Abstract
Expansion of “low complex” repeats of amino acids such as glutamine (Poly-Q) is associated with protein misfolding and the development of degenerative diseases such as Huntington's disease. The mechanism by which such regions promote misfolding remains controversial, the function of many repeat-containing proteins (RCPs) remains obscure, and the role (if any) of repeat regions remains to be determined. Here, a Web-accessible database of RCPs is presented. The distribution and evolution of RCPs that contain homopeptide repeats tracts are considered, and the existence of functional patterns investigated. Generally, it is found that while polyamino acid repeats are extremely rare in prokaryotes, several eukaryote putative homologs of prokaryote RCP—involved in important housekeeping processes—retain the repetitive region, suggesting an ancient origin for certain repeats. Within eukarya, the most common uninterrupted amino acid repeats are glutamine, asparagines, and alanine. Interestingly, while poly-Q repeats are found in vertebrates and nonvertebrates, poly-N repeats are only common in more primitive nonvertebrate organisms, such as insects and nematodes. We have assigned function to eukaryote RCPs using Online Mendelian Inheritance in Man (OMIM), the Human Reference Protein Database (HRPD), FlyBase, and Wormpep. Prokaryote RCPs were annotated using BLASTp searches and Gene Ontology. These data reveal that the majority of RCPs are involved in processes that require the assembly of large, multiprotein complexes, such as transcription and signaling.
Single amino acid repeats are regions within proteins that comprise a single homopolymeric tract of a particular amino acid. Uncontrolled genetic expansions of such regions have been shown to lead to the development of serious debilitating human diseases. For example, expanded poly-Q and poly-A tracts are associated with the development of neurological disorders such as Huntington disease and Oculopharyngeal Muscular Dystrophy (OPMD), respectively. Several studies have also demonstrated that many nondisease-linked polyamino acid tracts are toxic to cells and/or lead to protein aggregation or misfolding (Dorsman et al. 2002; Fandrich and Dobson 2002).
Of the polyamino acid repeats characterized to date, poly-Q repeats are the most extensively studied. Nine poly-Q-linked diseases have been identified, and the proteins believed to be responsible for the disease contain expanded poly-Q tracts that have been shown to possess an enhanced tendency to aggregate and form fibrils both in vitro and in vivo (Scherzinger et al. 1997; Skinner et al. 1997; Becher et al. 1998; Holmberg et al. 1998; Li et al. 1998; Warrick et al. 1998; Chow et al. 2004c). The pathogenic length of the poly-Q tract appears to be specific to each protein family (Cummings and Zoghbi 2000). For example, Huntington's disease only develops when the poly-Q repeat within the Huntingtin protein is 38 amino acids (generally encoded by 36 CAG repeats + CAA + CAG). In contrast, Machado-Joseph disease develops when the poly-Q repeat in ataxin-3 is 45 amino acids in length (Cummings and Zoghbi 2000; Chow et al. 2004a). The accumulation of the aggregated state in vivo appears to correlate with cell death and the onset of degenerative disease; however, the mechanism of poly-Q toxicity remains controversial. While the aggregated or fibrillized conformation of poly-Q tracts is postulated to be β-sheet rich, the precise structure and mechanism of fibril formation remains to be characterized (Wetzel 2002; Chow et al. 2004b). Furthermore, the function (if any) of a typical poly-Q tract remains to be determined. In contrast, studies focusing on the function of poly-Q RCPs have revealed interesting functional patterns; an investigation of Drosophila melanogaster poly-Q RCPs revealed that many of these proteins are transcription factors involved in development (Karlin and Burge 1996; Alba and Guigo 2004).
More recently, proteins containing expanded alanine tracts have been linked to several human diseases (Brown and Brown 2004). Like poly-Q RCPs, many of the disease-linked alanine RCPs are transcription factors (Brown and Brown 2004), and proteins containing lengthened alanine tracts (>10) also demonstrate an enhanced tendency to aggregate and form fibrils (Fan et al. 2001). Structural studies have revealed that while short polyalanine peptides form α-helices (Giri et al. 2003), the secondary structure of longer polyalanine tracts is predicted to be predominantly β-strand like (Giri et al. 2003). It is thus suggested that longer alanine-tracts possess an enhanced tendency to form β-sheet-rich fibrillar structures.
Several other repeat types have also been investigated. A polyglycine tract in the plant protein Toc-75 (a component of the protein import machinery in the chloroplast) has been shown to be important for targeting this protein to the outer envelope of the chloroplast (Inoue and Keegstra 2003) and several viral polyarginine-rich proteins have been shown to be involved in RNA binding (Calnan et al. 1991; Nam et al. 2001). Finally, a recent biochemical study (Oma et al. 2004) revealed that long amino acid tracts of almost all types possess a general tendency to aggregate.
To date, the role of many amino acid repeats and RCPs remains somewhat obscure, and it is likely that numerous disease-linked RCPs remain to be identified. To begin to address this problem, a global genome survey has been performed to identify all homopeptide RCPs; these data have been stored in an online database. Resources such as Online Mendelian Inheritance in Man (OMIM) and FlyBase were used to map function onto eukaryote RCPs. BLASTp and Gene Ontology were used to functionally annotate where possible prokaryote RCPs. When considered as a whole, striking functional patterns, independent of amino acid type, can be observed across all RCPs; these data reveal that the majority of RCPs perform roles in processes that require the assembly of large multiprotein or protein/nucleic acid complexes.
Results
A Web-accessible database of RCPs
We identified all homopeptide repeats in GENPEPT greater than six amino acids in length; these data are available at http://repeats.med.monash.edu.au. The homepage includes a table listing the number of RCPs for each amino acid type. A variety of search options are available (accession numbers, description of the protein, etc.) and searches based upon a repeat pattern (for example, poly-A followed by poly-Q) within a single RCP can be performed for multirepeat-containing proteins. A graphical display of the results is presented and all proteins are linked to their National Centre for Biotechnology Information (NCBI) record and an OMIM record, if applicable.
Within GENPEPT (2,677,049 proteins) 1.4% of proteins are RCPs; a total of 54,566 homopeptide repeats could be identified in 37,355 RCPs (Table 1; Fig. 1A). RCPs from environmental sequences (Venter et al. 2004) and viral sequences are listed in Table 1; however, these have not been further considered in this present study.
Table 1.
Number of homopeptide repeats and RCPs in GENPEPT, Eukaryotes, and Prokaryotes
|
GENPEPT
|
Eukaryote
|
Prokaryote
|
Other (viruses/environmental sequences)
|
|||||
|---|---|---|---|---|---|---|---|---|
| Repeats | Proteins | Repeats | Proteins | Repeats | Proteins | Repeats | Proteins | |
| Alanine | 6132 | 5045 | 5465 | 4425 | 251 | 250 | 416 | 370 |
| Valine | 149 | 117 | 94 | 83 | 9 | 9 | 46 | 25 |
| Leucine | 1638 | 1602 | 1446 | 1426 | 70 | 70 | 122 | 106 |
| Isoleucine | 57 | 56 | 34 | 33 | 3 | 3 | 20 | 20 |
| Proline | 4837 | 3931 | 4157 | 3333 | 217 | 184 | 463 | 414 |
| Methionine | 27 | 22 | 19 | 18 | 0 | 0 | 8 | 4 |
| Phenylalanine | 196 | 186 | 175 | 172 | 1 | 1 | 20 | 13 |
| Tryptophan | 3 | 3 | 3 | 3 | 0 | 0 | 0 | 0 |
| Glycine | 5981 | 5020 | 5002 | 4168 | 310 | 281 | 669 | 571 |
| Serine | 6383 | 5463 | 5424 | 4742 | 378 | 258 | 581 | 463 |
| Threonine | 2997 | 2415 | 2492 | 1984 | 63 | 59 | 442 | 372 |
| Cystine | 64 | 52 | 38 | 38 | 0 | 0 | 26 | 14 |
| Asparagine | 7126 | 3731 | 6962 | 3597 | 31 | 29 | 133 | 105 |
| Glutamine | 8334 | 5699 | 8022 | 5464 | 52 | 51 | 260 | 184 |
| Tyrosine | 56 | 51 | 39 | 38 | 4 | 4 | 13 | 9 |
| Aspartic Acid | 1835 | 1707 | 1554 | 1451 | 34 | 34 | 247 | 222 |
| Glutamic Acid | 4779 | 4302 | 4334 | 3912 | 67 | 61 | 378 | 329 |
| Lysine | 2081 | 1926 | 1920 | 1774 | 25 | 25 | 136 | 127 |
| Arginine | 751 | 714 | 462 | 443 | 60 | 57 | 229 | 214 |
| Histidine | 1140 | 1061 | 1049 | 971 | 32 | 32 | 59 | 58 |
| Total | 54,566 | 37,355 | 48,691 | 32,628 | 1607 | 1388 | 4268 | 3339 |
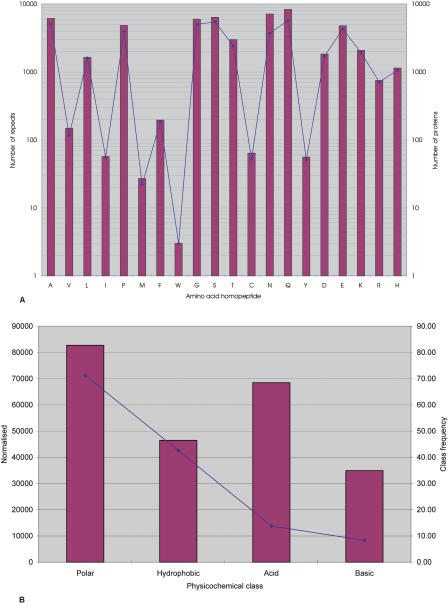
Figure 1.
