Abstract
In our continued effort to search for a Streptococcus suis protein(s) that can serve as a vaccine candidate or a diagnostic reagent, we constructed and screened a gene library with a polyclonal antibody raised against the whole-cell protein of S. suis type 2. A clone that reacted with the antibody was identified and characterized. Analysis revealed that the gene encoding the protein is localized within a 2.0-kbp EcoRI DNA fragment. The nucleotide sequence contained an open reading frame that encoded a polypeptide of 445 amino acid residues with a calculated molecular mass of 46.4 kDa. By in vitro protein synthesis and Western blot experiments, the protein exhibited an electrophoretic mobility of approximately 38 kDa. At the amino acid level the deduced primary sequence shared homology with sequences of unknown function from Streptococcus pneumoniae (89%), Streptococcus mutans (86%), Lactococcus lactis (80%), Listeria monocytogenes (74%), and Clostridium perfringens (64%). Except for strains of serotypes 20, 26, 32, and 33, Southern hybridization analysis revealed the presence of the gene in strains of other S. suis serotypes and demonstrated restriction fragment length differences caused by a point mutation in the EcoRI recognition sequence. We confirmed expression of the 38-kDa protein in the hybridization-positive isolates using specific antiserum against the purified protein. The recombinant protein was reactive with serum from pigs experimentally infected with virulent strains of S. suis type 2, suggesting that the protein is immunogenic and may serve as an antigen of diagnostic importance for the detection of most S. suis infections. Pigs immunized with the recombinant 38-kDa protein mounted antibody responses to the protein and were completely protected against challenge with a strain of a homologous serotype, the wild-type virulent strain of S. suis type 2, suggesting that it may be a good candidate for the development of a vaccine that can be used as protection against S. suis infection. Analysis of the cellular fractions of the bacterium by Western blotting revealed that the protein was present in the surface and cell wall extracts. The functional role of the protein with respect to pathogenesis and whether antibodies against the antigen confer protective immunity against diseases caused by strains of other pathogenic S. suis capsular types remains to be determined.
Streptococcus suis is an important swine pathogen that causes many pathological conditions, such as arthritis, endocarditis, meningitis, polyserositis, and bronchopneumonia (24). It is also an important zoonotic agent for people in contact with swine or their by-products and causes meningitis, permanent hearing loss, and septic shock (1, 22, 24). Thirty-five capsular types (types 1/2 and 1 to 34) are currently known. Type 2 is considered the type that is the most frequently associated with disease and is the type that is the most often isolated. Strains of other serotypes, such as serotypes 1/2, 7, 9, and 14, can also cause disease. Attempts to control the infection are hindered by a lack of thorough knowledge of the virulence factors and protective antigens of the bacterium, the existence of multiple serotypes with diverse genetic makeups, and the evolution of multidrug-resistant strains (3, 6, 13, 24).
Several protein components, including attenuated whole bacterial cells, have been evaluated as vaccines against S. suis. However, these studies did not achieve much success because the protection was either serotype or strain dependent, and in some instances the results were ambiguous (8, 9, 10, 27). For example, Jacobs et al. (10) evaluated a suilysin-based subunit vaccine and showed that it conferred complete protection. However, the absence of suilysin in a substantial number of isolates recovered from diseased pigs hampers the use of this vaccine (14, 19). Thus, identification of other antigenic factors will contribute to the development of a monovalent or a multivalent subunit vaccine that will protect pigs against infection by all capsular types.
In our effort to identify an S. suis gene(s) that may be involved in virulence and proteins that may be useful in the development of a reliable diagnostic reagent or vaccine to protect against infection with this bacterium, we identified a DNA region from a virulent strain of S. suis serotype 2 that encoded a polypeptide of 38 kDa. Of the 35 S. suis serotypes currently known, 31 contain and express the gene. The gene product was reactive with serum from pigs with S. suis infection, and the protein induced protective immunity in experimentally challenged pigs, making it a candidate for consideration in the development of a diagnostic reagent and vaccine.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
S. suis type 2 strain 1933, a virulent isolate (25), was used to construct the genomic library. Other S. suis isolates were recovered from pigs from diverse geographical locations. Plasmid pUC18, propagated in Escherichia coli DH5α, was used as the library expression vector; and pGEM (Promega, Madison, Wis.) was used for DNA sequencing. Luria-Bertani broth or agar was used to grow the E. coli strains. Todd-Hewitt medium supplemented with 0.6% yeast extract (Difco Laboratories, Detroit, Mich.) was used to grow the S. suis strains. When appropriate, ampicillin was used at 60 μg/ml for E. coli cultures. All cultures were incubated at 37°C.
Chemicals and enzymes.
Enzymes were purchased from Promega or New England Biolabs (Beverly, Mass.) and were used as recommended by the manufacturer. Chemicals were purchased from Sigma Chemical Co. (St. Louis, Mo.) or Fisher Scientific (Pittsburgh, Pa.). The digoxigenin-labeled DNA molecular-weight marker II and the digoxigenin-11-dUTP DNA-labeling kit and detection system were from Boehringer Mannheim (Indianapolis, Ind.). Serum samples were collected from seven pigs experimentally infected with virulent strains of S. suis type 2.
Construction and screening of a recombinant DNA library.
