Abstract
The biosynthesis of carbamoylphosphate is catalyzed by the heterodimeric enzyme carbamoylphosphate synthetase. The genes encoding the two subunits of this enzyme in procaryotes are normally transcribed as an operon, but the gene encoding the large subunit (carB) in Lactococcus lactis is shown to be transcribed as an isolated unit. Carbamoylphosphate is a precursor in the biosynthesis of both pyrimidine nucleotides and arginine. By mutant analysis, L. lactis is shown to possess only one carB gene; the same gene product is thus required for both biosynthetic pathways. Furthermore, arginine may satisfy the requirement for carbamoylphosphate in pyrimidine biosynthesis through degradation by means of the arginine deiminase pathway. The expression of the carB gene is subject to regulation at the level of transcription by pyrimidines, most probably by an attenuator mechanism. Upstream of the carB gene, an open reading frame showing a high degree of similarity to those of glutathione peroxidases from other organisms was identified.
In all organisms, pyrimidine metabolism is required in order to supply the cell with building blocks for the synthesis of DNA, RNA, and certain coenzymes needed in central metabolic pathways. In Lactococcus lactis, this requirement can be fulfilled either by the use of nucleosides and nucleobases present in the growth medium (36–38) or by means of the pyrimidine biosynthetic pathway, which seems to be universal in all prototrophic organisms investigated so far and consists of six enzymatic steps leading to the formation of UMP.
Many metabolic genes in L. lactis such as genes involved in the biosynthesis of amino acids (4, 10, 18) and glycolysis (6, 7, 31) have been identified and sequenced. The amino acid biosynthetic genes were found to be members of large operons, organized like the ones identified in Bacillus subtilis. Likewise, the genes encoding the pyrimidine biosynthetic enzymes in different gram-positive bacteria like Bacillus caldolyticus (17), B. subtilis (40), Lactobacillus plantarum (12), and Enterococcus faecalis (30) have been identified as members of a single operon. In contrast, it was recently shown that the genes of the pyrimidine biosynthetic pathway in L. lactis are organized differently. A pyr operon, which consists of only three biosynthetic genes, has been found in L. lactis (2). Two of the genes are the well-known pyr genes pyrD and pyrF, which encode dihydroorotate dehydrogenase and OMP decarboxylase, respectively. The third gene, pyrK, was identified as a new pyr gene encoding a protein which was shown to be necessary for the dihydroorotate dehydrogenase activity encoded by the adjacent pyrDb gene (2). The lactococcal pyrKDbF operon is highly homologous to the corresponding part of the much larger pyr operons found in other gram-positive bacteria. An interesting exception occurs with Lactobacillus plantarum, in which the pyrK analogue is absent from the operon (12). Another surprising feature of the pyrimidine biosynthesis pathway in L. lactis is the presence of two different genes, pyrDa and pyrDb, both of which encode a dihydroorotate dehydrogenase. The pyrDb gene belongs to the same family as the genes encoding dihydroorotate dehydrogenases in other gram-positive bacteria, whereas the pyrDa gene is closely related to that of the dihydroorotate dehydrogenase of Saccharomyces cerevisiae (1). Only pyrDb was shown to be part of the identified pyr operon (2).
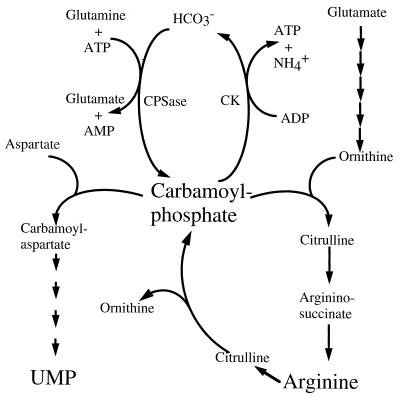
Carbamoylphosphate is formed from CO2, ATP, and glutamine and is used in the biosynthesis of both pyrimidine and arginine (Fig. 1). It is synthesized by the heterodimeric enzyme carbamoylphosphate synthetase (CPSase). The small subunit of the enzyme functions as a glutamine amidotransferase, whereas the large subunit has other catalytic properties. In all procaryotes described so far, CPSase activity is encoded by two genes commonly called carA and carB, and there is no reason to believe that this is not true for L. lactis. Procaryotes are characterized by having either a single set of genes that is responsible for all carbamoylphosphate synthesized and that encodes a single CPSase or two different sets of genes that encode CPSase (9). The two sets of genes differ in their regulatory features; one set is regulated by the level of pyrimidines in the cell, whereas the other responds to changes in the concentration of arginine (9). The genes encoding the two subunits have been sequenced for many procaryotes and have been found almost exclusively to be transcribed as an operon in the order carA-carB. Exceptions have, however, been reported. In Pseudomonas aeruginosa and Neisseria spp., sequences between carA and carB have been found (26, 27).