Distribution of RCPs in GENPEPT. (A) The distribution of the RCPs in GENPEPT and the total number of repeats in GENPEPT. The bars represent the total number of repeats and the solid diamonds the number of RCPs. (B) Distribution of the repeats based on physicochemical class (polar, hydrophobic, acidic, and basic). Red bars represent the number of repeats for the amino acid class normalized for the amino acid frequencies in GENPEPT for that amino acid class (i.e., number of repeats in class X/[amino acid frequency for class X, not including the RCPs]). The solid blue diamonds represent frequency of the amino acid class in the RCPs.
Several general trends are apparent across all the data. The vast majority (87%) of all RCPs are from eukaryotes; prokaryote RCPs are rare (4%) (Table 1). This is in agreement with previous studies (Karlin and Burge 1996; Marcotte et al. 1999; Huntley and Golding 2000). Within GENPEPT, poly-Q repeats are the most common (16%), whereas poly-W repeats are extremely rare (only three poly-W RCPs could be identified) (Fig. 1A). In eukaryotes, poly-Q, poly-N, poly-A, poly-S, and poly-G are the most common repeat types. In prokaryotes, poly-S, poly-G, poly-A, and poly-P are most common; however, poly-Q and poly-N repeats are relatively rare. In both eukaryotes and prokaryotes, Poly-I, Poly-M, Poly-W, Poly-C, and Poly-Y repeats are either absent or very rare.
When classified according to their physicochemical properties and normalized for the overall frequency of single amino acids within GENPEPT, there is an overrepresentation of polar repeats in comparison to hydrophobic repeats and of acidic repeats in comparison to basic repeats (Fig. 1B). These data are in agreement with a previous study that suggested that long stretches of hydrophobic residues possess greatly enhanced toxicity in comparison to similar stretches of hydrophilic residues (Dorsman et al. 2002; Oma et al. 2004) and are thus selected against.
Homopeptide length
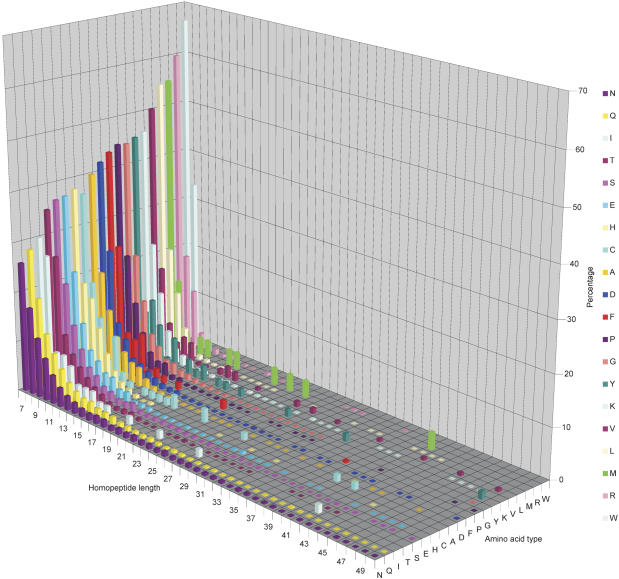
Figure 2 shows a graph of homopeptide repeat length against frequency for each amino acid type. Several general trends are apparent. Most hydrophobic or rare repeats (poly-I, poly-F, poly-Y, poly-V, poly-L, poly-M, poly-R, and poly-W) tend be short (<15 amino acids in length). In contrast, more extensive repeats of amino acids such as poly-Q, poly-N, poly-T, poly-S, and poly-E are common. Certain very long (>50 amino acids) repeats can be identified; however, these are very rare (148 repeats).
Figure 2.
Length of homopeptide repeats. Three-dimensional plot showing repeat length (x-axis) versus amino acid type (y-axis; also highlighted in key) versus percentage of each repeat class of a particular length limited to those repeats <51 amino acids in length. A blank square indicates that no repeat of that length and type exists in GENPEPT. There are repeats >50 amino acids in length; however, these are infrequent, with lengths up to 410 amino acids and a sporadic distribution.
Proteins with more than one homopeptide repeat
Within GENPEPT, 23% of all RCPs contain more than one repeat tract (Table 1). In eukaryotes, 24% of RCPs contain multiple repeat tracts and only 9% of proteins in prokaryotes (two archaeal and 113 bacterial proteins) are multirepeat-containing proteins.
The most common pattern in GENPEPT after a single amino acid repeat is the doublet GG (752) followed by QQ (736), PP (596), SS (505), and NN (485). The propensity of one repeat type to occur with another in the same protein was investigated. Repeat pairs were tallied according to the number of related sequence families in which they were found; Table 2 shows the frequency with which a repeat of one type occurs with another. Strikingly, for all repeats except poly-L, poly-R, and poly-V, the strongest association was with either poly-N or poly-Q tracts (excluding self–self pairs).
Table 2.
Frequency of repeat pairing
| Amino acid (no. of proteins) | % | Self | Decreasing frequency (%, first column) of pairs (<5% not shown) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A (1117) | 18.7 | A | 22.4 | Q | 15.4 | G | 12.6 | S | 7.8 | P | 5.0 | N | ||||||
| D (511) | 15.3 | D | 20.9 | N | 12.0 | E | 10.2 | Q | 8.0 | T | 7.6 | S | 7.6 | G | 6.5 | A | ||
| E (681) | 28.9 | E | 9.8 | Q | 9.6 | N | 9.0 | D | 8.1 | S | 6.3 | P | 6.0 | A | 5.6 | K | 5.5 | T |
| F (22) | 13.6 | F | 22.7 | N | 13.6 | K | 9.1 | D | 9.1 | L | 9.1 | P | ||||||
| G (1126) | 28.0 | G | 16.1 | Q | 15.2 | A | 11.3 | S | 7.6 | P | 5.8 | N | ||||||
| H (309) | 8.9 | H | 21.5 | Q | 14.2 | A | 12.4 | S | 12.3 | N | 10.4 | G | 7.2 | T | 5.2 | P | ||
| K (382) | 23.8 | K | 36.0 | N | 10.0 | E | 8.5 | S | 6.0 | D | ||||||||
| L (96) | 11.5 | L | 16.7 | A | 14.6 | E | 13.5 | Q | 9.4 | P | 7.3 | G | 6.2 | S | ||||
| N (2098) | 37.2 | N | 15.8 | Q | 12.6 | T | 9.9 | S | 6.6 | K | 5.1 | D | ||||||
| P (718) | 28.3 | P | 13.0 | Q | 12.1 | A | 11.9 | G | 10.0 | S | 6.0 | E | ||||||
| Q (2172) | 30.3 | Q | 15.2 | N | 11.5 | A | 11.0 | S | 8.8 | T | 8.8 | G | ||||||
| R (126) | 15.1 | R | 14.3 | G | 11.1 | P | 11.1 | E | 10.3 | S | 8.3 | A | 7.1 | T | 5.2 | D | ||
| S (1460) | 22.1 | S | 16.3 | Q | 14.3 | N | 10.8 | T | 9.7 | A | 8.7 | G | ||||||
| T (1064) | 20.3 | T | 24.8 | N | 17.9 | Q | 14.8 | S | ||||||||||
| V (21) | 28.6 | V | 19.0 | S | 9.5 | A | 9.5 | P | 9.5 | R | ||||||||
Distribution of repeats within eukaryote organisms
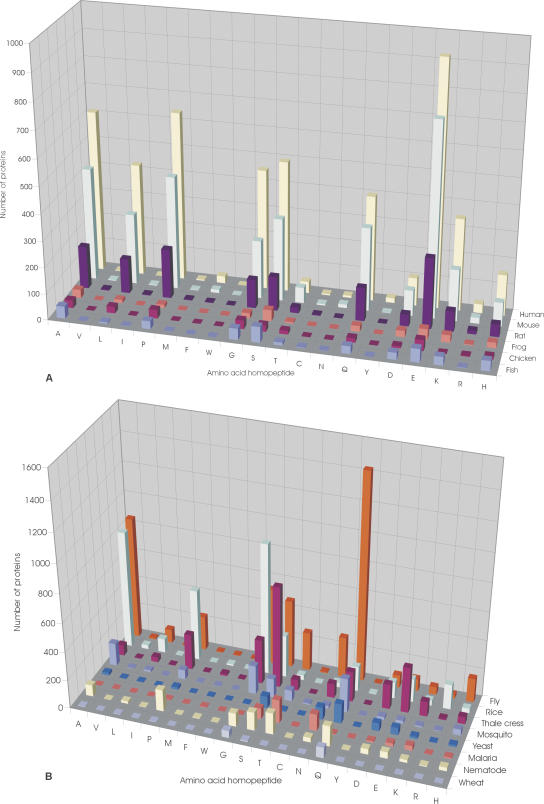
Figure 3, A and B, shows the distributions of RCPs in eukaryotes whose genomes are either complete or near completion. These data highlight several interesting anomalies. Drosophila melanogaster possesses an overabundance of poly-Q RCPs, >3.5-fold more than that of Homo sapiens and sixfold more than another insect, the mosquito Anopheles gambiae (Fig. 3B). In contrast, poly-Q repeats are extremely rare in Plasmodium falciparum; this organism instead possesses an overabundance of poly-N RCPs. Analysis of other complete eukaryote genomes revealed that poly-Q repeats are absent in Encephalitozoon cuniculi (an intracellular parasite). Another striking difference is in the distribution of poly-N RCPs. Nonvertebrate organisms all contain asparagine RCPs, whereas poly-N tracts are either absent or extremely rare in vertebrates (Fig. 3A). The human genome contains 233 poly-Q RCPs, but only eight poly-N RCPs, all of which are 8-residue repeats in the N terminus of the insulin receptor substrate 2. The genome of Mus musculus contains 170 poly-Q RCPs and only 13 poly-N RCPs (seven from thioredoxin interacting factor, one in the insulin receptor substrate 2, one in a transcription factor, and four in unknown proteins). The genomes of Gallus gallus and Xenopus laevis do not contain asparagine RCPs.