S. suis DNA, which was extracted by a previously described method (13), was digested with the EcoRI restriction endonuclease. Restriction fragments were then size fractionated by agarose gel electrophoresis. Fragments in the size range of 1 to 23 kb were excised from the gel, purified by electroelution, and ligated into the pUC18 plasmid cloning vector that had been digested with EcoRI. The recombinant plasmids were transformed into E. coli DH5α by electroporation. Transformed cells were plated unto Luria-Bertani agar containing 60 μg of ampicillin per ml, isopropyl-β-d-thiogalactopyranoside (4 μl of a 20% solution), and 5-bromo-4-chlor-3-indolyl-β-d-galactopyranoside (40 μl of a 20-mg/ml solution) and grown at 37°C overnight. The resulting white colonies were transferred to a fresh plate and were grown as described above. One loopful of each colony was solubilized in 100 μl of 1× sodium dodecyl sulfate (SDS) sample buffer by heating for 5 min at 100°C. The preparation was centrifuged for 2 min at 13,000 × g to remove the cellular debris; and the cell-free lysate was used for Western blot analysis with a 1:500 dilution of polyclonal antibody raised against the whole-cell protein of S. suis type 2 as the primary antibody, followed by incubation in a 1:1,000 dilution of anti-rabbit immunoglobulin G (IgG) conjugated with horseradish peroxidase. The blots were developed with hydrogen peroxide and 4-chloro-1-naphthol. A colony designated DH5α(pOT301) was identified and characterized.
Nucleotide sequence determination and bioinformatics.
The complete nucleotide sequences of both strands of the 2.0-kb EcoRI fragment in plasmid pOT301 were determined by the dideoxy-chain termination method (18) with an automated nucleotide sequencer (Applied Biosystems, Foster City, Calif.). The nucleotide sequences and the deduced amino acid sequences were analyzed with MacVector software (Oxford Molecular Group, Inc., Campbell, Calif.). Searches for the similarity of the sequences with the sequences in GenBank were performed by using the BLAST network service.
PCR.
Oligonucleotide primers were designed by using the 38-kDa gene sequence data. The sequences of the primers were 5′-ATGCCACGGATTACCTTCCC-3′(primer BAY46F) and 5′-CCGTCTCCTTAATGATCCGC-3′ (primer BAY46R). The primer pair was designed to amplify a 253-bp product from S. suis DNA. Amplification was performed with 100 ng of purified genomic DNA in a total volume of 50 μl containing 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 50 mM KCl, 0.001% gelatin, 200 μM each deoxynucleoside triphosphate (dATP, dCTP, dGTP, and dTTP), 1 μM each primer, and 2.5 U of Taq polymerase (Perkin-Elmer Corp., Norwalk, Conn.). The PCR assay was carried out in a Perkin-Elmer 2400 thermocycler, comprising 5 min of preincubation at 94°C, followed by 35 cycles of 1 min at 94°C, 1 min at 55°C, and 1 min at 72°C. A final extension was performed for 7 min at 72°C. The negative control was a reaction mixture containing all reagents but no DNA template. The PCR products were visualized by electrophoresis on a 0.8% agarose gel by standard procedures.
Electrophoresis and Southern blotting.
Three micrograms of genomic DNA was digested with EcoRI, and the fragments were separated on a 0.8% agarose gel (Promega) and transferred to a nylon membrane (Boehringer Mannheim) by the method of Southern (17, 23). After the fragments were transferred, the DNA was UV crossed-linked to the membrane (GS gene linker; Bio-Rad, Richmond, Calif.).
Probe preparation and hybridization.
A 1,170-bp EcoRV-HindIII internal DNA fragment from pOT301 was labeled with digoxigenin-11-dUTP, according to the specifications of the manufacturer (Genius System; Boehringer Mannheim). Hybridization, washes, and hybrid detection were done according to the instructions provided with the Genius II nonradioactive labeling and detection kit (Boehringer Mannheim).
Analysis of the regions flanking the gene.
To extend the known sequence beyond the EcoRI site flanking the gene, inverse PCR (IV-PCR) was performed with primers HP6 (5′-CTCGTCACGGGAAAACCATG-3′) and HP7 (5′-TGCTTCTTGGATACCTGCTG-3′). Chromosomal DNA from S. suis strain 1933 was restricted with HindIII and religated prior to IV-PCR. The PCR product was purified and sequenced with the same primers, and the nucleotide sequence was used to design primers that permitted analysis of DNA from isolates with different hybridization patterns.
Overexpression and purification of the recombinant 38-kDa protein.
A 1,626-bp PCR fragment containing the open reading frame of the gene encoding the 38-kDa protein was obtained by amplification of pOT301 DNA with primers BAY46-1 (5′-CTA CGG CTA GAG TAC TCG GC-3′) and BAY 46-2 (5′-CAG TCA ATA TCG GCT CGA CC-3′) and cloned into the pCR 2.1 vector (Invitrogen, Carlsbad, Calif.) to create pOT308. To clone the 1,626-bp fragment in frame for overexpression, pOT308 was digested with the XhoI and KpnI restriction enzymes in combination to release the fragment. The fragment was then purified and ligated into the XhoI and KpnI sites of the pBAD/Myc-HisA expression vector to create pOT312. Plasmid pOT312 was transformed into E. coli TOP10 competent cells and overexpressed with arabinose according to the protocol of the manufacturer (Invitrogen). Following arabinose induction, the protein was purified as described previously (15).
Antigen and polyclonal antibody preparation.
Polyclonal antibody against the recombinant protein was obtained by immunizing New Zealand White rabbits (Shelton's Bunny Barn Rabbits, Waverly Hall, Ga.) subcutaneously at multiple sites with approximately 200 μg of purified protein emulsified 1:1 with Freund complete adjuvant. The rabbits received one booster injection with the same antigen concentration emulsified 1:1 with Freund incomplete adjuvant 14 days later and were then bled 7 days after the booster was administered. The sera were filter sterilized and stored at −30°C until they were used.
