the following primer sequences: –20 kb, AGTCTTCTGTTGCCACTT GCCG and AGAGAAGGTGGCAGG TTGGCTG; +20 kb, ACACAGCAGTGA TAAGCCAGGG and TGCTTTACCTAT CCAGCACCC; and +40 kb, TGTGAG GAGGGAACCTCAAGGC and TGTCT CTTTTCCACTGTCTCCC. 32P-labeled probes were generated by in vitro transcription to detect the transcripts of the same direction as G430028I15. The sequences of the synthetic 30-mer DNA probes were CGAAGCCCAGAGGACAGGAGTTT GAGAGCC (FANTOM2 ID, 6330439J10), GGCTCTCAAACTCCT GTCCTCTGGGCTTCG (FANTOM2 ID, A230019L24), and AAT GACACCCTCTTTCCGCCTCTCCTTTTG (FANTOM2 ID, G430028I15). The sequences of 30-mer probes used for Fig. 3C are CTCCAAAACGATGAAGTTAACCCACCACCG (#1), CCCTC CAGGCCCTTGGCAACCCGAAACCCT (#2), GGCACAACCCGC GGGGCTCGTCCTAGCTGT (#3), GGCTCTCAAACTCCTG TCCTCTGGGCTTCG (#4), CCGGGGGTTGGGGGTGGGGGCT TGGAGACG (#5), and GCTGGAGGAGGGTGGGGAGGAGGG TGGATT (#6). These 30-mer DNAs were labeled with [32P]ATP and polynucleotide kinase (Takara) and hybridized using ULTRAhyb-Oligo hybridization solution (Ambion).
cDNA synthesis, PCR, and 5′ RACE
cDNA was synthesized with the SuperScript III RT System (Invitrogen) according to the manufacturer's instructions. 5′ RACE was performed with the GeneRacer Kit (Invitrogen) according to the manufacturer's instructions. EXTaq or LATaq (TaKaRa) was used in PCR reactions. The sequence used for the 3′ primer in the 5′ RACE reaction was GAGGCAAGGAGACCTCAGCAGG AAA, and its nested primer sequence was TGAGGCTAGTGTG GGGTTACTGTTCC (these sequences lie within the G430028I15 sequence).
Acknowledgments
We thank Misako Yuzuriha for her excellent technical assistance. We also thank Drs. Kazuo Shinozaki and Motoaki Seki for sharing A. thaliana sense and antisense gene data. Whole-rosette A. thaliana plants were kindly provided by Dr. Masatomo Kobayashi at RIKEN BRC. This work was supported in part by a Grant-in-aid of Ministry of Education, Science and Culture of Japan to H.K. and K.A. and by the Special Coordinating Funds for Promoting Science and Technology to K.A.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3155905. Article published online before print in March 2005.
Footnotes
[Supplemental material is available online at www.genome.org. The expression data from this study have been submitted to Gene Expression Omnibus (GEO) at the National Center for Biotechnology Information (NCBI) under accession no. GSE2185.]
References
- The Arabidopsis Genome Initiative. 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796-815. [DOI] [PubMed] [Google Scholar]
- Bornstein, P., McKinney, C.E., LaMarca, M.E., Winfield, S., Shingu, T., Devarayalu, S., Vos, H.L., and Ginns, E.I. 1995. Metaxin, a gene contiguous to both thrombospondin 3 and glucocerebrosidase, is required for embryonic development in the mouse: Implications for Gaucher disease. Proc. Natl. Acad. Sci. 92: 4547-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, M., Rojnuckarin, P., Zhu, Y.H., and Bornstein, P. 1998. A far upstream, cell type-specific enhancer of the mouse thrombospondin 3 gene is located within intron 6 of the adjacent metaxin gene. J. Biol. Chem. 273: 21816-21824. [DOI] [PubMed] [Google Scholar]
- The FANTOM Consortium, and The RIKEN Genome Exploration Research Group Phase I & II Team. 2002. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420: 563-573. [DOI] [PubMed] [Google Scholar]
- Hall, I.M., Shankaranarayana, G.D., Noma, K., Ayoub, N., Cohen, A., and Grewal, S.I. 2002. Establishment and maintenance of a heterochromatin domain. Science 297: 2232-2237. [DOI] [PubMed] [Google Scholar]
- Herbert, A. 2004. The four Rs of RNA-directed evolution. Nat. Genet. 36: 19-25. [DOI] [PubMed] [Google Scholar]
- Imamura, T., Yamamoto, S., Ohgane, J., Hattori, N., Tanaka, S., and Shiota, K. 2004. Non-coding RNA directed DNA demethylation of Sphk1 CpG island. Biochem. Biophys. Res. Commun. 322: 593-600. [DOI] [PubMed] [Google Scholar]
- Jackson, D.A., Pombo, A., and Iborra, F. 2000. The balance sheet for transcription: An analysis of nuclear RNA metabolism in mammalian cells. FASEB J. 14: 242-254. [PubMed] [Google Scholar]
- Kiyosawa, H. and Abe, K. 2002. Speculations on the role of natural antisense transcripts in mammalian X chromosome evolution. Cytogenet. Genome Res. 99: 151-156. [DOI] [PubMed] [Google Scholar]