Figure 3.
Distribution of RCPs in eukaryotes. (A) Distribution of the RCPs in vertebrate species, Homo sapiens (human), Mus musculus (mouse), Rattus norvegicus (rat), Xenopus laevis (frog), Danio rerio (fish), and Gallus gallus (chicken). (B) Distribution of RCPs in nonvertebrate species, Drosophila melanogaster (fly), Oryza sativa (rice), Arabidopsis thaliana (thale cress), Saccharomyces cerevisiae (yeast), Plasmodium falciparum (malaria), Anopheles gambiae str. PEST (mosquito), Caenorhabditis elegans (nematode), and Triticum aestivum (wheat). The genomes of the chosen species have been completed or are near completion.
In order to include an avian representative in our analysis, we also examined the distribution of RCPs in the chicken. These data reveal an apparent paucity of RCPs in G. gallus as compared with H. sapiens and M. musculus (Fig. 3A); however, we cannot exclude the possibility that this observation is a result of the preliminary nature of the available genomic data. Finally, we note that repeats are completely absent in the nucleomorph of Guillardia theta (Chromophyte algae).
Evolution of RCPs
In order to investigate the evolutionary context of RCPs, prokaryote RCPs were clustered into 1435 families (www.genome.org). Of these families, the majority (1056) are “orphan” RCPs (i.e., a single member). In order to search for eukaryote homologs of RCPs, PSI-BLAST searches of GENPEPT using probes from all clusters with more than five members (47) and eight randomly chosen clusters with more than five members (a total of 55 clusters) were performed (Table 3). The largest cluster considered contains 117 members from the xylanase family. Eukaryote putative homologs of this family were identified, and these putative homologs did not contain the repeat tract. Three other large families, the Ribonuclease E (25 members), membrane carboxypeptidase (25 members), and the β-propeller domain of methanol dehydrogenase (23 members), were also identified. Eukaryote homologs of the Ribonuclease family did not contain repeat tracts, and no convincing eukaryote putative homolog of methanol dehydrogenase could be identified. These results are summarized in Table 3. We were able to detect 20 prokaryote RCP families that extended into eukaryotes. Of these, only three families contained analogous repeat regions or an “interrupted” single amino acid-rich repeat tract in both prokaryote and eukaryote members, the heat-shock protein DnaJ (a molecular chaperone; HSP40) (Fig. 4), and the two ribosomal proteins L10 and L12 (Figs. 5 and 6).
Table 3.
RCP's representatives from the prokaryota clusters with more than four members and the seven randomly chosen clusters with less than five members
| gi accession | Species | Repeat type (length) | Number of members in the cluster | Protein/function | |
|---|---|---|---|---|---|
| 26987138 | Pseudomonas putida | G(7) | 5 | Hypothetical protein | Unknown | |
| 2983200 | Aquifex aeolicus | A(7) & R(9) | * | 25 | Ribonuclease E |
| 33602211 | Bordetella bronchiseptica | P(11) | 8 | Proline-rich inner membrane protein | Unknown | |
| 33331898 | Mycoplasma hyopneumoniae | Q(13) | 5 | P216 surface protein | Unknown | |
| 32473910 | Rhodopirellula baltica | G(7) | * | 6 | Polyribonucleotide nucleotidyltransferase | RNA/DNA binding |
| 32474384 | R. baltica | R(7) | 10 | Probable serine/threonine-protein kinase | Enzyme | |
| 27375632 | Bradyrhizobium japonicum | P(7) & P(8) & P(7) | 11 | Hypothetical protein blr0521 | Unknown | |
| 33864553 | Synechococcus sp. WH 8102 | G(7) | 9 | RNA-binding region RNP-1 | RNA binding | |
| 27378819 | B. japonicum | S(7) | 7 | Efflux protein | Transport | |
| 29830831 | Streptomyces avermitilis | G(7) | 6 | Putative single-stranded DNA-binding protein | DNA binding | |
| 34499217 | Chromobacterium violaceum | E(7) | 8 | RNA polymerase σ factor RpoD | Transcription | |
| 32041487 | Pseudomonas aeruginosa | S(17) | 23 | β-propeller domains of methanol dehydrogenase type | Enzyme | |
| 27367908 | Vibrio vulnificus | Q(89) | 5 | TPR repeat containing protein | Unknown | |
| 23015533 | Magnetospirillum magnetotacticum | G(7) | *+ | 9 | DnaJ-class molecular chaperone with C-terminal Zn finger domain |
| 23129192 | Nostoc punctiforme | G(8) | 13 | Periplasmic protein TonB, links inner and outer membranes | Structural | |
| 20094262 | Methanopyrus kandleri | E(7) & E(10) | *+ | 9 | Ribosomal protein L10 | Translation |
| 16124685 | Caulobacter crescentus | A(7) | 6 | Methyl-accepting chemotaxis protein McpA | Signaling | |
| 23027882 | Microbulbifer degradans | G(7) | 5 | Cation/multidrug efflux pump | Transport | |
| 23472931 | Pseudomonas syringae | A(7) & A(7) | * | 5 | Tfp pilus assembly protein FimV | Structural |
| 21226168 | Methanosarcina mazei | G(7) | 13 | Hypothetical protein MM0066 | Unknown | |
| 4887174 | Neisseria meningitidis | T(7) | 5 | Porin | |
| 22991761 | Enterococcus faecium | S(9) | * | 19 | Muramidase (flagellum-specific) | Enzyme |
| 15839665 | Mycobacterium tuberculosis | A(8) | 10 | PPE family protein | Unknown | |
| 399792 | Erwinia chrysanthemi | S(7) | 9 | PECTIC ENZYMES SECRETION PROTEIN OUTD | Transport | |
| 22968935 | Rhodospirillum rubrum | P(7) | * | 10 | Mg-chelatase subunit ChII | Enzyme |
| 22125459 | Yersinia pestis | G(7) & S(8) | 13 | Hypothetical | Unknown | |
