Abstract
The interaction of proteins to form macromolecular complexes is the basis for most biological processes. Approaches have been described that employ artificial constructs to promote such complexes and assess the consequences. For example, a protein interaction scheme has been described that examines the effects of a specific phosphorylation event catalyzed by a protein kinase via the provision of an artificial protein binding interface between a modified version of the kinase and a single substrate. We have generalized this type of approach to form the basis for a genetic selection to identify proteins that exert an activity when recruited to a target protein. The assay uses the leucine zipper domains from the mammalian transcription factors Fos and Jun to force the interaction of two proteins. With a target protein fused to the Jun zipper and a library of open reading frames fused to the Fos zipper, we demonstrate this approach in yeast with both a selection to identify membrane-associated proteins and a selection to identify candidate components of the filamentous growth MAP kinase pathway.
Protein complexes and signaling pathways comprise a physical network that underpins the activities of cells. Formation of these complexes is often highly regulated, with proteins recruited to specific cellular locations to participate in activities that include signal transduction, determination of cellular polarity, and the initiation of transcription or DNA replication. Unregulated recruitment, as can occur with constitutively activated receptors, may lead to oncogenesis (Porter and Vaillancourt 1998). Experimental strategies to artificially generate protein interactions and assess their effects include the use of chemical ligands that induce dimerization (Spencer et al. 1993). Alternatively, provision to two proteins of heterologous domains that interact with each other can be sufficient to reconstitute a process; in yeast, such domains added to a mutant Ste5 scaffolding protein and a downstream protein kinase restored activity of the pheromone signaling pathway (Park et al. 2003). In the approach of Mayer and colleagues (Sharma et al. 2003), complementary coiled coil domains were added to a substrate and to a protein kinase defective for substrate binding in order to ascertain the consequences of tyrosine phosphorylation of that single substrate. This previous use of this type of assay was designed to confirm hypotheses generated by traditional methods. Here, we have expanded this approach to use leucine zippers, which heterodimerize to form a parallel, doubled-stranded coiled-coil (O'Shea et al. 1989), in order to force the interaction of each member of a library with a protein of interest. A library-encoded protein that exerts a specific activity can thereby be identified in a genetic screen.
Results
Leucine zippers result in protein colocalization
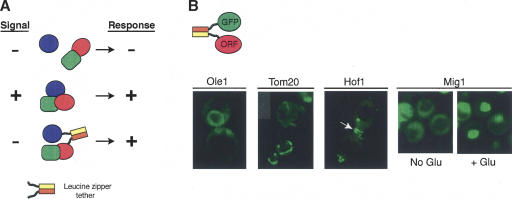
Our approach is described in Figure 1A, using as an example a signaling pathway in which the presence of a signal results in the formation of a protein complex and a cellular response. By placing complementary zippers on two proteins in this pathway, we hypothesize that in some cases a response will be observed in the absence of the relevant signal. The Fos and Jun zippers preferentially heterodimerize with a dissociation constant on the order of 1 nM (Heuer et al. 1996). We first identified the optimal leucine zipper domains and their arrangement in protein fusions by linking several versions of the Fos and Jun zippers to the Gal4 DNA-binding and activation domains, and using the two-hybrid assay as a measure of the relative strength of the interaction (data not shown). These experiments led us to place the Fos leucine zipper near the amino terminus followed by a short flexible linker (MRGS – Fos residues 159–202 – SGTGQFQLTTM – ORF) and the Jun leucine zipper at the carboxy terminus (ORF – GGGTG – Jun residues 277–334). To determine what fraction of the Fos and Jun fusion proteins associate within individual cells, we coexpressed a Fos–green fluorescent protein fusion (Fos-GFP) with proteins of known subcellular localization fused to Jun, with both fusion genes under the control of a tetracycline-regulated promoter. When the Fos-GFP was coexpressed with an empty Jun vector, GFP fluorescence was distributed throughout the cytoplasm and nucleus (data not shown). However, when the Fos-GFP was coexpressed with a protein fused to Jun, a large fraction of the GFP fluorescence was properly localized, as observed for Ole1, at the endoplasmic reticulum; Tom20, at the mitochondrial outer membrane; Hof1, at the septin ring at the bud neck; and Mig1, in the nucleus (Fig. 1B). Mig1 is a regulated transcription factor that rapidly moves from the cytoplasm into the nucleus upon addition of glucose (DeVit et al. 1997), as did the Fos-GFP when coexpressed with Jun-Mig1 (Fig. 1B). Coassociation was typically visible in 70%–90% of the cells in a culture. We constructed a library of all yeast ORFs fused to Fos (see Supplemental data) under tetracycline control and observed a variety of subcellular localizations in a large fraction of randomly isolated transformants that coexpressed a GFP-Jun fusion with a member of the library (data not shown). These results confirm that Fos and Jun leucine zippers can cause a strong coassociation of two proteins and can do so in a variety of subcellular localizations in a yeast cell.
