Abstract
The 46.4-kb nucleotide sequence of pSK41, a prototypical multiresistance plasmid from Staphylococcus aureus, has been determined, representing the first completely sequenced conjugative plasmid from a gram-positive organism. Analysis of the sequence has enabled the identification of the probable replication, maintenance, and transfer functions of the plasmid and has provided insights into the evolution of a clinically significant group of plasmids. The basis of deletions commonly associated with pSK41 family plasmids has been investigated, as has the observed insertion site specificity of Tn552-like β-lactamase transposons within them. Several of the resistance determinants carried by pSK41-like plasmids were found to be located on up to four smaller cointegrated plasmids. pSK41 and related plasmids appear to represent a consolidation of antimicrobial resistance functions, collected by a preexisting conjugative plasmid via transposon insertion and IS257-mediated cointegrative capture of other plasmids.
Staphylococcus aureus and, increasingly, coagulase-negative staphylococci are significant nosocomial pathogens, largely as a consequence of their propensity to develop antimicrobial resistance (37). Resistance determinants can be chromosomally encoded and/or carried by one or more plasmids commonly harbored by clinical staphylococcal strains. Several plasmid types have been identified in staphylococci (36, 37). The rolling-circle (RC) replicating plasmids are generally less than 5 kb in size and are cryptic or encode only a single resistance determinant (20). The staphylococcal RC plasmids have been subdivided into four families, exemplified by pT181, pC194, pSN2, and pE194, based on replication region sequence similarity. In contrast, the pSK639 family of trimethoprim resistance plasmids are thought to replicate via the theta mode (1). The pSK639-like plasmids, 8 to 13 kb in size, found to date contain two or three copies of IS257 (23). Because of their large size, (i.e., 15 to 40 kb) staphylococcal multiresistance plasmids are also presumed to replicate by a theta mechanism (18); the β-lactamase–heavy-metal resistance (46) and pSK1 (28) families are the most thoroughly characterized groups of such plasmids. The determinants carried by multiresistance plasmids are often located on one or more transposon-like structures and/or associated with insertion sequence (IS) elements.
The largest of the staphylococcal plasmids identified are the conjugative multiresistance plasmids. One structurally related group of these, typified by pSK41, pGO1, and pJE1 (see Fig. 1 below) (9, 11, 53), are capable of mediating their own conjugative transfer and the mobilization of coresident plasmids (1, 40). These plasmids, now designated the pSK41 family (37), were first detected in strains isolated in the mid 1970s and have subsequently been identified in geographically diverse isolates in both S. aureus and coagulase-negative staphylococci (2, 9, 19, 30).
FIG. 1.
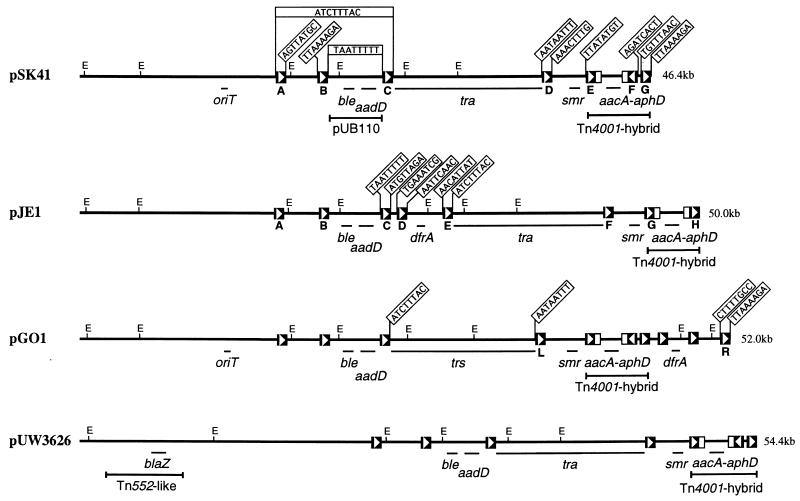
Maps of the staphylococcal conjugative plasmids, pSK41, pJE1, pGO1, and pUW3626 (3, 9, 11, 34); plasmid sizes are shown on the right. The genetic loci shown are aacA-aphD (Gmr Tmr Kmr), aadD (Nmr), ble (bleomycin resistance), dfrA (trimethoprim resistance), smr (antiseptic and disinfectant resistance), blaZ (penicillin resistance), tra/trs (conjugative transfer functions), and oriT (origin of conjugative transfer). The positions and extents of the cointegrated copy of the plasmid pUB110 (4), the Tn4001-IS257 hybrid structure (3), and the Tn552-like transposon (15) are indicated. Truncated copies of IS256 are represented by open boxes, whereas IS257 elements are represented as solid boxes containing an arrowhead indicating the direction of transposase transcription and hence the element’s orientation. IS257 element designations, following the nomenclature of Leelaporn et al. (23), are shown for pSK41 and pJE1, whereas designations for pGO1 are taken from Morton et al. (34). Where known, the 8 nt of sequence adjacent to each IS257 element is indicated; flanking sequences on pGO1 are from Morton et al. (33, 34). EcoRI restriction sites (E) are shown.
pSK41 family plasmids typically confer resistance to the aminoglycosides, gentamicin, tobramycin, and kanamycin (Gmr Tmr Kmr), via an aacA-aphD gene located on a Tn4001-IS257 hybrid structure (3) and neomycin (Nmr) via an aadD determinant encoded by a cointegrated copy of the small RC plasmid pUB110 (4), as well as multidrug resistance to antiseptics and disinfectants encoded by smr (formerly qacD) (25). pGO1 and pJE1 additionally confer dfrA-encoded trimethoprim resistance (Tpr), and other family members encode resistance to mupirocin or contain a Tn552-like β-lactamase transposon (15, 21, 34). Plasmids of the pSK41 family contain multiple copies of the insertion element IS257, flanking several of the resistance determinants and a transfer-associated region, tra (37). We have completed the nucleotide sequence of pSK41 to identify additional functions encoded by plasmids of this type and to gain insights into their evolution.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Escherichia coli DH5α (F− endA hsdR17 supE44 thi-1 λ− recA1 gyrA96 relA1 φ80dlacZΔM15; Bethesda Research Laboratories) was used as the host for all E. coli plasmids. S. aureus SK982 (resistant to rifampin and novobiocin) (27) and SK983 (resistant to streptomycin and fusidic acid) (51) were used as hosts of S. aureus plasmids. General culture conditions were 37°C on Luria agar (LA) plates or in Luria broth (LB). Plasmids used as sequencing templates were constructed by ligation of pSK41 and pJE1 restriction fragments into pUC (54) and pBluescript (Stratagene) series vectors; other plasmids used in this study are shown in Table 1. Where appropriate, antimicrobial agents were used at the following concentrations: ampicillin, 100 μg/ml; ethidium bromide, 40 μg/ml; fusidic acid, 10 μg/ml; gentamicin, 20 μg/ml; neomycin, 15 μg/ml; novobiocin, 2 μg/ml; rifampin, 20 μg/ml; and streptomycin, 100 μg/ml.
TABLE 1.
