Abstract
Type 10 17β-hydroxysteroid dehydrogenase (17β-HSD10) is the HSD17B10 gene product playing an appreciable role in cognitive functions. It is the main hub of exercise-upregulated mitochondrial proteins and is involved in a variety of metabolic pathways including neurosteroid metabolism to regulate allopregnanolone homeostasis. Deacetylation of 17β-HSD10 by sirtuins helps regulate its catalytic activities. 17β-HSD10 may also play a critical role in the control of mitochondrial structure, morphology and dynamics by acting as a member of the Parkin/PINK1 pathway, and by binding to cyclophilin D to open mitochondrial permeability pore. 17β-HSD10 also serves as a component of RNase P necessary for mitochondrial tRNA maturation. This dehydrogenase can bind with the Aβ peptide thereby enhancing neurotoxicity to brain cells. Even in the absence of Aβ, its quantitative and qualitative variations can result in neurodegeneration. Since elevated levels of 17β-HSD10 were found in brain cells of Alzheimer’s disease (AD) patients and mouse AD models, it is considered to be a key factor in AD pathogenesis. Since data underlying Aβ-binding-alcohol dehydrogenase (ABAD) were not secured from reported experiments, ABAD appears to be a fabricated alternative term for the HSD17B10 gene product. Results of this study would encourage researchers to solve the question why elevated levels of 17β-HSD10 are present in brains of AD patients and mouse AD models. Searching specific inhibitors of 17β-HSD10 may find candidates to reduce senile neurodegeneration and open new approaches for the treatment of AD.
Keywords: ABAD, Alzheimer’s disease, 17β-HSD10, mitochondria, multifunctional protein, neurosteroid metabolism
1. Introduction
The HSD17B10 gene (Gene ID: 3028—HSD17B10) was first cloned from human brain and mapped to Xp11.2 in 1997 by He et al. [1]. Its product, type 10 17β-hydroxysteroid dehydrogenase (17β-HSD10) (OMIM 300256—17beta-hydroxysteroid dehydrogenase X.) is a mitochondrial, homo-tetrameric protein composed of 1044 amino acid residues with a molecular weight of 108 kDa, also known as human brain short-chain L-3-hydroxyacyl-CoA dehydrogenase (SCHAD) [1,2,3,4,5] as a member of the short-chain dehydrogenase/reductase family [1,2,3,4,5,6] or 3-hydroxy-2-methylbutyryl-CoA dehydrogenase [7,8,9,10,11,12,13,14,15,16]. 17β-HSD10 is the main hub of exercise-upregulated mitochondrial protein [17]. It had also been named as the endoplasmic reticulum-associated Aβ-binding protein (ERAB) [18,19,20,21,22] and renamed again as Aβ-binding alcohol dehydrogenase (ABAD) based upon so-called generalized alcohol dehydrogenase activities (C2-C10) [21,22,23]. The ERAB/ABAD was well known for the mediating of Aβ neurotoxicity to destroy brain cells (see Figure 1 of Ref. [18]). ABAD was first reportedly associated with the endoplasmic reticulum same as the ERAB [18,19,20,21,22] because it reportedly had an ‘ER targeting signal’ [20], although human L-3-hydroxyacyl-CoA dehydrogenase type II (HADH2), the equivalent of ERAB/ABAD [21,22] was never isolated from microsomes the endoplasmic reticulum [18,19,20,21,22,23,24,25,26,27,28,29,30]. After a mitochondrial targeting signal was found in the N-terminal of 17β-HSD10/SCHAD [2,3,31] (see Figure 1), it was followed by a claim [23] that ABAD was a mitochondrial protein without 17β-HSD10/SCHAD literature being cited.
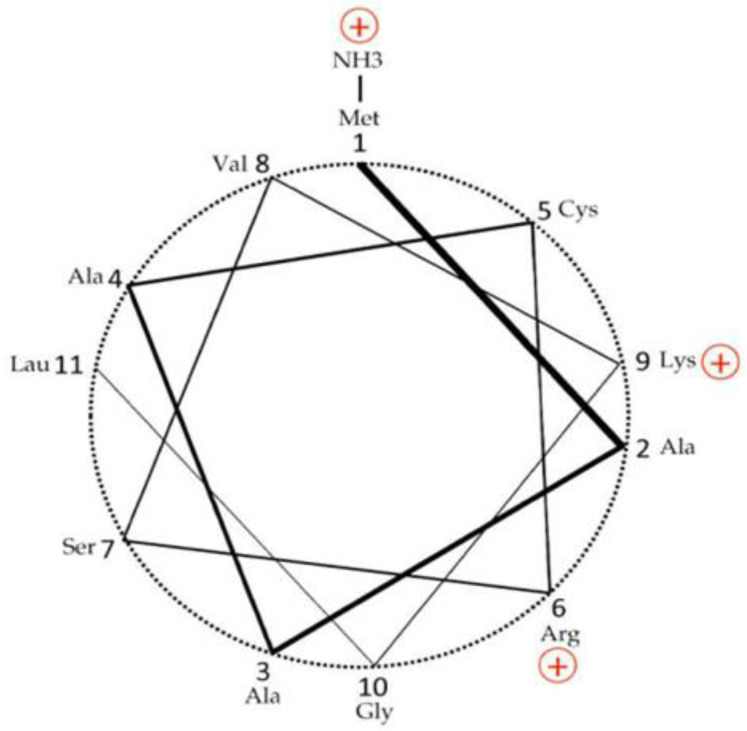
Figure 1.
N-terminal mitochondrial targeting sequence of 17β-HSD10 from human [1,2,3] and rat [32] displayed as ‘helical wheel’ (3.6 residues per turn), showing an amphiphilic helix. Adapted from Figure 5 of Ref. [31].
A mystery emerged—why human ABAD had beenrecognized as an equivalent to ERAB [21,22,23,24], a peptide with molecular weight 27 kDa only [18,19,20,21,22] even if rat ABAD was reportedly a tetramer later [30] same as rat 17β-HSD10 [32]. Unfortunately, there have been no corrigenda of ERAB or ABAD available in the prestigious journals JBC or Nature to date, although human 17β-HSD10, a multifunctional protein [33], was well recognized to be important to the pathogenesis of neurodegeneration [1,2,3,33,34,35,36].
2. Dehydration of Straight or Branched Chain Acyl-CoA Derivatives
The first catalytic function of the HSD17B10 gene product 17β-HSD10 was found to be its L-3-hydroxyacyl-CoA dehydrogenase (HAD) activity [1]. Since it can catalyze the β-oxidation of branched-chain fatty acyl-CoAs (see Figure 2), 17β-HSD10 belongs to the HADII family so it has an alternative name as SDR5C1 [37,38,39]. In contrast, L-3-Hydroxyacyl-CoA (HADH) dehydrogenase or medium-chain/short-chain HAD (EC 1.1.1.35) is a dimer with a molecular weight of 66 kDa. All HADHs including the membrane-associated long-chain HADH catalyze the third step of the fatty acid β-oxidation [40,41,42,43] (see Figure 1 of Ref. [43]) but have no role to play in the β-oxidation of the branched-chain fatty acyl-CoA derivatives [44,45] (see Figure 2). On the other hand, HADH deficiency known by clinicians as short-chain 3-hydroxyacyl-CoA dehydrogenase (SCHAD) deficiency (OMIM#231530) is a fatty acid oxidation disorder causing hyperinsulinism in pediatric patients [46], unrelated to 17β-HSD10 indeed even if both could catalyze the oxidation of acyl-CoA derivatives in mitochondria to provide energy.
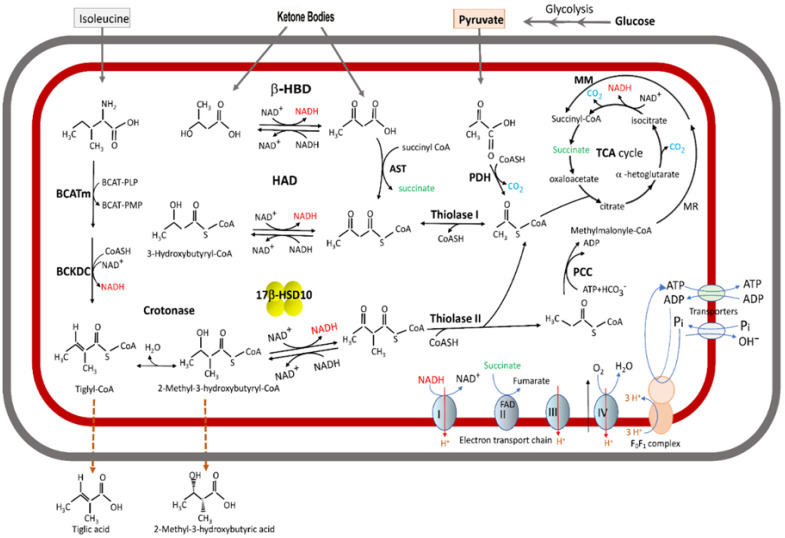
Figure 2.
Acyl thioester metabolism and oxidative phosphorylation in brain mitochondria. The fatty acid β-oxidation pathway is shown only the second half here. A cluster of four yellow balls represents 17β-HSD10/HADII. β-HBD rather than 17β-HSD10/HADII or HAD plays a key role in the ketone body metabolism. A missense mutation at 17β-HSD10/HADII would block the isoleucine catabolic pathway to result in the accumulation of isoleucine metabolites so that glucoronated tiglic acid and 2-methyl-3-hydroxybutyric acid are excreted from HSD10 deficiency patients’ urine. Adapted from Figure 1 of Ref. [47].
The HADII, a new kind of HAD, was first isolated from rat [7] and bovine sources [39]. Such HADII enzymes catalyze the oxidation of 2-methyl-3-hydroxyacyl-CoA. They belong to the HADII/SCHAD family [34], being homologs of human type 10 17β-hydroxysteroid dehydrogenase [38,45] (see Figure 2). The human brain SCHAD/17β-HSD10 [1] was later found to be a multifunctional protein [2,3,5,6,7,8,9,10,30,44,45], the same as other members of the SDR family [6,33,34,44], and therefore it was redesignated according to the international nomenclature system as type 10 17β-hydroxysteroid dehydrogenase [3,47,48,49].
3. 17β-HSD10 as a Multitask Enzyme Involved in Different Metabolic Pathways
Mitochondrial 17β-HSD10 not only catalyzes the oxidation of various acyl-CoA derivatives [33,34,47] (see Figure 2) but also plays an important role in steroid hormone and neurosteroid metabolism [2,3,47,50,51] (see Figure 3). It could modulate neuro-excitability via a two-enzyme molecular switch mechanism [51,52], namely 3α-HSD3/AKR1C2 [53] and 17β-HSD10 [52] (see Figure 4). In addition, it could act as cardiolipin phospholipase [54]. However, it needs more in vivo studies to determine [55] whether it plays a role in the metabolism of cardiolipin phospholipid, an important component of the mitochondrial inner membrane [56]. It also acts as a member of the Parkin/PINK1 pathway [57] to control mitochondrial structure and dynamics [58]. Furthermore, it is capable of binding to other proteins to carry out more physiological functions, such as mtRNA maturation [59,60,61,62,63,64,65,66,67].
