Abstract
Analysis of chlorobenzene-degrading transconjugants of Pseudomonas putida F1 which had acquired the genes for chlorocatechol degradation (clc) from Pseudomonas sp. strain B13 revealed that the clc gene cluster was present on a 105-kb amplifiable genetic element (named the clc element). In one such transconjugant, P. putida RR22, a total of seven or eight chromosomal copies of the entire genetic element were present when the strain was cultivated on chlorobenzene. Chromosomal integrations of the 105-kb clc element occurred in two different loci, and the target sites were located within the 3′ end of glycine tRNA structural genes. Tandem amplification of the clc element was preferentially detected in one locus on the F1 chromosome. After prolonged growth on nonselective medium, transconjugant strain RR22 gradually diverged into subpopulations with lower copy numbers of the clc element. Two nonadjacent copies of the clc element in different loci always remained after deamplification, but strains with only two copies could no longer use chlorobenzene as a sole substrate. This result suggests that the presence of multiple copies of the clc gene cluster was a prerequisite for the growth of P. putida RR22 on chlorobenzene and that amplification of the element was positively selected for in the presence of chlorobenzene.
Pseudomonas sp. strain B13 was the first described pseudomonad which could use 3-chlorobenzoate (3CBA) as a sole carbon and energy source (7). The degradation of 3CBA involves initial oxidation to chlorocatechols, which are subsequently converted to 3-oxoadipate by the action of four enzymes of the modified ortho cleavage pathway. This ortho cleavage pathway, also referred to as the chlorocatechol oxidative pathway, is encoded by the clcABDE genes (9). The clc genes can be transferred from strain B13 to many different recipient bacteria, thereby enabling the recipients to degrade chlorocatechols as well (20, 23, 29, 35). Although presumed to be a conjugative process, the transfer mechanisms and the nature of the transferred element have remained unclear. The isolation of a 110-kb plasmid carrying the clc genes in Pseudomonas sp. strain B13 was reported (3). However, other research groups were unable to isolate plasmid DNA from strain B13 (20, 35). Weisshaar et al. hypothesized the clc genes to be present on the chromosome of strain B13, but clear evidence for this hypothesis was not presented (33).
Strain B13 remained the basic organism for biochemical studies on the chlorocatechol oxidative pathway, but the clc genes were characterized initially from Pseudomonas putida AC827 (9). Indeed, in this strain a plasmid (pAC27) harboring the clc genes was discovered. The clc genes are organized in two clusters. One is formed by the clcABDE structural genes, comprising a 4-kb region (9). The other is formed by a 0.9-kb regulatory gene, clcR, oriented divergently from clcABDE and with a 0.3-kb spacing from clcA (5). It was long presumed that the clc genes of strain B13 would be very similar to those of P. putida AC827. Only recently was a DNA fragment containing the clcR gene of strain B13 characterized and shown to be identical to the pAC27 clcR gene (17).
To study the transmissible clc element of strain B13, we tested the possibility of trapping the element in a clear distinguishable form in another host. As a new recipient for the clc genes, we chose P. putida F1. This bacterium can metabolize toluene due to the catalytic activities of the enzymes encoded by the chromosomally located tod genes (36, 37). It had previously been demonstrated that the clc genes of Pseudomonas sp. strain B13 could be transferred to P. putida F1 (20, 22). The resulting F1 transconjugants expressed both the chlorocatechol oxidative pathway genes and those for toluene degradation and were therefore able to completely metabolize monochlorobenzene (MCB) and 1,4-dichlorobenzene (1,4-DCB).
In this report, we describe previously unknown features of the transmissible clc element of strain B13. Using pulsed-field gel electrophoresis (PFGE), cosmid mapping, and DNA-DNA hybridizations, we show that the element has a size of 105 kb and is integrated into the chromosome of strain F1. A physical map of the complete element and the sequences of its integration sites in strain F1 are presented. We demonstrate the unusual amplification and deamplification behaviors of the clc element in strain F1. To our knowledge, this is the first large chromosomal tandem amplification found in Pseudomonas spp.
MATERIALS AND METHODS
Bacterial strains and relevant characteristics.
Pseudomonas sp. strain B13 grows on 3CBA as a sole source of carbon and energy (20, 23). P. putida F1, which uses toluene as a sole carbon and energy source, was kindly obtained from Dave Gibson (36, 37). Strains P. putida RR1, RR3, RR4, RR6, RR7, RR8, RR21, RR22, RR28, and RR29 are transconjugants of strain F1 which have obtained the clc genes from strain B13. These transconjugants were isolated from matings between P. putida F1 and Pseudomonas sp. strain B13 on agar plate surfaces with 1,4-DCB as a sole substrate. They all use toluene, 3CBA, MCB, and 1,4-DCB as sole sources of carbon and energy. Strains RR1 to RR8 and strains RR21 to RR29 were obtained from two independent matings. P. aeruginosa rec1 (PAO rec1) is a recA mutant which was obtained from Michael Kertesz, Institute of Microbiology, Swiss Federal Institute for Technology, Zürich, Switzerland. PAO rec1 carries a Tn501::A7 insertion of the mercury resistance genes in recA as described by Ohman et al. (19). Escherichia coli DH5α (26) and E. coli XL1-Blue MR (Stratagene, La Jolla, Calif.) were used as host strains for recombinant DNA.
Media and culturing conditions.
For routine growth of the Pseudomonas strains and their enumeration by selective plating, Z3 minimal medium (30) to which the appropriate aromatic compounds were added was used; for strain B13, this compound was 3CBA, for strain F1, it was toluene, and for F1 transconjugants, it was MCB. Agar plates were incubated in gas-tight glass jars to which toluene or MCB was applied through the vapor phase. 3CBA was dissolved directly in the agar at a concentration of 5 mM. Ultrapure agar (Merck AG, Dietikon, Switzerland) was used to minimize background growth. For liquid cultures, toluene, MCB, and 1,4-DCB were dissolved in a secondary phase (2,2′,4,4′,6,8,8′-heptamethylnonane [HMN]; Sigma Chemical Co., St. Louis, Mo.). Relative to HMN, toluene was dissolved at a ratio of 0.1 (vol/vol), MCB was dissolved at 0.04 (vol/vol), and 1,4-DCB was dissolved at 0.02 (wt/vol). Per liter of Z3 minimal medium, 400 μl of toluene (346 mg), 400 μl of MCB (443 mg), or 400 mg of 1,4-DCB was added. For growth on aromatic substrates in liquid media, 1:100 volumes from an overnight preculture in nutrient broth (Biolife, Milano, Italy) or Luria-Bertani (LB) medium were used as inocula. PAO rec1 was grown in the presence of 6 μg (for Z3 minimal medium) or 20 μg (for LB medium) of mercury chloride per ml. For every experiment (unless otherwise stated), the bacteria were restreaked on selective agar plates from stock cultures which were maintained in 15% (vol/vol) glycerol at −80°C.
Prolonged cultivation of strain RR22 on specific media.
To determine the stability of the clc element in P. putida RR22, this strain was grown on LB medium and on Z3 medium with 3CBA for more than 200 generations. Cells were grown in batch cultures for approximately 20 generations each time. From each new batch culture, a fraction of 10−6 was inoculated into fresh medium. This procedure was repeated until 220 generations had passed. From every transferred culture, a sample was frozen in 15% (vol/vol) glycerol at −80°C. In addition, 10 individual cell lineages were derived from strain RR22 after cultivation on nonselective medium. Ten colonies were randomly picked from LB plates after 100 generations on LB medium. These lineages were designated RR2231 to RR2240. They all had different capabilities to grow on MCB as a sole carbon and energy source. Strain RR221, which could not use MCB anymore, was picked as a single colony after 200 generations of growth of RR22 on LB medium.
PFGE.