| 580740 | Azotobacter vinelandii | A(8) | 6 | Dihydrolipoyltransacetylase | Enzyme | |
| 8163676 | Streptococcus pneumoniae | S(7) | 7 | Hypothetical protein Rv3088 U | |
| 7481905 | Streptomyces sp. | A(7) | 7 | Polyketide synthase | Enzyme | |
| 14602144 | Aeropyrum pernix | G(7) | 10 | Hypothetical protein APE2556 | |
| 23050783 | Methanosarcina barkeri | E(14) | 10 | CO dehydrogenase/acetyl-CoA synthase β subunit | Enzyme | |
| 23051653 | M. barkeri | E(7) | *+ | 5 | Ribosomal protein L12E | Translation |
| 22968174 | R. rubrum | G(9) | * | 8 | Membrane protease subunits, stomatin/prohibitin homologs | Enzyme |
| 15604160 | Rickettsia prowazekii str. Madrid E | P(8) | 7 | VIRB10 protein (virB10) U | |
| 17549401 | Ralstonia solanacearum | G(7) | * | 117 | Xylanase | Enzyme |
| 23465971 | Bifidobacterium longum | A(7) | 7 | Cell division protein FtsK-DNA translocase | DNA binding | |
| 15806123 | Deinococcus radiodurans | P(8) | 7 | ABC transporter, ATP-binding protein, EF-3 family | Transport | |
| 20149845 | Archaeoglobus fulgidus | A(32) | 7 | Chain B, Reverse Gyrase | DNA binding | |
| 29840119 | Chlamydophila caviae | S(7) | * | 16 | ATP-dependent Clp protease, ATP-binding subunit ClpC | Enzyme |
| 11095653 | Treponema pallidum | A(8) | 15 | TprK U | Unknown | |
| 7442918 | Mycoplasma pneumoniae | S(7) | 8 | Adhesin P1, group 2 variant precursor | Adhesion | |
| 15639317 | Treponema pallidum subsp. pallidum str. Nichols | S(11) | * | 7 | Outer membrane protein | Unknown |
| 7443057 | Salmonella typhimurium | P(9) | 8 | Virulence-associated protein mkaA | Unknown | |
| 22979423 | Ralstonia metallidurans | H(7) | * | 7 | Putative GTPases | Enzyme |
| 23037072 | Oenococcus oeni | S(8) | 25 | Membrane carboxypeptidase | Enzyme | |
| 21231078 | Xanthomonas campestris pv. campestris str. ATCC 33913 | G(13) | * | 6 | Unknown acidic aa rich protein | Unknown |
| 19745410 | Streptococcus pyogenes | G(9) | * | 4 | Hypothetical protein spyM18_0265 | Unknown |
| 23108546 | Novosphingobium aromaticivorans | P(12) | * | 4 | Translation initiation factor 2 | Translation |
| 15596494 | Pseudomonas aeruginosa | H(14) | * | 2 | Probable metal transporter | Transport |
| 548937 | Clostridium thermocellum | G(19) | * | 2 | Cell surface glycoprotein 1 precursor | Adhesion |
| 22978658 | Ralstonia metallidurans | G(7) | * | 2 | Translation initiation factor 2 | Translation |
| 15606084 | Aquifex aeolicus | H(7) | * | 2 | Hydrogenase expression/formation protein B | Enzyme |
| 15922873 | Sulfolobus tokodaii | T(17) | * | 2 | Hypothetical protein ST2539 | Transport |
Putative eukaryote homologs, (+) the putative eukaryote homologs contain the repeat or an equivalent amino acid rich region
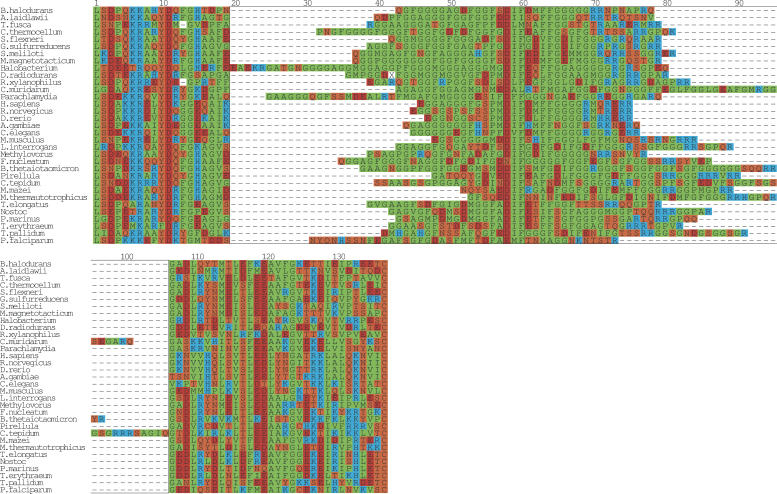
Figure 4.
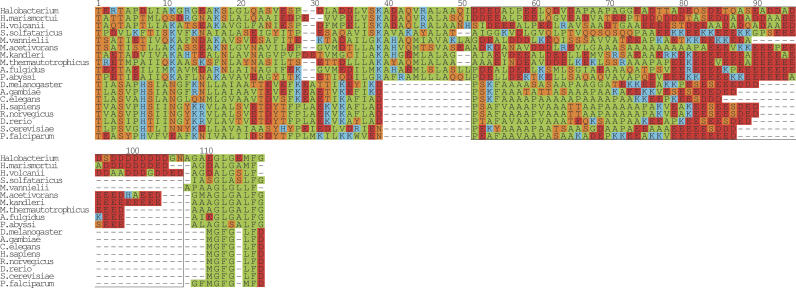
Multiple sequence alignment of DnaJ. A multiple alignment of the glycine repeat in DnaJ (Hsp40). From prokarya: Bacillus halodurans, Clostridium thermocellum, Sinorhizobium meliloti, Shigella flexneri, Magnetospirillum magnetotacticum, Geobacter sulfurreducens PCA, Leptospira interrogans serovar lai str. 56601, Thermosynechococcus elongatus BP-1, Nostoc sp. PCC 7120, Prochlorococcus marinus, Trichodesmium erythraeum IMS101, Methanosarcina mazei Goe1, Treponema pallidum, Rubrobacter xylanophilus DSM 9941, Fusobacterium nucleatum subsp. polymorphum, Methanothermobacter thermautotrophicus str. Delta H, Pirellula sp., Chlamydia muridarum, Parachlamydia sp. UWE25, Bacteroides thetaiotaomicron VPI-5482, Acholeplasma laidlawii, Chlorobium tepidum TLS, Thermobifida fusca, Halobacterium sp. NRC-1, and Deinococcus radiodurans. From eukarya: H. sapiens, R. norvegicus, M. musculus, D. rerio, A. gambiae str. PEST, C. elegans, and P. falciparum. The boxed region from positions 20–106 highlights the glycine/phenylalanine-rich region. The sequences were manually positioned with respect to the first instance of the highly conserved motif DxF (boxed, position 58–60). The regions to the left and right of the boxed region were aligned with T-COFFEE (Notredame et al. 2000) and the final figure was generated with ALSCRIPT (Barton 1993).
Figure 5.
Multiple sequence alignment of the ribosomal protein L12. A multiple sequence alignment of the glutamic acid repeat in L12 from the prokaryote species M. thermautotrophicus str. Delta H, Archaeoglobus fulgidus DSM 4304, Pyrococcus abyssi, M. kandleri AV19, Methanosarcina barkeri, Methanosarcina mazei Goe1, Halobacterium sp. NRC-1, Haloarcula marismortui, Haloferax volcanii, Nanoarchaeum equitans Kin4-M, Ferroplasma acidarmanus, Sulfolobus solfataricus, Thermoplasma acidophilum, and the eukaryote species M. musculus, H. sapiens, R. norvegicus, D. rerio, C. elegans (A) gi 25141400, (B) gi 17543850, S. cerevisiae (A) gi 171813, (B) gi 171815, (C) gi 236358, O. sativa, Eremothecium gossypii, P. falciparum. The boxed region from positions 98–144 highlights the two amino acid-rich regions, the N-terminal alanine-rich region and the C-terminal glutamic acid-rich region. Since no obvious alignment could be built of this region, the sequences were flushed left. The regions to the left and right of the boxed region was aligned with T-COFFEE (Notredame et al. 2000) and then manually adjusted. The final figure was generated with ALSCRIPT (Barton 1993).
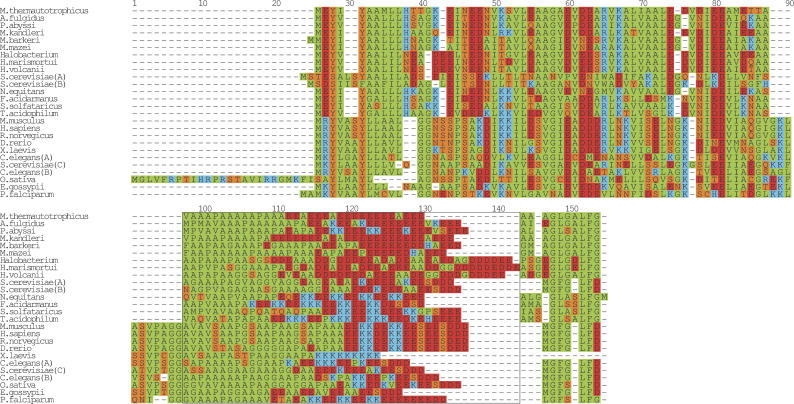
Figure 6.