Plasmids used in this study
| Plasmid | Descriptiona | Reference |
|---|---|---|
| pSK41 | 46.4-kb conjugative plasmid; Nmr EbQar Gmr Tmr Kmr Tra+ | 11 |
| pJE1 | 50.0-kb conjugative plasmid; Nmr EbQar Gmr Tmr Kmr Tpr Tra+ | 9, 11 |
| pUW3626 | 54.4-kb conjugative plasmid; Nmr EbQar Gmr Tmr Kmr Pcr Tra+ | 3 |
| pSK5093 | pSK41 deletion mutant lacking region between IS25741B and IS25741E; Nms EbQas Gmr Tmr Kmr Tra− | This study |
| pSK5094 | pSK41 deletion mutant lacking region between IS25741E and IS25741G; Nmr EbQar Gms Tms Kms Tra+ | This study |
Abbreviations: EbQar, resistance to ethidium bromide and quaternary ammonium compounds; Gmr Tmr Kmr, resistance to gentamicin, tobramycin, and kanamycin; Nmr, neomycin resistance; Pcr, penicillin resistance; Tpr, trimethoprim resistance; Tra, conjugative transfer. s = sensitive.
DNA manipulation.
Plasmid DNA was isolated from E. coli by using either the alkaline lysis method (45) or the Quantum Prep plasmid Miniprep Kit (Bio-Rad) and from S. aureus as described by Lyon et al. (26). Restriction endonucleases and T4 DNA ligase were used in accordance with the manufacturers’ instructions. DNA cloning was performed by standard techniques (45). PCR was carried out with Pfu DNA polymerase (Stratagene) or the Expand High-Fidelity PCR System (Boehringer Mannheim) according to the manufacturers’ recommendations. Primers for PCR and nucleotide sequencing were made with a Beckman Oligo-1000 synthesizer. PCR products to be sequenced were subjected to ammonium acetate precipitation and concentrated with a Microcon 100 microconcentrator (Amicon).
Nucleotide sequence determination and data analysis.
Nucleotide sequencing was performed with the SequiTherm cycle sequencing kit (Epicentre Technologies) according to the manufacturer’s instructions or by the Sydney University and Prince Alfred Macromolecular Analysis Centre with the ABI Ready Reaction kit. Double-stranded plasmids and PCR products amplified directly from pSK41 were utilized as sequencing templates; sequences from PCR products were derived from at least two independent amplifications. All restriction sites were crossed, and all novel sequences were determined on both DNA strands. Sequences were stored and assembled with the program SEQUENCHER (Gene Codes Corp.) and analyzed with the GCG package (8) maintained by the Australian National Genomic Information Service, University of Sydney. Sequence similarities were assessed by using pairwise alignments generated with the program GAP (8) with the Dayhoff 250 PAM matrix (7). Statistical significance (Z) was calculated as follows: Z = (a − m)/s, where a is the alignment score; m is the mean of 100 alignment scores where one of the sequences has been randomly shuffled; and s is the standard deviation of m (39). Z scores greater than 3, 6, or 10 were taken to be indicative of a “possible,” “probable,” or “highly probable” evolutionary relatedness, respectively (24). Phylogenetic analyses were performed by using programs in the PHYLIP package (10). Dot plots were generated with the program DOTTY PLOTTER (14). Potential transmembrane segments were identified with the program TOPPRED II (5).
Detection and characterization of deletion mutants.
Conjugative matings were performed with the plasmid pSK41 between S. aureus strains SK982 and SK983. Overnight cultures of donor and recipient strains grown in LB were diluted 1:10 in fresh medium and grown to an optical density at 600 nm (OD600) of 0.6. Portions (0.5 ml each) of donor and recipient cells were combined in 2 ml of LB and filtered through a 2.5-cm (0.45-μm pore size) Millipore filter. The filter was placed bacterium-side-up on LA and incubated upright overnight. Cells were removed from the filter by vortexing in 2 ml of LB, and transconjugants were selected on medium containing gentamicin, neomycin, or ethidium bromide, in addition to selection for the recipient strain, and then screened against the nonselected agents to reveal any loss of resistance. DNA was isolated from sensitive transconjugants, and PCR was used to confirm the loss of a resistance gene. Southern blot analysis of plasmid digests was performed to indicate the extent of a deletion. AccI-digested plasmids run on a 1% (wt/vol) agarose gel were transferred to Hybond N+ membranes as described elsewhere (45). The ECL direct nucleic acid labeling and detection system (Amersham) was used to label an IS257 probe (nucleotides [nt] 194 to 719; see Fig. 6) and to detect hybridizing fragments, according to the manufacturer’s directions. IS257 elements at the deletion junctions were sequenced with PCR products that entirely spanned the elements.
FIG. 6.
Comparison of IS257 elements from pSK41, pJE1, and pSK639. Element designations are given on the left, whereas sequence numbering is shown on the right. The pSK639 sequences are taken from GenBank entry U40259. Dots indicate identity to the top sequence, whereas dashes indicate the positions of indels. The terminal inverted repeats of IS257 are indicated by arrowed lines. The position of the proposed crossover sites in IS2575093B and IS2575094E can be localized to the regions between the brackets in these sequences.
Nucleotide sequence accession number.
The complete nucleotide sequence of pSK41 is available under the GenBank accession number AF051917. The nucleotide sequence of the additional segment in pJE1 is available under the GenBank accession number AF051916.
RESULTS AND DISCUSSION
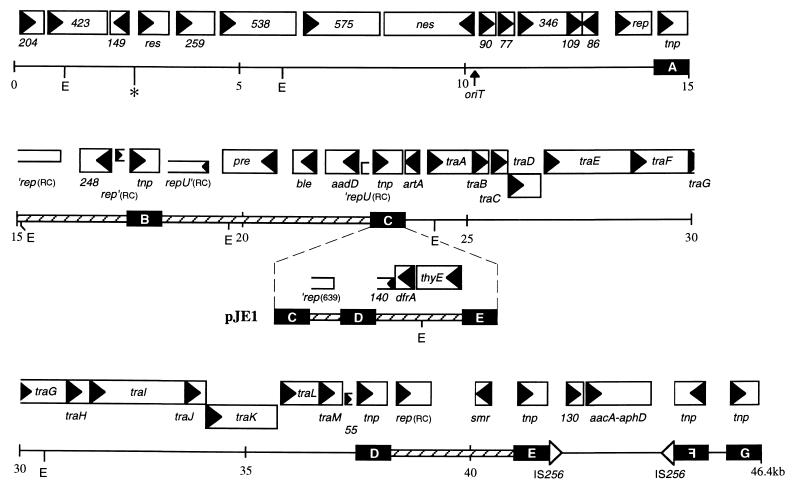
The physical and genetic maps of pSK41 and related plasmids are shown in Fig. 1. The 46,445-nt sequence of pSK41 was found to have an overall A+T content of 70.3%, which is consistent with a prolonged existence in low G+C content hosts, such as staphylococci. A total of 42 open reading frames (ORFs) likely to represent functional translated genes are evident in the sequence, and a further three truncated remnants of genes can also be identified; the genetic organization of the entire plasmid is illustrated in Fig. 2. The DNA segments flanked by IS25741G and IS25741A and by IS25741C and IS25741D (Fig. 1 and 2) probably represent the basic backbone of the plasmid into which other molecules have been inserted (see below). The latter of these corresponds to the previously reported tra region (11) and will therefore not be described in detail here. Characteristics of the deduced products of the 14 genes identified in the 14.2-kb segment between IS25741G and IS25741A are summarized in Table 2.
FIG. 2.
