Abstract
Pseudomonas aeruginosa is a major cause of nosocomial (hospital-derived) infections, is the predominant pathogen in chronic cystic fibrosis lung infections, and remains difficult to treat due to its high intrinsic antibiotic resistance. The completion of the P. aeruginosa PAO1 genome sequence provides the opportunity for genome-wide studies to increase our understanding of the pathogenesis and biology of this important pathogen. In this report, we describe the construction of a mini-Tn5-luxCDABE mutant library and a high-throughput inverse PCR method to amplify DNA flanking the site of insertion for sequencing and insertion site mapping. In addition to producing polar knockout mutations in nonessential genes, the promoterless luxCDABE reporter present in the transposon serves as a real-time reporter of gene expression for the inactivated gene. A total of 2519 transposon insertion sites were mapped, 77% of which were nonredundant insertions. Of the insertions within an ORF, -55% of total and unique insertion sites were transcriptional luxCDABE fusions. A bias toward low insertion-site density in the genome region that surrounds the predicted terminus of replication was observed. To demonstrate the utility of chromosomal lux fusions, we performed extensive regulatory screens to identify genes that were differentially regulated under magnesium or phosphate limitation. This approach led to the discovery of many known and novel genes necessary for these environmental adaptations, including genes involved in resistance to cationic antimicrobial peptides. This dual-purpose mutant library allows for functional and regulation studies and will serve as a resource for the research community to further our understanding of P. aeruginosa biology.
Genome-wide mutant libraries are collections of isogenic mutants in nonessential genes and are a powerful tool to understand the biology of an organism. Mutant libraries have been constructed systematically in Escherichia coli and Bacillus subtilis (Kobayashi et al. 2003; Kang et al. 2004) or by random transposon mutagenesis in Neisseria meningitidis, Mycoplasma, Pseudomonas aeruginosa, Helicobacter pylori, and the yeast Saccharomyces cerevisiae (Hutchison et al. 1999; Geoffroy et al. 2003; Jacobs et al. 2003; Salama et al. 2004; Scherens and Goffeau 2004). Saturating mutagenesis strategies enable the recovery of mutants in all nonessential genes, and therefore provide an estimate for the number of essential genes. In addition, mutant libraries allow the characterization of many known and unknown genes by standard phenotypic screening. Mutants can be generated with additional properties, depending on the features of the transposon used. For example, the transposon can encode a variety of promoterless reporter genes (phoA, lacZ, gfp, lux) to generate transcriptional or translational fusions to the inactivated gene. Transposons may contain outward-facing, inducible promoters that avoid polar mutations and permit identification of essential genes under noninducing conditions (Judson and Mekalanos 2000). Also, the presence of recombination sites within the transposon permits the use of epitope or hexahistidine tags for protein localization studies or protein purification, respectively (Ross-Macdonald et al. 1999; Jacobs et al. 2003).
P. aeruginosa is the primary cause of chronic lung infections in cystic fibrosis patients, one of the most frequently isolated pathogens from nosocomial infections and poses problems for immunocompromised AIDS and cancer patients (Govan and Deretic 1996; Quinn 1998; Hancock and Speert 2000). P. aeruginosa initiates infections at mucosal surfaces, but has the capacity to become invasive and therefore is frequently isolated from the lung, wounds, urinary tract, and blood. Infections are difficult to treat due to the high intrinsic antibiotic resistance of this organism attributed to low outer membrane permeability coupled with additional resistance mechanisms, such as active drug efflux and antibiotic modification (Hancock and Speert 2000). In addition to being a prominent human pathogen, P. aeruginosa is pathogenic to plants, animals, nematodes, and insects, and is therefore an excellent model for host–pathogen interaction studies, given the variety of experimental infection models (Hendrickson et al. 2001).
Between the original manual PAO1 genome annotation (http://www.pseudomonas.com) (Stover et al. 2000) and a subsequent reannotation (Weinel et al. 2003), up to 75% of the 5570 ORFs have been assigned probable functions. The aim of this research was to generate a library of multifunctional mutants that could be used for further studies to ultimately increase our understanding of the function of those genes that remain uncharacterized. We demonstrate the utility of this library by identifying differentially regulated genes from bacteria grown under magnesium and phosphate limitation and using this information to establish functions for selected genes involved in cationic antimicrobial peptide (CAP) resistance.
Results and Discussion
Mini-Tn5-lux transposon mutagenesis
Random mini-Tn5 transposon mutagenesis was used to generate mutants in P. aeruginosa PAO1. The mini-Tn5 derivative used contains the inner and outer transposase recognition sequences flanking a promoterless luxCDABE operon and a tetracycline (Tc) resistance cassette (Winson et al. 1998). This transposon was selected because insertion into a gene results in a knockout mutation and may lead to the production of a transcriptional luxCDABE fusion. The bioluminescence lux reporter is ideal for high-throughput gene expression studies because of the ability to monitor light production in real-time without the addition of enzyme substrates. All transposon insertions result in polar mutations of downstream genes in the same operon, due to the presence of transcriptional and translational termination signals that flank the tetracycline resistance marker.