Agarose-embedded DNA suitable for separations by field inversion gel electrophoresis (FIGE) or PFGE was prepared in accordance with instructions provided by the manufacturer of the FIGE Mapper electrophoresis system (Bio-Rad Laboratories AG, Glattbrugg, Switzerland) with some modifications. Bacterial cultures were grown to the late exponential phase. The cells were harvested by centrifugation, washed in 20 mM phosphate buffer (pH 7), and resuspended in a cell suspension buffer (10 mM Tris-HCl [pH 7.2], 20 mM NaCl, 5 mM EDTA) to a final density of 109 cells per ml. The cell suspension was mixed with the same volume of a 2% low-melting-temperature agarose solution (in distilled water) at 40°C, and the mixture was transferred into plug molds (Bio-Rad). After the agarose plugs had solidified, they were incubated in lysozyme buffer at 37°C for 30 min (lysozyme buffer contains 10 mM Tris-HCl [pH 7.2], 50 mM NaCl, 0.2% sodium deoxycholate, 0.5% sodium lauryl sarcosine, and 1 mg of lysozyme per ml). The lysozyme buffer was removed, and the plugs were rinsed once with wash buffer (20 mM Tris-HCl [pH 8.0], 50 mM EDTA) and incubated overnight at 50°C in proteinase K reaction buffer (100 mM EDTA [pH 8.0], 0.2% sodium deoxycholate, 1% sodium lauryl sarcosine, 1 mg of proteinase K per ml). Subsequently, the plugs were washed four times in wash buffer with gentle agitation for 1 h each time. In the second wash, phenylmethylsulfonyl fluoride was added to a final concentration of 1 mM to inactivate any residual proteinase K activity. After the fourth wash, the plugs were stored in wash buffer at 4°C.
Prior to restriction enzyme digestion of the DNA in the agarose plugs, the EDTA concentration in the plugs was lowered by incubation in 10-times-diluted wash buffer for 0.5 h. Each agarose plug was then incubated in an Eppendorf tube with 1 ml of the appropriate restriction enzyme buffer for 1 h. After replacement of this buffer with 0.3 ml of fresh restriction enzyme buffer, restriction digestion was performed by adding 50 U of restriction enzyme per 100 μl of plug volume and incubating the mixture overnight at the appropriate temperature for the enzyme. Before agarose plugs were loaded on the gels, they were equilibrated in electrophoresis buffer for 30 min. Separations of DNA molecules up to 500 kb were performed with the FIGE Mapper electrophoresis system, whereas for separations up to 2,000 kb, a CHEF DR II system (Bio-Rad) was used. Routinely, gels contained 1% agarose in 0.5× Tris-borate buffer (0.5× Tris-borate buffer is 45 mM Tris-borate and 1 mM EDTA); TBE buffer was also used as the running buffer for electrophoresis. Total run time and pulse times were set to obtain the desired separation range by following the instructions given by the manufacturers. Both FIGE and PFGE separations were performed at 4°C. After electrophoresis, the gels were stained with ethidium bromide.
DNA-DNA hybridizations.
DNA fractionated by agarose gel electrophoresis was blotted onto a Qiabrane nylon membrane (Qiagen AG, Basel, Switzerland) by the following procedure (modified from protocol 18-1023-07, Pharmacia Biotech, Uppsala, Sweden). The ethidium bromide-stained gel was irradiated by UV light (312 nm) for 45 s, denatured with a solution of 0.4 M NaOH plus 0.6 M NaCl for 30 min, and finally neutralized in a solution of 1.0 M Tris-HCl plus 1.5 M NaCl (pH 7.6) for 30 min. The DNA was transferred in 20× SSC (1× SSC is 150 mM NaCl plus 15 mM trisodium citrate at pH 7.0) to the nylon membrane over 45 to 60 min with a vacuum blotting device (VacuGene XL; Pharmacia). The membrane was removed from the gel and treated with the NaOH-NaCl solution for 30 s and with the Tris-HCl–NaCl solution for 1 min. The DNA was fixed on the membrane with UV light (254 nm, 120 mJ/cm2) by use of a model 1800 Stratalinker (Stratagene GmbH, Heidelberg, Germany). Hybridizations were performed with SDS/BSA hybridization buffer (0.5 M sodium phosphate [pH 7.0], 1 mM EDTA, 7% sodium dodecyl sulfate, 1% bovine serum albumin) at 62°C for approximately 16 h, followed by washing twice with a solution of 5× SSC and 1 mM EDTA for 2 min at room temperature and twice with a solution of 0.2× SSC and 0.1% sodium dodecyl sulfate for 30 min at 62°C. DNA fragments used in the hybridizations were labelled with [α-32P]dATP (3,000 Ci/mmol; Amersham, Buckinghamshire, United Kingdom) by use of a random-primer DNA labeling kit (Boehringer GmbH, Mannheim, Germany). As a DNA probe for the clc genes, we used a 4.2-kb BglII fragment of pDC100 (Table 1) containing the clcABD genes from P. putida(pAC27) (9). Hybridized membranes were exposed to Kodak X-Omat film. Hybridization signal intensities on exposed films were measured and analyzed on a computing densitometer with ImageQuant software (Molecular Dynamics, Sunnyvale, Calif.). Densitometric analyses were always performed on autoradiograms with relatively short exposure times to be in the linear range.
TABLE 1.
Plasmids used in this study
| Plasmid | Source of insert DNA | Relevant characteristics |
|---|---|---|
| pDC100 | 4.2-kb BglII fragment of pAC27 (9) | Contains the clcABD genes |
| pRR101 | 4.2-kb NheI-HindIII fragment of cosmid insert 3C9 cloned into pUC18Not | Contains the right junction (R1) of the integrated clc element at INT1 |
| pRR102 | 1.9-kb NotI-HindIII fragment of pRR101 cloned into pUC28 | Contains the right junction (R1) of the integrated clc element at INT1 |
| pRR104 | 4.2-kb EcoRI fragment of cosmid insert 3G3 cloned into pUC18Not | Contains the left junction (L1) of the integrated clc element at INT1 |
| pRR108 | 4.1-kb NheI-EcoRI fragment of cosmid insert 2B1 cloned into pUC18Not | Contains the right junction (R2) of the integrated clc element at INT2 |
| pRR123 | 871-bp PCR product obtained from strain F1 DNA with primers RR301 and RR303, cloned into pGEM-T Easy | Contains INT1 in strain F1 prior to integration |
| pRR148 | 984-bp iPCR product obtained from strain RR221 DNA with primers RR315 and RR316 after digestion with SphI and religation, cloned into pGEM-T Easy | Contains the left junction (L2) of the integrated clc element at INT2 |
| pRR157 | 1.1-kb PCR product obtained from strain F1 DNA with primers RR302 and RR325, cloned into pGEM-T Easy | Contains INT2 in strain F1 prior to integration |
Construction and analysis of a cosmid library from transconjugant strain RR221.
High-molecular-weight total DNA of strain RR221 was isolated from an LB medium overnight culture with an Easy-DNA kit (Invitrogen, Carlsbad, Calif.). From this DNA preparation, a SuperCos 1 cosmid library with DNA inserts ranging from 33 to 42 kb was constructed by Stratagene, La Jolla, Calif. The unamplified library was transfected into host strain E. coli XL1-Blue MR according to the recommendations given by Stratagene. At random, a total number of 768 individual colonies were picked and inoculated into eight 96-well microtiter plates. The bacteria were grown overnight at 37°C in 200 μl of LB medium (per well) supplemented with 50 μg of kanamycin per ml. For screening of the library by Southern hybridizations, 25 μl of bacterial culture from each well was transferred to a Qiabrane nylon membrane by use of a filtration manifold system (series 1055; Life Technologies, Gaithersburg, Md.). Cosmid DNA from positive clones was isolated and analyzed by restriction enzyme digestions by standard procedures (26). Some cosmid DNA fragments were subcloned in pUC18, pUC18Not, or pUC28 vectors (Table 1).
DNA sequencing, PCR, and sequence analysis.