Genetic organization of pSK41. Genes are represented by open boxes with names shown within or below and with arrowheads indicating the direction of transcription; smaller open boxes represent interrupted genes. Copies of IS257 are denoted by black boxes containing the element’s designation (A through G), whereas inverted copies of truncated IS256 elements are indicated by open triangles. Hatched segments indicate integrated small plasmids. RC plasmid replication-initiation genes are suffixed (RC) to differentiate them from the probable theta-mode rep gene of pSK41. Kilobase coordinates are shown below, as are the positions of EcoRI restriction sites (E) and oriT (vertical arrow). The position in pSK41 corresponding to the insertion site of the Tn552-like transposon in pUW3626 is indicated by an asterisk (∗). The position and genetic organization of the additional DNA segment present in pJE1 is also indicated; the tnp genes of the IS257 elements have been omitted for clarity. The pSK639-like rep gene remnant is suffixed (639) to differentiate it from pSK41 rep. Genetic nomenclature is as described for Fig. 1 and as follows: numbers, size in codons of deduced ORF of unknown function; res, resolvase; oriT, origin of conjugative transfer; nes, oriT nickase; rep/repU, replication initiation; tnp, transposase; pre, recombinase; artA/traA-M, transfer-associated genes; thyE, thymidilate synthetase.
TABLE 2.
Deduced products of pSK41 encoded by the region flanked by IS25741G and IS25741A
| Protein (amino acid size)a | Product size (kDa)b | Comments and predictionsc |
|---|---|---|
| Orf77 | 8.7 | Two transmembrane segments |
| Orf86 | 10.1 | Cytoplasmic; helix-turn-helix at amino acids 27 to 48 |
| Orf90 | 11.0 | Cytoplasmic |
| Orf109 | 12.9 | Cytoplasmic |
| Orf149 | 17.7 | Cytoplasmic |
| Orf204 | 24.1 | Cytoplasmic; UUG start codon |
| Orf259 | 30.1 | Cytoplasmic |
| Orf346 | 38.8 | Cytoplasmic |
| Orf423 | 49.6 | One transmembrane segment; potential signal peptide cleavage after amino acid 36 |
| Orf538 | 63.8 | Cytoplasmic |
| Orf575 | 66.3 | Cytoplasmic; similar to L. lactis pRS01 LtrC |
| Nes (665) | 78.9 | Homologous to oriT nickase from pGO1 |
| Rep (319) | 37.7 | Homologous to replication initiation proteins from other gram-positive plasmids |
| Res (185) | 21.4 | Member of the recombinase superfamily; helix-turn-helix at amino acids 161 to 182 |
Size in amino acids is indicated by protein name or is given in parentheses.
Product sizes were calculated from the deduced protein sequences.
Predicted cellular locations are based on the presence or absence of potential transmembrane segments (5).
Plasmid replication and maintenance functions.
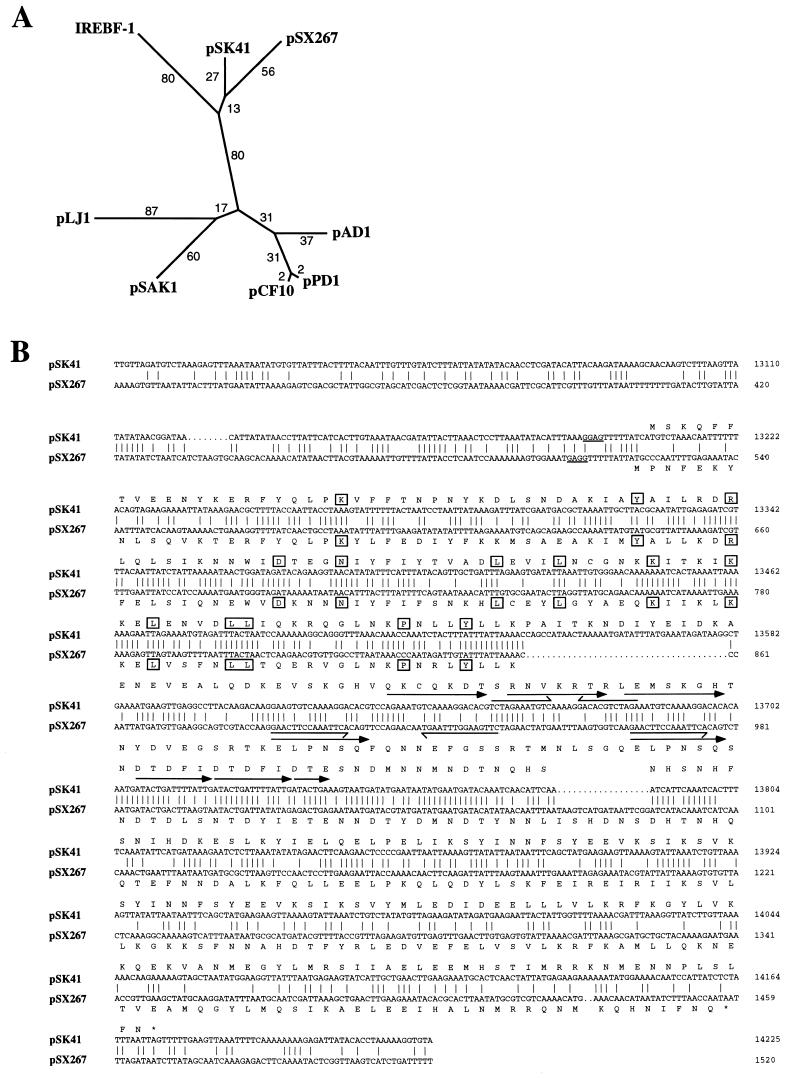
The segment of pSK41 between IS25741G and IS25741A was presumed to encode the plasmid’s replication functions since it is the only region conserved in all members of the plasmid family. Consistent with this notion, the gene immediately adjacent to IS25741A, now designated rep (Fig. 2), encodes a deduced product, Rep, which shares statistically significant sequence similarity to the replication-initiation proteins from several plasmids of gram-positive origin: viz., the staphylococcal plasmid pSX267 (Z = 48.5) (13); the enterococcal plasmids pAD1 (Z = 30.9) (56), pCF10 (Z = 31.9) (44), and pPD1 (Z = 37.5) (12); and the Lactobacillus plasmids pLJ1 (Z = 22.2) (50) and pSAK1 (Z = 35.1) (GenBank entry Z50862). The pSK41 rep product is most similar to the RepA protein of the arsenate resistance plasmid, pSX267, from Staphylococcus xylosus (Fig. 3A) (13), a member of the β-lactamase–heavy-metal resistance family of plasmids (17). This finding establishes an evolutionary link between the replication functions of two clinically significant groups of staphylococcal plasmids: viz., the pSK41 family of conjugative plasmids and the β-lactamase–heavy-metal resistance plasmids that emerged approximately three decades earlier (28).
FIG. 3.
(A) Phylogenetic analysis of the deduced pSK41 rep product and related proteins. The amino acid sequences were obtained from the following GenBank entries: pSX267, X92404; pCF10, L14285; pPD1, D78016; pAD1, L01794; pLJ1, J04240; pSAK1, Z50862; and IREBF-1, M55290. The unrooted tree was constructed by using the programs PROTDIST and NEIGHBOR (10) from a multiple alignment generated by PILEUP (8). An equivalent tree was obtained with the program PROTPARS (10). (B) Nucleotide sequence alignment of the replication regions of pSK41 and pSX267 (13); identities are indicated by vertical lines and indels are denoted by dots. The nucleotide sequences are numbered on the right. Ribosome binding sites for the rep genes are underlined, and the deduced amino acid sequences are shown above and below the nucleotide sequences, respectively. Directly repeated sequences are indicated by arrowed lines, whereas inverted repeats are shown by half-arrowed lines. Amino acids conserved in all proteins shown in panel A are boxed.