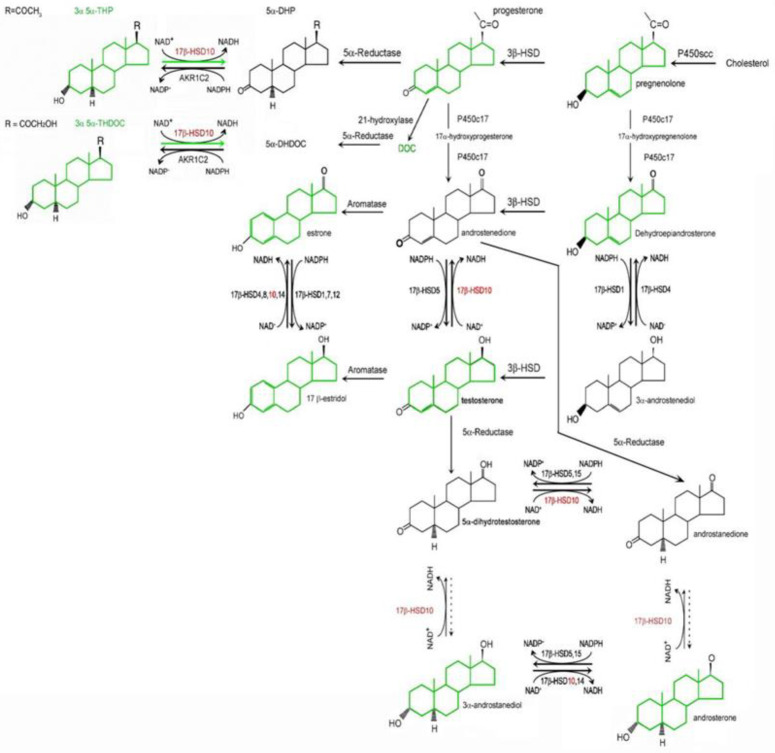
Figure 3.
Roles of 17β-hydroxysteroid dehydrogenases in neurosteroid metabolism. Neurosteroids including steroid hormones involved in brain-specific functions are shown in green. Type 10 17β-HSD shown in red, is localized in mitochondria. In contrast, types 2, 3, 6, 7 17β-HSD are in the endoplasmic reticulum while types 1 and 5 17β-HSD in cytosol and type 4 17β-HSD in the peroxisome. Reproduced from Figure 1 of Ref. [51].
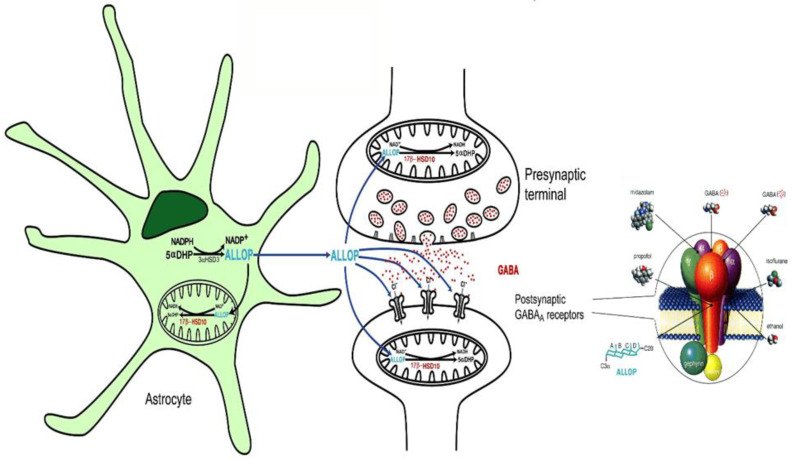
Figure 4.
Homeostasis of allopregnanolone (ALLOP) maintained by a dual enzyme molecular switch, consisting of 17β-HSD10 and 3α-HSD3 (AKR1C2). ALLOP is a positive modulator of GABAA receptors that potentiates GABA to increase the opening of Cl− channels. ―• indicates the binding sites of individual modulators on the GABAA receptor. The postsynaptic GABAA receptor was magnified and showed at the right side. Reproduced from Figure 2 of Ref. [51].
The structure of the HSD17B10 gene was found to be highly conserved across a broad evolutionary distance [65], as its gene product 17β-HSD10 is vital to life [47,52]. Studies on the properties of 17β-HSD10 are critical to the search for potential treatments of HSD17B10 gene-related disorders, including infantile neurodegeneration or HSD10 deficiency due to a missense mutation [52,58,66,67,68,69,70,71,72,73,74,75,76,77,78] and intellectual disabilities caused by a silent mutation [79,80] as well as a gene duplication [81]. Elevated levels of 17β-HSD10 were reportedly involved in the pathogenesis of Alzheimer’s disease [11,82,83,84,85,86,87]. Such studies are also important to the elucidation of mechanisms underlying its protective activity in Parkinson’s disease [88,89,90]. The Human Genome Organization (HUGO) announced in 2007 that HSD17B10 serves as the official symbol of this gene and thus the terminology of the gene product is type 10 17β-hydroxysteroid dehydrogenase [49].
4. Enzymatic Activities Regulated by Deacetylation
17β-HSD10 was found to be acetylated at lysine residues (K79, K99 and K105) by the acetyltransferase CREB-binding protein (CBP) and deacetylated by the NAD-dependent deacetylase Sirtuin 3 [91]. Its acetylation regulates its enzymatic activities and the formation of mitochondrial RNase P [91]. The regulation of its intracellular functions affects cell growth and cell resistance in response to stresses [47,88,91].
5. Additional Functions Irrelevant to Its Dehydrogenase Catalytic Activities
17β-HSD10 serves as an essential component of the mitochondrial RNase P [50]. It was also found to be a substrate of Parkin, the cytosolic E3 ubiquitin-protein ligase [88], and thus involved in mitochondrial quality control [58,92]. Furthermore, its interaction with cyclophilin D regulates the opening of a mitochondrial permeability transition pore [93].
17β-HSD10 binds to TRMT10C, a methyltransferase, to form a sub-complex catalyzing a conserved m1G/A methylation at position 9 of mitochondrial tRNAs [59]. This subcomplex can bind to a single-subunit protein-only RNase P enzyme (PRORP also known as MRPP3) to form mitochondrial RNase P [59,60]. Under these circumstances, 17β-HSD10 is renamed as MRPP2. Here, 17β-HSD10 acts as a platform to play a scaffolding role only as shown in the cryo-EM density map of the mtRNase P complex (see Figure 1 of Ref. [55]).
6. Re-Discovery of ABAD/ERAB in Mitochondria
As the nucleotide sequence of this gene (AF037438) and its cDNA (AF035555) were deposited into the GenBank in 1997 [1], an article appeared in which a 27 kDa Aβ-binding peptide with 262 amino acid residues was reported to be associated with the endoplasmic reticulum, and thus designated as the endoplasmic reticulum-associated Aβ-binding protein (ERAB) [18] with the attention of media and the focus of some research articles [19,20,21]. As the HSD17B10 gene product (17β-HSD10/SCHAD) was isolated and demonstrated to be a mitochondrial homo-tetrameric protein, where each subunit consisted of 261 amino acid residues [1,2,3,5,6,7], ERAB was re-designated as Aβ-binding alcohol dehydrogenase (ABAD) based on its so-called generalized alcohol dehydrogenase activity (C2-C10) [21,22]. At the end of Ref. [21], it was again emphasized that “…generalized alcohol dehydrogenase activity, in addition to HADH activity, lead us to propose the new name Aβ-binding protein alcohol dehydrogenase or ABAD to better describe the unusual properties of the enzyme previously referred to as ERAB”. Fortunately, it was found that data underlying such reports are not reproducible (see Figure 5 of Ref. [47]).
The ABAD was first reportedly associated with the endoplasmic reticulum (see Figure 2C of Ref. [22]). As a matter of fact, no reliable data to support those ABAD/ERAB reports including Refs. [21,22].
It is still a mystery why the intracellular localization of this protein could be altered from the ER (see Figures 6, 7 and 8a,b of Ref. [21], Figure 4 of Ref. [18] and Figure 2c of Ref. [22]) to mitochondria and then published in the Science journal [23] as a new discovery in 2004 by omitting all previous literature already showing the intra-mitochondrial localization of the human [1,2,3,4,5,31,92], mouse [86,87] and rat [32] HSD17B10 (HADII) gene product—17β-HSD10 or SCHAD.
7. An Erroneous Story of Aβ-Binding Alcohol Dehydrogenase
By making a comparison between Figure 5b,c (the reported immune-histological micrographs of ABAD and ERAB, respectively), the following questions emerged. Why are the immune-histological micrographs stained with guinea pig or mouse anti-ABAD [23] completely distinct from those stained with rabbit anti-ERAB/ABAD [18,19,20,21,22]? Since no specific controls [94] were applied in those studies, it is difficult to attribute the action [18,19,20,21,22] to be due to a technical error. More importantly, it was revealed that the subcellular fractionation data for the demonstration of the association of ERAB/HADII with the endoplasmic reticulum [21] were secured by the use of an unreasonable modification of the traditional procedure invented by the Nobel laureate Prof. Christian de Duve (see SM1 and SM2a of Ref. [35]).
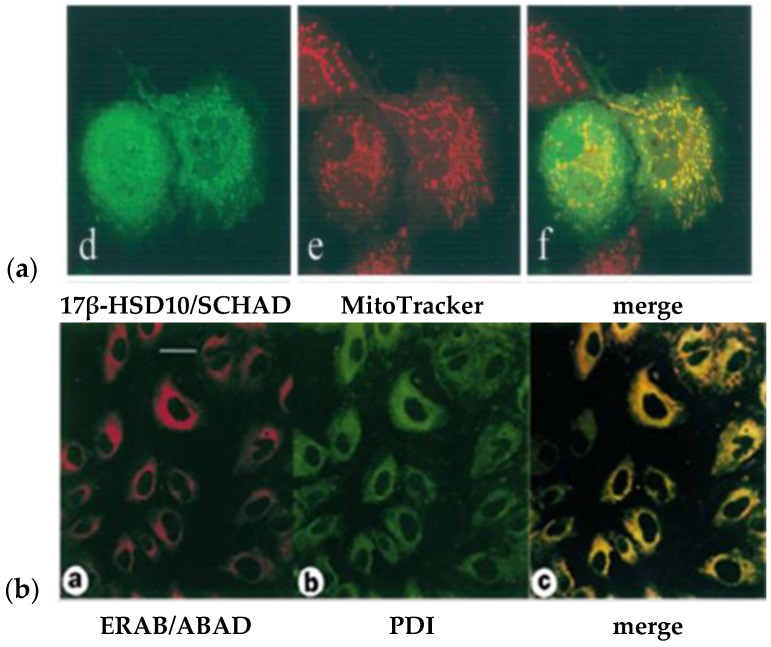
Figure 5.
Comparison of the reported intracellular localization data of 17β-HSD10 (a) with those of ERAB/ABAD (b). In the left column images ((d) green and (a) red) showed the protein staining; in the middle column, images ((e) red) and ((b) green) showed the mitochondrial and ER’s staining, respectively. In the right column, image in each low (a) and (b) showed the merge of the left and middle column images (yellow) in the same row. The low (a) is reproduced from Figure 1 of Ref [2]. The low (b) is reproduced from Figure 6 of Ref. [50].