Western immunoblotting and in vitro transcription-translation experiments.
Cell lysates, prepared as described above, were vacuum concentrated (15-fold), and 15 μl of sample was used for Western blot analysis (17). The proteins were reacted with a 1:500 dilution of the polyclonal antibody raised against the purified 38-kDa protein and then with a horseradish peroxidase-conjugated goat anti-rabbit IgG antibody (ICN) diluted 1:1,000. Bound antibodies were detected colorimetrically with hydrogen peroxide and 4-chloro-1-naphthol. To screen pig antisera for antibody against the 38-kDa antigen, a 1:100 dilution of the pig sera was used as the primary antibody, followed by the addition of a 1:1,000 dilution of alkaline phosphatase-conjugated affinity-purified anti-swine IgG (Rockland Immunochemicals, Gilbertsville, Pa.). The blots were developed with a 5-bromo-4-chloro-indolylphosphate-nitroblue tetrazolium salt mixture. For in vitro protein synthesis, 3 μg of purified plasmid DNA was added to an E. coli cell extract that contained [35S]methionine and that was capable of coupled transcription-translation of exogenous DNA (Promega). The resulting translation products were separated by SDS-polyacrylamide gel electrophoresis (PAGE). After electrophoresis, the gel was dried and exposed to X-ray film (Kodak X-OMAT AR) at −70°C for 2 days, and the film was developed in a Kodak film processor.
Cellular location of the 38-kDa protein.
The surface, cell wall, cytoplasmic, and periplasmic protein fractions of S. suis were prepared by previously described methods (12, 20, 26) and analyzed by Western blotting with a polyclonal antibody directed against the purified 38-kDa antigen.
Animals, immunization, and challenge.
Ten pigs (age, 3 weeks; average weight, 14 lb) were purchased from the Auburn University Swine Facility, Auburn, Ala.
The ears of the pigs were tagged with colored numbers for identification purposes and divided into two groups of five pigs each. The first group of pigs (red tag; identification numbers 43, 44, 45, 46, and 47) consisted of the vaccination group, and the second group (blue tag; identification numbers 92, 93, 94, 95, and 96) consisted of the control group. Prior to administration of the first dose of vaccine, serum (preimmune) was collected from all animals and screened to rule out the presence of serum antibodies against S. suis. The pigs in the first group were then vaccinated intramuscularly at two injection sites in the neck with 1 ml of vaccine preparations emulsified in Freund complete adjuvant. Each animal received approximately 100 μg of the purified recombinant 38-kDa protein-based vaccine, and the second group of pigs (pigs 92, 93, 94, 95, and 96) received a placebo composed of physiological saline in adjuvant (negative control). Two weeks after the initial vaccination, serum was collected from all pigs for antibody screening; and the pigs received a booster of the same vaccine preparations by the same route, but this time the vaccine was emulsified 1:1 with Freund incomplete adjuvant. Fourteen days after administration of the booster, serum was again collected from the pigs, followed by intravenous challenge in the ear vein with 1.5 × 106 CFU of an overnight culture of the homologous S. suis serotype 2 strain. The pigs were monitored twice daily for clinical signs of disease.
Nucleotide sequence accession number.
The GenBank accession number for the 38-kDa protein sequence reported in this paper is AF389083.
RESULTS
Identification of the 38-kDa protein.
The gene library yielded a colony designated E. coli DH5α(pOT301) that produced a 38-kDa protein which reacted faintly with a polyclonal antibody, which was raised in rabbit cells, against the whole-cell protein of S. suis type 2 (data not shown). Because of the poor reactivity of the antibody to the protein, an in vitro protein synthesis experiment was performed to verify gene expression and to confirm the size of the gene product. The result confirmed that the gene is expressed and that the size of the product is 38 kDa (Fig. 1). Restriction analysis localized the gene within a 2.0-kb EcoRI fragment, and expression was orientation dependent in the pUC18 cloning vector (data not shown).
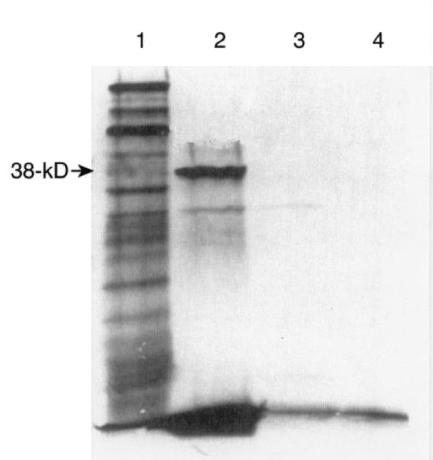
FIG. 1.
Translation products of the plasmid carrying the gene encoding the 38-kDa protein. An autoradiograph of the [35S]methionine-labeled products synthesized in an E. coli K-12 extract and separated by SDS-PAGE is shown. Lanes: 1, products of a positive control (Promega); 2, reactions of the gene template encoding the 38-kDa protein (pOT301); 3, reactions of the plasmid cloning vector pUC18 without the insert; 4, product of a control reaction with no template. The location of the ca. 38-kDa protein is indicated on the left. The molecular mass standard was very faint and as such was not labeled. The lower faint band in lane 2 was considered the plasmid cloning vector-coded protein since it is also present in lane 3.
Nucleotide and deduced amino acid sequence analysis.