Multiple sequence alignment of the ribosomal protein L10. A multiple alignment of the glutamic acid-rich region in L10 from the following prokaryote species: Halobacterium sp. NRC-1, Haloarcula marismortui, Haloferax volcanii, Sulfolobus solfataricus, M. vannielii, Methanosarcina acetivorans str. C2A, M. kandleri AV19, M. thermautotrophicus str. Delta H, Archaeoglobus fulgidus DSM 4304, P. abyssi, and the eukaryote species H. sapiens, R. norvegicus, D. rerio, A. gambiae str. PEST, C. elegans, P. falciparum. The boxed region from positions 52–117 highlights the acidic tail. Since no obvious alignment could be built of this region, the sequences were flushed left. The regions to the left and right of the boxed region was aligned with T-COFFEE (Notredame et al. 2000) and then manually adjusted. The final figure was generated with ALSCRIPT (Barton 1993).
Functional groups in H. sapiens, D. melanogaster, C. elegans, and prokaryote RCPs
We used the OMIM database and related resources to functionally group human RCPs. (Fig 7A). Sixty percent of the human RCPs have an OMIM record, and 120 diseases are associated with these records (Supplemental Table 1). In addition, all D. melanogaster RCPs were mapped onto FlyBase (FlyBase Consortium 2003). These were functionally grouped using Gene Ontology (GO) (Fig. 7B; Supplemental Table 2). These data reveal that the majority (85%) of D. melanogaster RCPs are intracellular proteins. Of the remainder, 13% are predicted to be membrane associated, and only 2% are predicted to be extracellular.
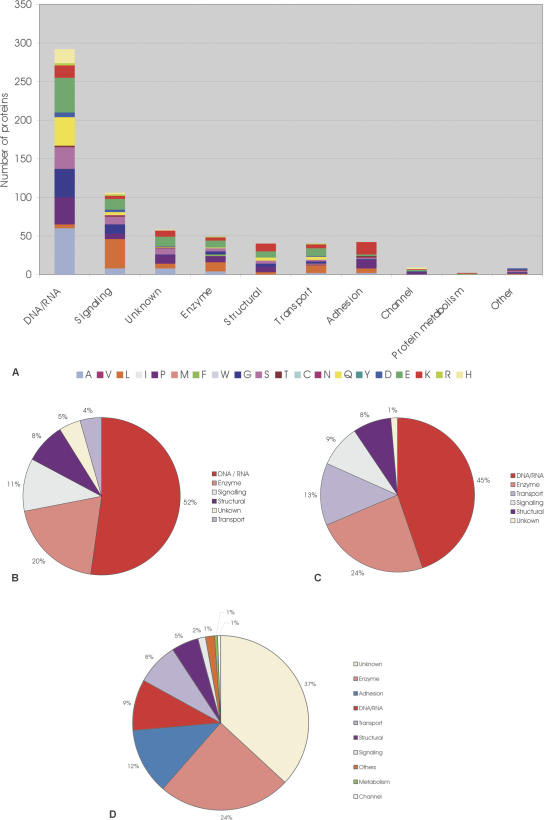
Figure 7.
Function of RCPs. (A) Bar graph showing the function of human RCPs, based upon OMIM. Amino acid types are represented by different colors (see key). On the x-axis, the functional classes DNA RNA (i.e., Transcription/chromatin binding/DNA binding/RNA binding/translation), signaling, unknown, Enzyme, structural, transport protein, adhesion, channel, metabolism, and other are shown. (B) Pie chart showing the function of D. melanogaster RCPs, (C) pie chart showing the function of C. elegans RCPs, (D) pie chart showing the function of prokaryote RCPs.
Interestingly, clear functional trends are apparent throughout the data set, the majority of both human and fruit-fly RCPs performing roles in transcription/translation and signaling processes. Enzymes, transport proteins, adhesion proteins, and structural proteins also commonly contain homopeptide repeats. We performed a similar analysis of C. elegans RCPs using Wormpep, and observed similar trends (Fig. 7C). Finally, we functionally annotated, where possible, prokaryote RCPs (Fig. 7D).
Discrete domains within RCPS
In order to investigate the functional context of homopeptide repeats within RCPs, we performed a survey of protein domains associated with repeat tracts, using the OMIM data set. Ninety-three percent of RCPs listed in OMIM contained a domain listed in pfam (Bateman et al. 2004) and/or SMART (Letunic et al. 2004). Of these, 43% contained an N-terminal repeat in relation to a characterized domain, 10% contained a repeat between domains, and 30% contained a C-terminal repeat in relation to a characterized domain. Also, 16% of RCPs contained a repeat within a characterized domain. Many RCPs are transcription factors or involved in transcriptional processes; hence, many repeat tracts are associated with a variety of nucleic acid-binding domains. However, no obvious pattern of association between repeat type and precise domain type could be detected.
Discussion
Our data reveal that RCPs are far more abundant in eukaryotes than in prokaryotes. In addition, based upon analysis of the D. melanogaster data set, the majority of eukaryote RCPs are predicted to be intracellular proteins. Furthermore, in agreement with the studies of Marcotte et al. (1999), there is an overrepresentation of polar repeats in comparison to hydrophobic repeats, and most hydrophobic repeats are relatively short in comparison to their polar counterparts. These data are consistent with previous studies that suggest hydrophobic RCPs aggregate and form toxic fibrils (Dorsman et al. 2002; Oma et al. 2004).
Glycine, serine, and proline repeats are common in both prokaryotes and eukaryotes; however, common eukaryote repeats such as glutamine, asparagines, and glutamic acid are relatively rare in prokaryote organisms; of 29 asparagine RCPs in prokaryotes, 11 are orphans, 12 do not have eukaryotic homologs, and six have putative eukaryote homologs. However, these homologs do not contain the repeat or an equivalent amino acid-rich region. Of the 51 glutamine RCPs in prokaryotes, 23 are orphans, 21 do not have eukaryotic homologs, and seven have putative eukaryote homologs, but again, these homologs do not contain a repeat or an equivalent amino acid-rich region.
Certain discrepancies are clearly apparent when considering repeat distribution within eukaryotes. For example, glutamine RCPs (the most common eukaryote repeat) are rare or absent in P. falciparum, E. cuniculi, and G. theta. Furthermore, our data reveal that while asparagine repeats are common in nonvertebrates such as insects and nematodes, such repeats are extremely rare in vertebrates. Kreil and Kreil (2000) previously observed the rarity of asparagine repeats in mammals. This is surprising, since glutamine and asparagine are chemically and structurally similar. We could not identify any function specific to poly-N repeats; indeed, an analysis of all D. melanogaster poly-N RCPs reveals that, like other RCPs (see below), a large proportion (>60%) are transcription factors or proteins involved in transcriptional processes (Table 2; Supplemental data). Interestingly, in most multirepeat-containing RCPs, glutamine or asparagine is the most likely associated partner polyamino acid (Table 2). The functional significance of these data remains to be understood.
Very rarely, repeats are conserved across entire protein families, and only three families (DnaJ and the ribosomal proteins L10 and L12) could be identified with repeat regions in eukaryote and prokaryote putative homologs. A sequence alignment of the DnaJ family (Fig. 4) reveals that an extensive glycine repeat is present in most putative homologs. However, in the majority of these, the repeat is interrupted (typically with an alanine or phenylalanine residue), and this region is contracted in many eukaryotic counterparts. DnaJ functions in complex with at least two other proteins (DnaK and GrpE) to control processes such as protein folding, apoptosis, and the degradation of misfolded proteins (Gragerov et al. 1992; Hendrick et al. 1993; Craig et al. 1994; Gotoh et al. 2004). Deletion of the glycine/phenylalanine-rich motif decreases the binding affinity of the substrate σ32 to DnaK (Wall et al. 1995), and it has been proposed that this region may act as a flexible linker to allow DnaJ/K complex formation and subsequent activation of DnaK (Wall et al. 1995).
Both archaeal and eukaryotic L10 and L12 proteins contain a C-terminal region that comprises an alanine-rich region, termed the hinge, followed by a glutamic acid-rich repeat (Figs. 5, 6). L10 and L12 form part of a complex in the large ribosomal subunit termed the stalk protuberance. The hinge, as well as the acidic region in both proteins, are postulated to function as flexible regions that mediate a variety of protein–protein interactions and are important for processes such as elongation (Remacha et al. 1995; Uchiumi et al. 2002a,b; Gonzalo and Reboud 2003). The acidic region in L10 and L12 is severely truncated in certain eukaryote orthologs; for example, this region is absent in L12e from X. laevis. Bacterial L12 and L10 proteins do not contain the acidic tail. The absence of this region has been observed previously (Ramirez et al. 1989), and it is suggested that the evolution of this region arose after the archaea/bacteria split, but prior to the emergence of eukaryotes.