Sequence similarity extends to the nucleotide level between the pSK41 and pSX267 rep genes, but is primarily confined to the 5′ ends of the coding sequences (Fig. 3B). The lack of sequence similarity beyond the extremities of the rep coding sequences is consistent with the finding that the origin of pSX267 replication appears to be located within the repA ORF, possibly associated with direct repeats present in the gene (13). Our analysis has revealed analogously located, albeit distinct, arrays of direct and inverted repeats in the sequences of both plasmids (Fig. 3B).
Like its homologous plasmid replication-initiation proteins, pSK41 Rep shares similarity with a protein identified as mouse interferon response element binding factor 1 (IREBF-1; Z = 37.1) (13, 56, 58). However, phylogenetic analysis (Fig. 3A) suggests that IREBF-1 may actually represent a contaminant of bacterial origin present in the mouse cDNA library from which this gene was obtained, since IREBF-1 is more similar to some of the plasmid rep products (e.g., pSK41) than some of the members of this plasmid replication-initiation protein family are to each other (e.g., pSK41 and pAD1); the high A+T content of the IREBF-1 nucleotide sequence (64%) supports this contention.
The deduced product of pSK41 orf86 contains a predicted helix-turn-helix domain, suggesting that it might be a DNA-binding protein. The location of this divergently transcribed gene immediately upstream of pSK41 rep (Fig. 2) raises the possibility that orf86 may be involved in the regulation of plasmid replication.
The pSK41 res gene was found to encode a predicted product sharing similarity to a superfamily of recombinases which includes plasmid resolvases. The regions of greatest similarity correspond to motifs diagnostic of the recombinase superfamily and, like the other members of this family, pSK41 Res is predicted to possess a helix-turn-helix DNA-binding domain. Such enzymes are thought to contribute to the segregational stability of plasmids by facilitating efficient partitioning through the conversion of plasmid multimers into monomers (49).
A resolvase-encoding gene, sin, has previously been detected on the staphylococcal plasmids pSK1 and pI9789, from the pSK1 and β-lactamase–heavy-metal resistance plasmid families, respectively, and Southern hybridization studies indicated that such genes were common on plasmids from these groups (38). However, the deduced pSK41 res product was found to be no more closely related to those of the sin genes than it is to resolvases from other genera (data not shown); indeed, the lack of nucleotide sequence similarity explains the previous failure of a pSK1 sin probe to hybridize to pSK41 (38). The related β-lactamase transposons, Tn552 (43) and Tn4002 (16), have been found to insert preferentially into both β-lactamase–heavy-metal resistance and pSK1 family plasmids at a site within an inverted repeat located immediately upstream of their respective sin genes (38). Site-specific insertion of Tn552-like β-lactamase transposons also appears to have occurred in pSK41-like conjugative plasmids (15, 57). Correlation of the pSK41 nucleotide sequence to restriction maps of the β-lactamase-encoding relatives pUW3626 (Fig. 1) and pCRG1600 (28) indicated that the Tn552-like transposons in these plasmids are likely to be located in the vicinity of genes equivalent to pSK41 res. To investigate this precisely, the junctions of the transposon in pUW3626 were amplified with primers corresponding to the ends of Tn552 and the pSK41 orf149 and res genes.
Sequence analysis revealed the transposon in pUW3626 to be flanked by a 6-nt target duplication, ATAGCG, corresponding to nt 2594 to 2599 of the pSK41 sequence; a duplication of this size was previously found flanking Tn552 in the β-lactamase–heavy-metal resistance plasmid, pS1 parent (pI9789::Tn552), whereas a 7-nt duplication was associated with Tn4002 in the pSK1 family plasmid pSK4 (38). The β-lactamase transposon in pUW3626 is located 185 nt upstream of this plasmid’s res allele; Tn552 and Tn4002 were found only 31 nt upstream of sin on pS1 parent and pSK4 (38). Furthermore, no obvious sequence similarity is discernible between the insertion sites in pUW3626 and pS1 parent or pSK4, and, in contrast to the large inverted repeat interrupted by the insertion of Tn552 and Tn4002 upstream of sin, only small repeats can be identified in the vicinity on pUW3626. However, the insertion sites are all located adjacent to canonical promoter-like sequences which may be responsible for the transcription of res and sin on their respective plasmids (38) (data not shown). The preference of Tn552-like transposons for insertion sites upstream of these resolvase genes is intriguing given the absence of nucleotide sequence similarity between them. It is conceivable that the insertion sites discussed here coincide with the target sites of the plasmids’ cognate resolvases and that it is a particular DNA-Res protein conformation that is being recognized by the Tn552-like transposons rather than a specific sequence per se.
Conjugative transfer functions.
The pSK41 DNA segment from nt 7982 to 10325 was found to be virtually identical to the equivalent region from the related plasmid pGO1. This segment of pGO1 encodes the origin of conjugative transfer, oriT, and the gene encoding the enzyme responsible for nicking at this site, nes (6). The nucleotide differences that are evident are confined to two small regions. The first of these corresponds to four “indels” (insertions-deletions), two in each plasmid, within a 24-nt segment in the central portion of nes, which results in a unique 8-amino-acid contiguous sequence within each product. The second region of divergence occurs downstream of nes and consists of a 22-nt stretch containing 5 indels (all nucleotides absent from the pSK41 sequence) and 2 nucleotide differences, which would be expected to result in a different and lengthened C terminus in the protein encoded by the pGO1 allele of pSK41 orf575.
Comparison of the pSK41-encoded conjugation-associated proteins to the sequence databases has revealed a relationship between this staphylococcal transfer system and that of the broad-host-range plasmid pIP501, which was originally identified in Streptococcus agalactiae but has also been shown to replicate in Staphylococcus, Clostridium, Pediococcus, and Listeria species (29). In addition to the previously described similarity between oriT nickases (6), amino acid similarity is evident between other proteins encoded by the pIP501 conjugation region A (55) and the pSK41 tra region (11). This conservation extends to the genetic organization of these regions (Fig. 4). The identification of any relationship between the remainder of the pSK41 tra genes and other pIP501 sequences, in particular a second segment associated with conjugative transfer, region B (29), awaits further characterization of the latter.
FIG. 4.
Relatedness of pSK41 and pIP501 conjugation systems. The genetic organization of the pIP501 conjugation region A (55) and pSK41 nes/oriT and artA-traG of the tra region (11) is illustrated. Genes (arrowed boxes) encoding homologous products are linked by shading; percent amino acid sequence identities and Z scores (in parentheses) are shown. The positions and strands of the oriT nick site of each plasmid are indicated by vertical arrows. A segment of approximately 11 kb between the two regions of pSK41 has been omitted for clarity.
