Abstract
The genome of Caenorhabditis elegans was the first animal genome to be sequenced. Although considerable effort has been devoted to annotating it, the standard WormBase annotation contains thousands of predicted genes for which there is no cDNA or EST evidence. We hypothesized that a more complete experimental annotation could be obtained by creating a more accurate gene-prediction program and then amplifying and sequencing predicted genes. Our approach was to adapt the TWINSCAN gene prediction system to C. elegans and C. briggsae and to improve its splice site and intron-length models. The resulting system has 60% sensitivity and 58% specificity in exact prediction of open reading frames (ORFs), and hence, proteins–the best results we are aware of any multicellular organism. We then attempted to amplify, clone, and sequence 265 TWINSCAN-predicted ORFs that did not overlap WormBase gene annotations. The success rate was 55%, adding 146 genes that were completely absent from WormBase to the ORF clone collection (ORFeome). The same procedure had a 7% success rate on 90 Worm Base “predicted” genes that do not overlap TWINSCAN predictions. These results indicate that the accuracy of WormBase could be significantly increased by replacing its partially curated predicted genes with TWINSCAN predictions. The technology described in this study will continue to drive the C. elegans ORFeome toward completion and contribute to the annotation of the three Caenorhabditis species currently being sequenced. The results also suggest that this technology can significantly improve our knowledge of the “parts list” for even the best-studied model organisms.
Caenorhabditis elegans, a soil nematode, is a major model organism for biomedical research and particularly for genomics. Its genome was the first genome of a multicellular organism to be sequenced (C. elegans Sequencing Consortium 1998) and continues to be a focus of intensive research. Expressed Sequence Tag (EST) and cDNA sequencing have been performed on C. elegans, a full-time staff curates its genome database (Stein et al. 2001; Harris et al. 2004), and the genome of a second soil nematode, C. briggsae, has been sequenced for comparison (Stein et al. 2003). A relatively accurate gene-prediction program called GENEFINDER was developed and optimized for C. elegans (P. Green, unpubl.), and an attempt has been made to amplify and clone all of the open reading frames (ORFs) in an early version of the C. elegans genome annotation (Reboul et al. 2003). Nonetheless, there are still thousands of genes in the standard annotation without any support from native EST/cDNA sequence. The annotations of Arabidopsis thaliana and Drosophila melanogaster are in a similar state.
To improve the completeness and accuracy of the C. elegans gene set, we adapted and extended the TWINSCAN gene-prediction algorithm (Korf et al. 2001) for C. elegans and C. briggsae. TWINSCAN combines the probabilistic Hidden Markov Model approach of programs like GENSCAN (Burge and Karlin 1997) with information derived from the alignment of the target genome to a second genome, called the informant. TWINSCAN was originally developed for the annotation of the human genome, using the sequence of the mouse genome as informant (Flicek et al. 2003). The predictions of this original system contributed to the discovery and verification of novel exons in human by RT–PCR and direct sequencing (Guigó et al. 2003). RT–PCR and sequencing of TWINSCAN predictions in rat, using human as the informant, amplified complete ORFs that were partially or completely absent from the standard annotation (Wu et al. 2004). More recently, TWINSCAN was adapted for the pathogenic fungus Cryptococcus neoformans, where 50% of the predicted ORFs tested were amplified and end-sequenced (Tenney et al. 2004).
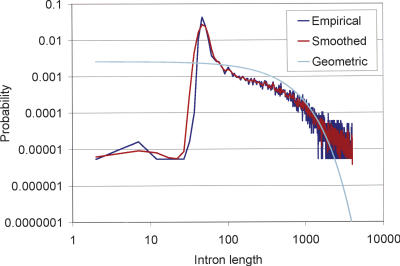
In adapting TWINSCAN for C. neoformans, we replaced the commonly used geometric model of intron lengths with a more accurate “smoothed empirical” model (for details, see Tenney et al. 2004). The latter is obtained by counting known introns of each length up to some maximum (400 for C. neoformans, 4000 for C. elegans), smoothing the counts, and then dividing each count by the total (Fig. 1). Nongeometric models have traditionally been avoided, because, in general, they require additional computing time that is proportional to the square of the maximum intron length. In this study, we demonstrate that smoothed empirical distributions are computationally feasible for C. elegans, despite the fact that it has a much larger genome and much larger introns than C. neoformans. The greater variability of intron lengths in C. elegans, as compared with C. neoformans, means that using an empirical distribution is less informative in principle, but we show that it is well worth the computational cost.