The total library consisted of -18,500 randomly picked mutants without discriminating by colony morphology or size. The first 9600 mutants were robotically arrayed in 96-well plates for long-term storage before any phenotypic screening. The remaining 8900 mutants were randomly picked and screened for differentially regulated lux fusions (see below) before selecting 384 mutants to be maintained, which were added to the initial batch of arrayed mutants. In addition, the library is available in the form of an unresolved pool of mutants to permit screening for additional phenotypes.
High-throughput inverse PCR and sequencing for insertion-site mapping
A variety of methods are available for mapping random transposon insertion sites, ranging from direct cloning of flanking DNA, direct sequencing of genomic DNA with transposon-specific primers (Slesarev et al. 2002), and various PCR methods to amplify DNA flanking the transposon for sequencing (Karlyshev et al. 2000; Geoffroy et al. 2003; Jacobs et al. 2003). We developed a high-throughput inverse PCR protocol that involved the isolation of genomic DNA, digestion with restriction enzymes that cut once within the transposon but numerous times within the genome, and ligation to circularize all linear genomic fragments, followed by PCR using two outward-facing, transposon-specific primers. All genomic DNA samples were NarI digested and processed through the inverse PCR protocol. If the PCR reaction failed, the process was repeated with SstII-digested genomic DNA. Restriction enzymes that cut more frequently in the genome, such as NarI, generate higher success rates for inverse PCR. This is likely due to the smaller average genomic fragment size after digestion, which facilitated circularization during ligation, and ultimately produced smaller PCR products.
An initial phase of random transposon mapping was carried out on mutants randomly distributed among the library, and a second phase of transposon mapping was performed on mutants screened for specific phenotypes. PCR products were obtained for -72% of all samples processed through two successive rounds of inverse PCR (3325/4608). All PCR-positive products were purified, sequenced, and mapped to the P. aeruginosa PAO1 genome. Sequencing primers were designed to sequence across the junction of the transposon and flanking genomic DNA where the transposon insertion site corresponded to the first nucleotide after the transposon-specific sequence. A summary of the sequencing and insertion-site mapping results are shown in Table 1.
Table 1.
Features of the mini-Tn5-luxCDABE mutant library in P. aeruginosa PAO1
| Feature | N |
|---|---|
| Random mini-Tn5-luxCDABE mutants isolated | 18,500 |
| Mutants arrayed and stored | 9984 |
| Samples processed through inverse PCR | 4608 |
| DNA sequencing reactions | 3325 |
| Total mapped insertion sites | 2519 |
| Insertions within a gene | 2275 |
| Intergenic insertions | 244 |
| Transcriptional luxCDABE fusions | 1253 |
| Nonredundant insertion sites | 1932 |
| Unique ORFs inactivated | 1284 |
| Unique intergenic insertions | 174 |
| Unique transcriptional luxCDABE fusions | 784 |
| Predicted PAO1 ORFs | 5570 |
In total, 2519 transposon insertion sites were mapped to the genome, both within predicted genes and in intergenic regions. Approximately 77% of all insertions were nonredundant (1932/2519), which was defined as insertions >50 bp apart. The exact insertion site was difficult to map for PCR products derived from SstII-digested genomic DNA, due to the proximity of the nested sequencing primer to the transposon junction. Mini-Tn5-lux transposons were mapped to 1284 unique predicted P. aeruginosa ORFs, representing knockouts in nearly 25% of nonessential P. aeruginosa genes (1284/5170) (Jacobs et al. 2003). A database was designed to facilitate searching for available mutants (http://pseudomutant.pseudomonas.com). These mutants will be made freely available to the community through the Pseudomonas Genome Database (http://www.pseudomonas.com).
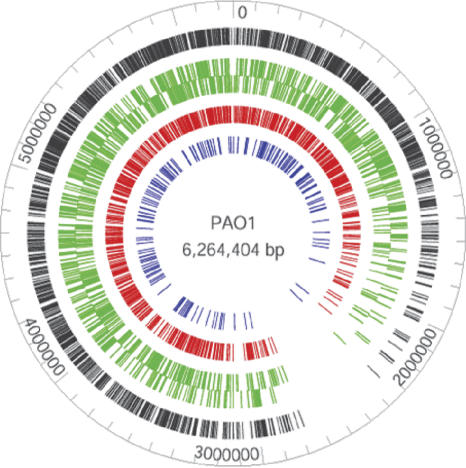
Transposon insertion sites are evenly distributed except for the predicted terminus of replication
The distribution of mini-Tn5-lux insertion sites in the PAO1 genome is shown in Figure 1. With the exception of a region between 2 and 3 Mb, transposon insertion sites were evenly distributed throughout the chromosome. The low-insertion density in this region has been previously reported by Jacobs et al. (2003); however, they did not propose an explanation for this distribution pattern. The transposon Tn5 inserts at a degenerate asymmetric 9-bp sequence, contributing to the randomness of transposition (Kang et al. 2004). We therefore searched the PAO1 genome for this sequence and concluded that this sequence is evenly distributed throughout the PAO1 genome (data not shown), indicating that the region of low-insertion site density is not due to a lack of transposition events. It is also worth nothing that putative essential genes are randomly distributed throughout the genome (Jacobs et al. 2003), suggesting that this particular region does not have an unusually high number of essential genes that cannot be inactivated.
Figure 1.