Figure 1.
Fos and Jun leucine zippers can be used to force proteins to associate. (A) A generalized example of the method. For a signal transduction pathway, a forced interaction might mimic the native interaction that occurs between components of the pathway and as a result cause the same downstream sequence of events, as in this example, activation of the pathway in the absence of signal. (B) Fos and Jun cause GFP to colocalize with various yeast proteins. GFP fluorescence was imaged by fluorescence microscopy of cells coexpressing a Fos-GFP fusion and Jun fused to the indicated protein. Cells expressing the Mig1 fusion were grown in the absence of glucose and then were placed on a thin agar pad containing glucose on a microscope slide. The cells were imaged immediately (no glucose) and then again in 2 min (+ glucose) without changing the field of view.
A screen for membrane-associated proteins
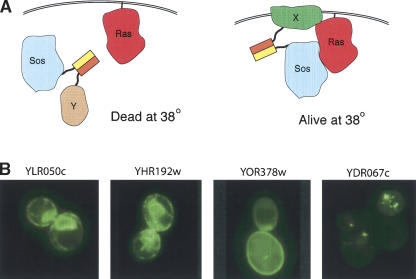
To demonstrate that this approach can be used as a genetic screening method, we carried out a screen to identify proteins that associate with the plasma membrane. We made use of the Sos-Ras recruitment system, which was originally designed as a method to identify protein–protein interactions (Aronheim et al. 1997). The yeast strain contains a temperature-sensitive mutant of Cdc25, an essential guanine–nucleotide exchange factor for Ras. The human homolog of Cdc25, Sos, can complement this mutant and permit growth at the restrictive temperature, but only if it is localized to the plasma membrane to be in close proximity with Ras. We sought to identify membrane-associated proteins in the Fos-ORF library by their rescue of the high-temperature growth defect of this strain in the presence of a Sos-Jun fusion (Fig. 2A). From 594 colonies able to grow at the restrictive temperature, we identified 215 Fos-fused genes (provided in Supplemental Table S1). Of these, 119 encode proteins known to be membrane-associated, as assessed by a gene ontology (GO) molecular component annotation in the Yeast Proteome Database (http://proteome.incyte.com) as plasma membrane, endoplasmic reticulum, Golgi, vesicles, vacuole, or nuclear membrane associated (Table 1). That we obtained proteins associated with various components of the endomembrane system is not surprising, given that they are continuously mixing as proteins are sorted, and that the Fos and Jun fusions are overexpressed. The rest of the ORFs identified are classified as having either an unknown or a nonmembrane molecular component (Table 1). We examined the localization of 54 of these by coexpressing them with a Jun-GFP fusion and found that 25, including seven characterized as having a nonmembrane localization, have an apparent membrane localization (examples shown in Fig. 2B), and another eight have a punctate appearance that suggests a potential association with vesicles (Table 1). Closer examination of the YPD annotations revealed that for an additional 17 ORFs classified as either uncharacterized or not membrane associated, there is some evidence of membrane association (e.g., they have roles in lipid biosynthesis or interact with known membrane proteins). Our results differ in some cases from published localization screens (Kumar et al. 2002; Huh et al. 2003) because we may be able to detect transient or weak membrane association that fulfills the genetic selection but is not sufficient to assign localization when GFP fluorescence is examined microscopically. In total, 169 (79%) of the ORFs isolated in the screen are known or likely membrane-associated proteins, and only 38 (17%) are likely to be false positives (Table 1). These results demonstrate this method works as a genomic screening method.
Figure 2.
A genetic screen that uses a Fos and Jun–mediated coassociation to identify membrane-localized proteins. (A) The screen is derived from the Sos-Ras recruitment system, which uses a yeast strain in which the Ras GTP exchange factor Sos, the human homolog of yeast Cdc25, replaces a temperature-sensitive Cdc25. Sos will function, and thus permit growth at the restrictive temperature, only if it is localized to the plasma membrane where it can interact with Ras. A plasma membrane-associated protein (protein X) is identified in the Fos-ORF library by its ability to bring the Sos-Jun fusion, via a leucine zipper interaction, into close proximity with Ras. (B) Localization of four proteins annotated in YPD as having an unknown localization (molecular component unknown). The Fos fusions isolated in the screen were coexpressed in yeast with a Jun-GFP fusion and the GFP imaged by fluorescence microscopy. The first three panels show proteins with likely plasma membrane and endoplasmic reticulum localization and the last panel shows a protein possibly localized in a transport vesicle.