Figure 1.
Empirical, smoothed empirical, and geometric intron length distributions up to 4000 nt, on a log–log scale. The smoothed empirical length distribution is very close to the distribution observed in the 3889 fully confirmed genes of WS100. The geometric distribution, in contrast, assigns far too much probability to very short introns, too little to introns of the most common lengths, too much to introns between 100 and 1000 nt, and too little to introns longer than 2000 nt.
We also added a probability model for GC splice donors, which are much rarer than those starting with GT and also less variable in the flanking splice site sequence. Specifically, GC donors were added to TWINSCAN's Maximum Dependency Decomposition model for splice donors (see Burge and Karlin 1997). The smoothed empirical intron length distribution, the model for GC splice donors, and the C. briggsae alignments are the major factors contributing to the accuracy of TWINSCAN's C. elegans predictions.
TWINSCAN for worms was tested both computationally, by comparison to known gene structures, and experimentally, by amplification and sequencing of predicted ORFs that do not overlap any ORF in the standard WormBase annotation. We also tested a sample of predicted ORFs from WormBase that did not overlap TWINSCAN predictions and a set of ORFs on which TWINSCAN and WormBase agreed on the start and stop codons, but not the internal structure. We conclude that, where there is no existing cDNA evidence, TWINSCAN is substantially more accurate than WormBase.
Results
Computational evaluation using known genes
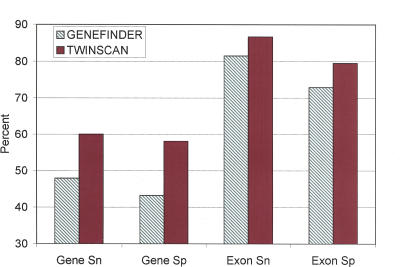
TWINSCAN 2.01 and GENEFINDER (release 980504) were both run on the C. elegans genome (see Methods). Their accuracy was evaluated by comparison to the 5569 transcripts at 4705 loci that are labeled “fully cDNA confirmed” in the WS130 version of WormBase (Stein et al. 2001; Fig. 2). In the figure, only predicted ORFs whose genomic extent overlaps that of a fully confirmed ORF by at least 1 nucleotide were used in order to avoid penalizing the prediction of real, but previously unknown genes; results for all predicted genes are given in Supplemental Table S1. TWINSCAN was substantially more accurate than GENEFINDER, especially in terms of its ability to predict the complete ORF, and hence, the protein product, exactly right. Although TWINSCAN was slightly more accurate than GENEFINDER in predicting internal exons, the bulk of its advantage is in predicting gene boundaries. Indeed, TWINSCAN predicted the annotated start codon for 75% of known genes, vs. 72% for GENEFINDER; the numbers for the stop codon were 86% vs. 81%, and for both start and stop, 64% vs. 55%.
Figure 2.
Accuracy of TWINSCAN and GENEFINDER on C. elegans estimated by comparison to 5569 fully confirmed ORFs (WS130). TWINSCAN uses alignments to the C. briggsae genome, an empirical intron-length model, and a model of GC splice donors. (Gene Sn) Percentage of loci with fully confirmed ORFs, at which TWINSCAN predicts one confirmed ORF exactly right. (Gene Sp) Percentage of TWINSCAN predictions that exactly match fully confirmed ORFs. Predictions that do not overlap any confirmed ORF are not counted. (Exon Sn and Exon Sp) Exact matches to coding regions of exons in fully confirmed ORFs.
In order to evaluate the effect of the C. briggsae genome alignment on prediction accuracy, we repeated this experiment using TWINSCAN in its nonconservation mode, which does not use genomic alignments. The results, shown in Table 1, indicate that comparison to C. briggsae yields clear, but modest improvements.
Table 1.
Comparison of TWINSCAN accuracy with and without alignments between the genomes of C. elegans and C. briggsae, calculated as in Figure 2
| Gene sensitivity | Overlap gene specificity | Exon sensitivity | Overlap exon specificity | |
|---|---|---|---|---|
| TS without briggsae | 57.0% | 55.8% | 85.5% | 77.5% |
| TWINSCAN with briggsae | 60.0% | 58.1% | 86.7% | 79.5% |
We also ran TWINSCAN with C. briggsae alignments but without the smoothed empirical intron-length model. A geometric intron-length model with the same mean as the empirical data was used instead (Fig. 1). The results indicate that the smoothed empirical intron-length model improved both exact gene prediction accuracy (4.7% Sn, 4.8% Sp) and exact exon prediction (1.4% Sn, 1.9% Sp) (Table 2). This comes at the cost of increased computing time—each 500-kb fragment takes about 10 min on a typical current machine when the intron length limit is 4000 nt, as compared with 47 sec using the geometric distribution. However, most of the accuracy improvement can be achieved with half the running time by using an empirical-length distribution up to 2000 nt and a geometric tail for longer introns (see Stanke and Waack 2003; data not shown).