A major question in repeat-related research fields is the role of RCPs and, in particular, the role of the repeat region itself. Evolutionary pressure on repeat regions is likely to include functional requirement, mutability of the underlying nucleotide sequence, and potential toxicity. Our analysis allows us to begin to address these questions from a functional perspective. Assigning function to protein sequences is nontrivial, since many proteins perform overlapping functions (for review, see Whisstock and Lesk 2003). However, using the OMIM database and other annotation resources, we have attempted to classify RCPs under as broad a class as possible. Most eukaryote RCPs are involved in transcription/translation or interact directly with DNA, RNA, or chromatin, irrespective of repeat type (Fig. 7A). Other common classes of RCPs include signaling molecules, structural proteins, transport molecules, and enzymes (Fig. 7). Previous research on smaller eukaryote data sets also note that repeat tracts are overrepresented in transcription factors and DNA-binding proteins (Karlin and Burge 1996; Mar Alba et al. 1999; Alba et al. 2002; Alba and Guigo 2004). The results of this study are consistent with these findings and identify other functional classes rich in RCPs.
We performed a functional analysis of RCPs from prokaryotes (Table 3; Fig. 7D). While 38% of these families perform as yet uncharacterized roles, several of the functional themes apparent in the eukaryote data set are also noticeable in prokaryotes. The most common functional class (accounting for 24% of classified molecules) are RCPs that perform roles as enzymes. Transport proteins, structural proteins, and transcription/translation-related RCPs are also common; however, in contrast to eukaryotes, the dramatic bias toward transcription/translation-related processes is not observed. One possible explanation for these data is that bacterial genomes are smaller in relation to eukaryotes and are not packaged and controlled in such a complex fashion. RCPs involved in signaling-related processes are also relatively rare in prokaryotes in comparison to eukaryotes. Again, we argue that this may be a result of the increased complexity of eukaryote processes; while bacteria utilize intracellular signaling processes to communicate intracellularly and with their environment, these processes are relatively modest in comparison to eukaryote-signaling cascades.
The repeats database provides a basis for understanding the function of RCPs and their associated repeats in all organisms. Strikingly, the majority of RCPs considered are involved in processes that require the assembly or association of large multiprotein and/or nucleic acid complexes (Fig. 7). For example, the ribosome (itself a large protein/RNA complex) requires a large number of additional factors (e.g., elongation factors) to properly function. Processes such as transcription involve the assembly of multiprotein complexes (e.g., RNA polymerase) and the binding of discrete sequences of DNA that may be kilobases apart (Tolhuis et al. 2002); chromatin condensation requires bundling and folding of nucleosome arrays by protein factors such as Nucleoplasmin (for review, see Akey and Luger 2003; Grigoryev 2004); signaling requires the assembly of a large number of proteins into a signalosome or transducisome (e.g., the MAP Kinase complex) (for review, see Burack and Shaw 2000) and ion channels such as the cGMP-gated channels in rod photoreceptors are involved in large protein complexes that link the plasma membrane with the outer segment disk (Korschen et al. 1999). In many of these processes, the involvement of repeats in mediating protein–protein interactions have been noted (for example, the glutamic acid-rich proteins of cGMP-gated channels and the acidic tails of ribosomal proteins) (Remacha et al. 1995; Korschen et al. 1999; Poetsch et al. 2001).
The data presented in this study reveal that the vast majority of the human repeat tracts present in the OMIM data set (83%) are located N-terminal to, as well as C-terminal to, or in-between discrete domains. Alba and Guigo (2004) have also noted that certain repeats (L, A, P, and Q) appear to be preferentially located at the N terminus. Structural studies reveal that most repeat regions do not adopt discrete well-ordered structures, and instead, are often disordered (Huntley and Golding 2002).
The function of RCPs, as well as the interdomain or terminal location of the repeat tract within these molecules and the disordered structure of these repeat sequences, supports the idea that repeats play roles as flexible spacer elements/“tethers” between individual folded domains in molecules that mediate protein–protein or protein–nucleic acid interactions (Karlin and Burge 1996).
Based upon this work, it is suggested that a general function of the majority of repeat sequences is to mediate the assembly of protein complexes, and that RCPs may act as molecular “fishing lines”, mediating interactions either through tethered distant domains, or indeed, through interactions with the repeat itself (e.g., Gerber et al. 1994; Korschen et al. 1999). Such a flexible structure would allow a single protein within a complex to recruit additional factors from the cytoplasmic or nuclear milieu (Fig. 8). A repeat tract would also be able to bridge large distances, such as is required in chromatin packaging or transcription. While many specialized proteins are able to achieve both distance and conformational mobility via an ordered three-dimensional fold (e.g., myosin) (Pollard 2000) or the serpin superfamily of proteinase inhibitors (Whisstock et al. 1998), a flexible unstructured region is a simple way of achieving a similar, albeit conformationally uncontrolled, outcome. A repeat does not require complicated molecular machinery to achieve flexibility and, because it is unstructured, appropriately sized repeats would be predicted to interfere minimally with other molecules in a complex.
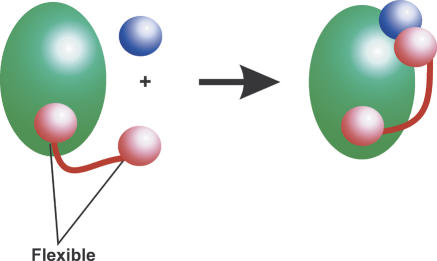
Figure 8.
General role of repeats regions. It is suggested that RCPs (red) function from within large mutiprotein and/or nucleic acid complexes (green circle). An example is shown where a two-domain protein (pink circles) functions via a flexible repeat to recruit an additional binding partner (blue circle).
Several studies have revealed that long stretches of hydrophobic amino acids are more toxic than hydrophilic counterparts (Dorsman et al. 2002; Oma et al. 2004). Thus, in order to bridge large distances, long tracts of polar repeats such as poly-Q, poly-N, and poly-E can be used as suggested by the distribution of repeat length and amino acid type (Fig. 2). The consequence of utilizing single amino acid repeats is that repeat expansion can result in the formation of ordered aggregates or fibrils. Such an event could result in cell death through a variety of mechanisms; the toxic nature of the fibril, loss or gain of function, the destruction of a large essential multiprotein complex, and/or the sequestering of nonspecific factors.
The majority of proteins sequenced to date do not contain repeats. While certain repeats are common throughout entire protein superfamilies (such as the DnaJ family), the data gleaned in this study reveals that repeat proteins are often “orphans.” We suggest that the putative role of repeats is ancient; however, the relatively sporadic distribution of these regions suggests that repeats often evolve to perform in specialized processes unique to a particular organism or set of organisms.
Methods
The March (2004) version of GENPEPT (from ftp://ftp.ncbi.nih.gov/blast/db) was used in this study.
Homopeptide searches
All homopeptides longer than 4 amino acids were discovered using the regular expression “([AaVvLlIiPpMmFfWwGgSsTtCc NnQqYyDdEeKkRrHh])\1+” across the GENPEPT database. This expression will find all occurrences of an amino acid repeat such as “aa,” “aaa,” and “gggg” (Friedl 2002). A minimum length threshold of 7 amino acids was set. The results from the study can be queried and visualized via the Web site http://repeats.med.monash.edu.au.
Evolution of RCPs
All RCPs from prokarya were used in an all-against-all BLASTp search with the following parameters: -f T (on), e = 0.001. Single-linkage clustering of the BLASTp results was then performed using a similar approach to the package GeneRAGE (Enright and Ouzounis 2000). That is, an NxN matrix was created from the BLASTp results and forced into symmetry. Clustering was performed across the matrix.
When considering eukaryote RCPs, only completed or near-complete genomes were used, so as to avoid potential bias due to overrepresentation of commonly studied protein families.
Thus, the following species were considered: Homo sapiens, Mus musculus, Rattus norvegicus, Gallus gallus, Danio rerio, Xenopus laevis, Drosophila melanogaster, Anopheles gambiae, Caenorhabditis elegans, Saccharomyces cerevisiae, Plasmodium falciparum, Oryza sativa, Triticum aestivum, and Arabidopsis thaliana.
The longest sequence from each of the prokaryote clusters was used as probes to search GENPEPT using PSI-BLAST. The following parameters were used: j = 5, b = 100,000, e = 0.001, and -F T. All sequences with significant expect scores (<0.001) (Park et al. 1998) from the sequenced eukaryote organisms (listed above) were selected. Sequences that were >98% identical were then removed using the nrdb90 algorithm (Holm and Sander 1998). For each eukaryote species, the top 10 putative homologs (with the lowest expect scores) were used for the multiple-sequence alignment. The alignments were initially generated using CLUSTALW (Thompson et al. 1994) with default parameters. Inspection of the alignments enabled identification of eukaryote putative homologs that contained a repeat or an equivalent amino acid-rich tract; these were subject to further analysis. The alignment shown in Figures 4, 5, 6 was initially generated with T-Coffee (Notredame et al. 2000), then manually curated.