Table 1.
Classification of ORFs identified in membrane screen (215 total)
| Membrane associated | |
| By GO annotation | 119 |
| By Jun-GFP colocalization | 25 |
| Total | 144 |
| Possible membrane associated | |
| By Jun-GFP colocalization | 8 |
| By other evidence | 17 |
| Total | 25 |
| Not membrane associated | |
| By GO annotation | 31 |
| By Jun-GFP colocalization | 7 |
| Total | 38 |
| Not enough information to assess | 8 |
A screen for proteins implicated in the filamentous growth pathway
As a second and more specific test, we screened for proteins that influence the filamentous growth MAP kinase pathway, which is required for the dimorphic transition to filamentation in nutrient-limiting conditions (Gimeno et al. 1992). The signal transduction cascade mediating this response includes an integral membrane protein, Msb2 (Cullen et al. 2004), at the head of a MAP kinase cascade (Liu et al. 1993). We used strains of the filamentous background, Σ1278b, and sought proteins that could activate the pathway when forced to associate with Msb2 (Fig. 3A). Yeast carrying Jun fused to the cytoplasmic tail of Msb2 were transformed with the Fos-ORF library and selected for growth by activation of an Msb2-dependent reporter (Cullen et al. 2004).
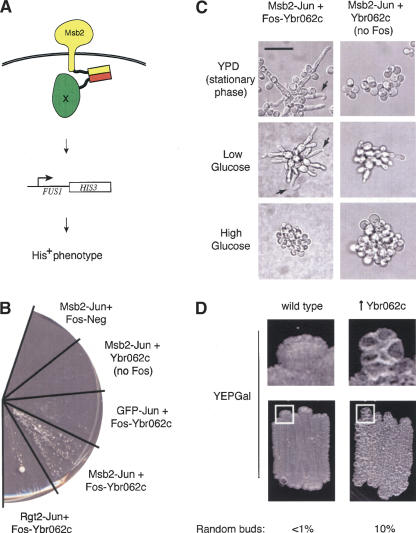
Figure 3.
A screen for proteins involved in the filamentous growth MAP kinase pathway. (A) The screen was designed to identify proteins that activate the filamentous growth pathway by forcing proteins to interact with Msb2 through a leucine zipper–mediated interaction. Msb2 is a plasma membrane protein required for activation of the filamentous growth MAP kinase cascade (Cullen et al. 2004). A candidate is identified as a protein that when forced to interact with Msb2 stimulates basal expression of the pathway, possibly by causing MAP kinase components to assemble a complex at the plasma membrane. Activation of the pathway induces expression of an Msb2-dependent reporter (Cullen et al. 2004) that allows the yeast to grow on media lacking histidine, provided that the filamentous growth pathway is intact. Proteins that are not part of the signaling pathway do not activate the pathway and thus do not confer growth on medium lacking histidine. (B) Induction of the filamentous growth pathway occurs when Msb2 and Ybr062c are forced to associate as leucine zipper fusions. Yeast coexpressing the indicated proteins were streaked on medium lacking histidine and grown at 25° for 9 d. The large colony in the Rgt2-Jun + Fos-Ybr062c sector is a likely revertant. Fast-growing revertants were seen with some combinations of proteins. (C) Forced interaction between Ybr062c and Msb2 results in morphologies associated with filamentous growth pathway hyperactivation. Cells containing the Msb2-Jun and Ybr062c-Fos fusions (left panels) or Msb2-Jun and Ybr062c alone (right panels, no Fos) were grown to stationary phase on SD minus uracil, leucine, and tryptophan solid-agar medium (top panels; stationary phase), or examined in the single cell invasive growth assay (Cullen and Sprague Jr. 2000) in glucose-limited (middle panels; low Glucose) or glucose-rich (bottom panels; high Glucose) medium. In addition to the elongated cell morphologies observed in cells associated with the Msb2–Ybr062c interaction, bent-cell morphologies were observed (arrows) indicative of hyperpolarized growth. As with hyperactive alleles of Msb2 (Cullen et al. 2004), glucose suppressed morphologies arising from the Msb2–Ybr062c interaction. Bar, 20 μm. (D) Phenotypes associated with overexpression of Ybr062c. Cells with wild-type level of Ybr062c (wild type; strain 538) and cells containing pGAL1-YBR062c (↑Ybr062c; strain 1492) were patched onto YEPGal media. The plate was incubated for 72 h at 30°C and photographed. An enlarged region of the colony border defined by a white square is also shown. A higher percentage of cells overexpressing Ybr062c also had a random budding pattern than wild-type cells, which was scored by direct visualization of cells scraped from YEPGal plates incubated for 24 h at 30°C.