Table 2.
Accuracy of TWINSCAN predictions with the commonly used geometric intron length distributions versus the smoothed empirical length distribution
| Gene sensitivity | Overlap gene specificity | Exon sensitivity | Overlap exon specificity | |
|---|---|---|---|---|
| Geometric intron length | 55.3% | 53.3% | 85.3% | 77.6% |
| Empirical intron length | 60.0% | 58.1% | 86.7% | 79.5% |
Both results use briggsae alignments and allow GC splice donors.
When TWINSCAN was run with C. briggsae alignments and the smoothed empirical intron-length distribution, but without the GC-AG intron model, exact gene sensitivity dropped by 0.7% and specificity by 0.2%. With the GC-AG intron model, a total of 913 genes were predicted to have GC-AG introns (4.2%); 53 of 142 known genes with GC-AG introns were predicted correctly (37%).
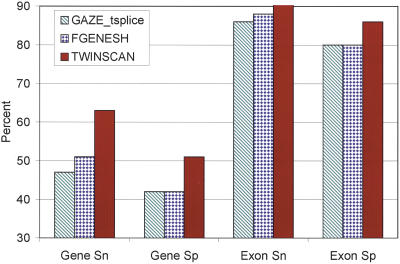
Finally, we compared TWINSCAN with two other gene-prediction systems that have recently been developed for nematodes–FGENESH (Salamov and Solovyev 2000; Stein et al. 2003) and GAZE (Howe et al. 2002). Figure 3 shows the results for FGENESH and GAZE on the GAZE data set (http://www.sanger.ac.uk/Software/analysis/GAZE) as reported by Howe et al. (2002), together with the result of running TWINSCAN on the same data set.
Figure 3.
Comparison of the accuracy of GAZE (with its trans-splicing model), FGENESH, and TWINSCAN on the GAZE test set. Numbers for GAZE and FGENESH are taken from Howe et al. (2002).
Computational annotation of the C. elegans genome
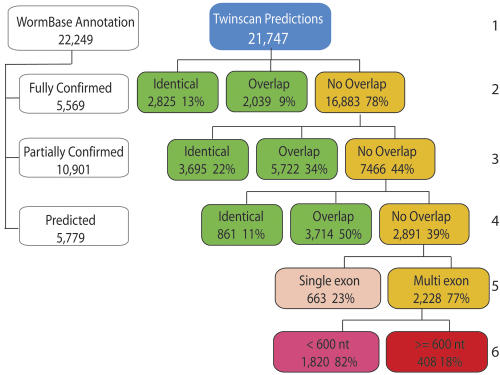
TWINSCAN 2.01 was run on the entire C. elegans genome (WS130) divided into 500-kb fragments. The 21,747 predicted ORFs were then compared with the annotations in WS130 by using the Eval software package (Keibler 2003; http://genes.cse.wustl.edu/eval/). The results are shown in Figure 4. Worm-Base contained 22,249 ORFs (including alternative splices) at 20,461 loci. Since the TWINSCAN ORFs are one per locus, TWINSCAN is predicting 1286 more gene loci than WormBase. WormBase transcripts are classified as confirmed, partially confirmed, or predicted. “Predicted” annotations are GENEFINDER predictions that have been manually reviewed, and in some cases, deleted or adjusted by experts in particular gene families (J. Spieth, pers. comm.). This comparison shows that there is both significant agreement between TWINSCAN and WormBase (7381 ORFs are identical) and substantial disagreement (7466 TWINSCAN predictions do not overlap either partially or fully confirmed WormBase annotations). Of the 7466 TWINSCAN predictions that do not overlap partially or fully confirmed Worm-Base annotations, 429 overlap repeats masked by the latest Rep-Base libraries (July, 2004) for at least 50% of the ORF, while 288 overlap pseudogenes annotated in WS130 for at least 50% of the ORF. When TWINSCAN predictions that overlap WormBase-predicted genes are factored out (Fig. 4, line 4), -9% of the remaining 2891 TWINSCAN predictions overlap repeats (261) and 10% overlap pseudogenes (276).