FIG. 1.
Carbamoylphosphate pathways in L. lactis. Pathways of carbamoylphosphate in the formation of arginine and pyrimidines and its synthesis and degradation to ammonia and carbonate with formation of ATP are shown. CK, carbamate kinase; CPSase, carbamoylphosphate synthetase.
In this study we have cloned and determined the nucleotide sequence of the carB region of the L. lactis chromosome. Surprisingly, the gene is shown to be transcribed as a monocistronic unit. Moreover it is shown that L. lactis has only one carB gene, regulated by the pyrimidine level in the cell.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and the plasmids used in this study are listed in Table 1. Plasmids pJS23 and pSJ24 were made in the following way. pKS2 was digested with HindIII and ligated into pSMA500 and pRC1, respectively. Competent L. lactis MG1363 cells were transformed with the Escherichia coli plasmids pSJ23 and pSJ24, which are unable to replicate in L. lactis but contain a selectable Emr marker and cloned pieces of the lactococcal chromosome. Transformants were selected and purified on plates containing 1 μg of erythromycin per ml. Only transformants in which plasmids have recombined into the chromosome will result in Emr colonies. Chromosomal DNA from strain MB35 was digested with SpeI, and the resulting linear molecules were circularized with T4 DNA ligase and transformed into E. coli DH5α. A plasmid (pSJ50) conferring erythromycin resistance and containing a 3,200-bp SpeI-HindIII lactococcal DNA fragment was obtained.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | F80lacZDM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17 supE44 thi-1 gyrA96 relA1 | Lab strain |
| L. lactis | ||
| MG1363 | Plasmid-free straina | 15 |
| MB35 | MG1363 carB::pSJ24 (Emr)a | This study |
| MB36 | MG1363 carB::lacLM (pSJ23) (Emr)a | This study |
| MB37 | MG1363 carB::lacLM (pSJ61) (Emr)a | This study |
| MB38 | MG1363 carB::lacLM (pSJ66) (Emr)a | This study |
| Plasmids | ||
| pRC1 | erm, L. lactis integration vector | 28 |
| pAK80 | erm lacLM, E. coli and L. lactis shuttle vector | 22 |
| pSMA500 | erm lacLM, L. lactis integration vector | 35 |
| pKS2 | amp, Sau3A fragments from L. lactis in pBR322 | 3 |
| pSJ23 | erm, 1,700-bp HindIII from pKS2 in pSMA500 | This study |
| pSJ24 | erm, 1,700-bp HindIII from pKS2 in pRC1 | This study |
| pSJ500 | erm, SpeI rescue from MB35 | This study |
| pSJ60 | erm lacLM, PCR, carB promoter in pAK80 | This study |
| pSJ61 | erm lacLM, PCR, carB promoter in pSMA500 | This study |
| pSJ66 | erm lacLM, PCR, carB terminator in pSMA500 | This study |
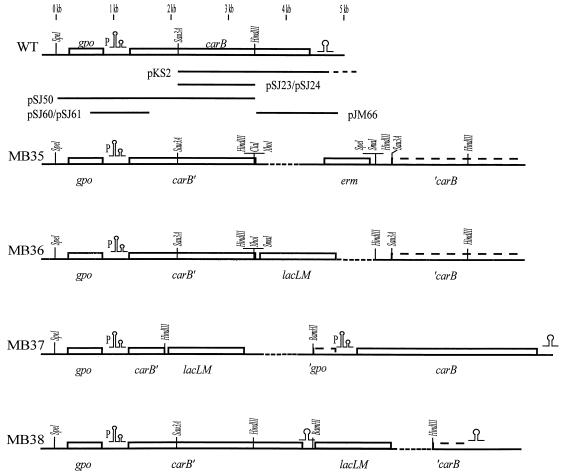
See Fig. 2 for a physical map.