Of 220 His+ colonies obtained from screening 230,000 transformants, only nine passed additional tests that showed that activation of the pathway was due to expression of a Fos-ORF protein. Sequencing revealed six proteins: Kss1, the MAP kinase for the filamentous growth pathway; Tpk2, a cAMP-dependent kinase subunit; Gis2, an uncharacterized protein involved in glucose sensing; Ypc1, a ceramidase enzyme; and two uncharacterized proteins, Yer158c and Ybr062c. To determine whether the Fos-ORFs activated the pathway by an association with Msb2, each was coexpressed with a GFP-Jun fusion and retested in the reporter strain. Only Ybr062c, which was independently identified twice, failed to activate the pathway in this test, indicating that it is the only protein that functioned via a forced association with Msb2. Ybr062c is conserved in other Ascomycete fungi, including Candida, Neurospora, and Aspergillus, and is predicted to contain a RING finger domain typically found in ubiquitin ligases (Lorick et al. 1999). Intriguingly, Ste5, the scaffold protein for the pheromone response pathway, contains such a domain (Inouye et al. 1997). When Fos-Ybr062c was coexpressed with another plasma membrane-localized protein (Rgt2) fused to Jun, the pathway was activated to a limited extent (Fig. 3B), possibly due to an increase in the localized concentration of Ybr062c at the plasma membrane. Coexpression of Fos-Ybr062c and Msb2-Jun in glucose-limited medium (Cullen and Sprague Jr. 2000) induced an extreme form of the elongated cell morphology associated with the shift to the filamentous growth pattern (Fig. 3C), indicative of a hyperactive signaling pathway (Cullen et al. 2004). Overexpression of Msb2 induces hyperpolarized growth (Cullen et al. 2004) but not to the same extent as when associated with Ybr062c, indicating that Ybr062c may influence the activity of Msb2 in a qualitatively similar but more striking manner as overexpression of Msb2 alone.
Overexpression of Ybr062c did not activate the filamentous growth pathway when not fused to Fos (Fig. 3B); thus, it would not have been obtained in a high copy screen. However, overexpression of this gene induced both colony ruffling and a budding defect, in which ∼10% of the cells formed buds in a random pattern as compared with <1% for control cells (Fig. 3D). The phenotypes manifested by overexpression of Ybr062c may not represent an accurate function of the protein, as overexpression of proteins can lead to mislocalization or nonphysiological protein interactions. However, colony ruffling is a phenotype associated with filamentation (Cullen and Sprague Jr. 2002), and budding pattern is reflective of effects on cell polarity, which under filamentous conditions requires Msb2 (Cullen et al. 2004). Deletion of the YBR062c gene did not disrupt the filamentous growth pathway (data not shown), and as such, it would not have been recovered in a screen for loss-of-function mutations. Thus, our approach may have identified a novel protein that would not have been recovered in conventional genetic screens. However, given the artificial nature of our screen, Ybr062c may not play a primary role in filamentous growth pathway activation but may influence another aspect of Msb2 function.
Discussion
We have shown that a method based on the forced association of two proteins can be used to identify a general class of proteins (membrane associated), as well as a specific component of a signaling process (Ybr062c). This method may be useful to identify proteins in any process or pathway that is activated when two or more proteins come into close physical proximity, provided that a conditional phenotype can be established. However, because these screens rely on protein tagging with leucine zipper domains, overexpression of these constructs, and forced association, any candidate proteins that emerge should be viewed with caution. The biological relevance of such proteins must be further established by additional experimentation.