Distribution of mini-Tn5-lux transposon insertions in the PAO1 genome. Total insertions are shown in the outer black ring. Green bars represent insertions within ORFs where the outer and inner rings correspond to genes encoded by the top and bottom strand. Red bars represent insertions within ORFs that result in transcriptional luxCDABE fusions. Blue bars represent insertions within intergenic regions.
Interestingly, analyses of nucleotide skews, including GC skew, AT skew, and purine excess indicated that the terminus of the P. aeruginosa genome is located around the 2.5-Mb position (Song et al. 2003; Hallin and Ussery 2004). Since the region between 2 and 3 Mb surrounds the predicted terminus of replication, we therefore propose that the low-insertion site density in this region may be due to the lower average number of targetgene copies in replicating cells.
Cationic antimicrobial peptide resistance genes are induced under Mg2+ starvation
Approximately 55% of all transposon mutants generated were transcriptional lux fusions (Table 1). To demonstrate the utility of this sensitive, real-time reporter of gene expression, mutants were screened in various growth conditions to identify genes that were differentially regulated. Two-component regulatory systems comprise a major class of regulatory proteins among the 10% of predicted P. aeruginosa ORFs that are annotated as regulatory proteins (Stover et al. 2000); however, few of the 64 two-component regulatory systems in P. aeruginosa have been well studied, and the environmental signals leading to their activation are largely unknown.
In P. aeruginosa, the PhoPQ and PmrAB two-component systems are known to respond to Mg2+ limitation and regulate the expression of genes necessary for resistance to cationic antimicrobial peptides and aminoglycosides (Macfarlane et al. 1999, 2000; McPhee et al. 2003). In an attempt to identify genes that are regulated by magnesium limitation and that are potentially members of the PhoPQ or PmrAB regulons, we screened a total of 18,500 randomly picked mutants for lux fusions that were strongly induced or repressed when grown in minimal medium supplemented with 20 μM MgSO4 (limiting) and 2 mM MgSO4 (sufficient). Mutants of interest were identified in a preliminary screen by visualizing the relative luminescence intensities of mid-log cultures grown in these two conditions. Initial phenotypes were then confirmed by quantitative measurement of light production throughout the growth cycle under both conditions.
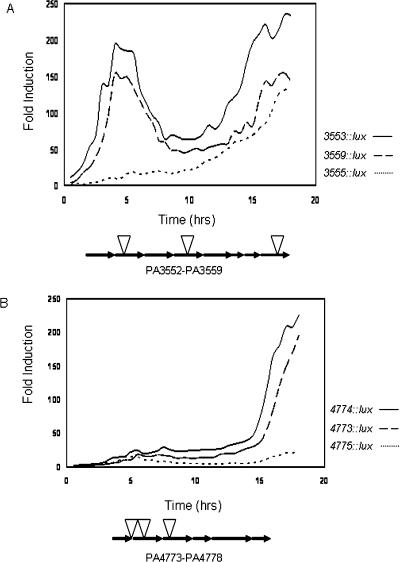
Table 2 lists the mutants with mapped insertion sites that were strongly induced during growth under Mg2+ limitation. The five-gene operon between PA4773 and PA4777 that encodes pmrAB, and the eight-gene operon between PA3552 and PA3559 that encodes a putative LPS modification operon were among the most highly induced operons (Table 2; Fig. 2). Interestingly, the putative LPS modification operon showed a biphasic pattern of induction during growth under limiting magnesium in two of three transcriptional lux fusions to this operon. This operon was massively induced by 134–235-fold throughout growth (Fig. 2A). Transcriptional lux fusions in the three genes upstream of pmrAB showed maximal expression in the later stages of growth with between 22- and 196-fold induction under magnesium limitation (Fig. 2B).
Table 2.
Genes induced under magnesium-limited growth
| Fold inductiona | Insertion site | Gene | Description | Mutant ID |
|---|---|---|---|---|
| 235 | PA3553 | pmrF | Glycosyl transferase, LPS modification | 53 D10 |
| 226 | PA4774 | Similar to polyamine synthesis protein | 38 F9 | |
| 196 | PA4773b | Similar to polyamine synthesis protein | 11 A9 | |
| 155 | PA4825 | mgtA | Mg2+ transport P-type ATPase | 25 E5 |
| 154 | PA3559 | pmrN | Nucleotide sugar dehydratase, LPS modification | 54 D3 |
| 134 | PA3555 | pmrj | Hypothetical protein, LPS modification | 146 H1 |
| 25 | PA3690 | Metal transport P-type ATPase | 12 F9 | |
| 23 | PA5398 | FMN oxidoreductase | 11 B7 | |
| 22 | PA4775 | Hypothetical protein | 50 F5 | |
| 20 | PA5400 | Electron transfer flavoprotein | 11 G6 | |
| 10 | PA4359 | feoA | Fe2+ transport protein | 39 G1 |
| 6 | PA5529 | Antiporter transport protein | 11 D7 | |
| 6 | PA4010 | DNA glycosylase | 19 C2 | |
| 3 | PA4496 | Periplasmic binding protein, ABC transporter | 21 E1 | |
| 3 | PA1048 | Outer membrane porin, OmpA-like | 36 H1 |
Ratio of normalized RLU/OD600 measurements from low and high magnesium conditions. The maximum fold induction values throughout growth are shown
Intergenic insertion between PA4773 and PA4774 (McPhee et al. 2003)
Figure 2.