Figure 4.
Breakdown of genome-wide predictions by TWINSCAN 2.01 in comparison to the WS130 annotations. (Row 1) Total number of WormBase annotations and TWINSCAN predictions. (Row 2) Breakdown of TWINSCAN predictions into those that are identical to fully confirmed WormBase predictions, those that overlap but are not identical, and those that do not overlap (orange). (Row 3) Breakdown of TWINSCAN predictions that do not overlap fully cDNA-confirmed ORFs by comparison to the partially cDNA confirmed WormBase ORFs. (Row 4) Breakdown of TWINSCAN predictions that do not overlap fully or partially confirmed WormBase ORFs by comparison to predicted WormBase ORFs. (Row 5) Breakdown of TWINSCAN predictions that do not overlap any of the above into single exon (beige) and multiexon (orange) predictions. (Row 6) Breakdown of novel multiexon TWINSCAN predictions into those that are shorter than 200 amino acids (pink) and those that are at least 200 amino acids (red). Analysis of predictions by an earlier and slightly less accurate version of TWINSCAN (2.0α), by comparison to WS100 ORFs, placed 265 novel ORFs of at least 200 amino acids in the red box, all of which were tested experimentally.
Amplifying, cloning, and sequencing predicted novel genes
These experiments were based on an earlier version of TWINSCAN (2.0α) that was less accurate than the one described above by about 4% in exact gene sensitivity and 2% in specificity (see Supplemental methods for differences). The first set of TWINSCAN predictions we targeted consisted of the 265 multi-exon ORFs that did not overlap any annotation in WormBase version WS100, nor anything in the ORFeome collection, and were at least 200 amino acids long. For each of these, we designed specific tailed PCR primers to anneal at the beginning and end of the predicted ORF (Hartley et al. 2000; Walhout et al. 2000a) and performed PCR on a highly representative C. elegans cDNA library (Walhout et al. 2000b). Tailed ORFs were cloned using the Gateway recombinational cloning system and the cloned inserts were end-sequenced using vector-specific primers. Of the 265 targets, nine (3%) yielded only low quality sequence, 33 (13%) yielded only vector sequence, 13 (5%) yielded nonvector sequence that did not match the targeted gene or matched another region better, 64 (24%) yielded sequence that matched the targeted gene without an intron gap, and 146 (55%) yielded sequences that matched the targeted gene and spanned an intron. Intron-spanning sequences are the most reliable indicators of transcription, although other sequences are not necessarily indicators of a wrong prediction. Some of the sequences amplified in these successful experiments may consist of two or more additional exons of known genes, but if so, then the ORF of the complete mRNA would necessarily be different from the one found by TWINSCAN. Estimating the frequency with which our amplicons are parts of larger known transcripts would require additional PCR experiments spanning the putative intergenic regions. Of the 504 introns whose boundaries we determined experimentally, 461 (92%) were predicted correctly by TWINSCAN; of the 1008 splice sites, 956 (95%) were predicted correctly. These numbers are higher than in computational comparison to known genes, because the predictions that fail PCR amplification and, hence, do not yield an experimentally determined intron, contain a disproportionate number of incorrectly predicted introns.
After the experiments were performed, we determined that 21 of the targets overlap the current pseudogene set by at least 50%, while three of them overlap the current set of interspersed repeats by at least 50%. The success rates for these targets were near zero (1/21 and 0/3, respectively). Thus, the success rate would have been higher had we been able to mask these pseudogenes and interspersed repeats prior to running TWINSCAN.