Signal transduction pathways may be particularly amenable to this approach, as ligand binding can cause proteins to interact at the plasma membrane with the cytoplasmic tail of a receptor, and a screen can be performed in the absence of ligand. Proteins that are modified by other proteins, e.g., substrates of a protein kinase, might be identified if the forced association increases the rate of phosphorylation sufficiently to activate a downstream response. Similarly, a protein kinase could be identified given a substrate. However, promiscuous specificities that result from forced association may occur that would need to be ruled out, and protein kinase–substrate pairs may not be detected if kinase activation is required. For other processes, a screen could be performed in a yeast strain modified such that normal protein interactions are disrupted. For example, a protein interaction domain could be deleted or a protein that acts as a bridging component could be deleted. Other screens to identify a class of proteins might also be possible. For example, we observed that no fluorescence is visible when GFP-Jun is coexpressed with several Fos-fused proteins. It seems likely that the association of GFP-Jun to the protein fused to Fos caused GFP degradation, which if so, suggests the possibility for a screen to identify proteins involved in protein degradation.
Although these proof of principle experiments have been carried out in yeast, it should be possible to carry out such screens in other organisms. As Sharma et al. (2003) have shown, a signal transduction cascade in mammalian cells can be activated by forcing substrates to associate with tyrosine kinase receptors. In fact, our approach may be particularly useful for mammalian cells, as it uses ectoptic expression of proteins and dominant gain-of-function phenotypes, although the presence of endogenous Fos and Jun interactions may require the use of different dimerization domains. Screens can be carried out without the need to create specially engineered mutant strains or to later identify mutated genes. The most limiting factor is the construction of a library of genes fused to Fos (or other small protein–protein interaction domain). However, the availability of complete genome sequences, which is leading to the construction of extensive gene sets, makes this method a powerful new tool for protein discovery studies.
Methods
Strains
Yeast were grown at 30°C in rich (YPD), or minimal (SD) medium lacking nutrients required to maintain selection of plasmids (Rose et al. 1990). Strain 839 used to observe GFP fluorescence was derived from strain DBY746 (MATa, ura3-52, his3Δ1, leu2-3,112, trp1-28). The tTA transactivator for tetracycline regulated promoters was integrated at the leu2 locus with plasmid pCM149 (Gari et al. 1997). The LEU2 marker of pCM149 was then removed by PCR-mediated gene replacement with a kanMX cassette (Guldener et al. 1996). Strain 843 used to screen for genes activating the filamentous growth pathway was created from PC1199 (Σ1278b, MATa ura3-52, leu2::NatMX, ste4, FUS1-HIS3 MSB2-HA). TRP1 was deleted by using plasmid pNKY1009 (Alani et al. 1987), which inserts the URA3 gene. After TRP1 disruption, the URA3 sequence was removed as the result of homologous recombination between repeated sequences that flank the URA3. Overexpression of the full-length YBR062c gene was performed by using primers 5′-AGGTCTATTTCAAAACTTTGGTAA CACCAGTGGTGAGGGTGATGCGAATTCGAGCTCGTTTAAAC-3′ and 5′-TATCCATTTCTTGCAACCATTCTTCCTGTAAT GATTCTGGAAGCATTTTGAGATCCGGGTTTT-3′ to amplify the GAL1 promoter by PCR from the pFA6a-kanMX6-PGAL1 template provided by John Pringle (Longtine et al. 1998). Integration was targeted to the YBR062c locus by homologous recombination (Baudin et al. 1993) in cells of the Σ1278b background in strain 538 (MATa ste4 FUS1-HIS3 FUS1-lacZ) to create strain 1492 (MATa ste4 FUS1-HIS3 FUS1-lacZ pGAL1-YBR062c::KanMX6).
Plasmids
Standard procedures for the manipulation of plasmid DNA and growth and transformation of Escherichia coli were followed (Sambrook et al. 1990). All plasmid constructs were made by recombinational cloning in yeast (Hudson Jr. et al. 1997) by linearization with a restriction enzyme that cuts at the position where the new DNA sequence was to be inserted. DNA sequences were amplified by PCR with primers that add flanking sequence such that homologous recombination targets the insert to the desired position in the recipient plasmid. The insert and linearized recipient plasmid DNAs were cotransformed into yeast (Gietz et al. 1992) followed by selection for the auxotrophic marker on the plasmid. Plasmids were recovered from extracts of yeast colonies by transformation into E. coli. Correct plasmids were identified by restriction enzyme digestion mapping and/or sequencing of inserts and fusion points and introduced back into yeast.