Double-stranded template sequencing was performed on plasmids by use of a Thermo Sequenase fluorescence-labelled primer cycle kit with 7-deaza-dGTP (Amersham). Primers labelled with the fluorescent dye IRD-800 at the 5′ end were purchased from MWG Biotech, Munich, Germany. An automated DNA sequencer (model 4000L; LI-COR Inc., Lincoln, Nebr.) was used for sequencing. Computer analysis of DNA sequences was done with DNASTAR software (DNASTAR Inc., Madison, Wis.). PCR was performed with Taq polymerase (Life Technologies). PCR primers used in this study (Table 2) were purchased from MWG Biotech or from Microsynth, Balgach, Switzerland. Amplified DNAs were cloned into pGEM-T Easy (Promega, Madison, Wis.).
TABLE 2.
PCR primers used in this study
| Primer | Nucleotide sequence | Position (5′ end)a |
|---|---|---|
| RR301 | 5′ GAG AAC GGA TTC AAC GCC ACC 3′ | 348 bp left of INT1 in strain F1 |
| RR302 | 5′ ATC GGC AAA CTG TGC CAT GAC TGG 3′ | 384 bp right of INT2 in strain F1 |
| RR303 | 5′ ACT GCA GCA GAG CAC GCC GTT CG 3′ | 523 bp right of INT1 in strain F1 |
| RR315 | 5′ TGC TCT CAG TTC CCG CAT CC 3′ | clc element, 209 bp from left end, directed inward |
| RR316 | 5′ GAT GAC GTT GTG ACG ACT GC 3′ | clc element, 178 bp from left end, directed outward |
| RR325 | 5′ AGA TAC TGC GCA GCG ACA ACA CCA 3′ | 295 bp left of INT2 in strain F1 |
Relative to the positions of the exact junctions between the clc element and chromosomal target sites in strain F1.
RESULTS
Transconjugants of P. putida F1 with multiple copies of a 100-kb element containing the clc genes.
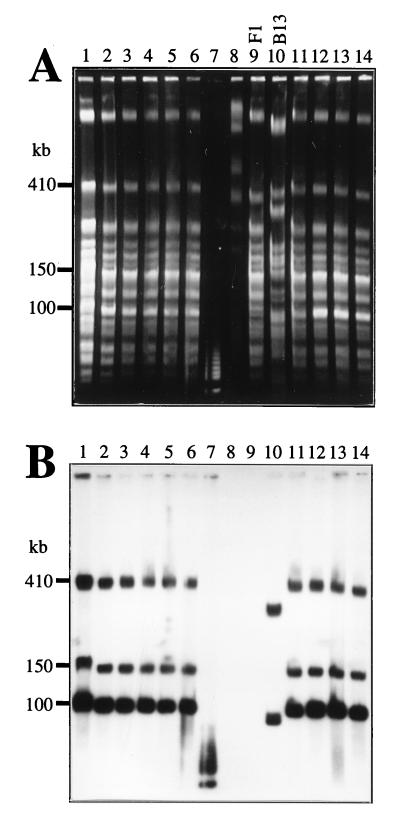
Transconjugants capable of mineralizing chlorobenzene were obtained in matings between Pseudomonas sp. strain B13 and P. putida F1 on agar plate surfaces incubated with 1,4-DCB as the sole substrate. These transconjugants were strain F1 derivatives having acquired the clc genes from strain B13 (22). To determine the location and presence of the clc genes, total DNA samples of the F1 transconjugants, F1, and B13 were analyzed by FIGE and hybridization with a clc gene probe. Total DNAs embedded in agarose plugs were prepared from LB medium-grown cultures. Among SpeI-digested DNAs, three similarly sized fragments of 100, 150, and 410 kb hybridized for all transconjugants (Fig. 1). As expected, no hybridizing fragments were observed in strain F1 DNA. In strain B13 DNA, two SpeI fragments of 90 and 320 kb hybridized to the clc gene probe. The 100-kb SpeI fragment in the transconjugants gave an approximately five-times-stronger hybridization signal than the other hybridizing fragments. Since the clc gene probe did not have internal SpeI restriction sites, the 90- and 320-kb fragments in B13 and the 150- and 410-kb fragments in the transconjugants were likely to each carry one copy of the clc genes. The more strongly hybridizing 100-kb fragment in the transconjugants suggested that multiple copies of the clc genes were present. The FIGE and hybridization mapping results also suggested that a DNA fragment carrying the clc genes had actually become integrated into the strain F1 chromosome, since the hybridizing 410-kb SpeI fragment in the transconjugants was clearly larger than the original 370-kb SpeI fragment in strain F1 (Fig. 1A).
FIG. 1.
Total DNAs digested with SpeI and separated by FIGE. (A) Gel stained with ethidium bromide. (B) Southern hybridization with a probe for the clc genes; the probe was a 4.2-kb BglII fragment containing the clcABD genes from P. putida(pAC27) (9). Lanes: 1 to 6, P. putida RR1, RR3, RR4, RR6, RR7, and RR8, respectively; 7, 5-kb marker (Bio-Rad); 8, Saccharomyces cerevisiae molecular size marker (225 to 2,200 kb; Bio-Rad); 9, P. putida F1; 10, Pseudomonas sp. strain B13; 11 to 14, P. putida RR21, RR22, RR28, and RR29, respectively. Note that the sample in lane 1 contained more DNA than the others, resulting in inaccurate migration velocity for this sample. The 5-kb marker consisted of pUC concatemers and hybridized with the gene probe since the probe contained traces of pUC vector.
Because all F1 transconjugants seemed identical, only one (RR22) was analyzed further. In RR22 total DNA digested with XbaI, fragments of 48, 75, and 100 kb hybridized to the clc gene probe (results not shown). Like the results observed with SpeI digests, the 100-kb XbaI fragment hybridizing with the clc gene probe appeared much more intense than the other two hybridizing fragments. In double digests with SpeI and XbaI, the more intense band appeared at 70 kb. This result indicated that both enzymes cut only once in this multicopy unit, that the distance between the SpeI and XbaI restriction sites was 70 kb, and that the total size of the unit was 100 kb. The multicopy unit was named the clc element. The largest common DNA fragment in strains B13 and RR22 hybridizing to the clc gene probe was a 28-kb NheI fragment (data not shown).
Construction of a physical map of the region containing the clc element.
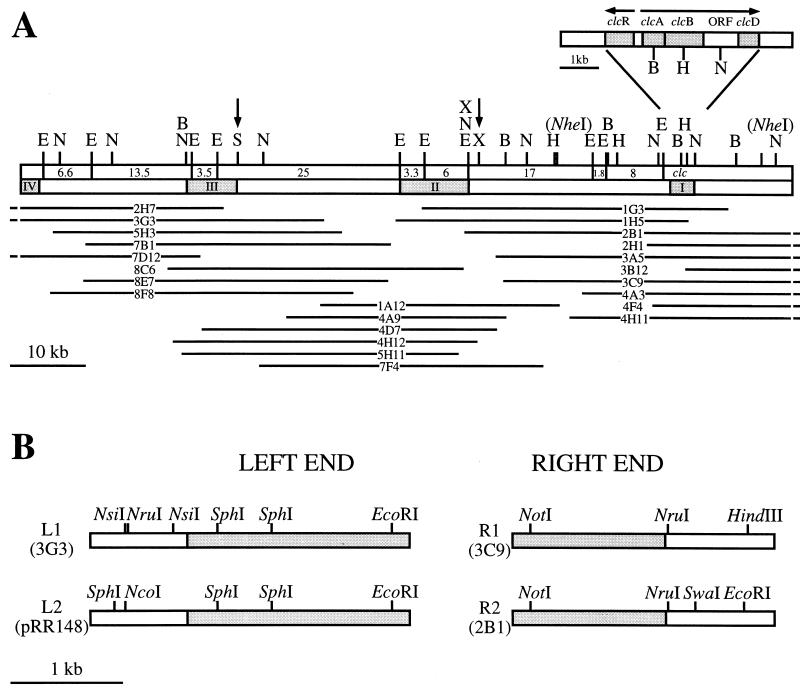
To derive a physical map of the clc element, we isolated overlapping cosmid clones from a library constructed for one particular F1 transconjugant. This transconjugant (RR221) contained only two copies of the clc element (see below). The first series of cosmids was isolated by hybridization with the clc gene probe (probe I; Fig. 2A). The DNAs of positive clones were mapped with restriction enzymes BamHI, EcoRI, HindIII, and NotI. A second gene probe (II) located at the left outer border of the farthest reaching cosmid insert was chosen for further screening of the library, with subsequent mapping of positive clones. This step was again repeated with a third gene probe (III). Finally, a physical map of the entire region containing the clc element could be constructed (Fig. 2A). The positions of the NheI sites flanking the clc gene cluster and the SpeI and XbaI sites were mostly confirmed with respect to the results from FIGE experiments. However, the higher resolution of cosmid insert mapping revealed two XbaI sites just 1.4 kb apart, instead of only one XbaI site (Fig. 2A).
