Abstract
With the availability of the human genome map and fast algorithms for sequence alignment, genome-based EST clustering became a viable method for gene modeling. We developed a novel gene-modeling method, ECgene (Gene modeling by EST Clustering), which combines genome-based EST clustering and the transcript assembly procedure in a coherent and consistent fashion. Specifically, ECgene takes alternative splicing events into consideration. The position of splice sites (i.e., exon–intron boundaries) in the genome map is utilized as the critical information in the whole procedure. Sequences that share any splice sites are grouped together to define an EST cluster in a manner similar to that of the genome-based version of the UniGene algorithm. Transcript assembly is achieved using graph theory that represents the exon connectivity in each cluster as a directed acyclic graph (DAG). Distinct paths along exons correspond to possible gene models encompassing all alternative splicing events. EST sequences in each cluster are subclustered further according to the compatibility with gene structure of each splice variant, and they can be regarded as clone evidence for the corresponding isoform. The reliability of each isoform is assessed from the nature of cluster members and from the minimum number of clones required to reconstruct all exons in the transcript.
Expressed sequence tag (EST) clustering has played a central role in finding unknown genes, as evidenced by the widespread use of NCBI's UniGene (Schuler et al. 1996; http://www.ncbi.nlm.nih.gov/UniGene). Even with their intrinsic shortcomings such as contamination by genomic DNA and limited sequence quality, EST clusters have been a major source of information for many academic laboratories and pharmaceutical companies in the identification of novel genes (Adams et al. 1991). Furthermore, qualitative or quantitative patterns of gene expression can be inferred by inspecting the tissue origin of the cDNA libraries comprising an EST cluster, as for example seen in the BodyMap project (Kawamoto et al. 2000).
Several well known EST clustering algorithms exist, most of which depend on pairwise alignment of ESTs. NCBI's UniGene is the most widely used such algorithm, whose original version was “transcript-based,” examining all pairwise alignments of mRNA and EST sequences. UniGene recently switched to a “genome-based” algorithm for the human genome since build number 162, which is quite similar to our algorithm (http://www.ncbi.nlm.nih.gov/UniGene). Its major strengths are the rapid update (releases generally take less than one month) and the extensive annotation providing ample external links to important resources such as LocusLink, OMIM, MapViewer, etc. TIGR Gene Indices (TGI) is another well known EST clustering procedure that combines EST clustering based on sequence similarity and the transcript assembly procedure (Quackenbush et al. 2001). Whereas the UniGene algorithm does not produce any consensus or contig sequences, each cluster in TGI has a consensus sequence, and the splice variants from the same gene belong to different clusters unlike the UniGene algorithm. STACKdb (developed at the South Africa National Bioinformatics Institute) also combines EST clustering with transcript assembly, and it is designed to examine transcript variations in the context of developmental and pathological states (Christoffels et al. 2001).
Alternative splicing (AS) is an important mechanism of modulating gene expression and function. Recent studies on AS estimated that 40%–70% of human genes have alternatively spliced transcripts, and AS thus is established as a major mechanism of expanding proteome diversity (Graveley 2001; Maniatis and Tasic 2002; Black 2003). Furthermore, many splice variants with missing motifs or domains are causally related to various diseases, thereby representing important targets of therapeutic drug development (Caceres and Kornblihtt 2002; Levanon and Sorek 2003). Attempts to identify transcript variations due to AS are mainly based on pairwise EST alignments with mRNA or known gene sequences (Mironov et al. 1999; Krause et al. 2002; Zavolan et al. 2002; Gopalan et al. 2004; Pospisil et al. 2004). For example, if an exon present in an mRNA sequence is missing in some of the EST sequences, this is considered strong evidence of an exon-skipping event. Notably, Heber et al. (2002) introduced the splicing graphs to represent AS patterns using the graph theory. However, predictive power based on comparing only the mRNA and EST sequences is rather limited, because it does not utilize information in the intronic part of the genome. The splicing mechanism during transcription is a carefully controlled process, as can be deduced from the fact that 98% of intron sequences have a consensus of 5′GT... AG3′ (represented as GT→AG hereafter).
Now that the genome map is available, it is possible to exploit the information hidden in the intronic part of the genome. Lee and coworkers at UCLA developed an algorithm that took advantage of the intronic information to detect splice variants (Modrek et al. 2001). Their algorithm analyzed the connection diagram of splice sites detected from the genomic-EST-mRNA multiple sequence alignments for selected UniGene clusters. A recent genome-wide analysis for 96,109 UniGene clusters identified 19,384 splice variants in 14,015 clusters (Lee et al. 2003). They further improved the assembly procedure by adopting a graph-theoretic method that is conceptually similar to our transcript assembly method (Xing et al. 2004). Kan et al. (2001) developed an innovative method called TAP (transcript assembly program) that delineates AS patterns inferred from the genomic alignment of ESTs. Their genome-wide analysis for 6400 RefSeq transcripts identified 11,011 AS patterns in 4032 genes (Kan et al. 2002). Sugnet et al. (2004) recently analyzed the connectivity of exon–intron boundaries using splicing graphs to find AS events conserved in both human and mouse transcriptomes.
There have been some efforts to combine sequence clustering and transcript assembly procedures. Eyras and coworkers (2004) at the European Bioinformatics Institute (EBI) developed an algorithm to extract a minimal set of clones compatible with the splicing structure of genomically aligned ESTs. Similarly, Ace-View at NCBI shows genes and alternative transcripts reconstructed from alignments of mRNAs and ESTs using the Acembly program (D. Thierry-Mieg, J. Thierry-Mieg, M. Potdevin, and M. Sienkiewicz, unpubl.; http://www.aceview.org).
Here we present a novel gene-modeling method, ECgene (gene modeling by EST Clustering), which combines genome-based EST clustering and transcript assembly procedure in a coherent and consistent fashion, taking alternative splicing events into account. Algorithmic details are described with genome-wide analyses of the human, mouse, and rat genomes.
Results
Algorithm overview
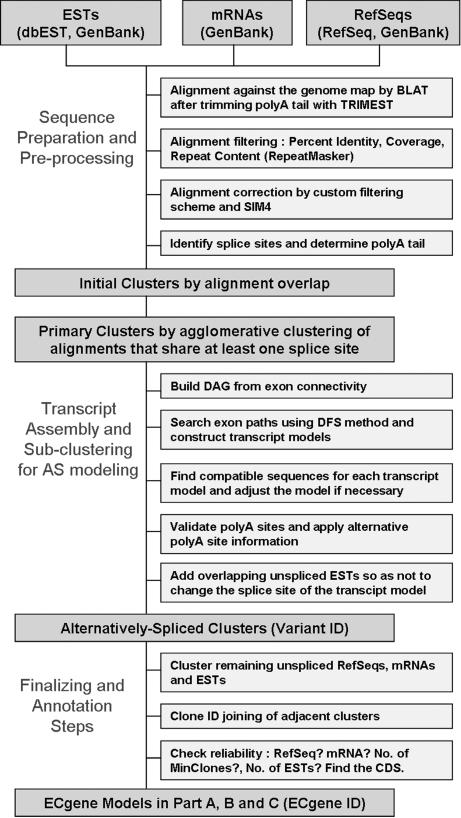
An overview of the ECgene algorithm is shown in Figure 1 as a flow chart. The ECgene algorithm was already implemented as a Web server application in ASmodeler (Kim et al. 2004). Several options including protein sequence-based gene prediction are available in ASmodeler, which predicts transcript models from user-supplied sequences. Even though the overview of the ECgene algorithm is described in the ASmodeler manuscript, we repeat the key portion of the summary here to give full algorithmic details and to describe the unique features in ECgene. Further details regarding each step are described in the Methods section.