The published erroneous data had been formally challenged since 2000 [4] (see SM1 of Ref. [35]), but it appears to be ignored completely by the responsible journal whatsoever. Since ERAB and ABAD both have been recognized as alternate names for the HSD17B10 gene product, i.e., mitochondrial 17β-HSD10/SCHAD [1,2,3,4,5,31,32,33,34,35,36,37,38,39,40,41,42,43,45,46,47,95], it would be necessary to clarify such erroneous nomenclature and experimental data published in prestigious journals without corrigenda [18,19,20,21,22,23,24,25,26,27,28] for scientific research to advance.
7.1. Kinetic Constants of ABAD/ERAB Not Based upon Experiments
Although the intracellular localization of Aβ-binding alcohol dehydrogenase has been transferred from the endoplasmic reticulum [18,19,20,21,22] to mitochondria in 2004 [23] after 17β-HSD10/SCHAD had been isolated from brain mitochondria in 1997 [1], the reported enzymatic properties of ABAD/ERAB [18,19,20,21,22,23,24,25] were always based upon unreliable data (see Figure 6 and Table 1).
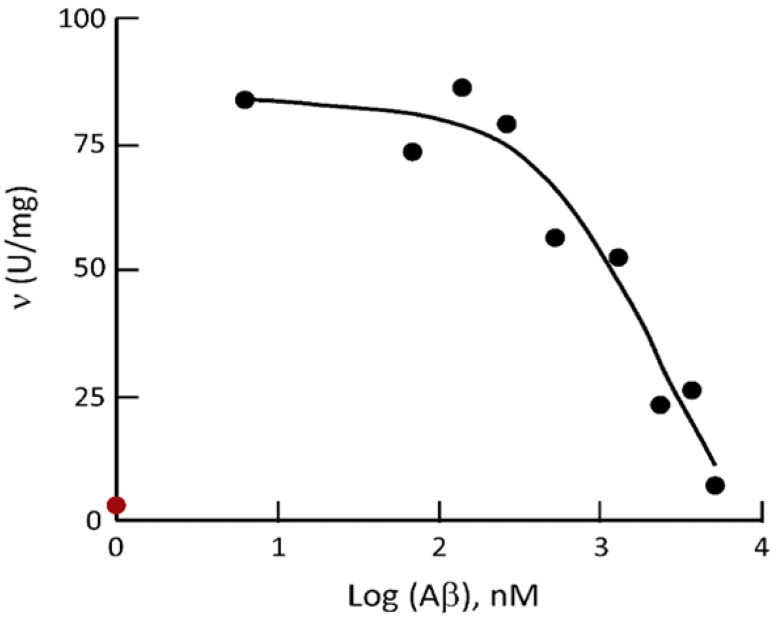
Figure 6.
Comparison of the ceiling of HAD activity of human 17β-HSD10 determined by use of the experimental procedure for ABAD/ERAB [21] in the absence of Aβ (red dot) to those reported for ABAD/ERAB under the Aβ1–40 inhibition (black dots). Reproduced from Figure 6 of Ref. [47].
Table 1.
Alternative designations of human HSD17B10 gene product ‡.
| Year | Accession Number | Name | Acronym | Comments | Ref. | |
|---|---|---|---|---|---|---|
| cDNA | Gene | |||||
| 1997–1998 |
AF035555 AF037438
[11/21/97] [12/9/97] Deposited into the Genbank respectively |
Short-chain 3-hydroxyacyl CoA dehydrogenase |
SCHAD | MW = 108 kDa, composed of 1044 residues. Homotetrameric enzyme exhibits HAD activity and proposed to reside in mitochondria | [1] | |
| U96132 n/a | Endoplasmic reticulum-associated Aβ-binding protein | ERAB | MW = 27 kDa, composed of 262 amino acid residues and associated with endoplasmic reticulum (ER) | [18] | ||
| 1999 | Novel 17β-Hydroxysteroid dehydrogenase |
Novel 17β-HSD | Mitochondrial, multifunctional protein inactivates 17β-estradiol to estrone | [2] | ||
| Amyloid β-peptide binding alcohol dehydrogenase | ABAD | Substitution for ERAB but it still associated with ER and to further convey incongruous data of generalized alcohol dehydrogenase (C2-C10) activities | [21] | |||
| 2000 | 2-methyl-3-hydroxyacyl-CoA dehydrogenase | MHBD | Appropriate for the isoleucine metabolism | [8] | ||
| 2001 |
OMIM300256:
17beta-Hydroxy- steroid dehydrogenase X |
Type 10 17β-Hydroxy-steroid dehydrogenase | 17β-HSD10 | Identification of its N-terminal mitochondrial targeting signal | [3] | |
| 2004 | Amyloid β-peptide binding alcohol dehydrogenase | ABAD | Claimed to change the ER-associated ABAD to be a mitochondrial ABAD without any citations of 17β-HSD10/SCHAD literature | [23] | ||
| 2007 | NM_004493, Gene symbol: HSD17B10 * | 3-Hydroxyacyl-CoA dehydrogenase type 2 | HADH2 | A silent mutation was found in MRXS10 ** patients | [80] | |
| 2008 | Mitochondrial RNase P protein 2 | MRPP2 | In RNA-free RNase P complex | [59] | ||
| 2013 | Short-chain dehydrogenase/reductase 5C1 | SDR5C1 | From a short-chain de-hydrogenase/reductase (SDR) evolution tree |
[37] | ||
Since the enzymatic activity first found in the HSD17B10 gene product is its 3-hydroxyacyl-CoA dehydrogenase (HAD) activity [1,2,3], human 17β-HSD10 was previously designated as short-chain 3-hydroxyacyl-CoA dehydrogenase (SCHAD) [1,2,3,4,5]. Although the HAD assay had already been well-established [96,97,98], a novel experimental procedure for the determination of ERAB/ABAD enzymatic activities was created by authors of Ref. [21] (see Figure 2 of Ref. [47]), and related experimental data were displayed in Table 2 and Figure 3 of Ref. [47]. Unfortunately, such an ABAD/ERAB experimental procedure was found to be nonsense for biomedical research at all (see Figure 2 of Ref. [47]). It was also revealed (see Figures 4 and 5 of Ref. [47]) that the reported kinetic constants (see the right column of Table 2 reproduced from Ref. [47]) are not based upon real assays. In other words, data are not secured by experimental studies, or no experiments had been performed according to the experimental conditions described in the same scientific report namely Ref. [21].
Table 2.
Comparison of catalytic constants of 17β-HSD10 and ABAD/ERAB.
| Substrate | 17β-HSD10 * | ABAD/ERAB ¶ | ||||
|---|---|---|---|---|---|---|
|
kcat (s −1) |
Km
(µM) |
kcat/Km (s−1/µM) |
kcat (S−1) |
Km (µM) |
kcat/Km (s−1/µM) | |
| Reduction by NADH: | ||||||
| S-Acetoacetyl-CoA | 37 | 89 | 0.42 | 190 | 68 | 2.8 |
| Oxidation by NAD+: | ||||||
| D-β-hydroxy-butyrate | Not detectable | ― | 0.40 × 10−6 | 4500 | 8.9 × 10−12 | |
| Ethanol | Not detectable | ― | 0.82 × 10−6 | 1.21 × 106 | 6.8 × 10−13 | |
| (−)-2-octanol ‡ | ND | ― | 1.3 × 10−3 | 43 × 103 | 3.0 × 10−8 | |
| 17β-Estradiol | 11 × 10−3 | 43 | 2.6 × 10−4 | 100 | 14 | 7.1 × 10−1 |
* Data from Refs. [1,3,35]. ¶ Data shown in Refs. [21,22] were recently revealed not secured obtainable from reported experimental studies. ‡ Not measurable by use of the experimental procedure reported in Refs. [21,22], because the actual saturated concentration of (−)-2-octanol is 8.6 mM only. ND, Not determined spectrophotometrically.
In that controversial publication, the reaction was reportedly catalyzed by ERAB (333 ng/mL). If the reported kinetic constants of ERAB/ABAD [21] are taken for granted, the enzymatic reaction would already be completed before the first data point was observed at 5 min. Although the change of A340 with time was observed at 5 min for the oxidation of 17β-estradiol catalyzed by 17β-HSD10/SCHAD (see Figure 3a of Ref. [35]), it is certainly not so in the reduction of S-acetoacetyl-CoA by NADH, because the Vmax of 17β-HSD10/SCHAD for catalyzing this hydrogenation reaction of S-acetoacetyl-CoA is much greater than that for its catalyzing of 17β-estradiol oxidation by about thirty-three hundred folds [3,4,5].
Although the v was proportional to [S] when [S-acetoacetyl-CoA] < 0.1 mM, it was certain that the v would have little increase with the increased [S-acetoacetyl-CoA] if it is greater than 0.1 mM because of the limitation of [NADH] in the assay system (see Figure 4 of Ref. [35]).
Although the assay designers of Ref. [13] appeared to disregard the basic enzymology concept of the initial velocity [99], a much more significant problem here is the inconsistency between published data and the reported experimental procedures, no matter whether the reported experimental procedures of the JBC article [21] are scientifically rational or not.
A comparison of the actual kinetic constants of the HSD17B10 gene product, 17β-HSD10, to those reported for ERAB/ABAD [21,22] was shown in Table 2. Although the ERAB problem had first been exposed two decades ago [4] (see SM1 of Ref. [35]), it was continuously covered by the related journals not making the necessary corrections. In this way, serious problems become ‘an old issue’ (see SM2 of Ref. [35]). The related editor took the so-called ‘old issue’ as an excuse for their omission, although an associate editor had previously promised that the editorial board would deal with the issue after the celebration of the 100th anniversary of the Journal.
7.2. ‘Competitive Inhibition’ of Aβ Defined by a Single Concentration of Substrate
It was reported [52] that a 17β-HSD10 mutant behaves as an allosteric enzyme with a Hill coefficient of about 1.3. Further investigation of the inhibition of the HSD17B10 gene product, 17β-HSD10, by Aβ is underway. It was reported to be a one-site competitive inhibition [21], and an equation for calculating the Ki value was recently released (see SM2 of Ref. [35]). However, it was found that only a single concentration of substrate and a single concentration of coenzyme, i.e., 0.18 mM S-acetoacetyl-CoA and 0.1 mM NADH as described in the legend for Figure 5 of Ref. [21], had been employed for the inhibition study on an ordered Bi-Bi reaction [99]. As a result, it is hard to know what kind of inhibition it could be (see Figure 5A of Ref. [35]). There is no reason to believe what was claimed by the authors of Ref. [21], a so-called ‘one-site model for competitive inhibition’, until more studies are completed as those recently reported for the inhibitor, benzothiazolyl ureas [100].