Induction of the PA3552–PA3559 and PA4773–PA4777 operons under Mg2+ limitation. The PA3552–PA3559 likely encodes an LPS modification operon that is strongly induced within hours of growth in Mg2+-limiting medium. (A) Transcriptional luxCDABE fusions to PA3553, PA3555, and PA3559 are shown. The PA4773–PA4777 operon that encodes pmrAB is strongly induced in the late stages of growth in Mg2+-limiting medium. (B) Transcriptional luxCDABE fusions to PA4773, PA4774, and PA4775 are shown. Luminescence (RLU) and OD620 were measured every 30 min throughout growth. All luminescence values were divided by the OD620 values to correct for growth and the ratio of normalized relative light units (RLU) in low to high Mg2+ conditions are plotted. All values shown are the mean of triplicate samples. Standard deviations were within 10% of the means.
Our laboratory previously reported that the pmrAB system was regulated by Mg2+ and was necessary for polymyxin B resistance (McPhee et al. 2003). It was subsequently shown that the pmrAB system regulates the addition of aminoarabinose to the lipid A moiety of P. aeruginosa LPS (Moskowitz et al. 2004). This species of modified LPS has been observed in minimally cultured CF isolates of P. aeruginosa (Ernst et al. 1999), and it is believed that the PA3552–PA3559 operon is responsible for the aminoarabinose modification, as it is homologous to a similar operon in Salmonella typhimurium (Gunn et al. 1998). MIC assays were performed to determine whether the PA3552–PA3559 operon plays a role in resistance to CAPs in P. aeruginosa. Mutations in both the pmrAB and PA3552–PA3559 operons resulted in significant increases in susceptibility to polymyxin B and other cationic antimicrobial peptides such as CP10A and CP11CN, which are improved variants of indolicidin, a cationic peptide from bovine neutrophils (Table 3) (Friedrich et al. 2000; Rozek et al. 2003).
Table 3.
Minimal inhibitory concentrations (MIC) of cationic antimicrobial peptides for P. aeruginosa strains grown in minimal BM2-glucose medium with limiting Mg2+
|
MIC (μg ml-1)
|
||||
|---|---|---|---|---|
| Strain | Genotype | Polymyxin B | CP10A | CP11CN |
| H103 | wt | 8 | 32 | >32 |
| 11 A9 | PA4773::lux | 4 | 2 | 8 |
| 38 F9 | PA4774::lux | 8 | 2 | 32 |
| 50 F5 | PA4775::lux | 1 | 2 | 16 |
| 53 D10 | PA3553::lux | <0.06 | 16 | 4 |
| 146 H1 | PA3555::lux | <0.06 | 16 | 4 |
| 54 D3 | PA3559::lux | <0.06 | 16 | 8 |
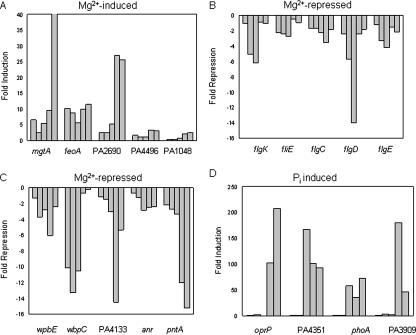
In addition to antibiotic-resistance genes, there were several transport proteins up-regulated under Mg2+ limitation, including a Mg2+-specific transport ATPase protein MgtA and a second P-type ATPase for metal transport (Table 2; Fig. 3A). Interestingly, an insertion into the first gene in an operon that is homologous to the Fe2+ iron transport operon feoAB was also induced under Mg2+ limitation, indicating that this iron transport pathway is coregulated by magnesium availability and may be promiscuous enough to mediate a certain level of divalent metal ion transport (Fig. 3A).
Figure 3.
Expression profiles of differentially regulated genes during growth in magnesium and phosphate limiting medium. (A) Genes involved in nutrient transport that were induced under Mg2+ limitation. (B) Flagellar genes that were repressed under Mg2+ limitation. (C) LPS biosynthesis and anaerobic genes that were repressed under Mg2+ limitation. (D) Phosphate acquisition and phospholipid synthesis genes that were induced under PO 3-4 limitation. Bars represent fold induction or repression at 2, 4, 6, 8, and 10 h of growth scanning from left to right for any given gene fusion. The y-axis represents the fold induction or fold repression. Standard deviations were within 10% of the means.
Screening for differentially regulated genes also led to the identification of a number of genes that were repressed when grown under magnesium limitation (Fig. 3; Supplemental data). There were five Mg2+-repressed genes involved in flagellar synthesis that fell into three operons (Fig. 3B). To confirm that Mg2+ starvation repressed flagella synthesis in P. aeruginosa, motility assays were conducted in motility agar supplemented with MgSO4 or the divalent cation chelator EDTA. Swimming motility (flagella-dependent; LB plus 0.3% agar) and swarming motility (type IV pili and flagella-dependent; BHI plus 0.5% agar) were increasingly inhibited as the concentration of magnesium decreased (data not shown).