Figure 1.
Flowchart of the ECgene algorithm. The primary cluster is a collection of spliced sequences sharing at least one splice site. The alternatively spliced clusters are subclusters of a primary cluster obtained from graph-theoretic analysis of exon connectivity. Unspliced ESTs are added to produce the final ECgene gene models, which are classified into three groups according to the transcript reliability.
Genomic alignment of mRNA and ESTs. The goal of this first step is to align input sequences against the genome. We chose the BLAT program developed by Dr. W.J. Kent (2002), mainly due to its speed and accuracy in determining the splice sites. The ability of the BLAT program to accurately align two sequences can be confounded by sequencing errors and polymorphisms. Erroneous BLAT alignments—small gaps, initial and terminal exons of low reliability, are corrected for valid splice sites. Alignments with introns not satisfying the intron consensus requirement (GT→AG or GC→AG, i.e., canonical introns) are further corrected with the SIM4 program (Florea et al. 1998). SIM4 uses a greedy algorithm to align two sequences and then forces the splice sites to be canonical if possible. This gives long unfragmented exons despite minor mismatches, which is a desirable feature for the 3′ EST region with low sequence quality. The combination of BLAT and SIM4 makes the genomic alignment process reliable and rapid, so that PC-based Linux clusters can handle genome-scale calculations. We keep only the significant hits that pass our filtering criteria, such as the minimum percent identity, alignment coverage, and the number of base pairs outside the repeat region.
Primary EST clustering. A major obstacle in EST data analysis is the genomic DNA contamination arising from mis-priming off of the polyA stretches present in the genomic DNA. Such contaminating sequences will turn out to be unspliced when aligned to the genome, since each contaminating sequence represents a part of contiguous genomic sequence (Sorek and Safer 2003). In an effort to resolve the problem of contamination by genomic DNA sequences, only the sequences with multiple exons (i.e., spliced sequences) are used in this step. Sequences that share any splice sites are grouped together to produce what we call the “primary” clusters. Primary clusters are equivalent to the UniGene cluster built by a genome-based algorithm, except that unspliced sequences are not included.
Transcript assembly procedure. The connectivity of exons in each primary cluster is represented as a directed acyclic graph (DAG). All possible paths along exons have been determined using the depth-first-search (DFS) method. Each path represents a putative transcript model, i.e., a splice variant. Sequences in the primary cluster are further clustered according to compatibility with each transcript model. Gene models without sufficient evidence are discarded or trimmed at thisstage. The presence of polyA tails, detected from detailed analyses of genomic alignment of mRNA and EST sequences, is specifically used to determine the gene boundary and direction. The direction of each gene model is determined by examining the direction of introns indicated by 5′GT... AG3′ consensus and presence of polyA tail. Details of the transcript assembly procedure are described below.
Addition of unspliced sequences. Unspliced sequences with correct orientation are added without altering the exon–intron boundaries of existing gene models. Extension of initial and terminal exons is allowed in this step. Sequences with opposite strand direction are not added, since they may represent genuine antisense transcripts even though the read direction information is believed to be wrong in many cases (Lavorgna et al. 2004). Primary clusters assembled thus far correspond to multi-exon genes, whose subclusters represent splice variants.
Clustering for unspliced genes. Remaining unspliced sequences are further clustered according to the overlap in the genomic loci and the sequence orientation. The resulting clusters, representing single-exon genes, are added to the list of primary clusters.
Clone-ID joining. Merging clusters by clone-based boundaries is useful in joining nonoverlapping 5′ and 3′ ESTs. We follow the UniGene procedure, which merges two clusters if clone-IDs link at least two 5′ ends from one cluster with at least two 3′ ends from the other neighboring cluster within 2 Mbp. This appears to be a reasonable assumption given that the largest intron is 825,779 base pairs long in the current RefSeq database for human.
Reliability check. Graph-theoretic approaches tend to overestimate the number of splice variants. We tested the reliability of the transcript models and divide the gene models into three groups. ECgene Part A represents transcripts of almost RefSeq quality. They should satisfy one of the following three requirements: (1) includes a RefSeq, (2) the minimal set of clones = 1, (3) includes mRNA and the number of clones ≥8 for singleexon genes. It should be noted that transcripts in Part A are not guaranteed to be full-length even though they have clone evidence for the presented model. It is quite possible that the actual full-length mRNA may be longer with additional exons. Part B is a collection of highly probable transcripts which satisfy one of the following three requirements: (1) includes an mRNA, (2) the minimal set of clones = 2, (3) the number of sequences ≥4 for single-exon genes. They do not show evidence of a single clone covering all exons of the transcript model. All other transcripts belong to the ECgene Part C. The method used to calculate the minimal set of clones is described in a later section.
Transcript assembly using graph theory—Splice variant analysis
The most critical part of the ECgene algorithm is the transcript assembly encompassing the analysis of splice variants. Other algorithmic details are given in the Methods section.
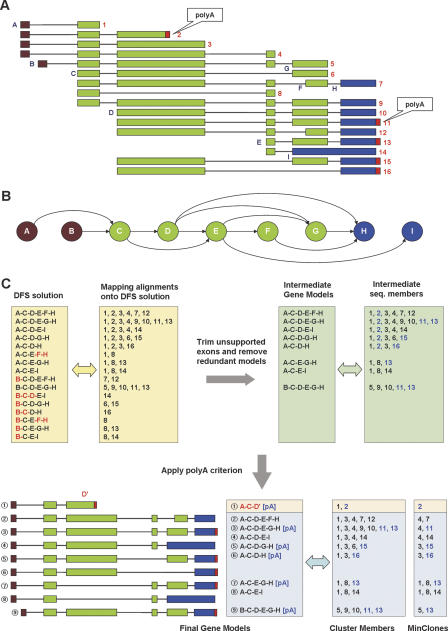
The hypothetical cluster in Figure 2A illustrates the principle. We have a cluster of 16 spliced sequences that share at least one splice site. This cluster includes examples of most frequently seen AS types—alternative promoters, exon skipping, alternative splice sites (5′ and 3′), and alternative polyA sites.
Figure 2.
Transcript assembly procedure based on the graph theory. (A) Example of genomic alignment of multi-exon sequences comprising an ECgene cluster. Exons are marked as A, B, C,..., and sequences are numbered as 1, 2, 3,... Exons A and B represent an example of alternative transcription start sites. Exons D, E, F, and G show exon-skipping events, whereas exons F and G occupy the same genomic loci with different 3′ splice sites (acceptor splice site variation). Sequence #14 shows an example of intron retention at exon I. PolyA tails are indicated as small red boxes; they do not align onto the genome. (B) Directed acyclic graph (DAG) representation of genomic alignment. Nodes and edges represent exons and introns, respectively. Exons are colored according to the type of nodes. Source nodes with outgoing arrows only are shown in brown, and terminal nodes with incoming arrows only are shown in blue. Internal nodes are colored green. (C) Transcript models and sequence members. Transcript models in the yellow boxes are the initial solutions from DFS (depth first search) that starts from one of the source nodes and ends with one of the terminal nodes. After mapping sequences onto the DFS solution, unsupported exons (indicated in red) are trimmed off and redundant transcript models are removed. This produces the intermediate gene models shown in green boxes. Then we examine sequences with a polyA tail (shown in blue letters) and ascertain that each transcript has only one polyA site. Truncation at the polyA site in sequence #2 creates a new exon, D′. Final transcript models and sequence members are shown with the MinClones. For example, the third transcript model (A-C-D-E-G-H) is a concatenation of ESTs #4 and #11, and the number of MinClones = 2.