Further investigation of the 146 confirmed novel predictions revealed that they are less conserved between elegans and briggsae than the known genes. By our methods of genome alignment, the confirmed novel predictions are, on average, 45% covered by briggsae genome alignments, as compared with 69% for all confirmed genes in WormBase. Within the aligned regions, the confirmed novel genes show only 75.4% nucleotide identity, as compared with 78.6% for WormBase confirmed. However, novel predictions that were not confirmed in this experiment showed even less conservation than those that were (26% aligned and 71.6% identity). Thus, highly conserved genes are likely to have been known already, whereas very poorly conserved predictions are likely to be false positives or at least difficult to confirm by our methods. When we started the experiments, only 25 of the 146 confirmed novel genes matched ESTs at 95% identity over 100 bp, and in the latest release of WormBase, there are an additional 13 that have such ESTs. Furthermore, this match criterion probably counts some ESTs that are not transcribed from the relevant locus. Overall, these numbers indicate that the majority of the 146 confirmed novel genes are expressed at levels below those that readily yield ESTs. Finally, the 146 show highly statistically significant differences in codon usage patterns as compared with known elegans genes for every amino with multiple codons except Histidine. For example, the two rarest codons for Leucine in known elegans genes are CTA (7.4%) and TTA (8.4%); in the confirmed novel genes, these rare codons are used more frequently (429 CTAs = 11.0% and 509 TTAs = 13.1%). Many of the codons whose frequency is greater in our genes are AT rich, consistent with the observed 3% increase in AT in our genes.
Seventy of the confirmed novel genes have <50% amino acid identity to the most similar gene in the WS130 release of WormBase, including predicted genes. Ninety-two of them have no PFAM hit. The other 54 have a total of 68 hits, of which the most common (six hits, two genes with three each) was the ShK toxin domain, found in a toxin from brown sea anemone, as well as several hypothetical C. elegans proteins. The next most common were WD40 (four hits in one gene) and F-Box (four hits in four genes). WD40 is found in β-transducin, a subunit of G proteins, which act as intermediaries in signal transduction, and F-Box is “present in numerous proteins and serves as a link between a target protein and a ubiquitin-conjugating enzyme.” The remaining PFAM hits showed no obvious patterns.
For comparison, we also targeted a random sample of 90 genes from the 1632 multiexon genes that are listed in Worm-Base as predicted, and which do not overlap any prediction by TWINSCAN 2.0α. Only six of these yielded sequences that matched the targeted gene and spanned at least one intron (7%). Unlike the TWINSCAN targets, these were not selected to have ORFs of at least 200 amino acids. However, the 53 ORFs longer than 200 amino acids had a lower success rate (3/53) than the 37 ORFs shorter than 200 amino acids (3/37). The difference between the PCR success rates for WormBase-predicted ORFs not overlapping TWINSCAN and TWINSCAN ORFs not overlapping WormBase is highly significant (χ2 = 64.4, P < 10-14). Of the 22 introns whose boundaries we determined experimentally, 17 (77%) were predicted correctly in WormBase; of the 44 splice sites, 37 (84%) were predicted correctly.
The fact that these WormBase targets had a low success rate is to be expected, given that many of them may already have been targeted for amplification and cloning in other experiments; if these experiments had succeeded, the targeted genes would no longer be considered predicted. Our results do not constitute an evaluation of GENEFINDER predictions or WormBase annotations in general, but they do constitute a fair evaluation of the 1632 predicted ORFs in WS100 that do not overlap TWINSCAN predictions.
Finally, we targeted a random sample of 96 multiexon ORFs on which TWINSCAN agreed with WormBase on the translation start and stop, but not the internal structure. Three of these experiments resulted in amplification of a related gene from a different locus (mispriming). Of the remaining 93, 31 yielded sequences that aligned to the target gene with at least one intron spliced out (33%). The fact that this success rate is lower than the 55% for TWINSCAN predictions not overlapping WormBase ORFs may be due to depletion of amplifiable ORFs from the predicted set in WS100. The fact that this success rate is higher than the 7% for WormBase predicted ORFs that do not overlap TWINSCAN predictions indicates that TWINSCAN has considerable power to discriminate good from bad annotations even within this depleted set. Once again, the targets with WormBase ORFs longer than 200 amino acids had a lower success rate (24/84) than those with WormBase ORFs shorter than 200 amino acids (7/12). Of the 139 introns whose boundaries we determined experimentally, TWINSCAN predicted 82% correctly, whereas WormBase predicted 76% correctly. Of 278 experimentally determined splice sites, TWINSCAN predicted 89% correctly, vs. 84% for WormBase.
All predictions, primers, experimental sequences and traces, and alignments to the genome can be found at http://genes.cse.wustl.edu/wei-2005/. This site also contains a link for visualizing the confirmed novel genes on the UCSC C. elegans genome browser. Traces have been submitted to the NCBI Trace Archive and assigned ID numbers 580100347–580100718.