FIG. 2.
Physical map of the region containing the 105-kb clc element and of its flanking chromosomal sites in P. putida RR221. (A) Restriction map of the complete clc element for the enzymes BamHI (B), EcoRI (E), HindIII (H), NotI (N), SpeI (S), and XbaI (X). Indicated with vertical arrows are the positions of SpeI and XbaI sites initially mapped by FIGE. Note that two XbaI sites just 1.4 kb apart are actually present. The two NheI sites flanking the clc genes are shown within parentheses, since other possible NheI sites were not mapped. For comparison with previous data on pB13 (4), the sizes of all EcoRI fragments within the clc element are given. The locations of DNA probes (I, II, and III) used for screening the cosmid library are indicated. Probe I was the 4.2-kb BglII fragment containing the clcABD genes from P. putida(pAC27) (9). The fourth DNA probe (IV) was used for hybridizations with total DNAs from P. aeruginosa transconjugants carrying the clc element. ORF, open reading frame. All of the overlapping cosmid clones (2H7, 3G3, and so forth) used for isolating and mapping the clc element are depicted below the physical map. The parts of the cosmid inserts extending beyond the right and left borders of the clc element are not shown. (B) Restriction maps of the left and right junctions of the two integrated elements in strain RR221. Grey shading indicates DNA which is part of the clc element. The remaining DNA is chromosomal DNA from parental strain F1.
Identification and characterization of the junctions between the integrated clc element in strain RR221 and the F1 chromosomal target sites.
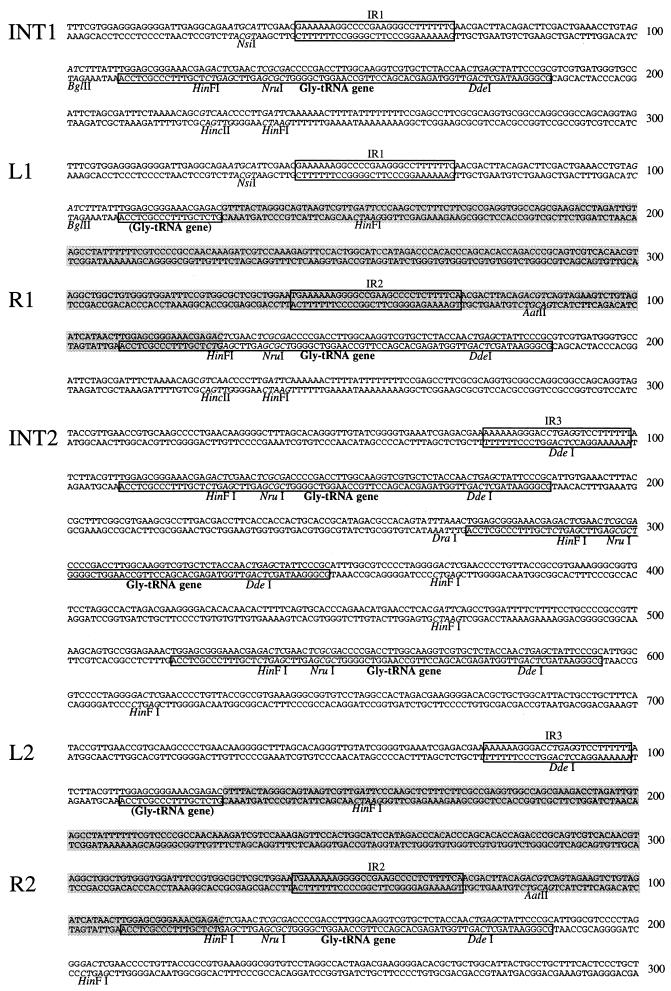
Since two copies of the clc element were present in strain RR221, two types of cosmid clones would be expected if the element were integrated at two locations in the chromosome. Inserts of cosmid clones near the clc genes indeed started to differ from an NruI site onward, suggesting that the right end of the clc element and the regions of integration on the chromosome were to be found here. These regions were subcloned (plasmids pRR101 [right end of first copy, R1] and pRR108 [right end of second copy, R2]), sequenced, and compared (Fig. 2B and 3). The two sequences were identical up to the point including a putative glycine-tRNA structural gene. From there on the sequences differed, suggesting that either the Gly-tRNA gene itself or a sequence within it formed the actual border of the clc element. The sequence of the Gly-tRNA gene in strain RR221 was 100% identical to that of the αGly-tRNA gene in E. coli K-12 (16).
FIG. 3.
Nucleotide sequences of the two chromosomal integration sites of the clc element in P. putida F1 (INT1 and INT2) and the junction regions of the integrated clc element copies in transconjugant RR221 (L1-R1 and L2-R2). The positions of relevant restriction sites, of a large conserved inverted repeat (IR1, IR2, and IR3), and of the Gly-tRNA gene copies (or parts thereof) are shown. The parts of the sequences belonging to the clc element are indicated with grey shading. The homologous sequences found at L1 and R1-R2 are located between nucleotides 37 and 127.
On the basis of the estimated total size of approximately 100 kb (based on FIGE), the left end of the element was already covered by the mapped cosmids, but three cosmid inserts at the left end (2H7, 3G3, and 7D12) did not differ with respect to restriction patterns. We tested the possibility that similar sequences were present at both outer ends of the integrated clc element. A DNA probe from the right end of the element (pRR102) was used to hybridize digested DNAs from cosmids covering the left end. Only very weak hybridization was observed for a 4.2-kb EcoRI fragment of cosmid 3G3. Sequencing of this fragment (pRR104) revealed the putative left end of the first copy (L1) of the clc element. Short homologous sequences between L1 and R1-R2 with 83% identity (in a 92-bp overlap) were present (Fig. 3). The 91-bp sequence at L1 contained the 18-bp 3′ end of the Gly-tRNA sequence at R1. This result indicated that the clc element had been integrated into the Gly-tRNA gene at this position. To confirm this hypothesis, the original integration site was isolated from strain F1. PCR was performed on F1 total DNA with primers located outside the borders of the clc element at the left (primer RR301) and right (primers RR302 and RR303) ends. With primers RR301 and RR303, a PCR product (insert pRR123) which contained a complete tRNA gene in the middle (named integration site INT1; Fig. 3) was obtained. The sequences to the left and right of the tRNA gene were identical to those adjacent to the integrated clc element in strain RR221 (Fig. 3). The exact ends of the clc element were now determined, and the total size of the element was estimated from restriction mapping to be 105 kb.
The left end of the second copy (L2) of the integrated clc element could be cloned after amplification by inverse PCR (iPCR). Total DNA from strain RR221 was digested with SphI, religated, and subjected to iPCR with primers RR315 and RR316. The obtained PCR product was cloned and sequenced, and again the 18-bp 3′ end of the Gly-tRNA gene was found (pRR148 [L2]). Beyond the tRNA gene the sequence of L2 started to differ from that of L1, confirming the left end of the element. The PCR primers RR302 and RR325 were then used to amplify and demonstrate the presence of the second integration site in strain F1(pRR157). This site was named INT2 (Fig. 3). Interestingly, the amplified DNA was 391 bp longer than expected and was found to contain sequences for three tandemly arranged Gly-tRNA genes (Fig. 3). The situation for strain RR221 suggested that the clc element had been integrated into one of these Gly-tRNA sequences. Perhaps the others disappeared during recombination between multiple copies of the clc element (see below).