P. aeruginosa smooth LPS is a heterogeneous mixture of two kinds of chemically and antigenically distinct molecules termed the A and B bands (Rivera et al. 1988). The B band LPS represents a major antigen that determines serological specificity in P. aeruginosa through the nature of the sugar residues in its O-antigen polysaccharide portion. Two genes in the B band O antigen biosynthesis cluster, wpbE and wbpC, were found to be repressed under growth in limiting magnesium (Fig. 3C). Unlike the mutants affecting lipid A modification described above, O-antigen mutants showed normal resistance to polymyxin B (data not shown). Interestingly, several genes that were associated with anaerobic or microaerobic growth, including the anaerobic transcriptional regulator anr, the pyridine nucleotide transhydrogenase pntA, and a cbb3-type cytochrome c oxidase (PA4133) were also repressed under Mg2+-limiting growth conditions (Fig. 3C).
Phosphate limitation induces phosphate acquisition genes
Genes regulated by environmental inorganic phosphate levels comprise the phosphate regulon. The PhoBR two-component system is required for the induction of genes necessary for phosphate acquisition (Filloux et al. 1988). Recent data indicated that the phosphate regulon is induced in P. aeruginosa after interaction with epithelial cells (Frisk et al. 2004). To identify genes that were regulated by phosphate limitation, and thus, potentially members of the PhoBR regulon, we screened a total of 12,000 mutants for lux fusions that were strongly induced when grown in minimal medium with limiting phosphate (0.2 mM). From this screen, we identified phosphate acquisition genes that were previously known to be phosphate regulated, including the major outer membrane phosphate-specific porin OprP (Siehnel et al. 1988) and alkaline phosphatase PhoA (PA3296) (Filloux et al. 1988; Table 4; Fig. 3D).
Table 4.
Genes induced under phosphate-limiting growth
| Fold inductiona | Insertion site | Gene | Description | Mutant ID |
|---|---|---|---|---|
| 207 | PA3279 | oprP | Phosphate-specific outer membrane porin | 81 C6 |
| 179 | PA3909 | Extracellular nuclease | 41 A5 | |
| 166 | PA4351 | olsA | Acyltransferase, ornithine lipid biosynthesis | 75 C4 |
| 72 | PA3296 | phoA | Alkaline phosphatase | 154 D1 |
| 15 | PA0688 | pstS | Phosphate-binding periplasmic protein, ABC transporter | 53 G1 |
| 6 | PA2645 | nuoJ | NADH dehydrogenase | 25 G2 |
| 4 | PA3965 | Transcriptional regulator | 14 B10 | |
| 3 | PA3690 | Metal-transport P-type ATPase | 12 F9 | |
| 3 | PA3682 | Carboxyphosphoenolpyruvate mutase | 145 C5 |
Ratio of normalized RLU measurements from low and high phosphate conditions. The maximum fold induction/repression values throughout growth are shown
A number of novel phosphate acquisition genes were also identified, including a protein with homology to the Escherichia coli PstS periplasmic-binding protein of a phosphate-specific ABC transporter (PA0688), and a protein with homology to an extracellular nuclease (PA3909) (Table 4, Fig. 3D). PA3909 may represent a novel phosphate acquisition mechanism in P. aeruginosa, whereby phosphate is obtained by degrading nucleic acids. Interestingly, a protein with homology to alkaline phosphatase (PA3910) is upstream and in the same operon as PA3909.
One interesting phosphate starvation-induced gene was PA4351, which encodes a protein with 50% similarity to an acyltransferase (olsA) involved in ornithine lipid synthesis in Sinorhizobium meliloti (Weissenmayer et al. 2002). Under phosphate-limiting conditions, bacteria can produce alternative phospholipids that replace the phosphate head group with the amino acid ornithine, or other basic amino acids, to produce phospholipids with a net positive charge. Pseudomonas aeruginosa is known to produce a small percentage of ornithine-derived lipids under growth in rich medium, but the biosynthetic genes have not been identified (Kawai et al. 1988).
Conclusion
We report the construction of a multifunctional mini-Tn5-luxCDABE mutant library in P. aeruginosa PAO1 that is useful for standard phenotypic screening and identifying differentially regulated genes. The utility of generating mutants with chromosomal luxCDABE fusions was illustrated by screening for genes differentially regulated by magnesium and phosphate-limiting conditions. Many of the Mg2+-regulated lux fusions mapped to genes required for resistance to cationic antimicrobial peptides. The database of available mutants has been integrated into the Pseudomonas Genome Database (http://www.pseudomonas.com).
There are multiple mutant library projects in P. aeruginosa, with each library constructed with various transposons and in several commonly used strains. A saturating mutagenesis was recently performed in P. aeruginosa PAO1 using a combination of mini-Tn5-phoA and lacZ transposons, which led to the identification of 300–400 candidate essential genes (Jacobs et al. 2003). Mutant libraries have also been constructed in strains PAK (Jacobs et al. 2003) and PA14 (F.M. Ausubel, pers. comm.). Signature-tagged mutagenesis has also been performed to create mutant libraries that permit the identification of genes required for virulence in vivo (Potvin et al. 2003). Each of these mutant libraries has its own assets, but the availability of such a large variety of mutants in multiple P. aeruginosa strains is of major benefit to Pseudomonas researchers and those performing comparative functional genomic analyses with other Pseudomonas species.