Discussion
The computational and molecular experiments described above all indicate that replacing the partially curated, “predicted” genes in WormBase with noncurated TWINSCAN predictions would improve the accuracy of the annotation. Two other gene-finding programs, GAZE (Howe et al. 2002) and FGENESH (Salamov and Solovyev 2000; Stein et al. 2003), also appear to be more accurate than GENEFINDER (Howe et al. 2002). The computational experiments reported above, however, indicate that the latest version of TWINSCAN is still more accurate, particularly in predicting complete ORFs (hence, proteins) exactly. We are not aware of any other system that, using genome sequences as its only inputs, can predict a correct protein product for 60% of known genes in a whole-genome annotation of a multicellular organism. The improvements that led to this high-level accuracy–the GC-AG intron model, briggsae alignments, and empirical intron-length distribution–could be incorporated into other comparative, hidden Markov model-based gene-finding programs.
In the C. briggsae genome paper, Stein et al. (2003) developed a procedure aimed at selecting the best gene model among those produced by several prediction programs (see Supplemental materials for details). When this procedure was tested on a set of elegans genes for which the WormBase annotation was known to be correct, the correct model was chosen 92% of the time. This number only bears on the sets of overlapping gene models that include at least one correct model. It provides no information about the overall accuracy of the “hybrid gene set” that the procedure produces, since we do not know what fraction of the overlapping predictions include a correct gene model. Thus, using this procedure on the latest TWINSCAN predictions, along with predictions from other programs, might improve on the accuracy of TWINSCAN alone, but that is not guaranteed–it depends on the number of genes for which the other programs have a correct model, but TWINSCAN does not.
Although the availability of the C. briggsae genome sequence was the original motivation for this work, we found that using it improved TWINSCAN's accuracy on C. elegans only modestly. This is almost certainly due to the high degree of divergence between elegans and briggsae (about 79% nucleotide identity in aligned coding regions, compared with 85% for mouse and human). For compact genomes like these, better results have been achieved at much closer evolutionary distances (Tenney et al. 2004). Three more species of Caenorhabditis are now being sequenced, but none are thought to be much closer to elegans than briggsae (Sternberg et al. 2003; Cho et al. 2004). It is not clear whether there is an extant organism anywhere near the optimal evolutionary distance for TWINSCAN prediction in elegans (probably 90%–95%). However, multigenome alignments based on several of these species may well yield substantial improvements in gene prediction accuracy (Siepel and Haussler 2004; Gross and Brent 2005).
The two other factors leading to the improved performance of TWINSCAN were modeling intron length accurately and allowing GC splice donors. The empirical intron-length model comes at the cost of increased computing demands, relative to other programs. However, we have shown that it is computationally feasible and worth the necessary investment of computing power. Modeling GC splice donors leads to a slight improvement in exact gene prediction, because, although only 0.54% of known worm introns begin with GC, about 2.6% of known transcripts contain at least one GC-AG intron.
C. elegans was the first multicellular organism to be fully sequenced, and its sequence is among the best annotated. Nonetheless, the latest version of TWINSCAN (2.01) predicted 7466 ORFs that do not overlap WormBase annotation with support from native cDNA sequence. Among these, 2891 do not even overlap predicted genes in WormBase. Using an earlier, slightly less-accurate TWINSCAN version (2.0α), we were able to amplify, clone, and sequence 146 previously unpredicted genes–55% of those targeted. Since short predicted ORFs tended to show a higher success rate, we feel safe in extrapolating this 55% rate to all 2228 multiexon TWINSCAN predictions that do not overlap any annotation in WormBase. Correcting for the 8.6% excess of repeats and pseudogenes in this set, as compared with the target set used for the experiments, yields an effective set size 2035. Multiplying by 55% yields an estimate of 1119 novel genes that can be confirmed in a single attempt in an organism with extensive experimental annotation and some curation. The 2891 TWINSCAN predictions that overlap, but do not agree with WormBase predictions, should yield about 1000 more confirmable targets. By amplifying, cloning, and sequencing the remainder of these targets, we expect to start closing in on the C. elegans ORFeome.
C. elegans is not unique. Other heavily studied model organisms, such as Arabidopsis thaliana, are also likely to contain well more than 1000 completely unannotated genes, and thousands more misannotated genes. Sequencing into cDNA libraries has reached saturation, but de novo gene prediction followed by RT–PCR and sequencing is providing a high yield of new, experimentally determined gene structures. This is largely the result of recent increases in the accuracy of gene structure prediction algorithms (Brent and Guigó 2004). Future improvements in prediction algorithms can be expected to lower the cost per confirmed gene, while bringing the experimental annotation of genomes ever closer to completion. Even with current technology, however, RT–PCR is a cost-effective approach to experimental annotation of eukaryotic genomes, from fungi to round worms to mammals.