RecA independence of integration.
Since no P. putida F1 recA mutant was available, PAO rec1 was used to test whether chromosomal integration of the clc element was dependent on RecA. Filter matings were performed between B13 and PAO rec1 on LB agar as described previously (22). PAO rec1 transconjugants having obtained the clc genes were selected on Z3 minimal medium supplied with HgCl2 and with 3CBA as the sole substrate. Several transconjugants were obtained and subjected to genetic characterization. Total DNAs of such transconjugants were hybridized with the clc gene probe. Among EcoRI-digested DNAs, a band of approximately 20 kb hybridized for all transconjugants. A second hybridization with a probe for the clc element left end (probe IV; Fig. 2) resulted in the hybridization of an EcoRI fragment of approximately 9 kb. If the clc element were present in P. aeruginosa as a closed circular molecule (e.g., a plasmid), both probes would have hybridized to the same 20-kb EcoRI fragment (see Discussion). These results showed that the clc element was integrated into the PAO rec1 chromosome in a single copy, suggesting a mechanism of integration independent of RecA activity. The sensitivity of the transconjugants to UV irradiation did not differ from that of PAO rec1, precluding the possibility of recA complementation by the clc element.
Deamplification of the 105-kb clc element in P. putida RR22 under nonselective growth conditions.
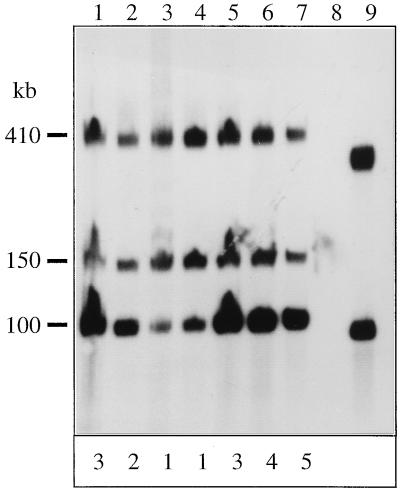
To investigate the nature of the multiple copies of the clc element in the F1 transconjugant RR22, we determined their stability during growth for 220 generations on LB medium. Isolated total DNAs from different generations were digested with SpeI, separated by FIGE, and hybridized to the clc gene probe; fragments of 150 and 410 kb were present at every stage (Fig. 4). Interestingly, the intensity of the 105-kb band decreased upon prolonged growth on LB medium, suggesting a decrease in the copy number of the clc element. Based on densitometric measurements of several autoradiograms, the 105-kb band in the DNA from RR22 cultures grown on MCB corresponded to an average clc element copy number of five or six. For this estimation, the intensities of the 150- and 410-kb bands served as single-copy standards. After 220 generations of growth on LB medium, the mean copy number had decreased to approximately one. The deamplification process was reversible. When cultures were grown again on MCB, the copy number of the 105-kb element was almost immediately restored to the original level (Fig. 4). In a control experiment with strain RR22 grown for 220 generations on 3CBA, the copy number of the 105-kb element did not change measurably (results not shown). This finding suggested that the copy number of the element in RR22 cultures decreased under nonselective conditions, whereas under selective conditions (i.e., a requirement for the clc genes), an approximate copy number of between five and six was maintained (not including the copies represented by the 150- and 410-kb SpeI fragments).
FIG. 4.
Autoradiogram of clc gene probe-hybridized total DNAs digested with SpeI and separated by FIGE. Total DNAs were prepared from strain RR22 after prolonged growth on LB medium or MCB. Lanes: 1, MCB grown; 2, 100 generations on LB medium; 3, 180 generations on LB medium; 4, 220 generations on LB medium; 5, 5 generations after inoculation from LB medium to MCB again; 6, 25 generations on MCB; 7, 45 generations on MCB; 8, P. putida F1; 9, Pseudomonas sp. strain B13. The intensities (determined by densitometric analysis) of the 100-kb band relative to the other two bands in the same lane are depicted at the bottom; each figure is the approximate copy number of the clc element represented by the 100-kb band. The densitometric analysis was performed on an autoradiogram with a shorter exposure time to be in the linear range.
Divergence of P. putida RR22 into a heterogenous population upon growth on nonselective medium.
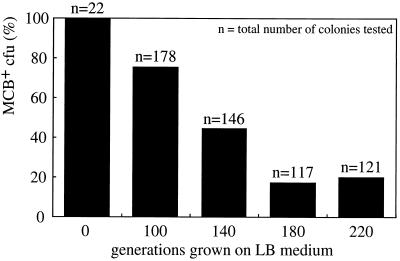
From samples of the RR22 culture after 100, 140, 180, and 220 generations on LB medium, individual colonies were tested for growth with MCB as the sole substrate on mineral agar plates. After 100 and 220 generations on LB medium, more than 20 and about 80% of the cells had lost their ability to mineralize MCB, respectively (Fig. 5). Prolonged cultivation of strain RR22 on LB medium for up to 300 generations did not further reduce the fraction able to grow on MCB (results not shown). The derivative strain RR221, used for the cosmid library and unable to grow on MCB, was picked as a single colony after 200 generations of growth on LB medium. Ten colonies obtained after 100 generations in liquid LB medium were picked and grown again on LB medium to obtain sufficient cells for DNA isolation. This procedure corresponded to another 30 generations of growth on nonselective medium. The 10 putative clones (named RR2231 to RR2240) were tested for their capability to grow on MCB by spotting 10 μl of an LB medium culture (from the one used for DNA preparation) on a selective agar plate (Table 3). To determine the copy number of the clc element in each putative clone, the total DNA was digested with XbaI, separated by FIGE, and hybridized to the clc gene probe. All strains still had the hybridizing 48- and 75-kb XbaI fragments, but four of the derived strains had lost the 100-kb XbaI fragment. This result correlated well with their capability to grow on MCB (Table 3). When strain RR221 (similar to RR2231) with only two copies of the clc element was again incubated with MCB, it was unable to grow. However, after prolonged incubation, single colonies that could use MCB appeared within the lawn of cells. When these single colonies were restreaked on MCB plates, they grew reasonably well, but when analyzed by FIGE and hybridization, they still contained only two copies of the clc element. These revertants were not further investigated.
FIG. 5.
Stability of the MCB growth phenotype of strain RR22 after prolonged growth on LB medium. Single colonies from a culture grown on MCB and from cultures after 100, 140, 180, and 220 generations on LB medium were tested for their ability to grow with MCB as the sole substrate on mineral agar plates.
TABLE 3.
Relationship between amplification level for the clc element and capability to metabolize MCB
| Derivative strain | Growth on MCBa | Mean copy no. of the clc element on XbaI restriction fragments ofb:
|
||
|---|---|---|---|---|
| 48 kb | 75 kb | 100 kb | ||
| RR221 | − | 1 | 1 | 0 |
| RR2231 | − | 1 | 1 | 0 |
| RR2232 | − | 1 | 1 | 0 |
| RR2233 | ++ | 1 | 1 | 5 |
| RR2234 | + | 1 | 1 | 1 |
| RR2235 | + | 1 | 1 | 1 |
| RR2236 | + | 1 | 1 | 1 |
| RR2237 | + | 1 | 1 | 1 |
| RR2238 | ++ | 1 | 1 | 3 |
| RR2239 | − | 1 | 1 | 0 |
| RR2240 | − | 1 | 1 | 0 |
| RR22 MCBc | +++ | 1 | 1 | 6 |
Relative capabilities to grow on MCB were tested by spotting 10 μl of overnight-grown LB medium cultures (except for strain RR22) on a mineral agar plate incubated in the presence of MCB vapor. −, no growth; +, moderate growth; ++, good growth; +++, excellent growth. It was not feasible to determine actual growth rates on MCB due to the unstable genotypes of these strains.