Analysis of the nucleotide sequence revealed that it contained three open reading frames (ORFs) of sufficient sizes to code for the 38-kDa polypeptide observed by the immunoblotting and the in vitro protein synthesis experiments (Fig. 1; see also Fig. 5). The first ORF starts at position 479, the second ORF starts at position 512, and the third ORF starts at position 551. All three ORFs end in the termination codon TAA at position 1814. Proximal to the 5′ end of the ORF are regions that resemble the consensus ribosome binding site and the −10 and −35 promoter sequences (2, 5, 21). The first ATG at nucleotide 479 is probably the start of translation, because it is preceded six nucleotides upstream by a hexanucleotide (AGGAGA) that is the putative ribosome binding site. The G+C content of the gene is 47.5 mol%, which is in close agreement with that reported for S. suis (38 to 42%) (11).
FIG. 5.
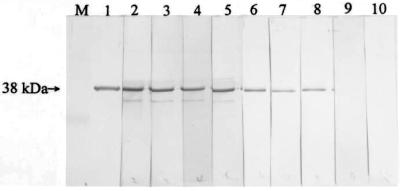
Immunoblot (Western blot) analysis with polyclonal antibody raised against the purified recombinant 38-kDa protein showing expression of the gene encoding the 38-kDa protein by strains of other serotypes. Lanes: M, rainbow molecular size marker (in kilodaltons; Amersham); 1, purified recombinant 38-kDa protein; 2, whole-cell lysate of S. suis serotype 2 strain 1933; 3, whole-cell lysate of a serotype 1 strain; 4, whole-cell lysate of a serotype 1/2 strain; 5, whole-cell lysate of a serotype 7 strain; and 6, whole-cell lysate of a serotype 9 strain. The location of the 38-kDa protein is indicated by the arrow on the left. Other reactive bands (lanes 2 to 6) were considered S. suis protein bands that cross-reacted with the antibody.
The sequence of the protein that is predicted from the DNA sequence and that starts at the first in-frame ATG codon (position 479) consists of 445 amino acid residues with a calculated molecular mass of 46.4 kDa, in contrast to the 38 kDa estimated in the Western blotting and in vitro experiments (Fig. 1; see also Fig. 5). No signal sequence was noted, as determined by a hydropathy plot (data not shown); and the protein had an estimated isoelectric point of 4.7. Several regions of high hydrophobicity, in comparison to regions of hydrophilicity, were observed on the hydropathy plots. Analysis of the deduced amino acid sequence of the protein revealed the absence of tryptophan.
A search of the GenBank database revealed that the sequences shared 83% identity at the nucleotide level to a gene of unknown function from the complete genome sequence of Streptococcus pneumoniae. At the amino acid level, the deduced primary sequence shared homology with those of unknown function from S. pneumoniae (89%), Streptococcus mutans (86%), Lactococcus lactis (80%), Listeria monocytogenes (74%), and Clostridium perfringens (64%).
PCR and hybridization.
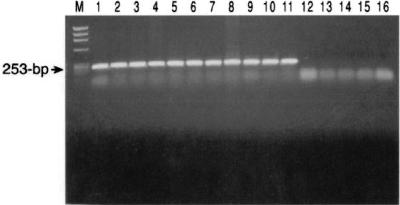
To demonstrate that the cloned fragment originated from S. suis and to examine the extent of conservation of the gene among the S. suis capsular types, PCR primers whose sequences were derived from the nucleotide sequence of the cloned fragment were used to amplify the DNA of the native S. suis strains encompassing all serotypes. The primers amplified the DNA from strains of most serotypes and produced a fragment of the expected size. DNA from strains belonging to serotypes 20, 26, 32, and 33 were nonreactive to the primers, as evidenced by the lack of an amplicon on agarose gels (Fig. 2). Because insufficient homology at the primer binding regions could result in the lack of amplification, we used hybridization experiments following EcoRI digestion to verify the PCR results. On the basis of the results of the analysis of our cloned fragment, digestion with the enzyme would produce a 2.0-kb fragment that would hybridize to the probe whose sequence was derived from within the gene. The results of the PCR and the hybridization methods were in agreement, suggesting that the cloned fragment originated from S. suis and that the gene is absent from strains of some serotypes. Unlike PCR, the hybridization studies revealed the existence of restriction fragment length differences that separated the strains into three genetic groups on the basis of the resulting fragment sizes of approximately 1.8, 2.0, and 5.0 kb, respectively (Fig. 3).
FIG. 2.
Example of ethidium bromide-stained agarose gels of PCR products with primers designed from the gene sequences encoding the 38-kDa protein. DNA from strains of serotypes 1/2 and 1 to 10 (lanes 1 to 11, respectively) and serotypes 20, 26, 32, and 33 (lanes 12 to 15, respectively) was used as the template. Lane M, HaeIII-digested φX174 DNA molecular mass markers (Promega); lane 16, a negative control (no template). The expected migration of the amplicons is indicated on the left.
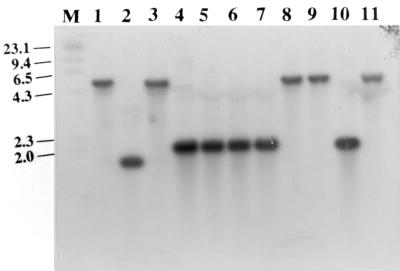
FIG. 3.