The X-ray diffraction study on rat ABAD (17β-HSD10) revealed its tetramer structure [30]. Information about the three-dimensional structure of the human protein is available in 2004 [23] (PDB:1U7T, see Figure 7c). However, it is uncertain whether binding of Aβ could lead to such radical changes of the 3D structure of 17β-HSD10, e.g., from what is seen in Figure 7c to that in Figure 7b displayed in Ref. [23] (PDB:1S08). A dramatic increase of the protein surface would need a supply of much energy and such a high energy status is very unstable. Since no electron density of Aβ was observed and described in that Science article [23], and since there is a clear difference between the reported 3D structures of Aβ-bound ERAB/ABAD if making a comparison between Figure 7a,b, the reliability of the reported Aβ-bound ERAB/ABAD structure [23] is questionable. In addition, the question of whether there is a large solvent channel in the center of ABAD [23] (see Figure 7a) also needs to be clarified by further studies.
Figure 7.
A comparison between three-dimensional structures of Aβ bound ABAD [23] (a,b) and that of 17β-HSD10/ABAD [50] (c).
As the pioneers in the research field of the human HSD17B10 gene and its product 17β-HSD10 [1,2,3,4,5,31,32,33,34], we eventually received an official reply from the JBC editorial board after about two decades (see SM2 of Ref. [35]), in which the so-called one-site competitive inhibition equation was shown below.
| (v/Vmax) = S/[Ks (1 + (I/Ki)) = S] or some derivative | (1) |
Surprisingly, it was found in this ‘rate equation’ that its left side, v/Vmax, was not equal to the right side. This rate equation cannot illustrate competitive, noncompetitive or uncompetitive inhibition (see SM2 of Ref. [35]). If careful corrections are made, it might be applicable to a competitive inhibition of the reaction catalyzed by a monomeric enzyme [99], but it is still useless for studies on the inhibition of 17β-HSD10 by Aβ (see Figure 5 of Ref. [21]) since substituting Log[I] for I as proposed by some JBC editorial board member is not warranted.
It was further stated in the questionable ABAD/ERAB report [21] that the data analysis was “using a one-site model by the method of Klotz and Hunston”. Since that literature’s title was found to be ‘Mathematical models for ligand-receptor binding. Real sites, Ghost sites’ [101], and the ABAD/ERAB researchers [18,19,20,21,22,23,24,25] should provide answers to the following questions:
7.3. Non-Reproducibility of Reported ABAD Assays
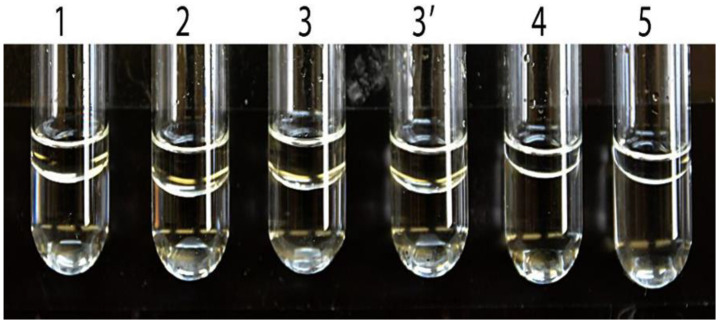
It is well known that alcohols with an alkyl >6 carbon have poor solubilities in water. (−)-2-Octanol, (+)-2-octanol and (±)-2-octanol are oils at 25 °C. Since the solubility of (−)-2-octanol and (+)-2-octanol are only 6 mM and 8.5 mM, respectively [103], the solubility of racemic (±)-2-octanol could not be greater than 15 mM. As shown in Figure 8, it is not possible to determine ABAD activity towards 2-octanols spectrophotometrically by following the published Experimental Procedures [21] (see SM3b and 3c of Ref. [35]), especially because the reported Km values of ABAD/ERAB for (−)-2-octanol, (+)-2-octanol and (±)-2-octanol were as high as 43, 86 and 87 mM, respectively.
Figure 8.
ABAD assay mixture prepared according to the published Experimental Procedure (see SM1 of [13]) with (−)-2-octanol, 160 mM (tube 1), 210 mM (tube 2), 210 mM and 1% Me2SO (tube 3), 210 mM mixed first with 1% Me2SO (tube 3′), or with 84 mM (+)-2-octanol ((tube 4), and 85 mM (±)-2-octanol (tube 5), respectively. All tubes were vortexed at high speed for 3 min and then incubated at room temperature (25 °C) for 72 h. The interface between two layers in the assay mixture is noticeable. Reproduced from Figure 5 of Ref. [47].
An interface between the layers does not disappear after the addition of 1% Me2SO to the assay system and being vortexed at high speed for 3 min and then incubated at room temperature (25 °C) for 72 h (see Tubes 3 and 3′ in Figure 8). The results are just opposite to the prediction by some members of the JBC editorial board who insisted on there being nothing wrong in those JBC articles [21,22]. Experimental details have been described in the SM3c of Ref. [35] for researchers who are interested in confirming the findings (see Figure 8). As a result, no one knows how the previously reported data and the published Figures 2B and 5B of Ref. [21] could have been obtained by assays performed under the reported experimental condition (see SM3c of Ref. [35]). Obviously, no reliable data support the conclusion that the HSD17B10 gene product exhibits generalized alcohol dehydrogenase activity (C2-C10) (see SM3b of Ref. [35]), which underlies the conversion of ABAD from ERAB [15,16,17,18,19,20,21,22,23,24,25,26,27]. This was the subject of a JBC report [22] and thereafter dozens of ABAD reports have appeared in various journals until the present time [26,27,28,29]. The term ABAD or ERAB, which probably originated from unreliable data, whenever used for the HSD17B10 gene product should be replaced by 17β-HSD10 without exception.
7.4. Importance of ABAD for Brain Cells’ Resistance to Oxidative and Nutritional Stress?
The ABAD/ERAB was reportedly contributing to the protective response to metabolic stress, especially in the setting of ischemia, because it could catalyze the oxidation of the ketone body D-3-hydroxybutyrate with Km of 4.5 mM and Vmax of 4 nmol/min/mg protein [21,22] (Only prokaryotes’ 3-hydroxyacyl-CoA dehydrogenase can catalyze the dehydration of both L- and D-isomers [104]). However, ABAD/ERAB was reportedly associated with the endoplasmic reticulum in the same JBC report (see Figure 2C of Ref. [22]). Most of the cell’s NAD+ is enclosed in mitochondria. It is hard to know whether such an ‘ER-associated NAD+-dependent dehydrogenase’ namely ERAB/ABAD could, indeed, play a physiological role in the ketone body metabolism. Furthermore, biochemists are generally aware that mitochondrial β-hydroxybutyrate dehydrogenase, β-HBD [105] rather than ABAD [21,22] or 17β-HSD10/HADII [31,32,33,34,35,36,37,38] is involved in ketone body metabolism (see Figure 2). Although data about D-3-hydroxybutyrate supporting the growth of COS cells were provided previously (see Figure 5 of ref. [22]), it could not serve as valid evidence for ABAD’s involvement in ketone body metabolism because β-HBD had not yet been knocked out from the COS cell line used by Yan et al. [22].
It had already been reported [1] that SCHAD/17β-HSD10 metabolizes only L-3-hydroxyacyl-CoA but not the D-isomer. Obviously, the report that ABAD catalyzes the oxidation of ketone bodies and, therefore, contributes to the protective response to metabolic stress, especially in the setting of ischemia [21,22] was also based upon non-reproducible data. Some possible mechanisms of 17β-HSD10, other than the oxidation of the ketone body, to play a role in the regulation of cell growth and cell resistance under oxidative and starvation stresses were reported recently [91]. After 17β-HSD10 proved to be an important mitochondrial enzyme (see Figure 2 and Figure 4), people are asking whether mitochondria are dysfunctioning in Alzheimer’s disease [47,58,106] and how we could find a way to deal with progressive neurodegeneration [58,107,108,109,110].
8. HSD17B10 Gene-Related Disorders (OMIM:#300438)
The expression of the HSD17B10 gene to generate an appropriate amount of 17β-HSD10 in different tissue cells, especially in different brain regions is critical to human health [111]. Either a mutation on this gene resulting in 17β-HSD10 mutants or the abnormal expression of this gene could cause human diseases such as infantile neurodegeneration [52] and senile neurodegeneration [112,113].
8.1. About Half Cases of HSD10 Deficiency Resulting from a p.R130C Mutation
About half of the cases of this disease are due to a missense C > T in exon 4 of the HSD17B10 gene, since the +2259 nucleotide from the ATG of this gene is >90% methylated in human X chromosome [114,115,116]. The 5-methylcytosine is prone to conversion to thymine by deamination. The substitution of arginine for cysteine eliminates several hydrogen bonds and reduces the van der Waals interaction between HSD10 subunits. HSD10 mutant could cause human diseases such as infantile neurodegeneration [52] and senile neurodegeneration [47,51,88]. Aβ levels in the cerebrospinal fluid of HSD10 deficiency patients are undetectable [117]. It indicates that the pathogenesis is most likely not due to the mediation of Aβ neurotoxicity by 17β-HSD10.
8.2. Few Female Cases of HSD10 Disease Because of X-Inactivation
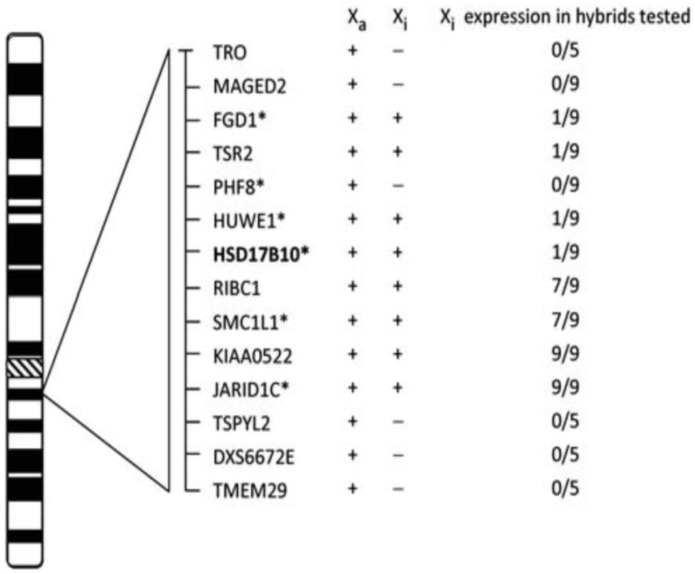
People who suffer from HSD10 disease are mostly male (see Figure 5 of Ref. [75]). Female cases with HSD10 deficiency were at least ten times less than male patients [72], because the female possesses two X-chromosomes. The random X-inactivation skewing mechanism [114,115] may randomly suppress the expression of the HSD10 mutant (see Figure 9). Clinical manifestations of some female HSD10 disease patients are also milder [52]. However, sons of the female carriers of HSD17B10 mutations are at high risk of suffering from this inherited metabolic disease.
Figure 9.
Expression of transcripts of HSD17B10 and surrounding genes from inactive X (Xi) hybrids. Samples scored as positive are expressed at least 10% of the Xa levels, and their number is shown as the numerator. The total number of hybrids tested is shown as the denominator. Gens with mutation(s) or copy number variation (CNV) causing mental retardation are marked with an asterisk. Reproduced from Figure 1 of Ref. [118].