Our gene model is based on the graph-theoretic analysis of exon connectivity. The exon connectivity can be represented as a DAG as shown in Figure 2B. Nodes and edges correspond to exons and introns, respectively. For example, connectivity in sequence #4 is A→C→D→E, and sequence #6 has a connectivity of C→D→G with the exon E skipped. We repeated the same procedure for all 16 sequences to construct a DAG for this cluster, as shown in Figure 2B.
Various ways of generating graphs have been reported. In the altSplice work by Sugnet et al. (2004), available as one of the gene prediction tracks in the UCSC genome browser, each node is the splice site and the edges are exons or introns connecting the splice sites. Each node in the ESTgenes of the Ensembl system represents the sequence itself, and two types of edge indicate the inclusion and extension relationships (Eyras et al. 2004). Nucleotides and sequences can be nodes and edges, respectively, as in Heber et al. (2002). In contrast to these existing systems, our model is conceptually simple, and extension towards clustering is straightforward.
Next, we search for all possible paths along the exons in the DAG, each path representing an inferred transcript model. The path should start from the source nodes (exons A and B) and end with the terminal nodes (exons H and I). The polyA tail in the middle of exon D is ignored for the moment. All possible paths along exons are found using the standard depth-first-search (DFS) method. Sixteen paths exist in this case, as shown in Figure 2C. For each DFS solution, we look for compatible sequences in the cluster. For example, the second path A→C→D→E→G→H has eight compatible sequences (1, 2, 3, 4, 9, 10, 11, 13) covering all six exons.
However, not all exons are necessarily covered by member sequences in every transcript model at this stage. For example, the path A→C→E→F→H has only two sequence members (1 and 8) that cover just three exons (A, C, E). This implies that the public sequence database does not have sufficient evidence for the proposed transcript model even though it is theoretically possible. Therefore, in order to reduce false positives, the program trims off the unsupported exons (F and H in red letters in Fig. 2C), leaving exons with transcript coverage (A→C→E) to reduce false positives. After trimming off unsupported exons from all 16 transcript models, some transcript models become redundant with all or part of other transcripts. We removed models that are part of longer transcript models. For example, the transcript model A→C→E is eliminated since it can be part of either A→C→E→G→H or A→C→E→I. After a redundancy check, only eight nonredundant transcript models remained.
The presence of a polyA tail is definite proof of the transcript end. Five sequences (2, 11, 13, 15, 16) show evidence of polyA tails. PolyA detection in the ECgene algorithm is carried out based on conservative criteria for examining the genomic DNA sequences, as described in the Methods section. Four of the five sequences with the identical genomic locus have polyA tails at the terminal exon. However, sequence #2 has a valid polyA tail in the middle of exon D. The transcript models with sequence #2 as a member should terminate at this site, and it cannot be part of longer transcript models. The program examines all transcript models with intermediate polyA tails, and creates separate shorter transcript models with an alternative polyA tail. Detailed descriptions regarding detection of polyA tails and criteria for termination of transcripts based on the presence of polyA tails are given in the Methods section.
In the end, we have nine transcript models for this example cluster, as shown in Figure 2C. Each gene model has cluster members and information on polyA tails as supporting evidence.
Minimal set of representative clones and ECgene reliability
It is important to note that some splice forms may not exist in vivo. Graphtheoretic approaches tend to overpredict the number of splice variants. If exon-skipping events are observed at two exons, the ECgene algorithm will predict four transcripts—with both exons present or missing, and with one of the two being skipped. If exon-skipping events occur at 10 different exons, we get 210 = 1024 splice variants, likely an overprediction.
The only direct evidence for validity of a gene model is to verify the existence of the full-length clone experimentally. As an aid to judging the reliability of a gene model, we calculated the minimal set of clones (“MinClones”) that are required to cover all exons in each transcript model. Alignments within a single exon (i.e., unspliced alignments) are not included in the calculation. Therefore, they represent a set of clones that reproduces the exon–intron structure of the transcript model. Transcripts whose number of MinClones = 1 can be regarded as gene models with experimentally verified full-length clones, being close to the RefSeq quality. Those transcripts are classified as the ECgene Part A, and transcripts with slightly lower quality (but still highly probable) are included in Part B.
Figure 2C shows MinClones for each transcript model. The first transcript has sequence #2 as the full-length clone. The second transcript needs two sequences (#4 and #7) to cover all exons. Those two sequences may be from different cDNA libraries and may not coexist in a single type of cell. This makes the second transcript less reliable than the first one. Isoforms that are possible only by joining two sequences are regarded as having less than sufficient evidence and are classified as belonging to ECgene Part B. Note that we have two transcript models with MinClones = 3, and that they belong to the ECgene Part C. Transcript structure in Part C may be questionable, since it requires concatenation of more than two clones. However, individual events of alternative splicing implied in the transcripts should be real unless the genomic alignment of mRNA/EST sequences is erroneous. There is a good chance that some of them will turn out to be real transcripts with more sequence data available in the future.
ECgene genome browser
Gene models from the ECgene algorithm are available via the genome browser at the UCSC Genome Center. Being one of the regular gene prediction tracks, ECgene can be easily combined with ample annotation data available at the Center. Information on putative transcription start sites, transcription factor binding sites, mRNA sequences from other organisms, and conserved regions between species can be valuable in determining the validity of the gene models. Other tools such as the table browser, gene sorter, and proteome browser are excellent utilities in inferring gene functions.
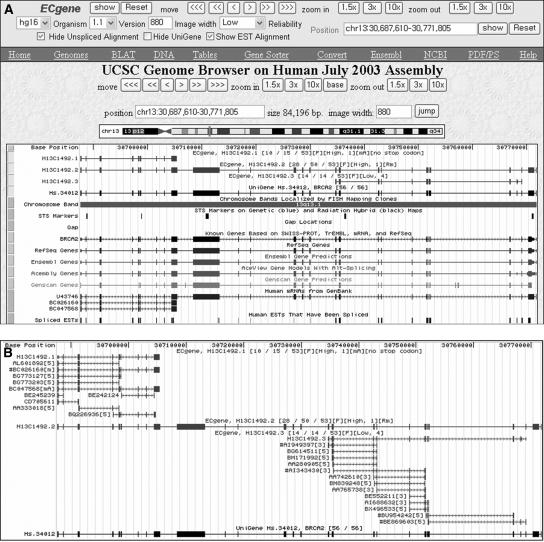
To provide more detailed information on the ECgene models, we created a utility program that shows ECgene models as custom tracks in the UCSC genome browser. Figure 3 shows the transcript structure of the BRCA2 gene using the ECgene genome browser available at http://genome.ewha.ac.kr/ECgene/gbr/. The GUI design is almost identical to that of the UCSC genome browser as shown in Figure 3A. The most useful feature would be the option of showing EST alignment that adds each transcript model and member sequences as a separate custom track. The title line includes a brief summary of the transcript model and clones. Clicking on the transcript or the title line expands the picture to show the alignment as in Figure 3B. We also provide the option of hiding unspliced alignments, since many of them are likely to be incomplete or artifactual. Furthermore, one can see the result of UniGene clustering for comparison even though they are just a collection of alignments without transcript models. For the BRCA2 gene, all 56 sequences in the UniGene cluster align in this genomic region. However, we often find that our clusters are substantially different from the UniGene clusters.