Methods
Mutant construction and transposon-insertion mapping
The mini-Tn5-lux transposon was delivered to P. aeruginosa PAO1 by conjugation with the donor strain E. coli S17-1 λ-pir (pUTmini-Tn5-luxCDABE-Tc) (Winson et al. 1998). P. aeruginosa recipient cells were grown at 42°C prior to biparental mating. Mixtures of the donor and recipient (1:2) were incubated on LB agar for 6–8 h at 37°C. To select for P. aeruginosa transconjugants, cells were resuspended in PBS and plated onto large 22 × 22-cm LB agar plates containing 50 μg/mL of tetracycline (Tc) to selectively kill the E. coli donor. Transconjugants were robotically picked into 96-well plates containing 100 μL of LB broth + 50 μg/mL of Tc, grown overnight, and frozen after the addition of DMSO (7% final concentration) for long-term storage at -80°C.
A high-throughput inverse PCR protocol was developed for the mapping of transposon sites, in which all steps were performed in a 96-well format with the use of a DNA Engine Tetrad gradient cycler (MJ Research). Cells were harvested from 24-h cultures (1.2 mL), and genomic DNA was isolated using the DNeasy Kit (Qiagen) according to manufacturer's recommendations. Genomic DNA was examined for purity and quantity on 1% agarose-Tris-acetate-EDTA (TAE) gels. For amplication of DNA flanking the left side of the transposon, SstII digestion, ligation, and inverse PCR were performed (SstII-IPCR), while for amplification of the right side of the transposon, NarI digestion, ligation, and inverse PCR (NarI-IPCR) were performed. Alternatively, SphI was used instead of SstII for amplification of the left side of the transposon. These restriction enzymes were chosen because their cut sites are present in the transposon, and they were likely to cut near the site of insertion due to the large number of cut sites in the PAO1 genome. Between 250 and 500 ng of genomic DNA was digested overnight with 2 U of NarI (NEB) or SstII (Invitrogen) (20 μL reaction volume) and heat inactivated at 65°C (SstII) or 85°C (NarI). T4 DNA Ligase (0.5 U) (Invitrogen) and its corresponding buffer was added to the digestion reaction to bring the volume to 25 μL and incubated overnight with temperatures cycling between 10 and 30°C. The ligation product was used as template for an inverse PCR reaction. (See Supplemental information for PCR conditions and primer sequences). The PCR reaction products were run on 1% agarose-TAE gels, and all samples yielding a single major band were selected for clean up with magnetic beads (Agencourt) and sequencing. Alternatively, a semi-random two-step (arbitrary) PCR method to amplify DNA flanking the transposon site was occasionally used as previously described (McPhee et al. 2003). PCR products were sequenced with nested primers at the University of Victoria Sequencing Centre or occasionally in our lab using Big Dye Terminator chemistry (Applied Biosystems) on a Basestation 51 Fragment Analyzer (MJ Research).
Transposon insertion sites in the PAO1 genome were determined using an automated process, whereby a PERL script parsed the top scoring BLASTN alignment to each query sequence. The coordinates for each insertion were then used to reference an annotation table of ORFs and intergenic regions. A second PERL script was used to identify transcriptional versus nontranscriptional fusions given the orientation of the gene where the insertion had occurred, the restriction enzyme used in the initial digestion, and the orientation of the transposon relative to the origin of replication in the PAO1 genome.
Global gene-expression assays
We developed an assay to screen thousands of mini-Tn5-lux mutants for differential expression during growth in various conditions. To identify genes induced or repressed under Mg2+ limitation, bacteria were grown in BM2-glucose minimal medium (Macfarlane et al. 2000) containing 20 μM (low) or 2 mM (high) MgSO4. To identify genes induced or repressed under PO 3-4 limitation, bacteria were grown in a defined phosphate minimal medium (Hancock et al. 1982) containing 200 μM (low) or 1 mM (high) PO 3-4. A 48-pin replicator stamp was used to subculture overnight LB cultures into the left and right half of 96-well plates containing “low” and “high” medium conditions, respectively. After growth for 4–6 h, the luminescence was observed using a ChemiGenius2 Bio-Imaging System (Syngene). From the preliminary screen, mutants were selected that were either strongly induced or repressed and were retested by quantitatively measuring luminescence (RLU) using a SPECTRAFluorPlus luminometer (Tecan) throughout growth. Luminescence was corrected for growth by simultaneously monitoring the absorbance at 620 nm.
Antibiotic susceptibility assays
The minimal inhibitory concentration (MIC) of various cationic antimicrobial peptides was determined using a modified microtitre broth dilution technique as previously described (Amsterdam 1991; Wu and Hancock 1999). Polypropylene plates were used to prevent aggregation of peptides on polystyrene and artificially high MICs. Susceptibility was assayed in BM2-glucose minimal medium containing 20 μM MgSO4. Growth was scored after 24-h incubation at 37°C.
Acknowledgments
S.L. was supported by a Canadian Cystic Fibrosis Foundation (CCFF) fellowship. J.B.M. was supported by a CCFF studentship. F.S.L.B. was a Michael Smith Foundation for Health Research Scholar. R.E.W.H. held a Canada Research Chair. We thank Glenn A. Cooper, Ross Gibbs, Marianne Beetz-Sargent, and Ben Koop for DNA sequencing performed at the Genome BC Sequencing Centre of the University of Victoria. Funding for this research was from the Functional Pathogenomics of Mucosal Immunity program supported by Genome Prairie and Genome BC with ancillary support from Inimex Pharmaceuticals Inc, the Canadian Institutes of Health Research (CIHR) and from the CCFF.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3513905.