Analysis of repeat pairs
In order to explore whether certain repeat types have a tendency to appear together within a given protein, the colocalization of repeat pairs was investigated. PSI-BLAST (three iterations, inclusion threshold E < 0.02, reporting threshold E <1 × 10–6) was used to obtain clusters of sequences from a database of multiple repeat-containing proteins, where each pairwise alignment spanned >50% of the length of the smaller sequence. For each unique repeat pair identified in a protein, a score was given as the reciprocal of the number of related proteins; i.e., a protein with the repeats TNNNNNK and related to four others would increment the scores for repeat pairs TN, TK, NN, and NK by 0.25. This was performed for all multiple repeat-containing proteins.
Functional annotation
Existing information within OMIM (http://www.ncbi.nlm.nih.gov/omim/), HPRD (http://www.hprd.org/), FlyBase (FlyBase Consortium 2003), and Wormpep (http://www.sanger.ac.uk/Projects/C_elegans/WORMBASE/current/wormpep.shtml) was used to annotate the human, Drosophila, and nematode data set, respectively. In order to annotate prokaryote sequences, the longest member from each of the 1435 clusters were used as probes in BLASTp searches of GENPEPT (using the default parameters). The results were manually analyzed, and putative general function assigned if a cluster shared significant similarity with a characterized family (only hits with an expect score <0.001 were considered). Literature searches were used to establish general function where required.
Acknowledgments
J.C.W. is a National Health and Medical Research Council of Australia Senior Research Fellow and Monash University Logan Fellow. S.P.B. is an NHMRC R.D. Wright Fellow and Monash University Logan Fellow. J.A.I. is an Anti-Cancer council of Victoria Fellow, Monash University Research Fund Fellow and NHMRC C.J. Martin Fellow. M.G.B. is a Monash University Logan Fellow. We thank the NHMRC, the Australian Research Council, the Victorian Partnership for Advanced Computing, and the State Government of Victoria for support. We thank Sophie Katsabanis for discussion and comment on the manuscript and Michael Cameron, Michelle Dunstone, Sheena McGowan, and Michelle Chow for helpful discussion.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3096505.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Akey, C.W. and Luger, K. 2003. Histone chaperones and nucleosome assembly. Curr. Opin. Struct. Biol. 13: 6-14. [DOI] [PubMed] [Google Scholar]
- Alba, M.M. and Guigo, R. 2004. Comparative analysis of amino acid repeats in rodents and humans. Genome Res. 14: 549-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alba, M.M., Laskowski, R.A., and Hancock, J.M. 2002. Detecting cryptically simple protein sequences using the SIMPLE algorithm. Bioinformatics 18: 672-678. [DOI] [PubMed] [Google Scholar]
- Barton, G.J. 1993. ALSCRIPT: A tool to format multiple sequence alignments. Protein Eng. 6: 37-40. [DOI] [PubMed] [Google Scholar]
- Bateman, A., Coin, L., Durbin, R., Finn, R.D., Hollich, V., Griffiths-Jones, S., Khanna, A., Marshall, M., Moxon, S., Sonnhammer, E.L., et al. 2004. The Pfam protein families database. Nucleic Acids Res. 32: D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becher, M.W., Kotzuk, J.A., Sharp, A.H., Davies, S.W., Bates, G.P., Price, D.L., and Ross, C.A. 1998. Intranuclear neuronal inclusions in Huntington's disease and dentatorubral and pallidoluysian atrophy: Correlation between the density of inclusions and IT15 CAG triplet repeat length. Neurobiol. Dis. 4: 387-397. [DOI] [PubMed] [Google Scholar]
- Brown, L.Y. and Brown, S.A. 2004. Alanine tracts: The expanding story of human illness and trinucleotide repeats. Trends Genet. 20: 51-58. [DOI] [PubMed] [Google Scholar]
- Burack, W.R. and Shaw, A.S. 2000. Signal transduction: Hanging on a scaffold. Curr. Opin. Cell. Biol. 12: 211-216. [DOI] [PubMed] [Google Scholar]
- Calnan, B.J., Tidor, B., Biancalana, S., Hudson, D., and Frankel, A.D. 1991. Arginine-mediated RNA recognition: The arginine fork. Science 252: 1167-1171. [DOI] [PubMed] [Google Scholar]
- Chow, M.K., Ellisdon, A.M., Cabrita, L.D., and Bottomley, S.P. 2004a. Polyglutamine expansion in Ataxin-3 does not affect protein stability: Implications for misfolding and disease. J. Biol. Chem. 279: 47643-47651. [DOI] [PubMed] [Google Scholar]
- Chow, M.K., Lomas, D.A., and Bottomley, S.P. 2004b. Promiscuous β-strand interactions and the conformational diseases. Curr. Med. Chem. 11: 491-499. [DOI] [PubMed] [Google Scholar]
- Chow, M.K., Paulson, H.L., and Bottomley, S.P. 2004c. Destabilization of a non-pathological variant of ataxin-3 results in fibrillogenesis via a partially folded intermediate: A model for misfolding in polyglutamine disease. J. Mol. Biol. 335: 333-341. [DOI] [PubMed] [Google Scholar]
- Craig, E.A., Weissman, J.S., and Horwich, A.L. 1994. Heat shock proteins and molecular chaperones: Mediators of protein conformation and turnover in the cell. Cell 78: 365-372. [DOI] [PubMed] [Google Scholar]
- Cummings, C.J. and Zoghbi, H.Y. 2000. Fourteen and counting: Unraveling trinucleotide repeat diseases. Hum. Mol. Genet. 9: 909-916. [DOI] [PubMed] [Google Scholar]
- Dorsman, J.C., Pepers, B., Langenberg, D., Kerkdijk, H., Ijszenga, M., den Dunnen, J.T., Roos, R.A., and van Ommen, G.J. 2002. Strong aggregation and increased toxicity of polyleucine over polyglutamine stretches in mammalian cells. Hum. Mol. Genet. 11: 1487-1496. [DOI] [PubMed] [Google Scholar]
- Enright, A.J. and Ouzounis, C.A. 2000. GeneRAGE: A robust algorithm for sequence clustering and domain detection. Bioinformatics 16: 451-457. [DOI] [PubMed] [Google Scholar]
- Fan, X., Dion, P., Laganiere, J., Brais, B., and Rouleau, G.A. 2001. Oligomerization of polyalanine expanded PABPN1 facilitates nuclear protein aggregation that is associated with cell death. Hum. Mol. Genet. 10: 2341-2351. [DOI] [PubMed] [Google Scholar]
- Fandrich, M. and Dobson, C.M. 2002. The behaviour of polyamino acids reveals an inverse side chain effect in amyloid structure formation. EMBO J. 21: 5682-5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FlyBase Consortium. 2003. The FlyBase database of the Drosophila genome projects and community literature. Nucleic Acids Res. 31: 172-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl, J.E.F. 2002. Mastering regular expressions. O'Reilly, Sebastopol, CA.