Southern blot of chromosomal DNA from different strains of S. suis type 2 (lanes 1 to 11) digested with EcoRI and hybridized to the 1,170-bp EcoRV-HindIII fragment derived from the gene encoding the 38-kDa antigen. Lane M, digoxigenin-labeled molecular weight marker II (Boehringer Mannheim).
Molecular basis for restriction fragment length differences.
To determine the genetic basis for the differences in the restriction fragment lengths, IV-PCR was performed to sequence the regions beyond the EcoRI fragment that contained the gene. The results revealed a base pair substitution (in boldface) at the EcoRI recognition sequence (GAATTC to GGATTC) at the 3′ end of the gene in isolates that gave a 5-kb band in the hybridization studies (Fig. 3, lanes 1, 3, 8, 9, and 11). The basis for the 1.8-kb fragment (Fig. 3, lane 2) was not determined. Because it is smaller than the expected 2.0-kb fragment, the fragment probably resulted from a deletion event.
Construction and overexpression of the 38-kDa protein.
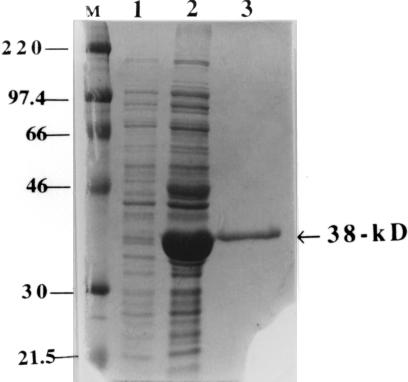
To produce a sufficient quantity of the protein for antibody production, the cloned fragment was inserted in frame into the pBAD/Myc-His version A expression vector (Invitrogen) as described in Materials and Methods, overexpressed, and gel purified. The purified protein gave a prominent 38-kDa band (Fig. 4) and was used to generate polyclonal antibody in rabbit cells.
FIG. 4.
Coomassie blue-stained SDS-polyacrylamide gel of the overexpressed and purified 38-kDa recombinant protein. The protein was overexpressed and purified as described previously (15). Lanes: M, rainbow molecular size marker (in kilodaltons); 1, whole-cell lysate of the uninduced pOT312 transformant of E. coli TOP10; 2, whole-cell lysate of the E. coli transformant with pOT312 following induction with arabinose; 3, recombinant protein purified from the pOT312 transformant of E. coli TOP10.
Reactivity of antibody produced against the 38-kDa recombinant protein.
In order for the recombinant protein to serve as a candidate for the development of a diagnostic reagent, it must be immunogenic and the antibody produced against the protein must be reactive with a similar protein from a wild-type S. suis strain(s). Figure 5 shows the reactivities of proteins from whole-cell lysates of wild-type S. suis strains to a polyclonal antibody raised against the purified recombinant protein. A protein from the wild-type strains reacted with the antibody and had a molecular mass identical to that of the recombinant protein. Other reactive bands were considered S. suis proteins that cross-reacted with the antibody. Figure 6 shows the reactivities of serum samples from pigs experimentally infected with S. suis type 2 strains to the 38-kDa recombinant protein. All samples produced a detectable signal against the antigen. These results demonstrate that the recombinant protein was not altered, that the gene encoding the protein is expressed in infected animals, and that the product is immunogenic. Thus, the results illustrate the potential use of the recombinant protein in the development of a serodiagnostic assay for the detection of S. suis infection. Isolates of the serotypes that were positive by the PCR and the hybridization studies expressed the protein, as determined by immunoblotting. The protein was not detected in the PCR- and hybridization-negative serotypes (serotypes 20, 26, 32, and 33) (Table 1). This result indicates the antigenic conservation of the protein among the strains of the serotypes that carry the gene.
FIG. 6.
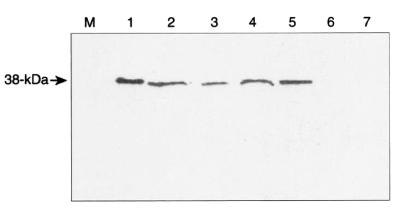
Immunoblot analysis of pig sera and their reactivities against the purified recombinant 38-kDa protein. Lanes: M, molecular size marker; 1 through 7, serum from pigs infected with S. suis type 2; 8, polyclonal antibody raised in a rabbit against the purified recombinant 38-kDa protein (positive control); 9, preimmune serum from the rabbit used to raise antibody against the purified protein (negative control); 10, preimmune serum from a pig prior to infection (negative control). Each lane is a strip cut from the membrane following Western transfer and prior to exposure to antibody.
TABLE 1.
Expression of the gene encoding the 38-kDa antigen by strains of different S. suis capsular types, as determined by immunoblot analysis
| S. suis serotype | No. of strains tested | No. of strains positive for:
|
|
|---|---|---|---|
| Gene by PCR and hybridization | 38-kDa protein by immunoblotting | ||
| 1 | 6 | 6 | 6 |
| 1/2 | 3 | 3 | 3 |
| 2 | 23 | 23 | 23 |
| 3 | 1 | 1 | 1 |
| 4 | 1 | 1 | 1 |
| 5 | 1 | 1 | 1 |
| 6 | 1 | 1 | 1 |
| 7 | 2 | 2 | 2 |
| 8 | 1 | 1 | 1 |
| 9 | 1 | 1 | 1 |
| 10 | 1 | 1 | 1 |
| 11 | 1 | 1 | 1 |
| 12 | 1 | 1 | 1 |
| 13 | 1 | 1 | 1 |
| 14 | 1 | 1 | 1 |
| 15 | 1 | 1 | 1 |
| 16 | 1 | 1 | 1 |
| 17 | 1 | 1 | 1 |
| 18 | 1 | 1 | 1 |
| 19 | 1 | 1 | 1 |
| 20 | 1 | 0 | 0 |
| 21 | 1 | 1 | 1 |
| 22 | 1 | 1 | 1 |
| 23 | 1 | 1 | 1 |
| 24 | 1 | 1 | 1 |
| 25 | 1 | 1 | 1 |
| 26 | 1 | 0 | 0 |
| 27 | 1 | 1 | 1 |
| 28 | 1 | 1 | 1 |
| 29 | 1 | 1 | 1 |
| 30 | 1 | 1 | 1 |
| 31 | 1 | 1 | 1 |
| 32 | 1 | 0 | 0 |
| 33 | 1 | 0 | 0 |
| 34 | 1 | 1 | 1 |
Cellular location of the 38-kDa protein.