8.3. Elevated Levels of 17β-HSD10 in Brain Cells of AD Patients or down Syndrome Patients with AD Pathology
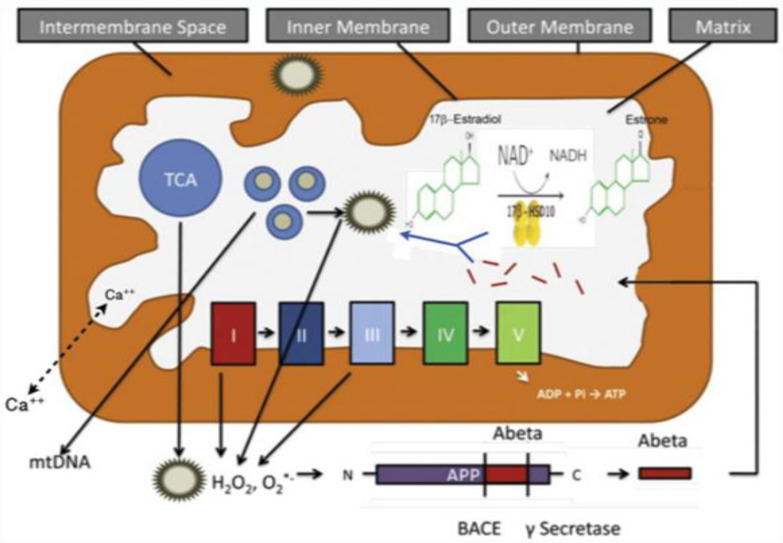
Most people with Down’s syndrome develop AD-like dementia by the fifth to sixth decade of life, a much younger age than is typically seen in sporadic, late-onset AD [119]. The HSD17B10 gene product, 17β-HSD10, catalyzes the oxidative inactivation of 17β-estradiol to estrone [2]. Elevated levels of the HSD17B10 gene product, 17β-HSD10, found in brain cells of Down’s syndrome patients with AD pathology (see Figure 1 of Ref. [120]), would accelerate the oxidative inactivation of the protective neurosteroid, 17β-estradiol (see Figure 10) and thus be harmful to the function and structure of brain mitochondria.
Figure 10.
Oxidative inactivation of neuroprotective neurosteroid 17β-estradiol by 17β-HSD10 in brain mitochondria. The I, II, III, IV and IV represent the complexes consisting of the electron-transport chain for the oxidative phosphorylation to generate ATP. Updated from Figure 1 of Ref. [58].
8.4. HSD10 Inhibitors as Potential Candidates for Treatment of Senile Neurodegeneration
There is no effective way to deal with increasing cases of the related neurodegeneration. Since 17β-HSD10 is involved in brain cells’ metabolism including neurosteroid metabolism and elevated levels of HSD10 were found in the brains of AD patients and AD mice models, inhibitors of this vital mitochondrial enzyme may be useful to alleviate the progress of neurodegeneration [106]. Whether such an inhibitor could effectively prevent mitochondrial dysfunction is considered an essential criteria for further investigation [107,108,109,110].
9. Perspective
Research in the field of the HSD17B10 gene product has been severely interfered with by ABAD/ERAB publications shown in prestigious scientific journals for two decades. No one should have the right to cover errors in their publications without corrigenda to prevent their readers from being misled. Nevertheless, consistent efforts of honest scientists in this field have resulted in significant advances such that ‘17beta-hydroxysteroid dehydrogenase X’ became the title of OMIM300256 and HSD17B10 has been approved by the human genome nomenclature committee (HGNC) as the gene symbol. It would encourage people to determine why elevated levels of 17β-HSD10/SCHAD, besides Aβ and phosphorylated Tau, are present in the brains of AD and some Down’s syndrome patients with AD pathology as well as mouse AD models. Such efforts may open new approaches to the finding of treatments for neurodegeneration seen in AD and Parkinson’s disease.
Acknowledgments
This work was supported in part by the New York State Office for People With Developmental Disabilities. We are in debt to I. H. Segel and H. H. Schulz for their invaluable advice, and to the late Nobel laureate Christin De Duve and members of the Academy of Science U.S.A. Salih J. Wakil and Victor A. McKusick for their kind support of scientific truth.
Abbreviations
| ABAD | Aβ-binding protein alcohol dehydrogenase |
| AD | Alzheimer’s disease |
| ERAB | Endoplasmic Reticulum-associated Aβ-binding protein |
| HADH | L-3-hydroxyacyl-CoA dehydrogenase |
| HADII | type II 3-hydroxyacyl-CoA dehydrogenase |
| HSD | hydroxysteroid dehydrogenase |
| 17β-HSD10 | 17β-hydroxysteroid dehydrogenase type 10 |
| Me2SO | dimethyl sulphoxide |
| mt | mitochondrial |
| MRPP | mitochondrial ribonuclease P protein |
| MRXS10 | mental retardation |
| X-linked | syndromic 10 |
| PD | Parkinson’s disease |
| PDI | protein disulfide isomerase (an ER marker) |
| PRORP | protein only RNase P |
| OMIM | Online Mendelian Inheritance in Man |
| SCHAD | short-chain 3-hydroxyacyl-CoA dehydrogenase |
| SM | Supplementary materials |
| TRMT10C | methyltransferase 10C |
| VDAC | anti-voltage-dependent anion channel (a mitochondrial marker) |
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research received no external funding.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.He X.Y., Schulz H., Yang S.Y. A human brain L-3-hydroxyacyl coenzyme A dehydrogenase is identical with an amyloid β-peptide binding protein involved in Alzheimer’s disease. J. Biol. Chem. 1998;273:10741–10746. doi: 10.1074/jbc.273.17.10741. [DOI] [PubMed] [Google Scholar]
- 2.He X.Y., Merz G., Mehta P., Schulz H., Yang S.Y. Human brain short chain L-3-hydroxyacyl coenzyme A dehydrogenase is a single-domain multifunctional enzyme. Characterization of a novel 17β-hydroxysteroid dehydrogenase. J. Biol. Chem. 1999;274:15014–15019. doi: 10.1074/jbc.274.21.15014. [DOI] [PubMed] [Google Scholar]
- 3.He X.Y., Merz G., Yang Y.Z., Schulz H., Yang S.Y. Characterization and localization of human type 10 17β-hydroxysteroid. dehydrogenase. Eur. J. Biochem. 2001;268:4899–4907. doi: 10.1046/j.0014-2956.2001.02421.2421.x. [DOI] [PubMed] [Google Scholar]
- 4.He X.Y., Yang Y.Z., Schulz H., Yang S.Y. Intrinsic alcohol dehydrogenase and hydroxysteroid dehydrogenase activities of human mitochondrial short chain L-3-hydroxyacyl-CoA dehydrogenase. Biochem. J. 2000;345:139–143. doi: 10.1042/bj3450139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He X.Y., Merz G., Yang Y.Z., Pullakart R., Mehta P., Schulz H., Yang S.Y. Function of human brain short chain L-3-hydroxyacyl coenzyme A dehydrogenase in androgen metabolism. Biochim. Biophys. Acta. 2000;1484:267–277. doi: 10.1016/S1388-1981(00)00014-7. [DOI] [PubMed] [Google Scholar]
- 6.Jornvall H., Persson B., Krook M., Atrian S., Gonzalez-Duarte R., Jeffery J. Short-chain dehydrogenase/reductases (SDR) Biochemistry. 1995;34:6003–6013. doi: 10.1021/bi00018a001. [DOI] [PubMed] [Google Scholar]
- 7.Luo M.J., Mao L.F., Schulz H. Short-chain 3-hydroxy-2-methylacyl-CoA dehydrogenase from rat liver: Purification and characterization of a novel enzyme of isoleucine metabolism. Arch. Biochem. Biophys. 1995;321:214–220. doi: 10.1006/abbi.1995.1388. [DOI] [PubMed] [Google Scholar]
- 8.Zschocke J., Ruiter J.P., Brand J., Lindner M., Hoffmann G.F., Wanders R.J., Mayatepek E. Progressive infantile neurodegeneration caused by 2-methyl-3-hydroxybutyryl-CoA dehydrogenase deficiency: A novel inborn error of branched-chain fatty acid and isoleucine metabolism. Pediatr. Res. 2000;48:852–855. doi: 10.1203/00006450-200012000-00025. [DOI] [PubMed] [Google Scholar]
- 9.Poll-The B.T., Duran M., Ruiter J.P.N., Wanders R.J.A., Barth P.G. Mild cerebral white matter disease associated with 2-methyl-3-hydroxybutyryl-CoA dehydrogenase deficiencccy. J. Inherit. Metab. Dis. 2001;24:59. [Google Scholar]
- 10.Olpin S.E., Pollitt R.J., McMenamin J., Manning N.J., Besley G., Ruiter J.P.N., Wanders R.J.A. 2-methyl-3-hydroxybutyryl-CoA dehydrogenase deficiency in a 23-year-old man. J. Inherit. Metab. Dis. 2002;25:477–482. doi: 10.1023/A:1021251202287. [DOI] [PubMed] [Google Scholar]
- 11.Ensenauer R., Niederhoff H., Ruiter J.P.N., Wanders R.J.A., Schwab O., Brandis M., Lehnert W. Clinical variability in 3-hydroxy-2-methylbutyryl-CoA dehydrogenase deficiency. Ann. Neurol. 2002;51:656–659. doi: 10.1002/ana.10169. [DOI] [PubMed] [Google Scholar]
- 12.Ofman R., Ruiter P.J., Feenstra M., Duran M., Zschocke J., Ensenauer R., Wanders J.R. 2-methyl-3-hydroxybutyryl-CoA dehydrogenase deficiency is caused by mutations in the HADH2 gene. Am. J. Hum. Genet. 2003;72:1300–1307. doi: 10.1086/375116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sutton V.R., O’Brien W.E., Clark G.D., Kim J., Wanders R.J.A. 3-Hydroxy-2-methylbutyryl-CoA dehydrogenase deficiency. J. Inherit. Metab. Dis. 2003;26:69–71. doi: 10.1023/A:1024083715568. [DOI] [PubMed] [Google Scholar]
- 14.Sass J.O., Forstner R., Sperl W. 2-Methyl-3-hydroxybutyryl-CoA dehydrogenase deficiency: Impaired catabolism of isoleucine presenting as neurodegenerative disease. Brain Dev. 2004;26:12–14. doi: 10.1016/S0387-7604(03)00071-8. [DOI] [PubMed] [Google Scholar]
- 15.Perez-Cerda C., García-Villoria J., Ofman R., Sala P.R., Merinero B., Ramos J., Ribes A. 2-Methyl-3-hydroxybutyryl-CoA dehydrogenase (MHBD) deficiency: An X-linked inborn error of isoleucine metabolism that may mimic a mitochondrial disease. Pediatr. Res. 2005;58:488–491. doi: 10.1203/01.pdr.0000176916.94328.cd. [DOI] [PubMed] [Google Scholar]
- 16.García-Villoria J., Navarro-Sastre A., Fons C., Pérez-Cerdá C., Baldellou A., Fuentes-Castelló M.Á., Ribes A. Study of patients and carriers with 2-methyl-3-hydroxybutyryl-CoA dehydrogenase (MHBD) deficiency: Difficulties in the diagnosis. Clin. Biochem. 2009;42:27–33. doi: 10.1016/j.clinbiochem.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 17.Amar D., Gay N.R., Jimenez-Morales D., Beltran P.M.J., Ramaker M.E., Raja A.N., MoTrPAC Study Group The mitochondrial multiomic response to exercise training across tissues. bioRxiv. 2023;13 doi: 10.1101/2023.01.13.523698. [DOI] [Google Scholar]
- 18.Du Yan S., Fu J., Soto C., Chen X., Zhu H., Al-Mohanna F., Stern D. An intracellular protein that binds amyloid-beta peptide and mediates neurotoxicity in Alzheimer’s disease. Nature. 1997;389:689–695. doi: 10.1038/39522. [DOI] [PubMed] [Google Scholar]