Figure 3.
ECgene genome browser. (A) Dense view showing the gene structure. The ECgene ID “H13C1492.1” indicates that the gene is the 1,492nd cluster located on human chromosome 13. The variant number is appended after the ECgene ID. The title line has additional information. “[10/15/53][F][High, 1][mA][no stop codon]” means that this cluster has 53 sequences. The first variant has 15 sequence members, 10 of which are multi-exon clones. The transcript is on the sense (+) strand. It contains mRNA sequence and has polyA evidence. [High, 1] means that the transcript belongs to the ECgene Part A, and the number of MinClones = 1. (B) Expanded view showing sequence alignment. The first variant has a polyA tail on BC047568 mRNA. The third variant belongs to the ECgene Part C (Low reliability) with the number of MinClones = 4. Representative clones belonging to the minimal set are indicated with the “#” sign in front of the accession number. Information on the EST read direction and the presence of mRNA or polyA is appended to the accession number. The browser supports an option of viewing unspliced alignments. If the option of showing EST alignment is unchecked, it will show just the transcript models in a single track. The navigating bars provided in the upper window should be used to make a query to our database. Otherwise, the data in the custom tracks do not change.
Analysis of human, mouse, and rat transcriptomes
We applied the ECgene algorithm to the human, mouse, and rat genomes. The total number of input sequences is summarized in Table 1; 92%–96% of RefSeq sequences align onto the genome with good quality. The aligned percentage decreases slightly for mRNA and EST sequences. The percentage of spliced sequences reflects the nature of EST clones, which are short sequences from single-pass reads. Note that the number of mRNA and EST sequences for rat is about one-tenth of those of the human and mouse genomes.
Table 1.
Summary of input sequences
|
Human
|
Mouse
|
Rat
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| RefSeq | mRNA | EST | RefSeq | mRNA | EST | RefSeq | mRNA | EST | |
| Raw data from GenBank | 25,975 | 133,271 | 5,426,061 | 40,568 | 113,526 | 3,918,650 | 21,937 | 11,779 | 538,134 |
| No. of aligned sequences onto the genome after initial filtering | 25,665 | 118,034 | 4,836,878 | 38,137 | 101,645 | 3,467,066 | 20,867 | 9996 | 487,771 |
| No. of sequences after removal of bad alignmentsa | 24,895 (96%) | 112,933 (84%) | 4,408,552 (81%) | 37,268 (92%) | 100,798 (89%) | 3,348,841 (85%) | 20,759 (94%) | 9871 (84%) | 471,043 (88%) |
| No. of spliced sequencesb | 22,649 (91%) | 86,897 (77%) | 2,076,217 (47%) | 29,948 (80%) | 68,912 (68%) | 1,315,511 (39%) | 18,289 (88%) | 8404 (85%) | 169,604 (36%) |
Sequences included in the final clustering of ECgene (percentage of aligned sequences)
Input sequences for transcript assembly procedure (percentage of multi-exon sequences out of all sequences in the ECgene)
Table 2 is a summary of the application of the ECgene algorithm on the human genome. Part A contains 57,172 genes of almost RefSeq quality, 37,497 (66%) of which are multi-exon genes. The portion of single-exon genes is rather high compared to the input RefSeq in Table 1, since our criterion requires an mRNA and the number of sequences ≥8 regardless of the availability of full-length clones. The percentage of alternatively spliced genes among multi-exon genes varies from 25% to 43% depending on the transcript reliability. The average numbers of isoforms for multi-exon genes range from 4.1 to 7.9. Approximately 80% of alternatively spliced genes have at least one splice variant being supported by EST sequences only. All of these numbers are in good agreement with previous reports.
Table 2.
Summary of ECgene for the human genome
| Part A | Part A + B | Part A + B + C | |
|---|---|---|---|
| No. of genes | 57,172 | 82,179 | 311,252 |
| No. of spliced genes (multi-exon genes) | 37,497 | 43,177 | 49,546 |
| No. of unspliced genes (single-exon genes) | 19,675 | 39,002 | 261,706 |
| No. of transcripts | 179,810 | 333,513 | 658,942 |
| No. of spliced transcripts | 154,741 | 287,934 | 389,778 |
| No. of protein-coding transcripts | 162,645 | 312,397 | 558,673 |
| No. of protein-coding transcripts with a polyA tail | 82,952 | 172,888 | 237,619 |
| No. of noncoding transcripts | 17,165 | 21,116 | 100,269 |
| No. of noncoding transcripts with a polyA tail | 5082 | 5454 | 5481 |
| No. of alternatively spliced genes | 9482 | 14,994 | 21,266 |
| Percentage of alternatively spliced genes among multi-exon genes | 25% | 35% | 43% |
| No. of alternative spliced genes with at least one EST-only splice variants | 7524 | 12,563 | 18,793 |
| Average number of isoforms for spliced genesa | 4.1 | 6.7 | 7.9 |
Average number of isoforms per gene for spliced genes = No. of spliced transcripts/No. of spliced genes
The total number of clusters is 311,252, a rather large number, but 55% (171,755 clusters) of those contain only one EST. Statistics regarding cluster size versus number of ECgene clusters are available in Supplemental Table S1. Clusters with an unusually large number of sequences are from the mitochondrial genome, except in one case. The statistics for coding versus non-coding transcripts are rather interesting. Whereas the number of coding transcripts shows a steady increase in the three groups in Table 2, the number of noncoding transcripts increases dramatically in Part C. Furthermore, we find that only 27 of the 79,153 noncoding transcripts in Part C have polyA tails. This strongly suggests that a substantial portion of noncoding transcripts in Part C might be artifacts, although we cannot rule out the possibility of noncoding RNAs being transcribed by different classes of RNA polymerase.
Summaries for the mouse and rat genomes are given in Tables 3 and 4, respectively. The trends in mouse are almost the same as in human. The extent of alternative splicing is slightly decreased, probably due to the smaller size of the EST database. The rat genome shows much less AS events, and the average number of isoforms does not increase by including less reliable transcripts, which is due to the limited number of EST sequences for rat, about one-tenth of those of the human and mouse genomes. We expect to observe more AS events with more data available.
Table 3.
Summary of ECgene for the mouse genome
| Part A | Part A + B | Part A + B + C | |
|---|---|---|---|
| No. of genes | 57,238 | 73,578 | 194,192 |
| No. of spliced genes (multi-exon genes) | 32,715 | 38,494 | 43,932 |
| No. of unspliced genes (single-exon genes) | 24,523 | 35,084 | 150,260 |
| No. of transcripts | 138,623 | 201,259 | 349,374 |
| No. of spliced transcripts | 100,707 | 149,979 | 180,480 |
| No. of protein-coding transcripts | 127,395 | 187,367 | 301,800 |
| No. of protein-coding transcripts with a polyA tail | 32,620 | 59,783 | 75,301 |
| No. of noncoding transcripts | 11,228 | 13,892 | 47,574 |
| No. of noncoding transcripts with a polyA tail | 1240 | 1327 | 1332 |
| No. of alternatively spliced genes | 7163 | 12,522 | 17,706 |
| Percentage of alternatively spliced genes among multi-exon genes | 22% | 33% | 40% |
| No. of alternative spliced genes with at least one EST-only splice variants | 5115 | 9655 | 14,633 |
| Average number of isoforms for spliced genesa | 3.1 | 3.9 | 4.1 |
Average number of isoforms per gene for spliced genes = No. of spliced transcripts/No. of spliced genes
Table 4.