Fos and Jun two hybrid constructs were derived by removal or addition of sequences to a Gal4 binding domain-Fos fusion (pPC76) and a Gal4 activation domain-Jun fusion (pPC79) (Chevray and Nathans 1992). Fos-BD and Jun-AD fusions were transformed into two-hybrid strain, PJ69-4a, and β-galactosidase assays were carried as previously described (Hudson Jr. et al. 1997). The following fusions were carried on a series of vectors created for tetracycline-regulated expression in yeast (Gari et al. 1997): Fos-GFP, GFP-Jun, Mig1-Jun, Tom20-Jun, Hof1-Jun, Ole1-Jun, Msb2-Jun, and Rgt2Δtail-Jun. Fos fusions were constructed in plasmid pCM181 that was modified so that the following amino acid sequence was added to the amino terminus of the desired protein: MRGS-Fos leucine zipper (amino acids 159–202)-SGTSQFQLTTM-ORF. Jun fusions were constructed in plasmid pCM187, so that the following sequence was added to the carboxy terminus of the desired protein: ORF-GGGTG-Jun leucine zipper (amino acids 277–334). A version of Ybr062c lacking the Fos leucine zipper was created by inserting a DNA segment containing the YBR062c ORF amplified from yeast genomic DNA into pCM187. Both wild-type and Fos-fused versions used here were based on an early ORF prediction in the Saccharomyces Genome Database (http://www.yeastgenome.org/). Since these experiments were completed, the sequence of the region upstream of YBR062c has been revised, resulting in a larger predicted ORF of 180 amino acids (see http://db.yeastgenome.org/cgi-bin/SGD/locusHistory.pl?sgdid=S0000266).
Library construction
A plasmid library of yeast proteins fused to Fos (Fos-ORF library) was created by inserting the sequence encoding the Fos leucine zipper in place of that encoding the Gal4 activation domain from a two-hybrid plasmid (GAD) library (Ito et al. 2000). This was carried out en masse by homologous recombination in yeast; the pooled GAD library, which was linearized by restriction enzyme digestion with enzymes that recognize sites in the GAL4 activation domain encoding sequence, was cotransformed with a DNA cassette containing the Fos encoding sequence, as well as the tet promoter that replaces the ADH1 promoter in the GAD plasmid and the kanMX gene as a marker to select for insertion of the cassette into the plasmid. The cassette also contains flanking sequence homologous to sequence in the pGAD plasmids so that homologous recombination causes the cassette to be inserted in the proper position and orientation: The 5′ homology targets the 5′ end of the cassette to be inserted upstream of the ADH1 promotor, and the 3′ homology targets the 3′ end of the cassette to be inserted just upstream of the sequence encoding for a yeast ORF. The homology was designed to cause the Fos coding sequence to be in the same reading frame as the yeast ORF and include an intervening linker. The resulting proteins have the following sequence: MRGS-Fos leucine zipper (amino acids 159–202)-ELGSGAPEFQLTT-ORF. For the recombination, the GAD library was linearized in separate reactions with three different restriction enzymes. The reactions were then combined and fractionated on 0.8% agarose gels to separate the linearized plasmids based on the size of the ORF. Pools of two size ranges were isolated by cutting the gel into two sections and electroeluting the DNA. Each size pool was cotransformed with the Fos cassette in separate reactions for the recombination. The transformations were plated on media lacking leucine and containing G418 to select for plasmids that have undergone a successful homologous recombination. The transformants were pooled by scraping colonies from all plates into one batch. Plasmids were recovered by transformation of purified extracts of aliquots of the pooled yeast into E. coli. The bacterial colonies were pooled by scraping the colonies into a single batch. Plasmids were purified from these bacteria. Sequencing of randomly picked bacterial colonies before pooling confirmed that the recombination successfully produced a library of Fos-fused yeast ORFs.
Microscopy
Bud site–selection phenotypes were scored by direct visualization of cells scraped from the plate at 1000× using DIC and were confirmed by staining with 1 μg/mL Calcofluor and analysis by using a UV filter at 1000× using established protocols (Chant and Pringle 1995; Pringle et al. 1995; Cullen and Sprague Jr. 2002). More than 200 cells were examined for budding pattern by each method. To visualize GFP fluorescence, yeast were grown to early log phase, concentrated, and imaged by fluorescence microscopy on a Zeiss Axiovert microscope with a 63Å × 1.4 NA objective.
Membrane screen
A cdc25 temperature-sensitive mutant yeast strain (MATα, cdc25-2, ura3, lys2, leu2, trp1, his3Δ200, ade101) carrying two plasmids, one expressing the tTA transactivator (pCM188) (Gari et al. 1997) and another expressing a Sos-Jun fusion (pSON8-Jun) (Aronheim et al. 1997), was transformed with the Fos-ORF library. The yeast were plated on SD media lacking uracil, tryptophan, and leucine to select for all three plasmids and incubated at 36°C. Aliquots were plated on the same media and incubated at the permissive temperature of 25°C to determine the total number of transformants. Plasmids were recovered from the yeast colonies that formed at 36°C, and the identity of the ORF fused to Fos was determined by sequencing with a primer that anneals in the Fos coding sequence.