Footnotes
[Supplemental information is available online at www.genome.org.]
References
- Amsterdam, G. 1991. Susceptibility testing of antimicrobials in liquid media, 3rd edition. Williams and Wilkins, Baltimore, MD.
- Ernst, R.K., Yi, E.C., Guo, L., Lim, K.B., Burns, J.L., Hackett, M., and Miller, S.I. 1999. Specific lipopolysaccharide found in cystic fibrosis airway Pseudomonas aeruginosa. Science 286: 1561-1565. [DOI] [PubMed] [Google Scholar]
- Filloux, A., Bally, M., Soscia, C., Murgier, M., and Lazdunsk, A. 1988. Phosphate regulation in Pseudomonas aeruginosa: Cloning of the alkaline phosphatase gene and identification of phoB- and phoR-like genes. Mol. Gen. Genet. 212: 510-513. [DOI] [PubMed] [Google Scholar]
- Friedrich, C.L., Moyles, D., Beveridge, T.J., and Hancock, R.E.W. 2000. Antibacterial action of structurally diverse cationic peptides on gram-positive bacteria. Antimicrob. Agents Chemother. 44: 2086-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisk, A., Schurr, J.R., Wang, G., Bertucci, D.C., Marrero, L., Hwang, S.H., Hassett, D.J., and Schurr, M.J. 2004. Transcriptome analysis of Pseudomonas aeruginosa after interaction with human airway epithelial cells. Infect. Immun. 72: 5433-5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoffroy, M.C., Floquet, S., Metais, A., Nassif, X., and Pelicic, V. 2003. Large-scale analysis of the meningococcus genome by gene disruption: Resistance to complement-mediated lysis. Genome Res. 13: 391-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govan, J.R. and Deretic, V. 1996. Microbial pathogenesis in cystic fibrosis: Mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60: 539-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunn, J.S., Lim, K.B., Krueger, J., Kim, K., Guo, L., Hackett, M., and Miller, S.I. 1998. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol. Microbiol. 27: 1171-1182. [DOI] [PubMed] [Google Scholar]
- Hallin, P.F. and Ussery, D.W. 2004. CBS Genome Atlas Database: A dynamic storage for bioinformatics results and sequence database. Bioinformatics 20: 3682-3686. [DOI] [PubMed] [Google Scholar]
- Hancock, R.E.W. and Speert, D.P. 2000. Antibiotic resistance in Pseudomonas aeruginosa: Mechanisms and impact on treatment. Drug Resist. Updat. 3: 247-255. [DOI] [PubMed] [Google Scholar]
- Hancock, R.E.W., Poole, K., and Benz, R. 1982. Outer membrane protein P of Pseudomonas aeruginosa: Regulation by phosphate deficiency and formation of small anion-specific channels in lipid bilayer membranes. J. Bacteriol. 150: 730-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson, E.L., Plotnikova, J., Mahajan-Miklos, S., Rahme, L.G., and Ausubel, F.M. 2001. Differential roles of the Pseudomonas aeruginosa PA14 rpoN gene in pathogenicity in plants, nematodes, insects and mice. J. Bacteriol. 183: 7126-7134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison, C.A., Peterson, S.N., Gill, S.R., Cline, R.T., White, O., Fraser, C.M., Smith, H.O., and Venter, J.C. 1999. Global transposon mutagenesis and a minimal Mycoplasma genome. Science 286: 2165-2169. [DOI] [PubMed] [Google Scholar]
- Jacobs, M.A., Alwood, A., Thaipisuttikul, I., Spencer, D., Haugen, E., Ernst, S., Will, O., Kaul, R., Raymond, C., Levy, R., et al. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. 100: 14339-14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson, N. and Mekalanos, J.J. 2000. TnAraOut, a transposon-based approach to identify and characterize essential bacterial genes. Nat. Biotechnol. 18: 740-745. [DOI] [PubMed] [Google Scholar]
- Kang, Y., Durfee, T., Glasner, J.D., Qiu, Y., Frisch, D., Winterberg, K.M., and Blattner, F.R. 2004. Systematic mutagenesis of the Escherichia coli genome. J. Bacteriol. 186: 4921-4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlyshev, A.V., Pallen, M.J., and Wren, B.W. 2000. Single-primer PCR procedure for rapid identification of transposon insertion sites. Biotechniques 28: 1078-1082. [DOI] [PubMed] [Google Scholar]
- Kawai, Y., Yano, I., Kaneda, K., and Yabuuchi, E. 1988. Ornithine-containing lipids of some Pseudomonas species. Eur. J. Biochem. 175: 633-641. [DOI] [PubMed] [Google Scholar]
- Kobayashi, K., Ehrlich, S.D., Albertini, A., Amati, G., Andersen, K.K., Arnaud, M., Asai, K., Ashikaga, S., Aymerich, S., Bessieres, P., et al. 2003. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. 100: 4678-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane, E.L., Kwasnicka, A., Ochs, M.M., and Hancock, R.E.W. 1999. PhoP-PhoQ homologues in Pseudomonas aeruginosa regulate expression of the outer-membrane protein OprH and polymyxin B resistance. Mol. Microbiol. 34: 305-316. [DOI] [PubMed] [Google Scholar]