Studies of pGO1 have indicated that the minimal conjugative unit of this class of plasmid consists of the oriT/nes and tra regions (6). However, analysis of transposon mutagenesis studies of the closely related plasmid, pJE1 (9), indicates that other functions encoded by the DNA segment between IS257JE1H and IS257JE1A (Fig. 1) might also play a role in conjugative processes. Sequence analysis of the equivalent region of pSK41, between IS25741G and IS25741A, lends credence to this possibility. The deduced product of pSK41 orf575 shares statistically significant sequence similarity (Z = 14.3) with that of ltrC from the lactococcal conjugative plasmid pRS01 (32). Like orf575 and nes, ltrC is located in proximity to a nickase gene, in this case ltrB. A transposon insertion in ltrC was found to slightly reduce conjugative transfer efficiency (32). It seems likely that ltrC might possess additional 5′ sequences since alignments reveal that similarity begins at approximately amino acid 200 of Orf575 at residues preceding the proposed N-terminal methionine residue of LtrC (data not shown); although a potential initiation codon has been identified for ltrC, the absence of a in-frame termination codon in the available upstream sequence is consistent with this proposal.
A 7-nt sequence, CATGACA, was found to overlap the −35 sequence of the three potential transcriptional promoters identified in the pSK41 tra region (11). Sharma et al. (47) have shown that for pGO1 the transcriptional repressor TrsN (equivalent to pSK41 ArtA) binds to sites overlapping this sequence in each of these promoters. Analysis of the DNA segment between IS25741G and IS25741A revealed three additional potential promoters with −35 boxes overlapped by CATGACA sequences. Two overlapping copies of this motif, one with a single mismatch, are found in a putative orf259 promoter; four overlapping copies, each with a single mismatch, are evident in the putative orf538 promoter; and two overlapping copies are associated with the promoter probably responsible for transcription of both orf346 and orf109. Although the significance of these sequences remains to be determined, their presence hints either that the genes transcribed by these promoters may play a role in conjugation or that they are at least coordinately regulated with transfer-associated functions.
IS257 and small plasmid cointegrates.
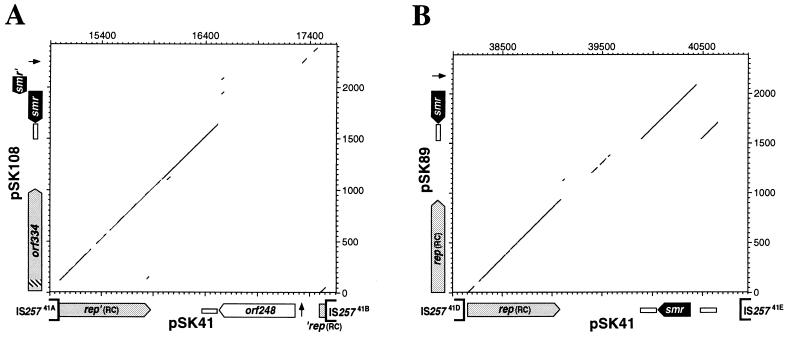
Restriction map similarity and limited nucleotide sequence data suggested that pSK41 contains a cointegrated copy of the small aminoglycoside and bleomycin resistance RC plasmid pUB110, which is flanked by IS25741B and IS25741C (Fig. 1 and 2) (4); the complete sequence data described here demonstrated identity between this segment of pSK41 and the published sequence of pUB110 (31). Analysis of the pSK41 sequence has revealed that two other segments, bounded by IS25741A and IS25741B and by IS25741D and IS25741E, also correspond to cointegrated copies of pC194 family RC plasmids (Fig. 2). The first of these segments, which is 2682 nt long, was found to be most similar to the Staphylococcus epidermidis plasmid pSK108 (22); their relationship is illustrated in a dot plot comparison (Fig. 5A). The extent of sequence similarity includes the remnants of a replication-initiation gene, rep′(RC) and ′rep(RC), at each end of the segment, a plus-strand-replication-origin nick site, and an SSOA (formerly palA) minus strand origin. Similarity breaks down at a region equivalent to that in pSK108 which encodes the smr multidrug resistance gene (Fig. 5A). In pSK41, this segment contains a gene encoding a deduced product of 248 amino acids which hydropathy analysis indicates is likely to be a membrane protein possessing up to eight transmembrane segments. Although no similar proteins were found in the sequence databases, it is tempting to speculate that this protein may be a transporter of unknown specificity. It would seem that in a stage of the process leading to cointegration of this plasmid, the replication gene, rep(RC), has been bisected by IS257 insertion (Fig. 5A). Duplications of 8 nt expected to result from IS257 transposition (35) are not found at the termini of the segment (Fig. 1). Flanking deletions, a previously described property of IS257 (21), may account for the absence of such repeats since comparison of the rep remnants with the homolog from pSK108 suggests that approximately 57 nt (encoding 19 amino acids) has been deleted from this pSK41 segment.
FIG. 5.
Dot plot comparisons between segments of pSK41 (x axes) and the small RC plasmids (y axes), pSK108 (A) and pSK89 (B). Dots indicate 20-nt stretches containing at least 17 identities. The genetic organization of the plasmids is illustrated beside the relevant axes. The extents and directions of genes are indicated by arrowed boxes; homologous genes are indicated by equivalent shading. The locations of the SSOA sequences are denoted by open boxes, and the positions of the putative replication nick sites are indicated by an arrow. The locations of the flanking IS257 elements in pSK41 are also shown. The segment of pSK108 orf334 not represented in the comparable pSK41 sequence is indicated by hatching. Sequence numbering for pSK89 and pSK108 starts at the first nucleotide their replication-initiation genes, rep and orf334, respectively. The pSK108 (22) and pSK89 (25) sequences are from the GenBank entries U15783 and M37889, respectively.
The nucleotide sequence of the smr gene of pSK41 has been reported previously (25), the sequence published representing one end of a segment flanked by IS25741D and IS25741E (Fig. 1 and 2). Analysis of this segment in its entirety revealed extensive similarity to another S. aureus plasmid of the pC194 family, pSK89 (Fig. 5B) (25). In addition to the previously described homologous smr multidrug resistance gene and SSOA sequences (25), this segment contains a homologous replication-initiation gene, rep(RC). No sequence resembling the replication nick site of pC194 family plasmids is evident in the sequence. Extrapolation based on its position in closely related plasmids indicates that the nick site would be expected to reside somewhere beyond the flanking IS257 elements. It is therefore likely that the nick site has been removed as a result of an IS257-mediated adjacent deletion, which would also explain the absence of flanking target duplications.
In pJE1, two additional IS257-flanked DNA segments are located between the integrated copy of pUB110 and the tra region (Fig. 1 and 2). The segment between IS257JE1D and IS257JE1E contains the trimethoprim resistance determinant, dfrA, and is virtually identical in sequence to an equivalent segment of the composite transposon-like element Tn4003 from pSK1 (42), except for a deletion flanking the IS257 copy upstream of dfrA (21). Tn4003 is now thought to be derived from a cointegrated copy of a pSK639 family trimethoprim resistance plasmid (1, 48). Consistent with the notion that this region of pJE1 is similarly derived, sequencing revealed that the segment between IS257JE1C and IS257JE1D (Fig. 2) is identical to a portion of pSK639. An equivalent segment was also found in Tn4003 (1), although the pSK639 rep gene remnant in pJE1 is 427 nt larger than that present in Tn4003 on pSK1. The incorporation of a pSK639-like plasmid into a pJE1 precursor appears to have occurred as the result of homologous recombination between an IS257 copy equivalent to IS25741C and an element equivalent to IS257639A (1, 23) rather than via IS257 transposition, since an additional element does not seem to have been generated. The identities of IS25741C, IS257639A, IS257JE1C, and IS257JE1E are consistent with this model (Fig. 6).