Filamentous growth pathway screen
The Fos-ORF library was transformed into strain 843, which carried MSB2-Jun on pCM187. Cells were plated on SD medium lacking uracil, tryptophan, leucine, and histine and containing 3 mM 3-aminotriazole (3AT). His+ colonies were restreaked on the same media to confirm growth. Each candidate colony was then streaked on the same media containing 25 μg/mL doxycycline, which represses expression of the Msb2-Jun and Fos-ORF fusions. Only candidates that did not grow, or grew much more slowly, on doxycycline were analyzed further. The candidates were recovered into bacteria, and the identity of the ORF was determined by sequencing with a primer complementary to the Fos coding sequence. Each unique Fos-ORF was retransformed into the test strain along with plasmids expressing Jun-Msb2, GFP-Jun, or Jun-Rgt2Δ tail and tested for its ability to confer growth on media lacking histidine and containing 3 mM 3AT. The single cell invasive growth assay was performed as described (Cullen and Sprague Jr. 2000). Cells containing fusion protein-bearing plasmids and control plasmids were also examined from colonies grown to saturation on SD minus uracil, leucine, and tryptophan solid-agar medium by microscopy at 100×. Protein overexpression was induced by serial passages of strains in YEP media supplemented with 2% galactose (YEPGal) followed by 24-h incubation at 30°C for bud site–selection assays and 72 h at 30°Cto examine colony ruffling phenotypes. Colony ruffling was observed by direct visualization of patches on YEPGal semisolid agar medium.
Acknowledgments
We thank Takashi Ito for providing the GAD library, and Enrique Hererro and John Pringle for providing plasmids. S.F. is an investigator of the Howard Hughes Medical Institute.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3259905.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Alani, E., Cao, L., and Kleckner, N. 1987. A method for gene disruption that allows for repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics 116: 541-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronheim, A., Zandi, E., Hennemann, H., Elledge, S.J., and Karin, M. 1997. Isolation of an AP-1 repressor by a novel method for detecting protein–protein interactions. Mol. Cell. Biol. 17: 3094-3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin, A., Ozier-Kalogeropoulos, O., Denouel, A., Lacroute, F., and Cullin, C. 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucl. Acids Res. 21: 3329-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chant, J. and Pringle, J.R. 1995. Patterns of bud-site selection in the yeast Saccharomyces cerevisiae. J. Cell Biol. 129: 751-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevray, P.M. and Nathans, D. 1992. Protein interaction cloning in yeast: Identification of mammalian proteins that react with the leucine zipper of Jun. Proc. Nat. Acad. Sci. 89: 5789-5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen, P.J. and Sprague Jr., G.F. 2000. Glucose depletion causes haploid invasive growth in yeast. Proc. Natl. Acad. Sci. 97: 13619-13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2002. The roles of bud-site–selection proteins during haploid invasive growth in yeast. Mol. Biol. Cell 13: 2990-3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen, P.J., Sabbagh Jr., W., Graham, E., Irick, M. van Olden, E.K., Neal, C., Delrow, J., Bardwell, L., and Sprague Jr., G.F. 2004. A signaling mucin at the head of the Cdc42- and MAPK-filamentous growth pathway in yeast. Genes & Dev. 18: 1695-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVit, M.J., Waddle, J.A., and Johnston, M. 1997. Regulated nuclear translocation of the Mig1 glucose repressor. Mol. Biol. Cell. 8: 1603-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gari, E., Piedrafita, L., Aldea, M., and Herrero, E. 1997. A set of vectors with a tetracycline-regulatable promotor system for modulated gene expression in Saccharomyces cerevisiae. Yeast 13: 837-848. [DOI] [PubMed] [Google Scholar]
- Gietz, D., St Jean, A., Woods, R.A., and Schiestl, R.H. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20: 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno, C.J., Ljungdahl, P.O., Styles, C.A., and Fink, G.R. 1992. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: Regulation by starvation and RAS. Cell 68: 1077-1090. [DOI] [PubMed] [Google Scholar]