Methods
TWINSCAN predictions
For the computational comparisons, TWINSCAN 2.01 was trained and tested on the 3889 genes that are labeled “fully cDNA confirmed” in the WS100 version of WormBase (Stein et al. 2001). Repetitive sequences were not masked out. The genome was divided into 200 segments of 500 kb each, and each segment was randomly assigned to one of eight groups. Each segment was then aligned to the genome of C. briggsae by using WU-BLAST (http://blast.wustl.edu), and the alignments were converted to conservation sequence (see Korf et al. 2001; Flicek et al. 2003). TWINSCAN was trained on the DNA and conservation sequences of known genes from seven of the eight groups and run on the 500-Kb segments from the eighth, in order to avoid training and testing on the same data. This was repeated eight times, holding out a different group each time, and the results were combined.
To compute the smoothed empirical distribution, we counted the introns of each length from 1 to 4000 in the training set and smoothed the counts using a discretized Gaussian filter with variance of five over a window of 10 nt to either side. At the boundaries where the window included lengths outside of the 1–4000 range, the counts were taken to be zero. The smoothed counts were then divided by their sum to yield a discrete distribution that sums to 1.
In both TWINSCAN runs, alignments between the genomes of C. elegans and C. briggsae (version cb25.agp8) were used. To prepare the C. briggsae database, sequences longer than 150 kb were cut into 150-kb fragments with 20 kb overlap. Each fragment was masked for low-complexity sequence by running NSEG with default parameters (Wootton and Federhen 1996). The C. elegans genome sequence was divided into 500-Kb query segments, which were aligned to the briggsae sequences by using nucleotide blast from the WU-BLAST package with parameters M = 1 N = -1 Q = 5 R = 1 B = 10000 V = 100 lcfilter filter = seg filter = dust topcomboN = 1.
In a subsequent repeat analysis, the July 2004 repeat libraries were downloaded from RepBase (http://www.girinst.org/server/RepBase/repeatmaskerlibraries/repeatmaskerlibrariesJuly2004.tar.gz).
PCR, cloning, and sequencing
PCR amplification, cloning and sequencing were performed as described in Reboul et al. (2003).
Analysis of experimental sequences
Sequence analysis was as described in Wu et al. (2004), except that reads were quality clipped before analysis, and quality values were not consulted thereafter.
Acknowledgments
We are grateful to Sean Eddy, John Spieth, and Jeltje van Baren for their insightful comments on early drafts. Thanks to the WormBase staff for their work in maintaining WormBase, as well as providing specific annotation data we requested. Thanks to Nansheng Chen for help with the pseudogene analysis. C. elegans work in the Brent lab was supported by NSF grant DBI-0132436. M.B. was also supported, in part, by NIH grant HG-02278. This work was also supported by grants 7 R33 CA81658-02 from the National Cancer Institute and 5R01HG01715-02 from the National Human Genome Research Institute and the National Institute of General Medical Sciences awarded to M.V.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3329005.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Brent, M.R. and Guigó, R. 2004. Recent advances in gene structure prediction. Curr. Opin. Struct. Biol. 14: 264-272. [DOI] [PubMed] [Google Scholar]
- Burge, C. and Karlin, S. 1997. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268: 78-94. [DOI] [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium. 1998. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science 282: 2012-2018. [DOI] [PubMed] [Google Scholar]
- Cho, S., Suk-Won, J., Cohen, A., and Ellis, R. 2004. A phylogeny of Caenorhabditis reveals frequent loss of introns during nematode evolution. Genome Res. 14: 1209-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek, P., Keibler, E., Hu, P., Korf, I., and Brent, M.R. 2003. Leveraging the mouse genome for gene prediction in human: From whole-genome shotgun reads to a global synteny map. Genome Res. 13: 46-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross, S.S. and Brent, M.R. 2005. Using multiple alignments to improve gene prediction. RECOMB 2005 (in press). [DOI] [PubMed]