Summary of ECgene for the rat genome
| Part A | Part A + B | Part A + B + C | |
|---|---|---|---|
| No. of genes | 30,267 | 38,566 | 94,673 |
| No. of spliced genes (multi-exon genes) | 23,707 | 26,215 | 27,406 |
| No. of unspliced genes (single-exon genes) | 6560 | 12,351 | 67,267 |
| No. of transcripts | 46,707 | 57,352 | 114,378 |
| No. of spliced transcripts | 39,441 | 44,082 | 46,083 |
| No. of protein-coding transcripts | 42,505 | 52,247 | 92,238 |
| No. of protein-coding transcripts with a polyA tail | 13,371 | 16,301 | 17,433 |
| No. of noncoding transcripts | 4202 | 5105 | 22,140 |
| No. of noncoding transcripts with a polyA tail | 879 | 1039 | 1040 |
| No. of alternatively spliced genes | 5294 | 7572 | 8699 |
| Percentage of alternatively spliced genes among multi-exon genes | 22% | 29% | 32% |
| No. of alternative spliced genes with at least one EST-only splice variants | 4476 | 6604 | 7722 |
| Average number of isoforms for spliced genesa | 1.7 | 1.7 | 1.7 |
Average number of isoforms per gene for spliced genes = No. of spliced transcripts/No. of spliced genes
Classification by AS type
Even though AS has been extensively studied in recent years, a truly genome-wide analysis of AS types is not available except for the mouse work using the FANTOM2 clones (Zavolan et al. 2003). We classified the AS types into six groups—exon skipping, donor (5′) splice site variation, acceptor (3′) splice site variation, alternative initiation, alternative termination, and intron retention. Examples of each case can be found in Figure 2A. They are not mutually exclusive, and several events may appear simultaneously in a single cluster of transcripts.
Table 5 is the summary of the results. Of all 21,266 alternatively spliced genes in human, 13,175 (62%) genes show an exon-skipping event. Genes with a variation of donor (acceptor) splice site are ∼50%. The cases of intron retention are rather infrequent, and a substantial portion of them likely result from incompletely processed mRNAs.
Table 5.
Analysis of AS types in ECgene
| Genes | Human | Mouse | Rat |
|---|---|---|---|
| Alternatively spliced genes | 21,266 | 17,706 | 8699 |
| with 5′ donor splice site variation | 10,471 (49%) | 7994 (45%) | 2570 (30%) |
| with 3′ acceptor splice site variation | 10,813 (51%) | 8019 (45%) | 2851 (33%) |
| with exon-skipping event | 13,175 (62%) | 9687 (55%) | 3833 (44%) |
| with intron retention event | 2895 (14%) | 1998 (11%) | 212 (2%) |
| with multiple transcription start sites | 6473 (30%) | 5486 (31%) | 1780 (20%) |
| with multiple transcription termination sites | 15,528 (73%) | 12,805 (72%) | 4872 (56%) |
| with multiple polyadenylation sites | 14,835 (70%) | 9123 (52%) | 3524 (41%) |
Alternative promoter usage is an important part of transcriptional regulation. A recent study by Landry et al. (2003) using the LocusLink database reported that ∼18% of all human genes (∼18,000 loci) show evidence of alternative promoter usage. Our analysis shows that 6473 genes have multiple transcription start sites (TSSs), which is almost twice as many. This increase seems to be due to the usage of all mRNA and EST sequences. However, the result should be examined critically for two reasons. First, the BLAT/SIM4 alignment for the first exon may not be correct, since detection of the small first exon is not a routine task, especially when the sequence quality is low. Second, the real transcript can turn out to be longer when more sequence data are available. The ECgene algorithm does not extend the transcript if it finds any exon–intron mismatches. For example, the first exon (node A in the graph) in the final gene models #7 and #8 in Figure 2C will disappear if the sequence #1 is missing in the example cluster. Even if exon A is present in sequences #2–#4, the final gene model will end up excluding exon A, since they would retain exon D. Without the sequence #1, these two gene models would start at exon C, which may not be the genuine first exon. This is an inherent problem in concatenating fragmented sequences to build the full-length model. To avoid this kind of pitfall, one should look for additional evidence of TSSs such as a CPG island, promoter site signatures, etc.
Alternative transcription termination and polyadenylation, producing mature transcripts with a 3′ end of variable length, are another important regulatory factor that affects mRNA stability and post-transcriptional regulation. It is rather striking that ∼73% of alternatively spliced genes show alternative transcription termination, which is more than double the number of alternative TSSs. Since the EST database is enriched with 3′ ESTs obtained from the oligo-dT primer, we do not have the problem of insufficient sequence data as in the transcription initiation site. Even when we consider just the polyA site—a definite sign of transcript end, ∼70% of alternatively spliced genes (30% of all multi-exon genes) show an alternative polyadenylation site. Our detection of polyA tails is very conservative, since we specifically filter it by cross-checking against the genomic sequence as described in the Methods section. In comparison, Gautheret and coworkers examined ∼13,000 human and 6000 mouse untranslated regions (UTRs) using the October 2000 release of dbEST (Beaudoing and Gautheret 2001). They found 5127 (40%) and 1296 (20%) UTRs with multiple polyA sites for human and mouse, respectively. This confirms that alternative polyadenylation is a much more common event in eukaryotes.
The numbers of each type of AS are slightly smaller in mouse and even smaller in rat than in human. This should be due to coverage of the EST database. Whereas we have 5.4 and 4 million ESTs for human and mouse, respectively, just 0.54 million ESTs are available for rat.
Discussion
The ECgene algorithm has many distinctive characteristics. Here we describe useful features other than the obvious merit—facile gene modeling of alternative splicing events.
Our genome-based clustering has advantages and disadvantages. The major weakness is that the algorithm can be applied only for organisms with a genome map. This may not be a major limitation, because at present there are completed genome sequences of most important organisms such as human, worm, fruit fly, and mouse, with many more genomes to be completed in the near future. Once the genome map is available, the genome-based approach has several significant advantages over the conventional EST clustering methods based on pairwise alignments.
First of all, the genomic alignment for each gene model is precisely defined in the genome-based approach. Therefore we can readily utilize ample information from the genome annotation, which includes promoter, transcription regulatory elements, intron sequences, sequence variations (e.g., single nucleotide polymorphisms), gene expression data from microarray and SAGE experiments, conserved regions across species, repeat sequences, and so on. The UCSC genome browser database is an excellent example of integrating genomic resources from the public sector (Karolchik et al. 2003). For example, even if an EST cluster represents only part of a gene due to insufficient data coverage, the structure of full-length mRNA can be inferred by examining the flanking genomic region, especially with the aid of ab initio gene predicting programs such as Genscan (Burge and Karlin 1997) and Fgenesh++ (Salamov and Solovyev 2000). The UCSC genome browser database contains precalculated results of many gene-finding programs. Comparative genomics tracks are also helpful to find conserved blocks of genome across species. Neighbor gene information, important in expression and pathway analyses, is also readily available in the genome browser. Furthermore, the consensus sequences for the gene models are of high quality because they are obtained from high-quality (99.99% accurate) genomic DNA sequence, not from the errorprone (up to 3% sequencing error) EST sequences.