- Guldener, U., Heck, S., Fiedler, T., Beinauer, J., and Hegemann, J.H. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24: 2519-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuer, K.H., Mackay, J.P., Podzebenko, P., Bains, N.P., Weiss, A.S., King, G.F., and Easterbrook-Smith, S.B. 1996. Development of a sensitive peptide-based immunoassay: Application to detection of the Jun and Fos oncoproteins. Biochemistry 35: 9069-9075. [DOI] [PubMed] [Google Scholar]
- Hudson Jr., J.R., Dawson, E.P., Rushing, K.L., Jackson, C.H., Lockshon, D., Conover, D., Lanciault, C., Harris, J.R., Simmons, S.J., Rothstein, R., et al. 1997. The complete set of predicted genes from Saccharomyces cerevisiae in a readily usable form. Genome Res. 7: 1169-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh, W.K., Falvo, J.V., Gerke, L.C., Carroll, A.S., Howson, R.W., Weissman, J.S., and O'Shea, E.K. 2003. Global analysis of protein localization in budding yeast. Nature 425: 686-691. [DOI] [PubMed] [Google Scholar]
- Inouye, C., Dhillon, N., and Thorner, J. 1997. Ste5 RING-H2 domain: Role in Ste4-promoted oligomerization for yeast pheromone signaling. Science 278: 103-106. [DOI] [PubMed] [Google Scholar]
- Ito, T., Tashiro, K., Muta, S., Ozowa, R., Chiba, T., Nishizawa, M., Yamamoto, K., Kuhara, S., and Sakaki, Y. 2000. Toward a protein–protein interaction map of the budding yeast: A comprehensive system to examine two-hybrid interactions in all possible combinations between the yeast proteins. Proc. Natl. Acad. Sci. 97: 1143-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, A., Agarwal, S., Heyman, J.A., Matson, S., Heidtman, M., Piccirillo, S., Umansky, L., Drawid, A., Jansen, R., Liu, Y., et al. 2002. Subcellular localization of the yeast proteome. Genes & Dev. 16: 707-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H., Styles, C.A., and Fink, G.R. 1993. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science 262: 1741-1744. [DOI] [PubMed] [Google Scholar]
- Longtine, M.S., McKenzie III, A., Demarini, D.J., Shah, N.G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J.R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953-961. [DOI] [PubMed] [Google Scholar]
- Lorick, K.L., Jensen, J.P., Fang, S., Ong, A.M., Hatakeyama, S., and Weissman, A.M. 1999. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. 96: 11364-11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shea, E.K., Rutkowski, R., and Kim, P.S. 1989. Preferential heterodimer formation by isolated leucine zippers from Fos and Jun. Science 245: 646-648. [DOI] [PubMed] [Google Scholar]
- Park, S.H., Zarrinpar, A., and Lim, W.A. 2003. Rewiring MAP kinase pathways using alternative scaffold assembly mechanisms. Science 299: 1061-1064. [DOI] [PubMed] [Google Scholar]
- Porter, A.C. and Vaillancourt, R.R. 1998. Tyrosine kinase receptor–activated signal transduction pathways which lead to oncogenesis. Oncogene 16: 1343-1352. [DOI] [PubMed] [Google Scholar]
- Pringle, J.R., Bi, E., Harkins, H.A., Zahner, J.E., De Virgilio, C., Chant, J., Corrado, K., and Fares, H. 1995. Establishment of cell polarity in yeast. Cold Spring Harb. Symp. Quant. Biol. 60: 729-744. [DOI] [PubMed] [Google Scholar]
- Rose, W.D., Winston, F., and Heiter, P. 1990. Methods in yeast genetics: A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
- Sambrook, J., Fritsch, E.F., and Maniatis, T. 1990. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
- Sharma, A., Antoku, S., Fujiwara, K., and Mayer, B.J. 2003. Functional interaction trap: A strategy for validating the functional consequences of tyrosine phosphorylation of specific substrates in vivo. Mol. Cell. Proteomics 2: 1217-1224. [DOI] [PubMed] [Google Scholar]
- Spencer, D.M., Wandless, T.J., Schreiber, S.L., and Crabtree, G.R. 1993. Controlling signal transduction with synthetic ligands. Science 262: 1019-1024. [DOI] [PubMed] [Google Scholar]
Web site references
- http://proteome.incyte.com; Yeast Protein Database.
- http://www.yeastgenome.org; Saccharomyces Genome Database.
- http://db.yeastgenome.org/cgi-bin/SGD/locusHistory.pl?sgdid=S0000266; description of YBR062c in the Saccharomyces Genome Database.