The relative levels of hybridization of the XbaI restriction fragments to the clc-gene probe were determined densitometrically. This determination provided an estimation of how many copies of the clc element were included in the 100-kb XbaI band.
Strain RR22 cultivated on MCB.
PFGE analysis of P. putida RR22 strains with various copy numbers of the clc element.
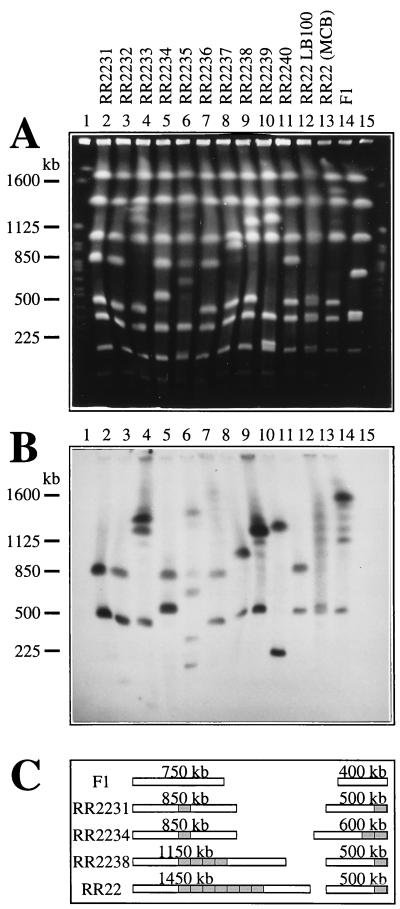
Digesting total DNA of P. putida F1 with SwaI resulted in only nine separate DNA fragments (30, 55, 200, 350, 400, 750, 1,050, 1,400, and 1,800 kb; Fig. 6). This result and the apparent absence of SwaI sites within the transferred clc element (see below) made it suitable for a large-scale genome analysis of the F1 transconjugants. Total DNAs of strains RR2231 to RR2240 were digested with SwaI, separated by PFGE, and hybridized to the clc gene probe. Similarly, DNA was prepared from strain RR22 grown on MCB, the “mixed” RR22 population after growth on LB medium for 100 generations, and strain F1 (Fig. 6). For RR22 grown on MCB, a ladder of hybridizing fragments between 1,100 and 1,500 kb could be seen, with the highest intensity being found for a new, 1,500-kb SwaI fragment (Fig. 6B, lane 13). The ladder with discrete 100-kb steps suggested that copies of the clc element were amplified and deamplified in tandem order. The DNA preparation from the RR22 culture after 100 generations on LB medium yielded a smeary ladder with some discrete steps still visible. At this stage, the bacterial culture was apparently quite heterogenous with respect to the copy number of the clc element.
FIG. 6.
Total DNAs of RR22 derivative strains obtained after growth on LB medium, digested with SwaI, and separated by PFGE. (A) Gel stained with ethidium bromide. (B) Southern hybridization with a probe for the clc genes; the probe was a 4.2-kb BglII fragment containing the clcABD genes from P. putida(pAC27) (9). Lanes: 2 to 11, strains RR2231, RR2232, RR2233, RR2234, RR2235, RR2236, RR2237, RR2238, RR2239, and RR2240, respectively; 12, P. putida RR22 after 100 generations on LB medium; 13, RR22 grown on MCB; 14, P. putida F1; 1 and 15, S. cerevisiae molecular size marker (225 to 2,200 kb; Bio-Rad). (C) Copy numbers and genetic organization of integrated clc elements on the two (originally) 400- and 750-kb SwaI fragments in some of the F1 transconjugant strains. The total size of each resulting SwaI fragment is indicated. No absolute location of the clc element integration on the 750-kb SwaI fragment is given.
The individual strains RR2231 to RR2240 appeared as “frozen” intermediate stages in the process of deamplification and loss of the clc element. RR2231, RR2232, and RR2240 were examples of strains without amplification. Two copies of the clc element were present, one on a 500-kb SwaI fragment (originally 400 kb in strain F1) and the other on an 850-kb SwaI fragment (originally 750 kb). This was also the case for strain RR221, used for the cosmid library (data not shown). RR2234 apparently had two copies on the 400-kb fragment and one on the 750-kb fragment, resulting in hybridizing bands at 600 and 850 kb. RR2237 had a duplication on the 750-kb fragment, resulting in hybridizing bands at 500 and 950 kb (Fig. 6B, lane 8). Some of the clones must have diverged to mixed populations even during the growth needed for DNA isolation (growth from one single cell to a colony and subsequent growth in liquid medium). For example, RR2233 showed hybridization with 500-, 1,150-, and 1,250-kb fragments, suggesting a heterogenous population of cells with four or five copies on the large SwaI fragment. As judged from the hybridization patterns of the 10 derivative strains and of RR22, multiple copies of the clc element seemed to appear preferentially on the (originally) 750-kb SwaI fragment of strain F1, rather than on the 400-kb SwaI fragment. Further hybridizations of the SwaI-digested DNAs of strain F1 indicated that INT1 is located on the 750-kb SwaI fragment, whereas INT2 is present on the 400-kb SwaI fragment (data not shown). INT2 is very close to one end of the 400-kb SwaI fragment (Fig. 2B), a fact which made it feasible to determine the size of the neighboring SwaI fragment. Hybridizations with a probe from outside the clc element right end (R2) (SwaI-EcoRI fragment; see Fig. 2B) indicated that this region is located on the 30-kb SwaI fragment of F1 (data not shown). As a consequence, the two integration sites in F1 are not located near one another.
Based on the PFGE hybridizations, a rough model of the physical presence of clc element copies on the two involved SwaI fragments in strains RR22, RR2231, RR2234, and RR2238 could be drawn (Fig. 6C). Deletions and/or larger DNA rearrangements were suspected in some of the other strains. For example, RR2235 and RR2239 (Fig. 6, lanes 6 and 10) showed hybridization with fragments smaller than 400 kb. Since the hybridization of XbaI restriction fragments had indicated that there were only two clc element copies in strain RR2239 (Table 3), the hybridizing 200- and 1,250-kb SwaI fragments indicated that a very large DNA rearrangement had taken place.
DISCUSSION
Genes encoding metabolic pathways for the degradation of aromatic compounds are often located on large, self-transmissible plasmids with a typical size of between 80 and 120 kb (6, 8, 11, 27, 31). Similarly, for Pseudomonas sp. strain B13, genes involved in the conversion of chlorocatechols were reported to be carried on the 110-kb plasmid pB13 (3). However, isolation of this plasmid could not be reproducibly performed; therefore, the location of the genes involved in chlorocatechol degradation remained obscure in strain B13 (20, 33, 35). Based on the present data, we propose the clc genes to be located on a transmissible element which can integrate site specifically into the chromosome with a Gly-tRNA structural gene as the target site.
The physical appearance of this element was detected by its tandem amplification in the new host strain, P. putida F1. Because of the presence of tandemly arranged multiple copies of the element, it was noted on hybridizations of total DNA digested with either SpeI or XbaI that the total size of the element was approximately 100 kb. The 410-kb hybridizing SpeI fragment gave an indication that the element had actually been integrated into the chromosome of strain F1. The F1 chromosomal fragments into which the element had been integrated could be observed in PFGE-separated DNA digested with SwaI. The actual increase in the size of the original F1 fragments was approximately 100 kb or multiples of 100 kb. The clc element actually had been integrated into two nonadjacent sites on the chromosome of strain F1. In addition, our data seemed to indicate that the clc element was present in two copies on the chromosome of strain B13 as well. The idea of tandem amplification of the clc element in F1 transconjugants was supported by (i) partial digests with SpeI and XbaI, resulting in a discrete stepwise ladder of 100-kb increases (21); (ii) SwaI digests of the different individual derivative strains with deamplified copies; and (iii) SwaI digests of the mixed RR22 population grown on MCB and LB medium. Amplification of the element could occur at both integration sites, with a preference for the site present on the (originally) 750-kb SwaI fragment (INT1).