Surface-exposed and cell wall-associated proteins of bacteria are, in general, targets for vaccine development or the production of a serodiagnostic reagent. Western blot analysis was performed with S. suis cell extracts to determine the structural location of the 38-kDa protein. The polyclonal antibody produced against the purified protein reacted to cell wall, surface, and cytoplasmic proteins; and its size was identical to that of the purified recombinant protein. No reactivity was observed in the periplasmic extract or the growth medium (Fig. 7).
FIG. 7.
Immunoblot analysis of proteins from cellular fractions of S. suis strain 1933 with polyclonal antibody raised against the purified 38-kDa recombinant protein. Lanes: M, molecular size standard; 1, the purified 38-kDa protein (positive control); 2, cell wall fractions; 3, surface fractions; 4 and 5, cytoplasmic fractions; 6, periplasmic fraction; 7, cell-free culture supernatant.
Protective value of the recombinant protein.
Because the protein was immunogenic in pigs and the antibody produced against the protein reacted with a protein of identical size from the wild-type strains of S. suis (Fig. 5), we wanted to determine its protective capability in an animal challenge model. None of the serum samples obtained from any of the experimental pigs reacted with the purified 38-kDa protein at the start of the experiment (day 0). The lack of reactivity with the protein indicated that the pigs had not previously been exposed to 38-kDa protein-positive strains of S. suis. In group 1 (vaccinated group), two of the five pigs (pigs 44 and 47) showed easily detectable antibodies against the protein 2 weeks postvaccination, while three pigs (pigs 43, 45, and 46) gave weak antibody responses (Table 2). Two weeks after administration of the booster dose (day 28), three of the five pigs (pigs 44, 45, and 47) had high titers of antibody against the protein, while two of the animals had moderate titers. Throughout this period no reactivity of the antibody with the protein was detected with sera from pigs in the control group (Table 2). These results confirm that the protein is immunogenic in pigs and that the level of antibody production is dependent on the particular animal.
TABLE 2.
Production of antibody against the 38-kDa antigen by individual pigs and antibody reactivity with the antigen, as determined by Western blot analysis
| Animal no. | Reactivity on daya:
|
Group | ||
|---|---|---|---|---|
| 0 | 14 | 28 | ||
| 43 | − | ± | + | Vaccinated |
| 44 | − | ++ | ++++ | Vaccinated |
| 45 | − | ± | +++ | Vaccinated |
| 46 | − | ± | + | Vaccinated |
| 47 | − | ++ | ++++ | Vaccinated |
| 92 | − | − | − | Control group |
| 93 | − | − | − | Control group |
| 94 | − | − | − | Control group |
| 95 | − | − | − | Control group |
| 96 | − | − | − | Control group |
±, weak reactivity; +, moderate reactivity; ++, good reactivity; +++, strong reactivity; ++++, very strong reactivity; −, no reactivity.
Three of the five pigs vaccinated with physiological saline in adjuvant (the control group) died as a result of the infection 2 to 4 days after the challenge. The remaining two animals were sick but recovered with time (Table 3). Specific clinical signs of disease, such as lameness, nervousness, and incoordination, were frequently recorded in the control group. Nonspecific clinical signs of disease, such as depression and a lack of appetite, were also observed. The pigs' body temperatures and leukocyte counts were also increased. In contrast, pigs in the vaccinated group were completely protected against challenge with a strain of a homologous serotype (Table 3). Only one of the pigs in this group showed temporary clinical signs, which consisted of arthritis and depression. These findings indicated that the S. suis recombinant 38-kDa protein could be a good candidate for consideration in the development of a recombinant subunit vaccine for the prevention of S. suis diseases in pigs.
TABLE 3.
Mortality rate among pigs immunized with the S. suis 38-kDa recombinant protein following challenge with a homologous S. suis type 2 strain
| Group no. | Treatment | No. of pigs sick/no. of pigs injected | No. of pigs dead/no. of pigs injected | % Mortality |
|---|---|---|---|---|
| 1 | Control | 5/5 | 3/5 | 60 |
| 2 | 38-kDa recombinant protein vaccine | 1/5 | 0/5 | 0 |
DISCUSSION
Several criteria can be used to identify antigens that can be useful in the development of a serodiagnostic reagent for the detection of S. suis infection or antigens which have the potential to elicit a cross-protective immune response against S. suis. First, the antigen should be present in at least all pathogenic strains of S. suis, regardless of their serotype. Second, antibody against the antigen should be present in infected animals, indicating that it is immunogenic and is expressed during infection.