Sequences corresponding to a pSK639-type rep gene are also present on pGO1, adjacent to IS257GO1R (34) within an IS257-flanked segment similarly neighboring that encoding dfrA (Fig. 1). The organization and restriction map of this region, with an additional three copies of IS257 in comparison to pSK41, suggests that cointegration of a pSK639-like plasmid has resulted from nonresolved replicative transposition of the element on the pGO1 precursor equivalent to IS25741G; IS257GO1R differs from IS25741G by a single nucleotide (C at position 107; Fig. 6). It should also be noted that for each of the small cointegrated plasmids described above, which include both RC and probable theta-mode replicons, the cognate replication function appears to have been inactivated either by interruption of the rep gene coding sequence by IS257 insertion or by the removal of essential sequences by IS257-flanking deletions.
The region of pSK41 from IS25741E to IS25741G (Fig. 1 and 2) is a Tn4001-IS257 hybrid structure essentially identical to that described previously for another pSK41-like plasmid, pSH6 (3), differing at only a single nucleotide within IS25741E. This structure represents a derivative of Tn4001 that has been immobilized by IS257 insertions and/or flanking deletions into its bounding IS256 elements. The DNA segment between IS25741F and IS25741G shares statistically significant similarity (data not shown) with a region of the Staphylococcus hominis chromosome distal to a gene of unknown function, smpB (41); the evolutionary implications of this relationship are not clear at this time.
Comparative analysis of the IS257 elements of pSK41 (Fig. 6) revealed that they can be grouped into several sequence types. IS25741A and IS25741B are identical and differ from IS25741C at only four positions. These elements are also very similar or identical to the two elements on pSK639, as are IS257JE1C, IS257JE1D, and IS257JE1E, a finding consistent with the notion that this portion of pJE1 is derived from such a plasmid. With the exception of two nucleotides, IS25741D and IS25741E are identical but differ from IS25741ABC-type at a minimum of 22 positions. Oriented in opposition to all other IS257 copies in pSK41 (Fig. 1 and 2), IS25741F differs markedly from the two aforementioned sequence types. IS25741G appears to be the product of multiple recombination events, since it possesses IS25741ABC-type ends and an IS25741F-like central portion. It can be seen that each of the three cointegrated plasmids within pSK41 are flanked by identical or nearly identical IS257 copies. Taken together, these observations, along with the likelihood that two of the cointegrated plasmids have resulted from IS257 insertions that directly inactivate their replication-initiation genes and the scarcity of naturally occurring RC plasmids carrying insertion sequences, argue strongly for nonresolved replicative transposition of IS257 as the basis for the cointegrative capture of these plasmids (23, 35, 48).
Site-specific deletions in pSK41 family plasmids.
Several researchers have described the generation of deletions within pSK41-type plasmids that correspond to DNA segments between copies of IS257 (30, 34). To investigate this phenomenon, we characterized two such deletion derivatives of pSK41, identified as exconjugant progeny that had lost one or more resistance phenotypes; of 1,200 transconjugants screened, only 2 (<0.2%) were found to have lost the capacity to confer aadD-encoded Nmr, smr-encoded ethidium bromide (Eb) resistance, and/or aacA-aphD-encoded Gmr. pSK5093 conferred a Nms Ebs Gmr phenotype, and restriction mapping demonstrated the loss of a DNA segment between IS25741B and IS25741E (Fig. 1), including three copies of IS257. pSK5094, which conferred an Nmr Ebr Gms phenotype, was found to have lost a DNA segment between IS25741E and IS25741G, including two IS257 copies. In both cases, nucleotide sequencing indicated that the IS257 elements remaining at the site of these deletions, IS2575093B and IS2575094E, are likely to represent chimeras of the elements that formerly bounded the deleted segments (Fig. 6). This finding strongly implicates homologous recombination as the basis of such deletions, as had been proposed previously (34, 52, 53). By using the available sequence variation, the crossover points can be localized to between nt 231 and 472 for IS2575093B and between nt 632 and 711 for IS2575094E (Fig. 6).
Concluding remarks.
The completion of the pSK41 sequence allows us to make some tentative conclusions about the evolution of pSK41-type conjugative plasmids. The transfer systems of these plasmids and that of the broad-host-range plasmid, pIP501, share a common evolutionary ancestry, although their replication systems appear to be distinct. Despite this and despite the relatedness of the pSK41 replication functions to those of staphylococcal β-lactamase–heavy-metal resistance plasmids, the degree of sequence divergence and the dispersion of conjugation-associated genes on both sides of the pSK41 rep region strongly suggest that pSK41-type plasmids have not arisen from the en bloc addition of conjugation functions to an existing, nontransmissible resistance plasmid. It would seem more likely that the antimicrobial resistance determinants on pSK41-like plasmids are contemporary additions. Probable target duplications adjacent to IS25741A and IS25741C (Fig. 1), still evident despite the propensity of deletion events adjacent to IS257, support this proposal. These duplications insinuate that the pSK41 rep and tra regions were juxtaposed prior to their separation by insertion of IS257 and cointegrated plasmids, including pUB110. The phenotype(s), if any other than conjugative transfer, associated with the primordial pSK41-type plasmid is not known but may still be encoded in the segment between IS25741G and IS25741A (Fig. 2); the availability of this sequence data should help to shed light on this possibility.
IS257 has clearly played a central role in the recent evolutionary history of pSK41 family plasmids, representing a catalyst for adaptation to a new niche, viz., an environment of widespread antimicrobial use. Shaped by the activities of this element, the structure of pSK41 illustrates the selfish nature of transmissible plasmids and its relevance to the development of antimicrobial resistance in staphylococci. For instance, the incorporation of small resistance plasmids presumably enhances the evolutionary fitness of the plasmid in subsequent hosts rather than benefiting the cell in which the cointegration event(s) occurred. The immobilization of the composite Tn4001-like aminoglycoside resistance transposon would seem to represent another manifestation of this phenomenon, presumably contributing to plasmid maintenance in hosts exposed to such antibiotics. The tight genetic linkage resulting from the accretion of resistance determinants on transmissible plasmids is of obvious consequence to the acquisition, dissemination, and maintenance of multiresistance. The capacity of plasmids such as pSK41 to collect activities as they move horizontally through bacterial populations highlights the importance of organisms, which may themselves be nonpathogenic, that act as reservoirs or transient hosts.
ACKNOWLEDGMENTS
We thank Melissa Brown, Ian Paulsen, and Carol Scaramuzzi for helpful discussions and Keith Dyke for pointing out the similarity between portions of the Tn4001-hybrid structure and the S. hominis chromosome.
This work was supported in part by a project grant from the National Health and Medical Research Council (Australia). T.B. was the recipient of an Australian Postgraduate Award. S.A. and A.L. were recipients of Australian International Development Assistance Bureau scholarships.