There are several examples of plasmid recombination in gram-negative bacteria. For example, the E. coli F episome is integrated into the chromosome (18), the TOL plasmid can recombine with specific chromosomal sites (14), and the pKA2 plasmid (carrying 2,4-dichlorophenoxyacetic acid degradation in Alcaligenes paradoxus) is integrated into and excised from the host chromosome (15). In most of these cases, insertion sequences seem to be targets for the recombination events, which are either RecA dependent or mediated by transposases. Since integration of the clc element was RecA independent in P. aeruginosa, it must have been actively mediated and not a result of general recombination. Apparently, the two chromosomal integration events for the clc element in P. putida F1 occurred orientation and site specifically with Gly-tRNA genes as target sites. Structural genes for tRNAs are common targets for insertions of bacteriophages, insertional actinomycete plasmids, and some conjugative transposons. This kind of insertion is mediated by site-specific recombinases of the integrase family (2, 4, 24, 25). The Gly-tRNA gene at INT1 (strain F1) was split in two parts by the integration of the clc element, inevitably leading to inactivation of the original gene copy. However, upon integration, the tRNA gene became restored at the right end of the elements (Fig. 3). As a rule, integrating genetic elements which use tRNA genes as targets create a duplication, restoring the tRNA gene (2). Our cosmids of the F1 transconjugant RR221 showed one anomaly, though. INT2 in strain F1 contained three tandemly arranged Gly-tRNA genes prior to integration, similar to their arrangement in E. coli K-12 (16). However, in strain RR221, two of these Gly-tRNA gene copies were absent. The R2-L2 junction sites were similar to the R1-L1 junction sites where integration into a single tRNA gene occurred. The apparent deletion at INT2 may have been a result of recombination between two integrated copies of the clc element.
The question remaining at the moment is whether the integrated clc element is identical to the previously described plasmid pB13 (3). We demonstrated that its present form in F1 resembles an integrative plasmid, which also seems to exist in two copies on the B13 chromosome. The former pB13 may at some time have lost its ability to replicate efficiently, or the determinant(s) necessary for chromosomal integration may have been acquired after its first characterization. However, deletion or accumulation of a substantial DNA fragment is not evident. Our EcoRI restriction map is basically identical to that published for pB13 (3), if circularization of the integrated element is accounted for. The size difference of approximately 5 kb (110 − 105 kb) seems to result only from inaccurate sizing of the larger EcoRI fragments. The two chromosomal copies in strain B13 may explain the low yield and irreproducible plasmid DNA isolation observed (20, 33, 35). When B13 was grown on 3CBA, the circular form was detected mainly in the stationary phase (results not shown), indicating that it was dispensable for growth on 3CBA.
To our knowledge, the tandem amplification of the clc element observed in strain F1 is rather unique for pseudomonads. The amplification led to an increase in the total DNA content of the cell of at least 10% (with an estimated genome size of 6 Mb and six copies of the clc element). Similar large amplifications have been observed for Streptomyces strains, where the tandem amplifications were assumed to have been caused by a rolling-circle replication mechanism (1, 34). The tandem amplifications in F1 transconjugants were mainly found at INT1; therefore, they cannot be associated with multiple integration sites. Only for derivative strain RR2234 was a duplication at INT2 evident. In this case, the duplication seemed to be a result of separate integrations into two adjacent Gly-tRNA gene copies (results not shown). Once formed, the amplified structures in the F1 transconjugants were quite unstable and were deamplified under nonselective conditions. Most likely, the disappearance of amplicons was caused by recombinational deletions between tandemly arranged copies. After deamplifications had finally led to two nonadjacent copies, no new amplification cycle occurred in derivative strains of RR22. This idea is similar to observations for Streptomyces, where at least two tandemly arranged copies of an amplifiable DNA element were required for amplification to occur (1). We found evidence for deletions and DNA rearrangements in some transconjugants after prolonged growth on LB medium, phenomena which are also known for Streptomyces.
The observations for strain F1 suggested that somehow amplification of the clc element occurs during transfer of the element into strain F1 and selection for growth on MCB. In a mixed population of cells, members which have the clc copy number necessary for growth on MCB will be the fastest to grow on MCB and will form the majority of the population (as observed for RR22 during switches from LB medium to MCB). This situation causes the apparent “amplification” of the clc element, reflecting the situation in a mixed population. Several reports have shown that chromosomal gene amplification can be advantageous for growth under specific conditions. For example, the amplification of chromosomally integrated plasmids bearing antibiotic resistance genes in Bacillus subtilis led to a higher level of antibiotic resistance (13, 32). Duplication of the permease genes in Salmonella typhimurium resulted in a dramatic increase in growth rate on certain substrates under carbon-limited conditions (12, 28). The advantage of these amplifications seemed to be an increased level of gene expression.
We found a positive correlation between the copy number of the clc element in RR22 derivative strains and their capability to grow on chlorobenzenes. Efficient conversion of chlorocatechols resulting from chlorobenzene metabolism is a critical step in the chlorobenzene degradation pathway (10, 20). Perhaps the F1 transconjugants achieved this conversion only by expressing the clc genes from multiple copies. The chlorocatechol 1,2-dioxygenase activities in strain B13 (with two chromosomal copies of the clc element and perhaps two or three plasmid copies) and in the F1 transconjugant RR1 (with approximately eight copies) grown on 3CBA were similar (22). Interestingly, the revertants of strain RR221, which were again capable of metabolizing MCB and 1,4-DCB, did so with two copies only. In these cases, a different mechanism, such as a simple mutation, might have enabled better expression of the clc genes in the F1 host. Since all the initially isolated chlorobenzene-degrading transconjugants of strain F1 had multiple chromosomal copies of the clc element, amplification seemed to occur at a frequency higher than that of other processes, such as mutation.
ACKNOWLEDGMENTS
We thank Jörg Hummerjohann, Frank Junker, and Michael Kertesz for the opportunity to perform experiments with the CHEF DR II PFGE equipment at the Institute of Microbiology, Swiss Federal Institute for Technology (ETH), Zürich, Switzerland.
This work was supported by grant 5002-038279 from the Swiss Priority Program Biotechnology.