- 19.Beyreuther K., Masters C.L. The ins and outs of amyloid-β. Nature. 1997;389:677–678. doi: 10.1038/39479. [DOI] [PubMed] [Google Scholar]
- 20.Sambamurti K., Lahiri D.K. ERAB contains a putative noncleavable signal peptide. Biochim. Biophys. Res. Commun. 1998;249:546–549. doi: 10.1006/bbrc.1998.9178. [DOI] [PubMed] [Google Scholar]
- 21.Du Yan S., Shi Y., Zhu A., Fu J., Zhu H., Zhu Y., Stern D.M. Role of ERAB/L-3-hydroxyacyl-coenzyme A dehydrogenase type II activity in Abeta-induced cytotoxicity. J. Biol. Chem. 1999;274:2145–2156. doi: 10.1074/jbc.274.4.2145. [DOI] [PubMed] [Google Scholar]
- 22.Du Yan S., Zhu Y., Stern E.D., Hwang Y.C., Hori O., Ogawa S., Ramasamy R. Amyloid β-peptide-binding alcohol dehydrogenase is a component of the cellular response to nutritional stress. J. Biol. Chem. 2000;275:27100–27109. doi: 10.1016/S0021-9258(19)61485-7. [DOI] [PubMed] [Google Scholar]
- 23.Lustbader J.W., Cirilli M., Lin C., Xu H.W., Takuma K., Wang N., Caspersen C., Chen X., Pollak S., Chaney M. ABAD directly links Abeta to mitochondrial toxicity in Alzheimer’s disease. Science. 2004;304:448–452. doi: 10.1126/science.1091230. [DOI] [PubMed] [Google Scholar]
- 24.Yan S.D., Stern D.M. Mitochondrial dysfunction and Alzheimer’s disease: Role of amyloid-β peptide alcohol dehydrogenase (ABAD) Int. J. Exp. Path. 2005;86:161–171. doi: 10.1111/j.0959-9673.2005.00427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vangavaragu J.R., Valasani K.R., Fang D., Williams T.D., Yan S.S. Determination of small molecule ABAD inhibitors crossing blood brain barrier and pharmacokinetics. J. Alzheimers Dis. 2014;42:333–344. doi: 10.3233/JAD-140252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.R Valaasani K., Sun Q., Hu G., Li J., Du F., Guo Y., Yan S. Identification of human ABAD inhibitors for rescuing Aβ-mediated mitochondrial dysfunction. Curr. Alzheimer Res. 2014;11:128–136. doi: 10.2174/1567205011666140130150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hroch L., Guest P., Benek O., Soukup O., Janockova J., Dolezal R., Musilek K. Synthesis and evaluation of frentizole-based indolyl thiourea analogues as MAO/ABAD inhibitors for Alzheimer’s disease treatment. Bioorg. Med. Chem. 2017;25:1143–1152. doi: 10.1016/j.bmc.2016.12.029. [DOI] [PubMed] [Google Scholar]
- 28.Morsy A., Trippier P.C. Amyloid-Binding Alcohol Dehydrogenase (ABAD) Inhibitors for the Treatment of Alzheimer’s Disease. J. Med. Chem. 2019;62:4252–4264. doi: 10.1021/acs.jmedchem.8b01530. [DOI] [PubMed] [Google Scholar]
- 29.Xiao X., Chen Q., Zhu X., Wang Y. ABAD/17β-HSD10 reduction contributes to the protective mechanism of huperzine a on the cerebral mitochondrial function in APP/PS1 mice. Neurobiol. Aging. 2019;81:77–87. doi: 10.1016/j.neurobiolaging.2019.05.016. [DOI] [PubMed] [Google Scholar]
- 30.Powell A.J., Read J.A., Banfield M.J., Gunn-Moore F., Yan S.D., Lustbader J., Brady R.L. Recognition of structurally diverse substrates by type II 3-hydroxyacyl-CoA dehydrogenase (HADII)/amyloid-β binding alcohol dehydrogenase (ABAD) J. Mol. Biol. 2000;303:311–327. doi: 10.1006/jmbi.2000.4139. [DOI] [PubMed] [Google Scholar]
- 31.Yang S.Y., He X.Y. Role of type 10 17beta-hydroxysteroid dehydrogenase in the pathogenesis of Alzheimer’s disease. In: Tolnay M., Probst A., editors. Neuropathology and Genetics of Dementia. Kluwer Academic/Plenum Publishers; New York, NY, USA: 2001. pp. 101–110. [DOI] [PubMed] [Google Scholar]
- 32.He X.Y., Merz G., Chu C.H., Lin D., Yang Y.Z., Mehta P., Yang S.Y. Molecular cloning, modeling, and localization of rat type 10, 17β-hydroxysteroid dehydrogenase. Mol. Cell. Endocrinol. 2001;171:89–98. doi: 10.1016/S0303-7207(00)00391-9. [DOI] [PubMed] [Google Scholar]
- 33.Yang S.Y., He X.Y., Schulz H. Multiple functions of type 10 17beta-hydroxysteroid dehydrogenase. Trends Endocrinol. Metab. 2005;16:167–175. doi: 10.1016/j.tem.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 34.Yang S.Y., He X.Y., Schulz H. 3-Hydroxyacyl-CoA dehydrogenase and short-chain 3-hydroxyacyl-CoA dehydrogenase in human health and disease. FEBS J. 2005;272:4874–4883. doi: 10.1111/j.1742-4658.2005.04911.x. [DOI] [PubMed] [Google Scholar]
- 35.He X.Y., Isaacs C., Yang S.Y. Roles of mitochondrial 17beta-hydroxysteroid dehydrogenase type 10 in Alzheimer’s disease. J. Alzhaimer Dis. 2018;62:665–673. doi: 10.3233/JAD-170974. [DOI] [PubMed] [Google Scholar]
- 36.Vinklarova L., Schmidt M., Benek O., Kuca K., Gunn-Moore F., Musilek K. Friend or enemy? Review of 17β-HSD10 and its role in human health or disease. J. Neurochem. 2020;155:231–249. doi: 10.1111/jnc.15027. [DOI] [PubMed] [Google Scholar]
- 37.Vilardo E., Rossmanith W. The amyloid-β-SDR5C1(ABAD) interaction does not mediate a specific inhibition of mitochondrial RNase P. PLoS ONE. 2013;8:e65609. doi: 10.1371/journal.pone.0065609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang S.Y., He X.Y. Hadh2 and 3-hydroxyacyl-CoA dehydrogenase. Am. J. Physiol. Endocrinol. Metab. 2008;295:E987. doi: 10.1152/ajpendo.90521.2008. [DOI] [PubMed] [Google Scholar]
- 39.Kobayashi A., Jiang L.L., Hashimoto T. Two mitochondrial 3-hydroxyacyl-CoA dehydrogenases in bovine liver. J. Biochem. 1996;119:775–782. doi: 10.1093/oxfordjournals.jbchem.a021307. [DOI] [PubMed] [Google Scholar]
- 40.Schulz H. Fatty acid oxidation. In: Lennarz W., Lane M.D., editors. Encyclopedia of Biological Chemistry. Elsevier; San Diego, CA, USA: 2004. pp. 90–94. [Google Scholar]
- 41.He X.Y., Yang S.Y. 3-Hydroxyacyl-CoA dehydrogenase (HAD) deficiency replaces short-chain hydroxyacyl-CoA dehydrogenase (SCHAD) deficiency as well as medium- and short-chain hydroxyacyl-CoA dehydrogenase (M/SCHAD) deficiency as the consensus name of this fatty acid oxidation disorder. Mol. Genet. Metab. 2007;91:205–206. doi: 10.1016/j.ymgme.2007.02.015. [DOI] [PubMed] [Google Scholar]
- 42.Birktoft J.J., Holden H.M., Hamlin R., Xuong N.C., Banaszak L.J. Structure of L-3-hydroxyacyl-CoA dehydrogenase. Preliminary chain tracing at 2.8 A resolution. Proc. Natl. Acad. Sci. USA. 1987;84:8262–8266. doi: 10.1073/pnas.84.23.8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He X.Y., Yang Y.X., Yang S.Y. Changes of the HSD17B10 gene expression levels in ulcerative colitis. Inflamm. Bowel Dis. 2013;19:E23–E24. doi: 10.1002/ibd.22882. [DOI] [PubMed] [Google Scholar]
- 44.Shafqat N., Marschall H.U., Filling C., Nordling E., Wu X.Q., Björk L., Oppermann U. Expended substrate screenings of human and Drosophila type 10 17β-hydroxysteroid dehydrogenase reveal multiple apecificities in bile acid and steroid hormone metabolism. Characterization of multifunctional 3α/7α/7β/20β/21-hydroxysteroid dehydrogenase. Biochem. J. 2003;376:49–60. doi: 10.1042/bj20030877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hiltunen J.K., Kastaniotis A.J., Autio K.J., Jiang G., Chen Z., Glumoff T. 17B-hydroxysteroid dehydrogenases as acyl thioester metabolizing enzymes. Mol. Cell. Endocrinol. 2019;489:107–118. doi: 10.1016/j.mce.2018.11.012. [DOI] [PubMed] [Google Scholar]
- 46.Zhang W., Sang Y.M. Genetic pathogenesis, diagnosis, and treatment of short-chain 3-hydroxyacyl-coenzyme A dehydrogenase hyperinsulinism. Orphanet. J. Rare Dis. 2021;16:467. doi: 10.1186/s13023-021-02088-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He X.Y., Dobkin C., Brown W.T., Yang S.Y. 3-Hydroxyacyl-CoA and alcohol dehydrogenase activities of mitochondrial type 10 17beta-hydroxysteroid dehydrogenase in neurodegeneration study. J. Alzheimer Dis. 2022;88:1487–1497. doi: 10.3233/JAD-220481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adamski J., Jacob F.J. A guide to 17β-hydroxysteroid dehydrogenases. Mol. Cell. Endocrinol. 2001;171:1–4. doi: 10.1016/S0303-7207(00)00383-X. [DOI] [PubMed] [Google Scholar]
- 49.Korman S.H., Yang S.Y. HSD17B10 replaces HADH2 as the approved designation for the gene mutated in 2-methyl-3-hydroxybutyryl-CoA dehydrogenase deficiency. Mol. Genet. Metab. 2007;91:115. doi: 10.1016/j.ymgme.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 50.Yang S.Y., He X.Y., Isaacs C., Dobkin C., Miller D., Philipp M. Roles of 17β-hydroxysteroid dehydrogenase type 10 in neurodegenerative disorders. J. Steroid Biochem. Mol. Biol. 2014;143:460–472. doi: 10.1016/j.jsbmb.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 51.He X.Y., Dobkin C., Yang S.Y. 17beta-Hydroxysteroid dehydrogenases and neurosteroid metabolism in the central nervous system. Mol. Cell. Endocrinol. 2019;489:92–97. doi: 10.1016/j.mce.2018.10.002. [DOI] [PubMed] [Google Scholar]
- 52.Yang S.Y., He X.Y., Olpin S.E., Sutton V.R., McMenamin J., Philipp M., Malik M. Mental retardation linked to mutations in the HSD17B10 gene interfering with neurosteroid and isoleucine metabolism. Proc. Natl. Acad. Sci. USA. 2009;106:14820–14824. doi: 10.1073/pnas.0902377106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trauger J.W., Jiang A., Stearns B.A., LoGrasso P.V. Kinetics of allopregnanolone formation catalyzed by human 3α-hydroxysteroid dehydrogenase type III (AKR1C2) Biochemistry. 2002;41:13451–13459. doi: 10.1021/bi026109w. [DOI] [PubMed] [Google Scholar]