Another major advantage is that genome-wide identification of AS events is naturally incorporated in our EST clustering algorithm. Most genome-based methods for detecting splice variants depend on the UniGene EST clusters (Modrek et al. 2001). In the early stage of the UniGene algorithm, “anchored clusters” based on polyA information are used as seed clusters (http://www.ncbi.nlm.nih.gov/UniGene). This is useful in discriminating the genomic DNA contamination, but the resulting cluster tends to miss many 5′ ESTs even after the clone-ID joining step. This is a substantial drawback considering that polyA tail is present in only ∼70% of transcripts and that we have more 5′ ESTs than 3′ ESTs in the dbEST database. To make things worse, polyA determination is not reliable if only mRNA or EST sequences are investigated without cross-checking against the genomic sequence specifically. Furthermore, when splice variants contain alternative polyA tails, the UniGene clustering leads to inappropriately fragmented clusters; i.e., splice variants from the same gene with different polyA tails belong to different UniGene clusters. In our approach, both EST clustering and analysis for AS utilize splice site information in a coherent and consistent way. Alternative promoters as well as alternative polyA can be analyzed within a single cluster.
Third, the UTRs are substantially longer since the terminal exons are extended by overlapping ESTs with correct orientation. Our analysis on ∼25,000 transcripts in the Ensembl database showed that the average lengths of 3′ UTRs were 550, 970, and 1130 base pairs for Ensemble genes, the UTRdb (Pesole et al. 2002), and ECgene, respectively. This is significant since the 3′ UTR, being the target region of siRNAs or microRNAs, is a critical determinant of mRNA stability and post-transcriptional regulation (Lewis et al. 2003). We were also able to develop an algorithm that predicts microRNA targets with a longer 3′ UTR database and can find many novel potential targets (S. Nam, Y. Kim, P. Kim, V.N. Kim, and S. Lee, in prep.).
Fourth, gene expression pattern can be inferred at the individual isoform level either by examining the cDNA library source of ESTs or by extracting SAGE tags from the transcript models. For example, Xu and Lee found many tissue-specific (Xu et al. 2002) and cancer-specific (Xu and Lee 2003) alternative splicing events by classifying cDNA libraries according to tissue and cancer types of origin. However, SAGE tags provide more direct and quantitative description of gene expression patterns, even though the number of publicly available libraries is rather small compared to cDNA libraries (Lash et al. 2000). Our initial results from exploring gene expression using EST and SAGE databases are available at http://genome.ewha.ac.kr/ECgene/ECexpress/ as part of the ECgene annotation project (Kim et al. 2005). A more detailed analysis is in progress.
Methods
Data sets
Genome-based EST clustering requires the genome map and transcript sequences. We used the July 2003 human reference sequence (UCSC version hg16) that is based on NCBI Build 34. The genome sequence was downloaded from the UCSC Genome Center (ftp://hgdownload.cse.ucsc.edu/goldenPath/). Genome maps for mouse and rat are still in the draft stage. We used the most recent releases, which are October 2003 assembly (UCSC version mm4) for mouse and June 2003 assembly (UCSC version rn3) for rat. ESTs and mRNA sequences were obtained from NCBI GenBank release 138 (October 24, 2003) (ftp://ftp.ncbi.nlm.nih.gov/genbank/). GenBank flat files were parsed to obtain organism-specific sequences. ESTs in gbestNN.seq and mRNAs in gbhtcNN.seq, gbpriNN.seq, and gbrodNN.seq were extracted. We also added the reference sequences to our mRNA catalog, which were obtained from NCBI RefSeq release 2 (November 4, 2003, vertebrate mammalian) (ftp:/ftp.ncbi.nlm.nih.gov/refseq/release/). The UniGene database was obtained from NCBI UniGene #163 (Genome-based method, September 22, 2003) (http://www.ncbi.nlm.nih.gov/UniGene).
Mapping sequences against the genome map
All EST, mRNA, and RefSeq sequences were mapped against the human genome using the C/S version of the BLAT program (Kent 2002). We used the BLAT version 23 downloaded from Dr. W.J. Kent's homepage (http://www.soe.ucsc.edu/∼kent/src). BLAT output contains many suboptimal alignments. Only the best alignment with the highest BLAT score was kept unless we had multiple hits of the same quality. Statistics showed that 1.0%, 0.4%, and 0.8% of EST, mRNA, and RefSeq sequences, respectively, were multiply aligned on the genome.
Sequences of poor quality were filtered out based on several criteria. Minimum percent identities were 93% for ESTs, 96% for mRNA and RefSeq. Aligned parts should be over half of the sequence length (i.e., minimum alignment coverage = 50%). Many mRNA and EST sequences have long polyA tails, which affect the percent identity and alignment coverage values. Putative polyA tails were identified by the TRIMEST program in the EMBOSS package (http://www.emboss.org), and removed from the sequence before aligning against the genome. Furthermore, the aligned regions should contain more than 100 base pairs outside the repeat regions. We used the RepeatMasker track (chrNN_rmsk files) in the UCSC genome center to find the repeat regions (http://genome.ucsc.edu). We also discarded sequences that had unaligned sequences (>30 base pairs) in the middle while neighboring sequences showed good alignment along both directions. This can be justified by the assumption that no parts of mRNA or EST sequences should be missing in the genome map. These filtering steps ensure that only high-quality sequences are used in clustering. Total numbers of sequences used in ECgene are summarized in Table 1.
BLAT alignments include many defects and are not quite ready for genome-based clustering. Erroneous alignments were corrected in several steps. BLAT alignment tends to create many small gaps due to the low sequence quality of ESTs (up to 3% of sequencing error). Therefore, we joined adjacent exons separated by very small introns that are shorter than 32 base pairs. Furthermore, if an alignment contained introns that did not satisfy the intron consensus signature (GT→AG or GC→AG), the SIM4 program (Florea et al. 1998) was used to find long unfragmented exons accepting minor mismatches.
Recent versions of the BLAT program are designed to identify additional exons at both ends of the transcript. However, we found that many small initial and terminal exons from the EST sequences were not reliable, probably due to low sequence quality near the sequence ends. In an effort to avoid improper extension of transcripts, we removed any initial and terminal exons from the alignment if the exons were smaller than 20 base pairs and if the connecting introns were not canonical.
Primary EST clustering
Many ESTs originating from genomic DNA are included in the dbEST. The original UniGene algorithm, a transcript-based clustering, solved this problem of genomic contaminants by using “anchored” clusters with polyA signal or tail (http://www.ncbi.nlm.nih.gov/UniGene). In the genome-based clustering, genomic contaminants can be filtered by using sequences with introns only, i.e., spliced sequences, since most contaminated ESTs are expected to be unspliced (Sorek and Safer 2003). Therefore, we divided the sequences in two groups—spliced and unspliced sequences. Only the spliced sequences are used in the primary clustering and gene modeling (transcript assembly) procedures. Unspliced sequences were added and clustered after transcript assembly had been completed.
Primary clustering is based on the assumption that sequences from the same gene should share at least one splice site. Such sequences were grouped together to generate the primary clusters. However, exact determination of exon–intron boundary was often problematic since the splice sites in EST sequences are often different by a few nucleotides from the true sites due to the low sequence quality of ESTs. Based on several numerical experimentations, we made an assumption that neighboring splice sites are identical if they are within ±16 base pairs. Therefore, our gene model can not distinguish splice variants whose splice sites are less than 16 base pairs apart due to this allowance. Among the splice sites within the 32-base pair range, the site supported by most sequences was chosen to be the representative splice site of the group, and was used in subsequent steps of gene modeling.
At this point, primary clusters were equivalent to the genome-based UniGene clusters except that unspliced sequences were missing and that clones with the same library ID were not joined. These steps were postponed until the transcript assembly procedure.