REFERENCES
- 1.Altenbuchner J, Eichenseer C, Brüderlein M. DNA amplification and deletion in Streptomyces lividans. Proc Biol Actinomycetes. 1988;7:139–144. [Google Scholar]
- 2.Campbell A M. Chromosomal insertion sites for phages and plasmids. J Bacteriol. 1992;174:7495–7499. doi: 10.1128/jb.174.23.7495-7499.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chatterjee D K, Chakrabarty A M. Genetic homology between independently isolated chlorobenzoate degradative plasmids. J Bacteriol. 1983;153:532–534. doi: 10.1128/jb.153.1.532-534.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheetham B F, Katz M E. A role for bacteriophages in the evolution and transfer of bacterial virulence determinants. Mol Microbiol. 1995;18:201–208. doi: 10.1111/j.1365-2958.1995.mmi_18020201.x. [DOI] [PubMed] [Google Scholar]
- 5.Coco W M, Rothmel R K, Henikoff S, Chakrabarty A M. Nucleotide sequence and initial functional characterization of the clcR gene encoding a LysR family activator of the clcABD chlorocatechol operon in Pseudomonas putida. J Bacteriol. 1993;175:417–427. doi: 10.1128/jb.175.2.417-427.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Don R H, Pemberton J M. Genetic and physical map of the 2,4-dichlorophenoxyacetic acid degradative plasmid pJP4. J Bacteriol. 1985;161:466–468. doi: 10.1128/jb.161.1.466-468.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dorn E, Hellwig M, Reineke W, Knackmuss H-J. Isolation and characterization of a 3-chlorobenzoate degrading pseudomonad. Arch Microbiol. 1974;99:61–70. doi: 10.1007/BF00696222. [DOI] [PubMed] [Google Scholar]
- 8.Franklin F C H, Bagdasarian M, Bagdasarian M M, Timmis K N. Molecular and functional analysis of the TOL plasmid pWW0 from Pseudomonas putida and cloning of genes for the entire regulated aromatic ring-cleavage pathway. Proc Natl Acad Sci USA. 1981;78:7458–7462. doi: 10.1073/pnas.78.12.7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frantz B, Chakrabarty A M. Organization and nucleotide sequence determination of a gene cluster involved in 3-chlorocatechol degradation. Proc Natl Acad Sci USA. 1987;84:4460–4464. doi: 10.1073/pnas.84.13.4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fritz H, Reineke W, Schmidt E. Toxicity of chlorobenzene on Pseudomonas sp. strain RHO1, a chlorobenzene-degrading strain. Biodegradation. 1991;2:165–170. doi: 10.1007/BF00124490. [DOI] [PubMed] [Google Scholar]
- 11.Fulthorpe R R, Wyndham R C. Transfer and expression of the catabolic plasmid pBRC60 in wild bacterial recipients in a freshwater ecosystem. Appl Environ Microbiol. 1991;57:1546–1553. doi: 10.1128/aem.57.5.1546-1553.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haack K R, Roth J R. Recombination between chromosomal IS200 elements supports frequent duplication formation in Salmonella typhimurium. Genetics. 1995;141:1245–1252. doi: 10.1093/genetics/141.4.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janniere L, Niaudet B, Pierre E, Ehrlich S D. Stable gene amplification in the chromosome of Bacillus subtilis. Gene. 1985;40:47–55. doi: 10.1016/0378-1119(85)90023-x. [DOI] [PubMed] [Google Scholar]
- 14.Jeenes D J, Williams P A. Excision and integration of degradative pathway genes from TOL plasmid pWW0. J Bacteriol. 1982;150:188–194. doi: 10.1128/jb.150.1.188-194.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ka J O, Tiedje J M. Integration and excision of a 2,4-dichlorophenoxyacetic acid degradative plasmid in Alcaligenes paradoxus and evidence of its natural intergeneric transfer. J Bacteriol. 1994;176:5284–5289. doi: 10.1128/jb.176.17.5284-5289.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Komine Y, Adachi T, Inokuchi H, Ozeki H. Genomic organization and physical mapping of the transfer RNA genes in Escherichia coli K12. J Mol Biol. 1990;212:579–598. doi: 10.1016/0022-2836(90)90224-A. [DOI] [PubMed] [Google Scholar]
- 17.Leveau J H J, van der Meer J R. The tfdR gene product can successfully take over the role of the insertion element-inactivated TfdT protein as a transcriptional activator of the tfdCDEF gene cluster, which encodes chlorocatechol degradation in Ralstonia eutropha JMP134(pJP4) J Bacteriol. 1996;178:6824–6832. doi: 10.1128/jb.178.23.6824-6832.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Low K B. Hfr strains of Escherichia coli K-12. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1987. pp. 1134–1137. [Google Scholar]
- 19.Ohman D E, West M A, Flynn J L, Goldberg J B. Method for gene replacement in Pseudomonas aeruginosa used in construction of recA mutants: recA-independent instability of alginate production. J Bacteriol. 1985;162:1068–1074. doi: 10.1128/jb.162.3.1068-1074.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oltmanns R H, Rast H G, Reineke W. Degradation of 1,4-dichlorobenzene by enriched and constructed bacteria. Appl Microbiol Biotechnol. 1988;28:609–616. [Google Scholar]
- 21.Ravatn, R. Unpublished results.
- 22.Ravatn R, Zehnder A J B, van der Meer J R. Low-frequency horizontal transfer of an element containing the chlorocatechol degradation genes from Pseudomonas sp. strain B13 to Pseudomonas putida F1 and to indigenous bacteria in laboratory-scale activated-sludge microcosms. Appl Environ Microbiol. 1998;64:2126–2132. doi: 10.1128/aem.64.6.2126-2132.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reineke W, Wessels S W, Rubio M A, Latorre J, Schwien U, Schmidt E, Schlömann M, Knackmuss H J. Degradation of monochlorinated aromatics following transfer of genes encoding chlorocatechol catabolism. FEMS Microbiol Lett. 1982;14:291–294. [Google Scholar]
- 24.Reiter W D, Palm P, Yeats S. Transfer RNA genes frequently serve as integration sites for prokaryotic genetic elements. Nucleic Acids Res. 1989;17:1907–1914. doi: 10.1093/nar/17.5.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salyers A A, Shoemaker N B, Stevens A M, Li L Y. Conjugative transposons: an unusual and diverse set of integrated gene transfer elements. Microbiol Rev. 1995;59:579–590. doi: 10.1128/mr.59.4.579-590.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 27.Shields M S, Reagin M J, Gerger R R, Campbell R, Somerville C. TOM, a new aromatic degradative plasmid from Burkholderia (Pseudomonas) cepacia G4. Appl Environ Microbiol. 1995;61:1352–1356. doi: 10.1128/aem.61.4.1352-1356.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sonti R V, Roth J R. Role of gene duplications in the adaptation of Salmonella typhimurium to growth on limiting carbon sources. Genetics. 1989;123:19–28. doi: 10.1093/genetics/123.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thiem S M, Krumme M L, Smith R L, Tiedje J M. Use of molecular techniques to evaluate the survival of a microorganism injected into an aquifer. Appl Environ Microbiol. 1994;60:1059–1067. doi: 10.1128/aem.60.4.1059-1067.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van der Meer J R, Roelofsen W, Schraa G, Zehnder A J B. Degradation of low concentrations of dichlorobenzenes and 1,2,4-trichlorobenzene by Pseudomonas sp. strain P51 in nonsterile soil columns. FEMS Microbiol Ecol. 1987;45:333–341. [Google Scholar]
- 31.van der Meer J R, van Neerven A R, de Vries E J, de Vos W M, Zehnder A J B. Cloning and characterization of plasmid-encoded genes for the degradation of 1,2-dichloro-, 1,4-dichloro-, and 1,2,4-trichlorobenzene of Pseudomonas sp. strain P51. J Bacteriol. 1991;173:6–15. doi: 10.1128/jb.173.1.6-15.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vazquez-Cruz C, Ochoa-Sanchez J C, Olmedo-Alvarez G. Pulsed-field gel-electrophoretic analysis of the amplification and copy-number stability of an integrational plasmid in Bacillus subtilis. Appl Microbiol Biotechnol. 1996;46:55–60. doi: 10.1007/s002530050782. [DOI] [PubMed] [Google Scholar]
- 33.Weisshaar M P, Franklin F C, Reineke W. Molecular cloning and expression of the 3-chlorobenzoate-degrading genes from Pseudomonas sp. strain B13. J Bacteriol. 1987;169:394–402. doi: 10.1128/jb.169.1.394-402.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Young M, Cullum J. A plausible mechanism for large-scale chromosomal DNA amplification in streptomycetes. FEBS Lett. 1987;212:10–14. doi: 10.1016/0014-5793(87)81547-8. [DOI] [PubMed] [Google Scholar]
- 35.Zhou J Z, Tiedje J M. Gene transfer from a bacterium injected into an aquifer to an indigenous bacterium. Mol Ecol. 1995;4:613–618. doi: 10.1111/j.1365-294x.1995.tb00261.x. [DOI] [PubMed] [Google Scholar]
- 36.Zylstra G J, Gibson D T. Toluene degradation by Pseudomonas putida F1. Nucleotide sequence of the todC1C2BADE genes and their expression in Escherichia coli. J Biol Chem. 1989;264:14940–14946. [PubMed] [Google Scholar]
- 37.Zylstra G J, McCombie W R, Gibson D T, Finette B A. Toluene degradation by Pseudomonas putida F1: genetic organization of the tod operon. Appl Environ Microbiol. 1988;54:1498–1503. doi: 10.1128/aem.54.6.1498-1503.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]