REFERENCES
- 1.Apisiridej S, Leelaporn A, Scaramuzzi C D, Skurray R A, Firth N. Molecular analysis of a mobilizable theta-mode trimethoprim resistance plasmid from coagulase-negative staphylococci. Plasmid. 1997;38:13–24. doi: 10.1006/plas.1997.1292. [DOI] [PubMed] [Google Scholar]
- 2.Archer G L, Johnston J L. Self-transmissible plasmids in staphylococci that encode resistance to aminoglycosides. Antimicrob Agents Chemother. 1983;24:70–77. doi: 10.1128/aac.24.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Byrne M E, Gillespie M T, Skurray R A. Molecular analysis of a gentamicin resistance transposonlike element on plasmids isolated from North American Staphylococcus aureus strains. Antimicrob Agents Chemother. 1990;34:2106–2113. doi: 10.1128/aac.34.11.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Byrne M E, Gillespie M T, Skurray R A. 4′,4" adenyltransferase activity on conjugative plasmids isolated from Staphylococcus aureus is encoded on an integrated copy of pUB110. Plasmid. 1991;25:70–75. doi: 10.1016/0147-619x(91)90008-k. [DOI] [PubMed] [Google Scholar]
- 5.Claros M G, von Heijne G. TopPred II: an improved software for membrane protein structure predictions. Comput Appl Biosci. 1994;10:685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- 6.Climo M W, Sharma V K, Archer G L. Identification and characterization of the origin of conjugative transfer (oriT) and a gene (nes) encoding a single-stranded endonuclease on the staphylococcal plasmid pGO1. J Bacteriol. 1996;178:4975–4983. doi: 10.1128/jb.178.16.4975-4983.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dayhoff M O, Schwartz R M, Orcutt B C. A model of evolutionary changes in proteins. In: Dayhoff M O, editor. Atlas of protein sequence and structure. Washington, D.C: National Biomedical Research Foundation; 1978. pp. 324–352. [Google Scholar]
- 8.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans J, Dyke K G. Characterization of the conjugation system associated with the Staphylococcus aureus plasmid pJE1. J Gen Microbiol. 1988;134:1–8. doi: 10.1099/00221287-134-1-1. [DOI] [PubMed] [Google Scholar]
- 10.Felsenstein J. PHYLIP: phylogeny inference package. Cladistics. 1989;5:164–166. [Google Scholar]
- 11.Firth N, Ridgway K P, Byrne M E, Fink P D, Johnson L, Paulsen I T, Skurray R A. Analysis of a transfer region from the staphylococcal conjugative plasmid pSK41. Gene. 1993;136:13–25. doi: 10.1016/0378-1119(93)90442-6. [DOI] [PubMed] [Google Scholar]
- 12.Fujimoto S, Tomita H, Wakamatsu E, Tanimoto K, Ike Y. Physical mapping of the conjugative bacteriocin plasmid pPD1 of Enterococcus faecalis and identification of the determinant related to the pheromone response. J Bacteriol. 1995;177:5574–5581. doi: 10.1128/jb.177.19.5574-5581.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gering M, Gotz F, Bruckner R. Sequence and analysis of the replication region of the Staphylococcus xylosus plasmid pSX267. Gene. 1996;182:117–122. doi: 10.1016/s0378-1119(96)00526-4. [DOI] [PubMed] [Google Scholar]
- 14.Gilbert D G. Dot plot sequence comparisons on Macintosh computers. Comput Appl Biosci. 1990;6:117. doi: 10.1093/bioinformatics/6.2.117. [DOI] [PubMed] [Google Scholar]
- 15.Gillespie M T, Lyon B R, Skurray R A. Structural and evolutionary relationships of β-lactamase transposons from Staphylococcus aureus. J Gen Microbiol. 1988;134:2857–2866. doi: 10.1099/00221287-134-11-2857. [DOI] [PubMed] [Google Scholar]
- 16.Gillespie M T, May J W, Skurray R A. Antibiotic resistance in Staphylococcus aureus isolated at an Australian hospital between 1946 and 1981. J Med Microbiol. 1985;19:137–147. doi: 10.1099/00222615-19-2-137. [DOI] [PubMed] [Google Scholar]
- 17.Gotz F, Zabielski J, Philipson L, Lindberg M. DNA homology between the arsenate resistance plasmid pSX267 from Staphylococcus xylosus and the penicillinase plasmid pI258 from Staphylococcus aureus. Plasmid. 1983;9:126–137. doi: 10.1016/0147-619x(83)90015-x. [DOI] [PubMed] [Google Scholar]
- 18.Helinski D R, Toukdarian A E, Novick R P. Replication control and other stable maintenance mechanisms of plasmids. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella. Washington, D.C: American Society for Microbiology; 1996. pp. 2295–2324. [Google Scholar]
- 19.Jaffe H W, Sweeney H M, Weinstein R A, Kabins S A, Nathan C, Cohen S. Structural and phenotypic varieties of gentamicin resistance plasmids in hospital strains of Staphylococcus aureus and coagulase-negative staphylococci. Antimicrob Agents Chemother. 1982;21:773–779. doi: 10.1128/aac.21.5.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khan S A. Rolling-circle replication of bacterial plasmids. Microbiol Mol Biol Rev. 1997;61:442–455. doi: 10.1128/mmbr.61.4.442-455.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leelaporn A, Firth N, Byrne M E, Roper E, Skurray R A. Possible role of insertion sequence IS257 in dissemination and expression of high- and low-level trimethoprim resistance in staphylococci. Antimicrob Agents Chemother. 1994;38:2238–2244. doi: 10.1128/aac.38.10.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leelaporn A, Firth N, Paulsen I T, Hettiaratchi A, Skurray R A. Multidrug resistance plasmid pSK108 from coagulase-negative staphylococci; relationships to Staphylococcus aureus qacC plasmids. Plasmid. 1995;34:62–67. doi: 10.1006/plas.1995.1034. [DOI] [PubMed] [Google Scholar]
- 23.Leelaporn A, Firth N, Paulsen I T, Skurray R A. IS257-mediated cointegration in the evolution of a family of staphylococcal trimethoprim resistance plasmids. J Bacteriol. 1996;178:6070–6073. doi: 10.1128/jb.178.20.6070-6073.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lipman D J, Pearson W R. Rapid and sensitive protein similarity searches. Science. 1985;227:1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- 25.Littlejohn T G, DiBerardino D, Messerotti L J, Spiers S J, Skurray R A. Structure and evolution of a family of genes encoding antiseptic and disinfectant resistance in Staphylococcus aureus. Gene. 1991;101:59–66. doi: 10.1016/0378-1119(91)90224-y. [DOI] [PubMed] [Google Scholar]
- 26.Lyon B R, May J W, Skurray R A. Analysis of plasmids in nosocomial strains of multiple-antibiotic-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1983;23:817–826. doi: 10.1128/aac.23.6.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lyon B R, May J W, Skurray R A. Tn4001: a gentamicin and kanamycin resistance transposon in Staphylococcus aureus. Mol Gen Genet. 1984;193:554–556. doi: 10.1007/BF00382099. [DOI] [PubMed] [Google Scholar]
- 28.Lyon B R, Skurray R. Antimicrobial resistance of Staphylococcus aureus: genetic basis. Microbiol Rev. 1987;51:88–134. doi: 10.1128/mr.51.1.88-134.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Macrina F L, Archer G L. Conjugation and broad host range plasmids in streptococci and staphylococci. In: Clewell D B, editor. Bacterial conjugation. New York, N.Y: Plenum Press, Inc.; 1993. pp. 313–329. [Google Scholar]