Determination of polyA tail and transcript ends
The determination of polyA tail by examining the end of mRNA or EST sequences could be erroneous because the same sequence may appear in the genomic sequence. The prediction accuracy of polyADQ (Tabaska and Zhang 1999), which detects polyA tails using quadratic discriminant functions, is about 50%. The most definite evidence of a polyA tail is to verify that the suspected A-rich region is not present next to the terminal exon in the genomic DNA. We examined the putative polyA sequences trimmed by the TRIMEST program in the EMBOSS program package. Substantial fractions (59% for human, 72% for mouse, 56% for rat) of presumed polyA tails align onto the genome, thereby not representing genuine polyA tails. This confirms that the reliable determination of polyA tails should be based on cross-checking with the genomic sequence specifically.
We developed our own empirical rules for identifying polyA tails; the rules depend on the length of polyA sequence and the quality of alignment onto the genomic DNA. The shortest polyA is five consecutive A's that do not align onto the genome. As the trimmed sequence gets longer, we gradually allowed matches between the putative polyA sequence and the genomic sequence. For 3′ EST sequences, polyT was assumed to be present instead of polyA tail.
The presence of polyA/T sites plays an important role in our gene modeling procedure, as described in the previous section. As a conservative approach, we acknowledged the transcript ends inferred from polyA tails only when we found polyA tails in one mRNA sequence or two spliced EST sequences or four unspliced EST sequences. The polyA attachment site should be identical in EST sequences. It should be noted that this conservative criterion could miss potentially genuine polyA tails. Transcript models with multiple polyA sites were split into many transcripts with only one polyA tail as described earlier.
Determination of gene direction
Determining the direction of transcription is not a trivial task for sequence clusters whose members have conflicting orientations. Our method is based on the presence of polyA tail and the intron consensus sequences and on the read direction of EST sequences. It was assumed that mRNA and 5′ ESTs have polyA tails and that 3′ ESTs have polyT tails. Possible polyA/T tails that do not satisfy this criterion were ignored. For sequences with introns, the strand direction can be inferred from the intron sequences, since approximately 99% of introns contain the consensus element, GT→AG. Therefore, we took the direction inferred from spliced sequences into account and determined the gene direction before adding unspliced sequences for multi-exon genes.
For each spliced sequence, we assigned the direction by counting the number of introns with GT→AG and CT→AC sequences corresponding to sense and antisense strands, respectively. If these two numbers were equal, we examined other spliced sequences with definite directions that share the exon–intron boundaries. Read directions inconsistent with the assigned strand direction were reversed. If the strand direction was still undecided, we used the read direction—the direction of 3′ ESTs was reversed. PolyA/T tails that do not agree with the strand direction were discarded. At this point, strand direction was assigned for most of the spliced sequences even though sequences in one cluster may have conflicting directions.
The next step was deciding the direction of gene models. We took a stepwise approach with different weighting factors for mRNA and ESTs, namely 3 and 1, respectively. Initially, we collected mRNA and EST sequences with polyA/T tails. Sums of weighting factors in the sense and antisense strands were compared to decide the gene direction. If the gene direction was still ambiguous, we examined sequences without polyA/T tails. Up to this stage, we used ESTs with known read directions only. If the gene direction was still undecided, ESTs without known read directions were collected. Weighting factors for ESTs with and without polyA/T tails were 2 and 1, respectively. Once the gene direction was decided, we assigned the result to all sequence members and inconsistent polyA/T tails were discarded.
The gene direction of clusters for single-exon genes was decided in a similar fashion. Sequence direction was decided from the read direction and polyA/T tails only, since no information from intron sequence was available.
Genes spanning multiple genomic loci
When a similar gene is present in the flanking region of the genome, some sequence alignment may span multiple genes. This happens occasionally in the human genome since many genes of a given family appear next to each other. It then becomes difficult to distinguish splice variants from different genes. In an effort to reduce false merging of two separate genes, we did not allow EST sequences merging two nonoverlapping mRNA or RefSeq sequences unless the EST alignment was highly reliable. Our criterion requires that the merging EST has at least two overlapping exons with both mRNAs or that the terminal exon should be longer than 50 base pairs connected by a canonical intron.
ORF and CDS determination
The transcript sequences in ECgene are extracted from the highly reliable genomic DNA sequences. Furthermore, all transcripts in our gene model have definite gene directions obtained as described above. Therefore, we considered only three reading frames and chose the longest open reading frame (ORF). However, simple implementation fails frequently in ECgene, since the 3′ UTR usually present inside the last exon is significantly extended by overlapping ESTs. ORFs in the last exon can be longer than alternative ORFs spanning multiple exons.
Our rule for ORF and the coding sequence (CDS) determination considered the number of exons, the ORF length, presence of the start codon (Met), and the CDS length. We classified ORFs (defined as the region between two adjacent stop codons) into four groups: (1) spliced ORFs with Met, (2) spliced ORFs without Met, (3) single-exon ORFs with Met, and (4) single-exon ORFs without Met. Initially, we searched the first group for the ORF with the longest CDS. We accepted the coding sequences that are longer than 30 amino acids (93 base pairs) or identical to one of the SWISS-PROT proteins excluding fragmented entries. If we could not find such an ORF in the first group, other groups were examined sequentially for the presence of ORFs using the same criteria. Genes lacking apparent ORFs were defined as non-coding RNA genes.
Acknowledgments
We thank the UCSC Genome Center for making such a wonderful resource available to the public. We would also like to thank Prof. Jaesang Kim for helpful comments and editing of the manuscript. This work was supported by the Ministry of Science and Technology of Korea through the bioinformatics research program of MOST NRDP (M1-0217-00-0027) and the Korea Science and Engineering Foundation through the center for cell signaling research at Ewha Womans University.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3030405.
Footnotes
[Supplemental material is available online at www.genome.org. Gene models from genome-wide analyses for the human, mouse, and rat genomes are available at the ECgene Web site (http://genome.ewha.ac.kr/ECgene) or may be viewed through the UCSC genome browser.]