- 54.Boynton T.O., Shimkets L.J. Myxococcus CsgA, Drosophila Sniffer, and human HSD10 are cardiolipin phospholipases. Genes Dev. 2015;29:1903–1914. doi: 10.1101/gad.268482.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wohlfarter Y., Eidelpes R., Yu R.D., Sailer S., Koch J., Karall D., Keller M.A. Lost in promiscuity? An evolutionary and biochemical evaluation of HSD10 function in cardiolipin metabolism. Cell. Mol. Life Sci. 2022;79:562. doi: 10.1007/s00018-022-04579-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haines T.H. A new look at cardiolipin, editorial. Biochim. Biophys. Acta. 2009;1788:1997–2002. doi: 10.1016/j.bbamem.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 57.Bertolin G., Jacoupy M., Traver S., Ferrando-Miguel R., Saint Georges T., Grenier K., Corti O. Parkin maintains mitochondrial levels of the protective Parkinson’s disease-related enzyme 17-β hydroxysteroid dehydrogenase type 10. Cell Death Differ. 2015;22:1563–1576. doi: 10.1038/cdd.2014.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.He X.Y., Wegiel J., Kooy R.F., Brown W.T., Yang S.Y. HSD17B10 gene-related disorders are associated with abnormalities of mitochondrial function, morphology, dynamics and clearance. Ann. Genet. Genom. 2021;1:1005. [Google Scholar]
- 59.Holzmann J., Frank P., Löffler E., Bennett K.L., Gerner C., Rossmanith W. RNase P without RNA: Identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell. 2008;135:462–474. doi: 10.1016/j.cell.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 60.Yang S.Y., He X.Y., Miller D. Hydroxysteroid (17β) dehydrogenase X in human health and disease. Mol. Cell. Endocrinol. 2011;343:1–6. doi: 10.1016/j.mce.2011.06.011. [DOI] [PubMed] [Google Scholar]
- 61.Oerum S., Roovers M., Leichsenring M., Acquaviva-Bourdain C., Beermann F., Gemperle-Britschgi C., Yue W.W. Novel patient missense mutations in the HSD17B10 gene affect dehydrogenase and mitochondrial tRNA modification functions of the encoded protein. Biochim. Biophys. Acta. 2017;1863:3294–3302. doi: 10.1016/j.bbadis.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 62.Oerum S., Roovers M., Rambo R.P., Kopec J., Bailey H.J., Fitzpatrick F., Yue W.W. Structural insight into the human mitochondrial tRNA purine N1-methyltransferase and ribonuclease P complexes. J. Biol. Chem. 2018;293:12862–12876. doi: 10.1074/jbc.RA117.001286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bhatta A., Dienemann C., Cramer P., Hillen H.S. Structural basis of RNA processing by human mitochondrial RNase P. Nat. Struct. Mol. Biol. 2021;28:713–723. doi: 10.1038/s41594-021-00637-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vilardo E., Nachbagauer C., Buzet A., Taschner A., Holzmann J., Rossmanith W. A subcomplex of human mitochondrial RNase P is a bifunctional methyltransferase--extensive moonlighting in mitochondrial tRNA biogenesis. Nucleic Acid Res. 2012;40:11583–11593. doi: 10.1093/nar/gks910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marques A.T., Antunes A., Fernandes P.A., Ramos M.J. Comparative evolutionary genomics of the HADH2 gene encoding Abeta-binding alcohol dehydrogenase/17beta-hydroxysteroid dehydrogenase type 10 (ABAD/HSD10) BMC Genom. 2006;7:202. doi: 10.1186/1471-2164-7-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Richardson A., Berry G.T., Garganta C., Abbott M.A. Hydroxysteroid 17-Beta Dehydrogenase Type 10 Disease in Siblings. JIMD Rep. 2017;32:25–32. doi: 10.1007/8904_2016_547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rauschenberger K., Schöler K., Sass J.O., Sauer S., Djuric Z., Rumig C., Zschocke J. A non-enzymatic function of 17beta-hydroxysteroid dehydrogenase type 10 is required for mitochondrial integrity and cell survival. EMBO Mol. Med. 2010;2:51–62. doi: 10.1002/emmm.200900055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fukao T., Akiba K., Goto M., Kuwayama N., Morita M., Hori T., Hasegawa Y. The first case in Asia of 2-methyl-3-hydroxybutyryl-CoA dehydrogenase deficiency (HSD10 disease) with atypical presentation. J. Hum. Genet. 2014;59:609–614. doi: 10.1038/jhg.2014.79. [DOI] [PubMed] [Google Scholar]
- 69.Seaver L.H., He X.Y., Abe K., Cowan T., Enns G.M., Sweetman L., Yang S.Y. A novel mutation in the HSD17B10 gene of a 10-year-old boy with refractory epilepsy, choreoathetosis and learning disability. PLoS ONE. 2011;6:e27348. doi: 10.1371/journal.pone.0027348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zschocke J. HSD10 disease: Clinical consequences of mutations in the HSD17B10 gene. J. Inherit. Metab. Dis. 2012;35:81–89. doi: 10.1007/s10545-011-9415-4. [DOI] [PubMed] [Google Scholar]
- 71.Upadia J., Walano N., Noh G.S., Liu J., Li Y., Deputy S., Andersson H.C. HSD10 disease in a female: A case report and review of literature. JIMD Rep. 2021;62:35–43. doi: 10.1002/jmd2.12250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.He X.Y., Dobkin C., Brown W.T., Yang S.Y. Infantile neurodegeneration results from mutants of 17β-hydroxysteroid dehydrogenase type 10 rather than Aβ-binding alcohol dehydrogenase. Int. J. Mol. Sci. 2023;24:8487. doi: 10.3390/ijms24108487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vilardo E., Rossmanith W. Molecular insights into HSD10 disease: Impact of SDR5C1 mutations on the human mitochondrial RNase P complex. Nucleic Acids Res. 2015;43:5112–5119. doi: 10.1093/nar/gkv408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Akagawa S., Fukao T., Akagawa Y., Sasai H., Kohdera U., Kino M., Kaneko K. Japanese male siblings with 2-methyl-3-hydroxybutyryl-CoA dehydrogenase deficiency (HSD10 disease) without neurological regression. JIMD Rep. 2017;32:81–85. doi: 10.1007/8904_2016_570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Falk M.J., Gai X., Shigematsu M., Vilardo E., Takase R., McCormick E., Hou Y.M. A novel HSD17B10 mutation impairing the activities of the mitochondrial RNase P complex causes X-linked intractable epilepsy and neurodevelopmental regression. RNA Biol. 2016;13:477–485. doi: 10.1080/15476286.2016.1159381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Su L., Li X., Lin R., Sheng H., Feng Z., Liu L. Clinical and molecular analysis of 6 Chinese patients with isoleucine metabolism defects: Identification of 3 novel mutations in the HSD17B10 and ACAT1 gene. Metab. Brain Dis. 2017;32:2063–2071. doi: 10.1007/s11011-017-0097-y. [DOI] [PubMed] [Google Scholar]
- 77.Hochberg I., Demain L.A., Richer J., Thompson K., Urquhart J.E., Rea A., Newman W.G. Bi-allelic variants in the mitochondrial RNase P subunit PRORP cause mitochondrial tRNA processing defects and pleiotropic multisystem presentations. Am. J. Hum. Genet. 2021;108:2195–2204. doi: 10.1016/j.ajhg.2021.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Waters P.J., Lace B., Buhas D., Gravel S., Cyr D., Boucher R.M., Maranda B. HSD10 mitochondrial disease: P.Leu122Val variant, mild clinical phenotype, and founder effect in French-Canadian patients from Quebec. Mol. Genet. Genom. Med. 2019;7:e1000. doi: 10.1002/mgg3.1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Reyniers E., Van Bogaert P., Peeters N., Vits L., Pauly F., Fransen E., Kooy R.F. A new neurological syndrome with mental retardation, choreoathetosis, and abnormal behavior maps to chromosome Xp11. Am. J. Hum. Genet. 1999;65:1406–1412. doi: 10.1086/302638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lenski C., Kooy F., Reyniers R.F., Loessner E.D., Wanders R.J., Winnepenninckx B., Hellerand H., Engert S., Schwartz C.E., Meindl A., et al. The reduced expression of the HADH2 protein causes X-linked mental retardation, choreoathetosis, and abnormal behavior. Am. J. Hum. Genet. 2007;80:372–377. doi: 10.1086/511527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang Q., Chen P., Liu J., Lou J., Liu Y., Yuan H. Xp11.22 duplications in four unrelated Chinese families: Delineating the genotype-phenotype relationship for HSD17B10 and FGD1. BMC Med. Genom. 2020;13:66. doi: 10.1186/s12920-020-0728-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shafik A.M., Zhou H., Lim J., Dickinson B., Jin P. Dysregulated mitochondrial and cytosolic tRNA m1A methylation in Alzheimer’s disease. Hum. Mol. Genet. 2022;13:ddab357. doi: 10.1093/hmg/ddab357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Oppermann U.C., Salim S., Tjernberg L.O., Terenius L., Jornvall H. Binding of amyloid β-peptide to mitochondrialhydroxyacyl-CoA dehydrogenase (ERAB): Regulation of an SDR enzyme activity with implication for apoptosis in Alzheimer’s disease. FEBS Lett. 1999;451:238–242. doi: 10.1016/S0014-5793(99)00586-4. [DOI] [PubMed] [Google Scholar]
- 84.Aitken L., Quinn S.D., Perez-Gonzalez C., Samuel I.D.W., Penedo J.C., Gunn-Moore F.J. Morphology-specific inhibition of β-amyloid aggregates by 17β-hydroxysteroid dehydrogenase type 10. ChemBioChem. 2016;17:1029–1037. doi: 10.1002/cbic.201600081. [DOI] [PubMed] [Google Scholar]