- Kiyosawa, H., Yamanaka, I., Osato, N., Kondo, S., and Hayashizaki, Y. 2003. Antisense transcripts with FANTOM2 clone set and their implications for gene regulation. Genome Res. 13: 1324-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara, T., Hsieh, J., Nakashima, K., Taira, K., and Gage, F.H. 2004. A small modulatory dsRNA specifies the fate of adult neural stem cells. Cell 116: 779-793. [DOI] [PubMed] [Google Scholar]
- Misra, S., Crosby, M.A., Mungall, C.J., Matthews, B.B., Campbell, K.S., Hradecky, P., Huang, Y., Kaminker, J.S., Millburn, G.H., Prochnik, S.E., et al. 2002. Annotation of the Drosophila melanogaster euchromatic genome: A systematic review. Genome Biol. 3: research0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuya, K., Meguro, M., Lee, M.P., Katoh, M., Schulz, T.C., Kugoh, H., Yoshida, M.A., Niikawa, N., Feinberg, A.P., and Oshimura, M. 1999. LIT1, an imprinted antisense RNA in the human KvLQT1 locus identified by screening for differentially expressed transcripts using monochromosomal hybrids. Hum. Mol. Genet. 8: 1209-1217. [DOI] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
- Nelson, P., Kiriakidou, M., Sharma, A., Maniataki, E., and Mourelatos, Z. 2003. The microRNA world: Small is mighty. Trends Biochem. Sci. 28: 534-540. [DOI] [PubMed] [Google Scholar]
- Nikaido, I., Saito, C., Mizuno, Y., Meguro, M., Bono, H., Kadomura, M., Kono, T., Morris, G.A., Lyons, P.A., Oshimura, M., et al. 2003. Discovery of imprinted transcripts in the mouse transcriptome using large-scale expression profiling. Genome Res. 13: 1402-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osato, N., Yamada, H., Satoh, K., Ooka, H., Yamamoto, M., Suzuki, K., Kawai, J., Carninci, P., Ohtomo, Y., Murakami, K., et al. 2003. Antisense transcripts with rice full-length cDNAs. Genome Biol. 5: R5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salditt-Georgieff, M. and Darnell Jr., J.E. 1982. Further evidence that the majority of primary nuclear RNA transcripts in mammalian cells do not contribute to mRNA. Mol. Cell. Biol. 2: 701-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki, M., Satou, M., Sakurai, T., Akiyama, K., Iida, K., Ishida, J., Nakajima, M., Enju, A., Narusaka, M., Fujita, M., et al. 2005. Full-length cDNAs for the discovery and annotation of genes in A. thaliana. In Plant functional genomics (ed. D. Leister), pp. 3-22. The Haworth Press, Inc., Binghamton, NY.
- Shinagawa, T. and Ishii, S. 2003. Generation of Ski-knockdown mice by expressing a long double-strand RNA from an RNA polymerase II promoter. Genes & Dev. 17: 1340-1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen, T. and Plasterk, R.H. 2003. Transposon silencing in the Caenorhabditis elegans germ line by natural RNAi. Nature 426: 310-314. [DOI] [PubMed] [Google Scholar]
- Sleutels, F., Zwart, R., and Barlow, D.P. 2002. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 415: 810-813. [DOI] [PubMed] [Google Scholar]
- Tada, M., Takahama, Y., Abe, K., Nakatsuji, N., and Tada, T. 2001. Nuclear reprogramming of somatic cells by in vitro hybridization with ES cells. Curr. Biol. 11: 1553-1558. [DOI] [PubMed] [Google Scholar]
- Vance, V. and Vaucheret, H. 2001. RNA silencing in plants—Defense and counterdefense. Science 292: 2277-2280. [DOI] [PubMed] [Google Scholar]
- Volpe, T.A., Kidner, C., Hall, I.M., Teng, G., Grewal, S.I., and Martienssen, R.A. 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833-1837. [DOI] [PubMed] [Google Scholar]
- Yamada, K., Lim, J., Dale, J.M., Chen, H., Shinn, P., Palm, C.J., Southwick, A.M., Wu, H.C., Kim, C., Nguyen, M., et al. 2003. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302: 842-846. [DOI] [PubMed] [Google Scholar]
- Yelin, R., Dahary, D., Sorek, R., Levanon, E.Y., Goldstein, O., Shoshan, A., Diber, A., Biton, S., Tamir, Y., Khosravi, R., et al. 2003. Widespread occurrence of antisense transcription in the human genome. Nat. Biotechnol. 21: 379-386. [DOI] [PubMed] [Google Scholar]
Web site references
- http://www.ncbi.nlm.nih.gov/geo/; National Center for Biotechnology Information.
- http://genome.ucsc.edu/; The UCSC Bioinformatics site.