- Macfarlane, E.L., Kwasnicka, A., and Hancock, R.E.W. 2000. Role of Pseudomonas aeruginosa PhoP-PhoQ in resistance to antimicrobial cationic peptides and aminoglycosides. Microbiology 146: 2543-2554. [DOI] [PubMed] [Google Scholar]
- McPhee, J.B., Lewenza, S., and Hancock, R.E.W. 2003. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol. Microbiol. 50: 205-217. [DOI] [PubMed] [Google Scholar]
- Moskowitz, S.M., Ernst, R.K., and Miller, S.I. 2004. PmrAB, a two-component regulatory system of Pseudomonas aeruginosa that modulates resistance to cationic antimicrobial peptides and addition of aminoarabinose to lipid A. J. Bacteriol. 186: 575-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potvin, E., Lehoux, D.E., Kukavica-Ibrulj, I., Richard, K.L., Sanschagrin, F., Lau, G.W., and Levesque, R.C. 2003. In vivo functional genomics of Pseudomonas aeruginosa for high-throughput screening of new virulence factors and antibacterial targets. Environ. Microbiol. 5: 1294-1308. [DOI] [PubMed] [Google Scholar]
- Quinn, J.P. 1998. Clinical problems posed by multiresistant nonfermenting gram-negative pathogens. Clin. Infect. Dis. 27: S117-S124. [DOI] [PubMed] [Google Scholar]
- Rivera, M., Bryan, L.E., Hancock, R.E.W., and McGroarty, E.J. 1988. Heterogeneity of lipopolysaccharides from Pseudomonas aeruginosa: Analysis of lipopolysaccharide chain length. J. Bacteriol. 170: 512-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Macdonald, P., Coelho, P.S., Roemer, T., Agarwal, S., Kumar, A., Jansen, R., Cheung, K.H., Sheehan, A., Symoniatis, D., Umansky, L., et al. 1999. Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature 402: 413-418. [DOI] [PubMed] [Google Scholar]
- Rozek, A., Powers, J.P., Friedrich, C.L., and Hancock, R.E.W. 2003. Structure-based design of an indolicidin peptide analogue with increased protease stability. Biochemistry 48: 14130-14138. [DOI] [PubMed] [Google Scholar]
- Salama, N.R., Sheperd, B., and Falkow, S. 2004. Global transposon mutagenesis and essential gene analysis of Helicobacter pylori. J. Bacteriol. 186: 7926-7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherens, B. and Goffeau, A. 2004. The uses of genome-wide yeast mutant collections. Genome Biol. 5: 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siehnel, R.J., Worobec, E.A., and Hancock, R.E.W. 1988. Cloning of the Pseudomonas aeruginosa outer membrane porin protein P gene: Evidence for a linked region of DNA homology. J. Bacteriol. 170: 2312-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slesarev, A.I., Mezhevaya, K.V., Makarova, K.S., Polushin, N.N., Shcherbinina, O.V., Shakhova, V.V., Belova, G.I., Aravind, L., Natale, D.A., Rogozin, I.B., et al. 2002. The complete genome of hyperthermophile Methanopyrus kandleri AV19 and monophyly of archaeal methanogens. Proc. Natl. Acad. Sci. 99: 4644-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, J., Ware, A., and Liu, S.L. 2003. Wavelet to predict bacterial ori and ter: A tendency towards a physical balance. BMC Genomics 4: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover, C.K., Pham, X.Q., Erwin, A.L., Mizoguchi, S.D., Warrener, P., Hickey, M.J., Brinkman, F.S., Hufnagle, W.O., Kowalik, D.J., Lagrou, M., et al. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406: 959-964. [DOI] [PubMed] [Google Scholar]
- Weinel, C., Ermolaeva, M.D., and Ouzounis, C. 2003. PseuRECA: Genome annotation and gene context analysis for Pseudomonas aeruginosa PAO1. Bioinformatics 19: 1457-1460. [DOI] [PubMed] [Google Scholar]
- Weissenmayer, B., Gao, J.L., Lopez-Lara, I.M., and Geiger, O. 2002. Identification of a gene required for the biosynthesis of ornithine-derived lipids. Mol. Microbiol. 45: 721-733. [DOI] [PubMed] [Google Scholar]
- Winson, M.K., Swift, S., Hill, P.J., Sims, C.M., Griesmayr, G., Bycroft, B.W., Williams, P., and Stewart, G.S. 1998. Engineering the luxCDABE genes from Photorhabdus luminescens to provide a bioluminescent reporter for constitutive and promoter probe plasmids and mini-Tn5 constructs. FEMS Microbiol. Lett. 163: 193-202. [DOI] [PubMed] [Google Scholar]
- Wu, M. and Hancock, R.E.W. 1999. Interaction of the cyclic antimicrobial cationic peptide bactenecin with the outer and cytoplasmic membrane. J. Biol. Chem. 274: 29-35. [DOI] [PubMed] [Google Scholar]
Web site references
- http://www.pseudomonas.com; Pseudomonas Genome Database.
- http://pseudomutant.pseudomonas.com; Pseudomutant Database.