- 85.Kristofikova Z., Springer T., Gedeonova E., Hofmannova A., Ricny J., Hromadkova L., Homola J. Interaction of 17β-hydroxysteroid dehydrogenase type 10 and cyclophilin D in Alzheimer’s disease. Neurochem. Res. 2020;45:915–927. doi: 10.1007/s11064-020-02970-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wen G.Y., Yang S.Y., Kaczmarski W., He X.Y., Pappas K.S. Presence of hydroxysteroid dehydrogenase type 10 in amyloid plaques (Aps) of Hsiao’s APP-Sw transgenic mouse brains, but absence in Aps of Alzheimer’s disease brains. Brain Res. 2002;954:115–122. doi: 10.1016/S0006-8993(02)03354-1. [DOI] [PubMed] [Google Scholar]
- 87.He X.Y., Wen G.Y., Merz G., Lin D., Yang Y.Z., Mehta P., Schulz H., Yang S.Y. Abundant type 10 17β-hydroxysteroid dehydrogenase in the hippocampus of mouse Alzheimer’s disease model. Mol. Brain Res. 2002;99:46–53. doi: 10.1016/S0169-328X(02)00102-X. [DOI] [PubMed] [Google Scholar]
- 88.Yang S.Y., He X.Y., Dobkin C., Isaacs C., Brown W.T. Mental retardation and isoleucine metabolism. In: Rajendram R., Preedy V.R., Patel V.B., editors. Branched Chain Amino Acids in Clinical Nutrition: Volume I, Nutrition and Health. Springer; New York, NY, USA: 2015. pp. 157–170. [Google Scholar]
- 89.Seshadri A., Alladi P.A. Divergent expression patterns of Drp1 and HSD10 in the nigro-striatum of two mice strains based on their MPTP susceptibility. Neurotox. Res. 2019;36:27–38. doi: 10.1007/s12640-019-00036-8. [DOI] [PubMed] [Google Scholar]
- 90.Tieu K., Perier C., Vila M., Caspersen C., Zhang H.P., Teismann P., Przedborski S. L-3-hydroxyacyl-CoA dehydrogenase II protects a model of Parkinson’s disease. Ann. Neurol. 2004;56:51–60. doi: 10.1002/ana.20133. [DOI] [PubMed] [Google Scholar]
- 91.Liu L., Chen S., Yu M., Ge C., Ren M., Liu B., Luo J. Deacetylation of HSD17B10 by SIRT3 regulates cell growth and cell resistance under oxidative and starvation stresses. J. Cell Death Dis. 2020;11:563. doi: 10.1038/s41419-020-02763-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.He X.Y., Yang Y.Z., Peehl D.M., Lauderdale A., Schulz H., Yang S.Y. Oxidative 3α-hydroxysteroid dehydrogenase activity of human type 10 17β-hydroxysteroid dehydrogenase. J. Steroid Biochem. Mol. Biol. 2003;87:191–198. doi: 10.1016/j.jsbmb.2003.07.007. [DOI] [PubMed] [Google Scholar]
- 93.Hemmerova E., Springer T., Kristofikova Z., Homola J. In vitro study of interaction of 17β-hydroxysteroid dehydrogenase type 10 and cyclophilin D and its potential implications for Alzheimer’s disease. Sci. Rep. 2019;9:16700. doi: 10.1038/s41598-019-53157-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Burry R.W. Specificity controls for immunocytochemical methods. J. Histochem. Cytochem. 2000;48:163–165. doi: 10.1177/002215540004800201. [DOI] [PubMed] [Google Scholar]
- 95.Fišar Z., Musílek K., Benek O., Hroch L., Vinklářová L., Schmidt M., Raboch J. Effects of novel 17β-hydroxysteroid dehydrogenase type 10 inhibitors on mitochondrial respiration. Toxicol. Lett. 2021;339:12–19. doi: 10.1016/j.toxlet.2020.12.012. [DOI] [PubMed] [Google Scholar]
- 96.Binstock J.F., Schulz H. Fatty acid oxidation complex from Escherichia coli. Methods Enzymol. 1981;71:403–411. doi: 10.1016/0076-6879(81)71051-6. [DOI] [PubMed] [Google Scholar]
- 97.He X.Y., Yang S.Y., Schulz H. Assay of L-3-hydroxyacyl-coenzyme A dehydrogenase with substrates of different chain lengths. Anal. Biochem. 1989;180:105–109. doi: 10.1016/0003-2697(89)90095-X. [DOI] [PubMed] [Google Scholar]
- 98.Yang S.Y., Schulz H. Kinetics of coupled enzyme kinetics. Biochemistry. 1987;26:5579–5584. doi: 10.1021/bi00391a054. [DOI] [PubMed] [Google Scholar]
- 99.Segel I.H. Enzyme Kinetics, Behavior and Analysis of Rapid Equilibrium and Steady-State Enzyme System. Wiley-Interscience; New York, NY, USA: 1975. [Google Scholar]
- 100.Schmidt M., Benek O., Vinklarova L., Hrabinova M., Zemanova L., Chribek M., Musilek K. Benzothiazolyl Ureas are Low Micromolar and Uncompetitive Inhibitors of 17β-HSD10 with Implications to Alzheimer’s Disease Treatment. Int. J. Mol. Sci. 2020;21:2059. doi: 10.3390/ijms21062059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kissinger C.R., Rejto P.A., Pelletier L.A., Thomson J.A., Showalter R.E., Abreo M.A., Villafranca J.E. Crystal structure of human ABAD/HSD10 with a bound inhibitor: Implications for design of Alzheimer’s disease therapeutics. J. Mol. Biol. 2004;342:943–952. doi: 10.1016/j.jmb.2004.07.071. [DOI] [PubMed] [Google Scholar]
- 102.Klotz I.M., Hunston D.L. Mathematical models for ligand-receptor binding. Real sites, Ghost sites. J. Biol. Chem. 1984;259:10060–10062. doi: 10.1016/S0021-9258(18)90927-0. [DOI] [PubMed] [Google Scholar]
- 103.Windholz M. The Merk Index. 10th ed. Merk & Co. Inc.; Rahway, NJ, USA: 1983. [Google Scholar]
- 104.Yang S.Y., He X.Y., Schulz H. Glutamate 139 of the large α-subunit is the catalytic base in the dehydration of both D- and L-3-hydroxyacyl-coenzyme A but not in the isomerization of Δ3, Δ2- enoyl coenzyme A catalyzed by the multienzyme complex of fatty acid oxidation from Escherichia coli. Biochemistry. 1995;34:6441–6447. doi: 10.1021/bi00130a009. [DOI] [PubMed] [Google Scholar]
- 105.Churchill P., Hempel J., Romovacek H., Zhang W.W., Churchill S., Brennan M. Primary structure of rat liver D-beta-hydroxybutyrate dehydrogenase from cDNA and protein analyses: A short-chain alcohol dehydrogenase. Biochemistry. 1992;31:3793–3799. doi: 10.1021/bi00130a009. [DOI] [PubMed] [Google Scholar]
- 106.Ayan D., Maltais R., Poirier D. Identification of a 17β-hydroxysteroid dehydrogenase type 10 steroidal inhibitor: A tool to investigate the role of type 10 in Alzheimer’s disease and prostate cancer. ChemMedChem. 2012;7:1181–1184. doi: 10.1002/cmdc.201200129. [DOI] [PubMed] [Google Scholar]
- 107.Xie J., Liang R., Wang Y., Huang J., Cao X., Niu B. Progress in Target Drug Molecules for Alzheimer’s Disease. Curr. Top. Med. Chem. 2020;20:4–36. doi: 10.2174/1568026619666191203113745. [DOI] [PubMed] [Google Scholar]
- 108.Lim J.W., Lee J., Pae A.N. Mitochondrial dysfunction and Alzheimer’s disease: Prospects for therapeutic intervention. BMB Rep. 2020;53:47–55. doi: 10.5483/BMBRep.2020.53.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Benek O., Vaskova M., Miskerikova M., Schmidt M., Andrys R., Rotterova A., Musilek K. Development of Submicromolar 17β-HSD10 inhibitors and their in vitro and in vivo evaluation. Eur. J. Med. Chem. 2023;258:115593. doi: 10.1016/j.ejmech.2023.115593. [DOI] [PubMed] [Google Scholar]
- 110.Schmidt M., Vaskova M., Rotterova A., Fiandova P., Miskerikova M., Zemanova L., Musilek K. Physiologically relevant fluorescent assay for identification of 17β-hydroxy-steroid dehydrogenase type 10 inhibitors. J. Neurochem. 2023;167:154–167. doi: 10.1111/jnc.15917. [DOI] [PubMed] [Google Scholar]
- 111.He X.Y., Wegiel J., Yang S.Y. Intracellular oxidation of allopregnanolone by human brain type 10 17β-hydroxysteroid dehydyrogenase. Brain Res. 2005;1040:29–35. doi: 10.1016/j.brainres.2005.01.022. [DOI] [PubMed] [Google Scholar]
- 112.Frackoviak J., Mazur-Kolecka B., Kazmarski W., Dickson D. Deposition of Alzheimer’s vascular amyloid-beta is associated with decreased expression of brain L-3-hydroxyacyl-coenzyme A dehydrogenase (ERAB) Brain Res. 2001;907:44–53. doi: 10.1016/S0006-8993(01)02497-0. [DOI] [PubMed] [Google Scholar]
- 113.Akwa Y. Steroid and Alzheimer’s disease: Changes associated with pathology and therapeutic potential. Int. J. Mol. Sci. 2020;21:4812. doi: 10.3390/ijms21134812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yang S.Y., Dobkin C., He X.Y., Brown W.T. Transcription start sites and epigenetic analysis of the HSD17B10 proximal promoter. BMC Biochem. 2013;14:17. doi: 10.1186/1471-2091-14-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yang S.Y., Dobkin C., He X.Y., Philipp M., Brown W.T. A 5-methylcytosine hotspot responsible for the prevalent HSD17B10 mutation. Gene. 2013;515:380–33384. doi: 10.1016/j.gene.2012.12.064. [DOI] [PubMed] [Google Scholar]
- 116.Miller A.P., Willard H.F. Chromosomal basis of X chromosome inactivation: Identification of a multigene domain in Xp11.21-p.11.22 that escapes X inactivation. Proc. Natl. Acad. Sci. USA. 1998;95:8709–8714. doi: 10.1073/pnas.95.15.8709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ortez C., Villar C., Fons C., Duarte S.T., Pérez A., García-Villoria J., García-Cazorla A. Undetectable levels of CSF amyloid-beta peptide in a patient with 17β-hydroxysteroid dehydrogenase deficiency. J. Alzheimer Dis. 2011;27:253–257. doi: 10.3233/JAD-2011-110647. [DOI] [PubMed] [Google Scholar]
- 118.He X.Y., Dobkin C., Yang S.Y. Does the HSD17B10 gene escape from X-inactivation? Eur. J. Hum. Genet. 2011;19:123–124. doi: 10.1038/ejhg.2010.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zigman W.B., Schupf N., Devenny D.A., Miezejeski C., Ryan R., Urv T.K., Silverman W. Incidence and prevalence of dementia in elderly adults with mental retardation without down syndrome. Am. J. Ment. Retard. 2004;109:126–141. doi: 10.1352/0895-8017(2004)109<126:IAPODI>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 120.Yang S.-Y., He X.-Y., Olpin S.E., Sutton V.R., McMenamin J., Philipp M., Denman R.B., Malik M. Mental retardation linked to mutations in the HSD17B10 gene. Proc. Natl. Acad. Sci. USA. 2009;106:14735–14736. doi: 10.1073/iti3509106. [DOI] [PMC free article] [PubMed] [Google Scholar]