- Gerber, H.P., Seipel, K., Georgiev, O., Hofferer, M., Hug, M., Rusconi, S., and Schaffner, W. 1994. Transcriptional activation modulated by homopolymeric glutamine and proline stretches. Science 263: 808-811. [DOI] [PubMed] [Google Scholar]
- Giri, K., Ghosh, U., Bhattacharyya, N.P., and Basak, S. 2003. Caspase 8 mediated apoptotic cell death induced by β-sheet forming polyalanine peptides. FEBS Lett. 555: 380-384. [DOI] [PubMed] [Google Scholar]
- Gonzalo, P. and Reboud, J.P. 2003. The puzzling lateral flexible stalk of the ribosome. Biol. Cell 95: 179-193. [DOI] [PubMed] [Google Scholar]
- Gotoh, T., Terada, K., Oyadomari, S., and Mori, M. 2004. hsp70-DnaJ chaperone pair prevents nitric oxide- and CHOP-induced apoptosis by inhibiting translocation of Bax to mitochondria. Cell Death Differ. 11: 390-402. [DOI] [PubMed] [Google Scholar]
- Gragerov, A., Nudler, E., Komissarova, N., Gaitanaris, G.A., Gottesman, M.E., and Nikiforov, V. 1992. Cooperation of GroEL/GroES and DnaK/DnaJ heat shock proteins in preventing protein misfolding in Escherichia coli. Proc. Natl. Acad. Sci. 89: 10341-10344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoryev, S.A. 2004. Keeping fingers crossed: Heterochromatin spreading through interdigitation of nucleosome arrays. FEBS Lett. 564: 4-8. [DOI] [PubMed] [Google Scholar]
- Hendrick, J.P., Langer, T., Davis, T.A., Hartl, F.U., and Wiedmann, M. 1993. Control of folding and membrane translocation by binding of the chaperone DnaJ to nascent polypeptides. Proc. Natl. Acad. Sci. 90: 10216-10220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm, L. and Sander, C. 1998. Removing near-neighbour redundancy from large protein sequence collections. Bioinformatics 14: 423-429. [DOI] [PubMed] [Google Scholar]
- Holmberg, M., Duyckaerts, C., Durr, A., Cancel, G., Gourfinkel-An, I., Damier, P., Faucheux, B., Trottier, Y., Hirsch, E.C., Agid, Y., et al. 1998. Spinocerebellar ataxia type 7 (SCA7): A neurodegenerative disorder with neuronal intranuclear inclusions. Hum. Mol. Genet. 7: 913-918. [DOI] [PubMed] [Google Scholar]
- Huntley, M. and Golding, G.B. 2000. Evolution of simple sequence in proteins. J. Mol. Evol. 51: 131-140. [DOI] [PubMed] [Google Scholar]
- ———. 2002. Simple sequences are rare in the Protein Data Bank. Proteins 48: 134-140. [DOI] [PubMed] [Google Scholar]
- Inoue, K. and Keegstra, K. 2003. A polyglycine stretch is necessary for proper targeting of the protein translocation channel precursor to the outer envelope membrane of chloroplasts. Plant J. 34: 661-669. [DOI] [PubMed] [Google Scholar]
- Karlin, S. and Burge, C. 1996. Trinucleotide repeats and long homopeptides in genes and proteins associated with nervous system disease and development. Proc. Natl. Acad. Sci. 93: 1560-1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korschen, H.G., Beyermann, M., Muller, F., Heck, M., Vantler, M., Koch, K.W., Kellner, R., Wolfrum, U., Bode, C., Hofmann, K.P., et al. 1999. Interaction of glutamic-acid-rich proteins with the cGMP signalling pathway in rod photoreceptors. Nature 400: 761-766. [DOI] [PubMed] [Google Scholar]
- Kreil, D.P. and Kreil, G. 2000. Asparagine repeats are rare in mammalian proteins. Trends Biochem. Sci. 25: 270-271. [DOI] [PubMed] [Google Scholar]
- Letunic, I., Copley, R.R., Schmidt, S., Ciccarelli, F.D., Doerks, T., Schultz, J., Ponting, C.P., and Bork, P. 2004. SMART 4.0: Towards genomic data integration. Nucleic Acids Res. 32: D142-D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M., Miwa, S., Kobayashi, Y., Merry, D.E., Yamamoto, M., Tanaka, F., Doyu, M., Hashizume, Y., Fischbeck, K.H., and Sobue, G. 1998. Nuclear inclusions of the androgen receptor protein in spinal and bulbar muscular atrophy. Ann. Neurol. 44: 249-254. [DOI] [PubMed] [Google Scholar]
- Mar Alba, M., Santibanez-Koref, M.F., and Hancock, J.M. 1999. Amino acid reiterations in yeast are overrepresented in particular classes of proteins and show evidence of a slippage-like mutational process. J. Mol. Evol. 49: 789-797. [DOI] [PubMed] [Google Scholar]
- Marcotte, E.M., Pellegrini, M., Yeates, T.O., and Eisenberg, D. 1999. A census of protein repeats. J. Mol. Biol. 293: 151-160. [DOI] [PubMed] [Google Scholar]
- Nam, Y.S., Petrovic, A., Jeong, K.S., and Venkatesan, S. 2001. Exchange of the basic domain of human immunodeficiency virus type 1 Rev for a polyarginine stretch expands the RNA binding specificity, and a minimal arginine cluster is required for optimal RRE RNA binding affinity, nuclear accumulation, and trans-activation. J. Virol. 75: 2957-2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame, C., Higgins, D.G., and Heringa, J. 2000. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302: 205-217. [DOI] [PubMed] [Google Scholar]
- Oma, Y., Kino, Y., Sasagawa, N., and Ishiura, S. 2004. Intracellular localization of homopolymeric amino acid-containing proteins expressed in mammalian cells. J. Biol. Chem. 279: 21217-21222. [DOI] [PubMed] [Google Scholar]
- Park, J., Karplus, K., Barrett, C., Hughey, R., Haussler, D., Hubbard, T., and Chothia, C. 1998. Sequence comparisons using multiple sequences detect three times as many remote homologues as pairwise methods. J. Mol. Biol. 284: 1201-1210. [DOI] [PubMed] [Google Scholar]
- Poetsch, A., Molday, L.L., and Molday, R.S. 2001. The cGMP-gated channel and related glutamic acid-rich proteins interact with peripherin-2 at the rim region of rod photoreceptor disc membranes. J. Biol. Chem. 276: 48009-48016. [DOI] [PubMed] [Google Scholar]
- Pollard, T.D. 2000. Reflections on a quarter century of research on contractile systems. Trends Biochem. Sci. 25: 607-611. [DOI] [PubMed] [Google Scholar]
- Ramirez, C., Shimmin, L.C., Newton, C.H., Matheson, A.T., and Dennis, P.P. 1989. Structure and evolution of the L11, L1, L10, and L12 equivalent ribosomal proteins in eubacteria, archaebacteria, and eucaryotes. Can. J. Microbiol. 35: 234-244. [DOI] [PubMed] [Google Scholar]
- Remacha, M., Jimenez-Diaz, A., Bermejo, B., Rodriguez-Gabriel, M.A., Guarinos, E., and Ballesta, J.P. 1995. Ribosomal acidic phosphoproteins P1 and P2 are not required for cell viability but regulate the pattern of protein expression in Saccharomyces cerevisiae. Mol. Cell. Biol. 15: 4754-4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherzinger, E., Lurz, R., Turmaine, M., Mangiarini, L., Hollenbach, B., Hasenbank, R., Bates, G.P., Davies, S.W., Lehrach, H., and Wanker, E.E. 1997. Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell 90: 549-558. [DOI] [PubMed] [Google Scholar]
- Skinner, P.J., Koshy, B.T., Cummings, C.J., Klement, I.A., Helin, K., Servadio, A., Zoghbi, H.Y., and Orr, H.T. 1997. Ataxin-1 with an expanded glutamine tract alters nuclear matrix-associated structures. Nature 389: 971-974. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis, B., Palstra, R.J., Splinter, E., Grosveld, F., and de Laat, W. 2002. Looping and interaction between hypersensitive sites in the active β-globin locus. Mol. Cell 10: 1453-1465. [DOI] [PubMed] [Google Scholar]
- Uchiumi, T., Honma, S., Endo, Y., and Hachimori, A. 2002a. Ribosomal proteins at the stalk region modulate functional rRNA structures in the GTPase center. J. Biol. Chem. 277: 41401-41409. [DOI] [PubMed] [Google Scholar]
- Uchiumi, T., Honma, S., Nomura, T., Dabbs, E.R., and Hachimori, A. 2002b. Translation elongation by a hybrid ribosome in which proteins at the GTPase center of the Escherichia coli ribosome are replaced with rat counterparts. J. Biol. Chem. 277: 3857-3862. [DOI] [PubMed] [Google Scholar]
- Venter, J.C., Remington, K., Heidelberg, J.F., Halpern, A.L., Rusch, D., Eisen, J.A., Wu, D., Paulsen, I., Nelson, K.E., Nelson, W., et al. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304: 66-74. [DOI] [PubMed] [Google Scholar]
- Wall, D., Zylicz, M., and Georgopoulos, C. 1995. The conserved G/F motif of the DnaJ chaperone is necessary for the activation of the substrate binding properties of the DnaK chaperone. J. Biol. Chem. 270: 2139-2144. [DOI] [PubMed] [Google Scholar]
- Warrick, J.M., Paulson, H.L., Gray-Board, G.L., Bui, Q.T., Fischbeck, K.H., Pittman, R.N., and Bonini, N.M. 1998. Expanded polyglutamine protein forms nuclear inclusions and causes neural degeneration in Drosophila. Cell 93: 939-949. [DOI] [PubMed] [Google Scholar]
- Wetzel, R. 2002. Ideas of order for amyloid fibril structure. Structure 10: 1031-1036. [DOI] [PubMed] [Google Scholar]
- Whisstock, J.C. and Lesk, A.M. 2003. Prediction of protein function from protein sequence and structure. Q Rev. Biophys. 36: 307-340. [DOI] [PubMed] [Google Scholar]
- Whisstock, J., Skinner, R., and Lesk, A.M. 1998. An atlas of serpin conformations. Trends Biochem. Sci. 23: 63-67. [DOI] [PubMed] [Google Scholar]
Web site references
- http://repeats.med.monash.edu.au; A database of homopeptide repeats.
- http://www.hprd.org/; Human Protein Reference Database.
- ftp://ftp.ncbi.nih.gov/blast/db/; NCBI ftp site of available databases.
- http://www.ncbi.nlm.nih.gov/omim/; Online Mendelian Inheritance in Man, OMIM. McKusick-Nathans Institute for Genetic Medicine, Johns Hopkins University (Baltimore, MD) and National Center for Biotechnology Information, National Library of Medicine (Bethesda, MD), 2000.
- http://www.sanger.ac.uk/Projects/C_elegans/WORMBASE/current/wormpep.shtml; Wormpep.