- 30.McDonnell R W, Sweeney H M, Cohen S. Conjugational transfer of gentamicin resistance plasmids intra- and interspecifically in Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob Agents Chemother. 1983;23:151–160. doi: 10.1128/aac.23.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKenzie T, Hoshino T, Tanaka T, Sueoka N. The nucleotide sequence of pUB110: some salient features in relation to replication and its regulation. Plasmid. 1986;15:93–104. doi: 10.1016/0147-619x(86)90046-6. [DOI] [PubMed] [Google Scholar]
- 32.Mills D A, McKay L L, Dunny G M. Splicing of a group II intron involved in the conjugative transfer of pRS01 in lactococci. J Bacteriol. 1996;178:3531–3538. doi: 10.1128/jb.178.12.3531-3538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morton T M, Eaton D M, Johnston J L, Archer G L. DNA sequence and units of transcription of the conjugative transfer gene complex (trs) of Staphylococcus aureus plasmid pGO1. J Bacteriol. 1993;175:4436–4447. doi: 10.1128/jb.175.14.4436-4447.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morton T M, Johnston J L, Patterson J, Archer G L. Characterization of a conjugative staphylococcal mupirocin resistance plasmid. Antimicrob Agents Chemother. 1995;39:1272–1280. doi: 10.1128/aac.39.6.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Needham C, Noble W C, Dyke K G H. The staphylococcal insertion sequence IS257 is active. Plasmid. 1995;34:198–205. doi: 10.1006/plas.1995.0005. [DOI] [PubMed] [Google Scholar]
- 36.Novick R P. Staphylococcal plasmids and their replication. Ann Rev Microbiol. 1989;43:537–565. doi: 10.1146/annurev.mi.43.100189.002541. [DOI] [PubMed] [Google Scholar]
- 37.Paulsen I T, Firth N, Skurray R A. Resistance to antimicrobial agents other than β-lactams. In: Archer G L, Crossley K B, editors. The staphylococci in human disease. New York, N.Y: Churchill Livingstone; 1997. pp. 175–212. [Google Scholar]
- 38.Paulsen I T, Gillespie M T, Littlejohn T G, Hanvivatvong O, Rowland S J, Dyke K G, Skurray R A. Characterisation of sin, a potential recombinase-encoding gene from Staphylococcus aureus. Gene. 1994;141:109–114. doi: 10.1016/0378-1119(94)90136-8. [DOI] [PubMed] [Google Scholar]
- 39.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Projan S J, Archer G L. Mobilization of the relaxable Staphylococcus aureus plasmid pC221 by the conjugative plasmid pGO1 involves three pC221 loci. J Bacteriol. 1989;171:1841–1845. doi: 10.1128/jb.171.4.1841-1845.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ross J I, Eady E A, Cove J H, Baumberg S. Identification of a chromosomally encoded ABC-transport system with which the staphylococcal erythromycin exporter MsrA may interact. Gene. 1995;153:93–98. doi: 10.1016/0378-1119(94)00833-e. [DOI] [PubMed] [Google Scholar]
- 42.Rouch D A, Messerotti L J, Loo L S, Jackson C A, Skurray R A. Trimethoprim resistance transposon Tn4003 from Staphylococcus aureus encodes genes for a dihydrofolate reductase and thymidylate synthetase flanked by three copies of IS257. Mol Microbiol. 1989;3:161–175. doi: 10.1111/j.1365-2958.1989.tb01805.x. [DOI] [PubMed] [Google Scholar]
- 43.Rowland S J, Dyke K G. Characterization of the staphylococcal β-lactamase transposon Tn552. EMBO J. 1989;8:2761–2773. doi: 10.1002/j.1460-2075.1989.tb08418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruhfel R E, Manias D A, Dunny G M. Cloning and characterization of a region of the Enterococcus faecalis conjugative plasmid, pCF10, encoding a sex pheromone-binding function. J Bacteriol. 1993;175:5253–5259. doi: 10.1128/jb.175.16.5253-5259.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 46.Shalita Z, Murphy E, Novick R P. Penicillinase plasmids of Staphylococcus aureus: structural and evolutionary relationships. Plasmid. 1980;3:291–311. doi: 10.1016/0147-619x(80)90042-6. [DOI] [PubMed] [Google Scholar]
- 47.Sharma V K, Johnston J L, Morton T M, Archer G L. Transcriptional regulation by TrsN of conjugative transfer genes on staphylococcal plasmid pGO1. J Bacteriol. 1994;176:3445–3454. doi: 10.1128/jb.176.12.3445-3454.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skurray R A, Firth N. Molecular evolution of multiply-antibiotic-resistant staphylococci. Ciba Found Symp. 1997;207:167–183. doi: 10.1002/9780470515358.ch11. [DOI] [PubMed] [Google Scholar]
- 49.Swinfield T J, Janniere L, Ehrlich S D, Minton N P. Characterization of a region of the Enterococcus faecalis plasmid pAMβ1 which enhances the segregational stability of pAMβ1-derived cloning vectors in Bacillus subtilis. Plasmid. 1991;26:209–221. doi: 10.1016/0147-619x(91)90044-w. [DOI] [PubMed] [Google Scholar]
- 50.Takiguchi R, Hashiba H, Aoyama K, Ishii S. Complete nucleotide sequence and characterization of a cryptic plasmid from Lactobacillus helveticus subsp. jugurti. Appl Environ Microbiol. 1989;55:1653–1655. doi: 10.1128/aem.55.6.1653-1655.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tennent J M, Lyon B R, Gillespie M T, May J W, Skurray R A. Cloning and expression of Staphylococcus aureus plasmid-mediated quaternary ammonium resistance in Escherichia coli. Antimicrob Agents Chemother. 1985;27:79–83. doi: 10.1128/aac.27.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thomas W D, Jr, Archer G L. Mobility of gentamicin resistance genes from staphylococci isolated in the United States: identification of Tn4031, a gentamicin resistance transposon from Staphylococcus epidermidis. Antimicrob Agents Chemother. 1989;33:1335–1341. doi: 10.1128/aac.33.8.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thomas W J, Archer G L. Identification and cloning of the conjugative transfer region of Staphylococcus aureus plasmid pGO1. J Bacteriol. 1989;171:684–691. doi: 10.1128/jb.171.2.684-691.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vieira J, Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- 55.Wang A, Macrina F L. Characterization of six linked open reading frames necessary for pIP501-mediated conjugation. Plasmid. 1995;34:206–210. doi: 10.1006/plas.1995.0006. [DOI] [PubMed] [Google Scholar]
- 56.Weaver K E, Clewell D B, An F. Identification, characterization, and nucleotide sequence of a region of Enterococcus faecalis pheromone-responsive plasmid pAD1 capable of autonomous replication. J Bacteriol. 1993;175:1900–1909. doi: 10.1128/jb.175.7.1900-1909.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weber D A, Goering R V. Tn4201, a β-lactamase transposon in Staphylococcus aureus. Antimicrob Agents Chemother. 1988;32:1164–1169. doi: 10.1128/aac.32.8.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yan C, Tamm I. Molecular cloning and characterization of interferon alpha/beta response element binding factors of the murine (2′-5′)oligoadenylate synthetase ME-12 gene. Proc Natl Acad Sci USA. 1991;88:144–148. doi: 10.1073/pnas.88.1.144. [DOI] [PMC free article] [PubMed] [Google Scholar]