This report describes the identification and characterization of the gene encoding a 38-kDa protein from a virulent strain of S. suis type 2, strain 1933. Characterization of the gene product showed that it is immunogenic in swine infected with pathogenic strains of S. suis type 2 as well as in swine vaccinated with the purified protein and provided protection against challenge with a strain of a homologous serotype. The protein was detected in S. suis cell wall and surface extracts and shares properties with the native protein. For example, a polyclonal antibody raised against the recombinant protein recognized the native protein from S. suis type 2, and the recombinant and native proteins had identical molecular masses (Fig. 5). The size similarity of the recombinant and native proteins and the reactivity between the recombinant and native proteins indicate that the recombinant protein was not altered in E. coli.
In an in vitro gene expression system, the cloned gene directed the production of a ca. 38-kDa polypeptide. This molecular mass is consistent with that observed by Western blotting with polyclonal antibody directed against the purified recombinant protein (Fig. 1, 5, and 7) but different from the calculated molecular mass of 46.4 kDa derived from the deduced amino acid sequence. The size disparity may be due to protein processing or anomalous migration of this primarily hydrophobic protein in SDS-polyacrylamide gels. It is also possible that the lack of a bulky amino acid residue such as tryptophan may influence the mobility of the protein in the gel.
The expression of the cloned gene in E. coli in the plasmid cloning vector pUC18 is orientation dependent, because in the reverse orientation the protein could not be detected on Western blots or in an in vitro gene expression system (data not shown). Thus, the cloned gene is probably not expressed from its own promoter, or it may contain a promoter that did not function in E. coli. In this case, expression is probably under the control of the ampicillin resistance gene promoter in the pUC18 cloning vector.
Surface-exposed and cell wall-associated proteins of bacteria are, in general, targets for vaccine development or the production of serodiagnostic reagents. In this study, the 38-kDa antigen was detected in the cell wall, surface, and cytoplasmic fractions (Fig. 7). We were therefore unable to conclude where the protein is located by the techniques that we used (12, 20, 26). However, cross contamination between fractions cannot be ruled out. A different approach is therefore needed to solve this problem.
The GenBank database is a useful source for the prediction of protein function. A search of the database revealed that the deduced primary sequence of the 38-kDa protein shared homology with sequences of unknown function in some gram-positive bacteria. We were therefore unable to use the database to assign a putative function to the protein. Nonetheless, it is safe to assume that the protein serves a common function in gram-positive bacteria. Work is under way in an effort to determine its function.
A recombinant subunit vaccine (suilysin) that confers protection in pigs has been reported previously (10). However, the usefulness of the vaccine was limited because the target protein is absent from a large number of S. suis strains isolated from diseased pigs (10, 14). In another study, purified muramidase-released protein and extracellular factor reportedly conferred protection in experimentally challenged pigs (27). However, most of the S. suis serotype 2 strains isolated from diseased pigs in Canada were negative for muramidase-released protein and extracellular factor (4). In this study, we tested strains from various geographical locations, including the United States, Canada, and Europe. Except for strains of serotypes 20, 26, 32, and 33, which lack the gene and which, as a result, do not produce the protein, strains of all other S. suis serotypes, regardless of their origins, contain the gene and express the 38-kDa protein. Strains of S. suis serotypes 1/2, 1, 2, 7, 9, and 14 are the most commonly associated with disease, with type 2 being the most significant (7, 16, 24). Strains of serotypes 20, 26, 32, and 33 are clinically insignificant and as such are not of major concern at present. Thus, the 38-kDa antigen has the potential for use in the development of a mono- or multivalent vaccine to protect against S. suis diseases.
Genetic heterogeneity can be a result of a mutation, insertion of a genetic element, or a deletion. Because genetic heterogeneity has previously been demonstrated in S. suis strains (13), we were interested in determining the molecular basis for the restriction fragment length differences observed in this study. Although point mutations are rare events, we noted that a point mutation in the EcoRI recognition sequence located 190 bp from the stop codon of the gene encoding the 38-kDa antigen was the basis for the differences. This mutation likely resulted in EcoRI cutting at the next site downstream in the isolates that yielded the 5.0-kb fragment.
In conclusion, we have identified a DNA region from a virulent strain of S. suis type 2 that encodes a protein with a molecular mass of 38 kDa. The gene and its product are present in strains belonging to the pathogenic serotypes of S. suis and most other S. suis serotypes. The protein was detected in the surface and cell wall fractions, is immunogenic in swine infected with strains of S. suis type 2 as well as in swine vaccinated with the purified protein, and provided protection against challenge with a strain of a homologous serotype. The biological functions of the protein antigen with respect to pathogenesis and the ability of antibody against the antigen to confer protective immunity against infections caused by other S. suis capsular types are the subjects of further research.
Acknowledgments
We thank Kevin Shuller, Noel Bennette, and Jeannine Bellamy for technical assistance.
This work was supported by the National Pork Producers Council (grant NPPC 00-131).
REFERENCES
- 1.Arends, J. P., and H. C. Zanen. 1988. Meningitis caused by Streptococcus suis in humans. Rev. Infect. Dis. 10:131-137. [DOI] [PubMed] [Google Scholar]
- 2.Ferretti, J. J., and R. Curtis III (ed.). 1987. Compilation of nucleotide sequences that signal the initiation of transcription and translation of streptococci, p. 293. In Streptococcal genetics. American Society for Microbiology, Washington, D.C.