- Guigó, R., Dermitzakis, E.T., Agarwal, P., Ponting, C., Parra, G., Reymond, A., Abril, J.F., Keibler, E., Lyle, R., Ucla, C., et al. 2003. Comparison of mouse and human genomes followed by experimental verification yields an estimated 1,019 additional genes. Proc. Natl. Acad. Sci. 100: 1140-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, T.W., Chen, N., Cunningham, F., Tello-Ruiz, M., Antoshechkin, I., Bastiani, C., Bieri, T., Blasiar, D., Bradnam, K., Chan, J., et al. 2004. WormBase: A multi-species resource for nematode biology and genomics. Nucleic Acids Res. 32: D411-D417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley, J.L., Temple, G.F., and Brasch, M.A. 2000. DNA cloning using in vitro site-specific recombination. Genome Res. 10: 1788-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe, K.L., Chothia, T., and Durbin, R. 2002. GAZE: A generic framework for the integration of gene-prediction data by dynamic programming. Genome Res. 12: 1418-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keibler, E. and Brent, M.R. 2003. Eval: A software package for analysis of genome annotations. BMC Bioinformatics 4: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf, I., Flicek, P., Duan, D., and Brent, M.R. 2001. Integrating genomic homology into gene structure prediction. Bioinformatics 17: S140-S148. [DOI] [PubMed] [Google Scholar]
- Reboul, J., Vaglio, P., Rual, J.F., Lamesch, P., Martinez, M., Armstrong, C.M., Li, S., Jacotot, L., Bertin, N., Janky, R., et al. 2003. C. elegans ORFeome version 1.1: Experimental verification of the genome annotation and resource for proteome-scale protein expression. Nat. Genet. 34: 35-41. [DOI] [PubMed] [Google Scholar]
- Salamov, A.A. and Solovyev, V.V. 2000. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 10: 516-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel, A.C. and Haussler, D. 2004. Computational identification of evolutionarily conserved exons. In RECOMB. ACM, San Diego, CA.
- Stanke, M. and Waack, S. 2003. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics 19: II215-II225. [DOI] [PubMed] [Google Scholar]
- Stein, L., Sternberg, P., Durbin, R., Thierry-Mieg, J., and Spieth, J. 2001. WormBase: Network access to the genome and biology of Caenorhabditis elegans. Nucleic Acids Res. 29: 82-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, L.D., Bao, Z., Blasiar, D., Blumenthal, T., Brent, M.R., Chen, N., Chinwalla, A., Clarke, L., Clee, C., Coghlan, A., et al. 2003. The genome sequence of Caenorhabditis briggsae: A platform for comparative genomics. PLoS Biol. 1: E45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg, P.W., Waterston, R.H., Spieth, J., Eddy, S.R., and Wilson, R.K. 2003. Genome sequence of additional Caenorhabditis species: Enhancing the utility of C. elegans as a model organism. National Human Genome Research Institute.
- Tenney, A., Brown, R.H., Vaske, C., Lodge, J.K., Doering, T.L., and Brent, M.R. 2004. Gene prediction and verification in a compact genome with numerous small introns. Genome Res. 14: 2330-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walhout, A.J., Sordella, R., Lu, X., Hartley, J.L., Temple, G.F., Brasch, M.A., Thierry-Mieg, N., and Vidal, M. 2000a. Protein interaction mapping in C. elegans using proteins involved in vulval development. Science 287: 116-122. [DOI] [PubMed] [Google Scholar]
- Walhout, A.J., Temple, G.F., Brasch, M.A., Hartley, J.L., Lorson, M.A., van den Heuvel, S., and Vidal, M. 2000b. GATEWAY recombinational cloning: Application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol. 328: 575-592. [DOI] [PubMed] [Google Scholar]
- Wootton, J.C. and Federhen, S. 1996. Analysis of compositionally biased regions in sequence databases. Methods Enzymol. 266: 554-571. [DOI] [PubMed] [Google Scholar]
- Wu, J.Q., Shteynberg, D., Arumugam, M., Gibbs, R.A., and Brent, M.R. 2004. Identification of rat genes by TWINSCAN gene prediction, RT-PCR, and direct sequencing. Genome Res. 14: 665-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
Web site references
- http://www.girinst.org/server/RepBase/repeatmaskerlibraries/repeatmaskerlibrariesJuly2004.tar.gz; Repeat libraries used in the foregoing analysis.
- http://www.sanger.ac.uk/Software/analysis/GAZE; GAZE data set.
- http://genes.cse.wustl.edu/eval/; Eval software.
- http://genes.cse.wustl.edu/wei-2005/; Predictions, primers, experimental sequences and traces, and genome alignments.
- http://blast.wustl.edu; Washington University BLAST archives.