References
- Adams, M.D., Kelley, J.M., Gocayne, J.D., Dubnick, M., Polymeropoulos, M.H., Xiao, H., Merril, C.R., Wu, A., Olde, B., Moreno, R.F., et al. 1991. Complementary DNA sequencing: Expressed sequence tags and human genome project. Science 252: 1651-1656. [DOI] [PubMed] [Google Scholar]
- Beaudoing, E. and Gautheret, D. 2001. Identification of alternate polyadenylation sites and analysis of their tissue distribution using EST data. Genome Res. 11: 1520-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black, D.L. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. BioChem. 72: 291-336. [DOI] [PubMed] [Google Scholar]
- Burge, C. and Karlin, S. 1997. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268: 78-94. [DOI] [PubMed] [Google Scholar]
- Caceres, J.F. and Kornblihtt, A.R. 2002. Alternative splicing: Multiple control mechanisms and involvement in human disease. Trends Genet. 18: 186-193. [DOI] [PubMed] [Google Scholar]
- Christoffels, A., van Gelder, A., Greyling, G., Miller, R., Hide, T., and Hide, W. 2001. STACK: Sequence Tag Alignment and Consensus Knowledgebase. Nucleic Acids Res. 29: 234-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyras, E., Caccamo, M., Curwen, V., and Clamp, M. 2004. ESTGenes: Alternative splicing from ESTs in Ensembl. Genome Res. 14: 976-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florea, L., Hartzell, G., Zhang, Z., Rubin, G.M., and Miller, W. 1998. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 8: 967-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalan, V., Tan, T.W., Lee, B.T., and Ranganathan, S. 2004. Xpro: Database of eukaryotic protein-encoding genes. Nucleic Acids Res. 32: D59-D63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley, B.R. 2001. Alternative splicing: Increasing diversity in the proteomic world. Trends Genet. 17: 100-107. [DOI] [PubMed] [Google Scholar]
- Heber, S., Alekseyev, M., Sze, S.H., Tang, H., and Pevzner, P.A. 2002. Splicing graphs and EST assembly problem. Bioinformatics (Suppl.) 18: S181-S188. [DOI] [PubMed] [Google Scholar]
- Kan, Z., Rouchka, E.C., Gish, W.R., and States, D.J. 2001. Gene structure prediction and alternative splicing analysis using genomically aligned ESTs. Genome Res. 11: 889-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan, Z., States, D., and Gish, W. 2002. Selecting for functional alternative splices in ESTs. Genome Res. 12: 1837-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik, D., Baertsch, R., Diekhans, M., Furey, T.S., Hinrichs, A., Lu, Y.T., Roskin, K.M., Schwartz, M., Sugnet, C.W., Thomas, D.J., et al. 2003. The UCSC Genome Browser Database. Nucleic Acids Res. 31: 51-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto, S., Yoshii, J., Mizuno, K., Ito, K., Miyamoto, Y., Ohnishi, T., Matoba, R., Hori, N., Matsumoto, Y., Okumura, T., et al. 2000. BodyMap: A collection of 3′ ESTs for analysis of human gene expression information. Genome Res. 10: 1817-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent, W.J. 2002. BLAT—The BLAST-like alignment tool. Genome Res. 12: 656-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, N., Shin, S., and Lee, S. 2004. ASmodeler: Gene modeling of alternative splicing from genomic alignment of mRNA, EST and protein sequences. Nucleic Acids Res. 32: W181-W186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, P., Kim, N., Lee, Y., Kim, B., Shin, Y., and Lee, S. 2005. ECgene: Genome annotation for alternative splicing. Nucleic Acids Res. 33: D75-D79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause, A., Haas, S.A., Coward, E., and Vingron, M. 2002. SYSTERS, GeneNest, SpliceNest: Exploring sequence space from genome to protein. Nucleic Acids Res. 30: 299-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry, J.R., Mager, D.L., and Wilhelm, B.T. 2003. Complex controls: The role of alternative promoters in mammalian genomes. Trends Genet. 19: 640-648. [DOI] [PubMed] [Google Scholar]
- Lash, A.E., Tolstoshev, C.M., Wagner, L., Schuler, G.D., Strausberg, R.L., Riggins, G.J., and Altschul, S.F. 2000. SAGEmap: A public gene expression resource. Genome Res. 10: 1051-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavorgna, G., Dahary, D., Lehner, B., Sorek, R., Sanderson, C.M., and Casari, G. 2004. In search of antisense. Trends Biochem Sci. 29: 88-94. [DOI] [PubMed] [Google Scholar]
- Lee, C., Atanelov, L., Modrek, B., and Xing, Y. 2003. ASAP: The Alternative Splicing Annotation Project. Nucleic Acids Res. 31: 101-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levanon, E.Y. and Sorek, R. 2003. The importance of alternative splicing in the drug discovery process. Targets 2: 109-114. [Google Scholar]
- Lewis, B.P., Shih, I.H., Jones-Rhoades, M.W., Bartel, D.P., and Burge, C.B. 2003. Prediction of mammalian microRNA targets. Cell 115: 787-798. [DOI] [PubMed] [Google Scholar]
- Maniatis, T. and Tasic, B. 2002. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature 418: 236-243. [DOI] [PubMed] [Google Scholar]
- Mironov, A.A., Fickett, J.W., and Gelfand, M.S. 1999. Frequent alternative splicing of human genes. Genome Res. 9: 1288-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrek, B., Resch, A., Grasso, C., and Lee, C. 2001. Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res. 29: 2850-2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesole, G., Liuni, S., Grillo, G., Licciulli, F., Mignone, F., Gissi, C., and Saccone, C. 2002. UTRdb and UTRsite: Specialized databases of sequences and functional elements of 5′ and 3′ untranslated regions of eukaryotic mRNAs. Update 2002. Nucleic Acids Res. 30: 335-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pospisil, H., Herrmann, A., Bortfeldt, R.H., and Reich, J.G. 2004. EASED: Extended Alternatively Spliced EST Database. Nucleic Acids Res. 32: D70-D74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quackenbush, J., Cho, J., Lee, D., Liang, F., Holt, I., Karamycheva, S., Parvizi, B., Pertea, G., Sultana, R., and White, J. 2001. The TIGR Gene Indices: Analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic Acids Res. 29: 159-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salamov, A.A. and Solovyev, V.V. 2000. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 10: 516-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler, G.D., Boguski, M.S., Stewart, E.A., Stein, L.D., Gyapay, G., Rice, K., White, R.E., Rodriguez-Tome, P., Aggarwal, A., Bajorek, E., et al. 1996. A gene map of the human genome. Science 274: 540-546. [PubMed] [Google Scholar]
- Sorek, R. and Safer, H.M. 2003. A novel algorithm for computational identification of contaminated EST libraries. Nucleic Acids Res. 31: 1067-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugnet, C.W., Kent, W.J., Ares Jr., M., and Haussler, D. 2004. Transcriptome and genome conservation of alternative splicing events in humans and mice. Pac. Symp. Biocomput. 66-77. [DOI] [PubMed]
- Tabaska, J.E. and Zhang, M.Q. 1999. Detection of polyadenylation signals in human DNA sequences. Gene 231: 77-86. [DOI] [PubMed] [Google Scholar]
- Xing, Y., Resch, A., and Lee, C. 2004. The multiassembly problem: Reconstructing multiple transcript isoforms from EST fragment mixtures. Genome Res. 14: 426-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Q. and Lee, C. 2003. Discovery of novel splice forms and functional analysis of cancer-specific alternative splicing in human expressed sequences. Nucleic Acids Res. 31: 5635-5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Q., Modrek, B., and Lee, C. 2002. Genome-wide detection of tissue-specific alternative splicing in the human transcriptome. Nucleic Acids Res. 30: 3754-3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavolan, M., van Nimwegen, E., and Gaasterland, T. 2002. Splice variation in mouse full-length cDNAs identified by mapping to the mouse genome. Genome Res. 12: 1377-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavolan, M., Kondo, S., Schonbach, C., Adachi, J., Hume, D.A., Hayashizaki, Y., and Gaasterland, T. 2003. Impact of alternative initiation, splicing, and termination on the diversity of the mRNA transcripts encoded by the mouse transcriptome. Genome Res. 13: 1290-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
Web site references
- http://genome.ewha.ac.kr/ECgene; ECgene Web site.
- ftp://ftp.ncbi.nlm.nih.gov/genbank/; GenBank FTP site.
- ftp://hgdownload.cse.ucsc.edu/goldenPath/; Genome Browser FTP site at the UCSC Genome Center.
- ftp:/ftp.ncbi.nlm.nih.gov/refseq/release/; RefSeq FTP site.
- http://genome.ucsc.edu; UCSC Genome Bioinformatics Home.
- http://www.emboss.org; EMBOSS: The European Molecular Biology Open Software Suite. [DOI] [PubMed]
- http://www.aceview.org; Identification and functional annotation of cDNA-supported genes in higher organisms using AceView.
- http://www.ncbi.nlm.nih.gov/UniGene; UniGene.
- http://www.soe.ucsc.edu/∼kent/src; Dr. W.J. Kent's homepage.