- 3.Gottschalk, M., P. Turqeon, R. Higgins, M. Beaudoin, and A. M. Bourgault. 1991. Susceptibility of Streptococcus suis to penicillin. J. Vet. Diagn. Investig. 3:170-172. [DOI] [PubMed] [Google Scholar]
- 4.Gottschalk, M., A. Lebrun, H. J. Wisselink, J. D. Dubreuil, H. E. Smith, and U. Vecht. 1998. Production of virulence related-proteins by Canadian strains of Streptococcus suis capsular type 2. Can. J. Vet. Res. 62:75-79. [PMC free article] [PubMed] [Google Scholar]
- 5.Graves, M. C., and J. C. Rabinowitz. 1986. In vivo and in vitro transcription of the Clostridium pasteurianum ferrodoxin gene. J. Biol. Chem. 261:11409-11415. [PubMed] [Google Scholar]
- 6.Hampson, D. J., D. T. Trott, I. L. Clarke, C. G. Mwaniki, and I. D. Robertson. 1993. Population structure of Australian isolates of Streptococcus suis. J. Clin. Microbiol. 31:2895-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Higgins, R., M. Gottschalk, M. Beaudoin, and S. A. Rawluk. 1992. Distribution of Streptococcus suis capsular types in Quebec and western Canada. Can. Vet. J. 33:27-30. [PMC free article] [PubMed] [Google Scholar]
- 8.Holt, M. E., M. R. Enright, and T. J. L. Alenxander. 1988. Immunization of pigs with live cultures of Streptococcus suis type 2. Res. Vet. Sci. 45:349-352. [PubMed] [Google Scholar]
- 9.Holt, M. E., M. R. Enright, and T. J. L. Alexander. 1990. Protective effect of sera raised against different fractions of Streptococcus suis type 2. J. Comp. Pathol. 103:85-94. [DOI] [PubMed] [Google Scholar]
- 10.Jacobs, A. A. C., A. J. G. van den Berg, and P. L. W. Loeffen. 1996. Protection of experimentally infected pigs by suilysin, the thiol-activated haemolysin (suilysin) of Streptococcus suis. Vet. Rec. 139:225-228. [DOI] [PubMed] [Google Scholar]
- 11.Kilper-Balz, R., and K. H. Schleifer. 1987. Streptococcus suis sp. nov., nom. rev. Int. J. Syst. Bacteriol. 37:160-162. [Google Scholar]
- 12.Mercurio, A., and P. A. Manning. 1985. Cellular localization and export of the soluble haemolysin of Vibrio cholerae El Tor. Mol. Gen. Genet. 200:472-475. [DOI] [PubMed] [Google Scholar]
- 13.Okwumabua, O., J. Staats, and M. M. Chengappa. 1995. Detection of genomic heterogeneity in Streptococcus suis isolates by DNA restriction fragment length polymorphisms of rRNA genes (ribotyping). J. Clin. Microbiol. 33:968-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okwumabua, O., O. Abdelmagid, and M. M. Chengappa. 1999. Hybridization analysis of the gene encoding a hemolysin (suilysin) of Streptococcus suis type 2: evidence for the absence of the gene in some isolates. FEMS Microbiol. Lett. 181:113-121. [DOI] [PubMed] [Google Scholar]
- 15.Okwumabua, O., J. S. Persaud, and P. G. Reddy. 2001. Cloning and characterization of the gene encoding the glutamate dehydrogenase of Streptococcus suis serotype 2. Clin. Diagn. Lab. Immunol. 8:251-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robertson, I. D., and D. K. Blackmore. 1989. Prevalence of Streptococcus suis types 1 and 2 in domestic pigs in Australia and New Zealand. Vet. Rec. 124:391-394. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 18.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Segers, R. P. A. M., T. Kenter, L. A. M. Haan, and A. A. C. Jacobs. 1998. Characterisation of the gene encoding suilysin from Streptococcus suis and expression in field strains. FEMS Microbiol. Lett. 167:255-261. [DOI] [PubMed] [Google Scholar]
- 20.Shimoji, Y., Y. Mori, and V. A. Fishetti. 1999. Immunological characterization of a protective antigen of Erysipelothrix rhusiopathiae: identification of the region responsible for protective immunity. Infect. Immun. 67:1646-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shine, J., and J. Dalgarno. 1974. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementary to non-sense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. USA 71:1342-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shnerrson, J. M., B. Chattopadhyay, M. F. G. Murphy, and I. W. Fawcett. 1980. Permanent perceptive deafness due to Streptococcus suis type II infection. J. Laryngol. Ontol. 94:425-427. [DOI] [PubMed] [Google Scholar]
- 23.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 24.Staats, J. J., I. Feder, O. Okwumabua, and M. M. Chengappa. 1997. Streptococcus suis: past and present. Vet. Res. Commun. 21:381-407. [DOI] [PubMed] [Google Scholar]
- 25.Staats, J. J., B. L. Plattner, J. Nietfield, S. Dritz, and M. M. Chengappa. 1998. Use of ribotyping and hemolysin activity to identify highly virulent Streptococcus suis type 2 isolates. J. Clin. Microbiol. 36:15-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tavares, F., and A. Sellstedt. 2000. A simple, rapid and non-destructive procedure to extract cell wall-associated proteins from Frankia. J. Microbiol. Methods 30:171-178. [DOI] [PubMed] [Google Scholar]
- 27.Wisselink, H. J., U. Vecht, N. Stockhofe-Zurwieden, and H. E. Smith. 2001. Protection of pigs against challenge with virulent Streptococcus suis serotype 2 strains by a muramidase released protein and extracellular factor vaccine. Vet. Rec. 148:473-477. [DOI] [PubMed] [Google Scholar]