Two plasmids fusing the carB promoter to lacLM were constructed as follows. With pSJ50 as the template and the primers 5′-CCCAAGCTTACAGCCAGTAAATGTGGT-3′ and 3′-CGCGGATCCCATAGTAAAAGCTG-5′, a 1,250-bp PCR product was obtained and subsequently digested with HindIII and BamHI. This fragment was inserted in the lacLM promoter fusion vectors pAK80 (22) and pSMA500 (35), which had been digested with HindIII and BamHI. The resulting plasmids were termed pSJ60 and pSJ61, respectively.
Plasmid pJM66 was constructed as follows. With pKS2 as the template and the primers 5′-CTTAGGAACTCAAGTCG-3′ and 3′-ACCGGATCCCTTCAAATACTTATTAAC-5′, a 1,300-bp PCR product was obtained. After digestion with HindIII and BamHI, the 1,000-bp fragment was inserted in the lacLM vector pSMA500 (35). Competent L. lactis MG1363 cells were transformed with plasmid pJM66, which is unable to replicate in L. lactis but contains a selectable Emr marker and a piece of the lactococcal chromosome. Transformants were selected and purified on plates containing 2 μg of erythromycin per ml. Only strains in which homologous recombination between plasmids and chromosomal DNA has occurred after transformation will result in Emr colonies. The carB regions in all the strains used in this work were mapped by Southern blot or PCR analysis.
Growth conditions and enzyme assay.
Lactococcal cultures were grown either on M17 glucose broth (48) or on synthetic media that were based on MOPS (morpholinepropanesulfonic acid), contained seven vitamins and either 19 (SA) or 8 (BIV) amino acids (23), and were supplied with 1% glucose. E. coli cultures were grown on Luria-Bertani broth. L. lactis was cultured at 30°C in filled culture flasks without aeration. E. coli in batch cultures was grown at 37°C with vigorous shaking. For all plates, agar was added to 15 g/liter. When needed, the following compounds were added to the different media: arginine at 200 μg/ml, uracil at 20 μg/ml, erythromycin at 1 μg/ml for lactococci and 150 μg/ml for E. coli, and ampicillin at 100 μg/ml. For enzyme assays, the cells were grown in SA or BIV glucose medium and aliquots were harvested at different times during exponential growth between optical densities at 450 nm (OD450s) of 0.2 and 0.8. The amount of β-galactosidase in the cells was assayed as previously described (22), but the cell density was measured at 450 nm. Specific enzymatic activity was determined as follows: OD420/(OD450 × min × ml of culture).
Transformation.
L. lactis was transformed by electroporation (21). E. coli cells were transformed as described previously (42).
DNA isolation, manipulations, and sequencing.
Chromosomal lactococcal DNA was prepared as described by Johansen and Kibenich (24). The methods described by Sambrook et al. (42) were used for general DNA methods in vitro. DNA sequences were determined from plasmid DNA by the dideoxy-chain termination method (44) with a Thermo Sequenase-radiolabelled-terminator cycle sequencing kit (product no. US 79750; Amersham) in accordance with the protocol of the manufacturer.
Southern blot analysis.
Southern blot analysis was performed with GeneScreen nylon membranes (New England Nuclear) and the digoxigenin system (Boehringer Mannheim) for colorimetric detection of hybridized products in accordance with the protocols of the manufacturers.
PCR amplification of DNA.
L. lactis chromosomal DNA was amplified by PCR with 1 μg of DNA in a final volume of 100 μl containing deoxyribonucleoside triphosphates (0.25 mM each), oligonucleotides (10 μM), and 2.5 U of AmpliTaq DNA polymerase (Perkin-Elmer). Amplification was performed with 30 cycles of 95°C for 1 min and 55°C for 1 min, followed by 3 min at 72°C.
RNA extraction.
L. lactis RNA was harvested from strain MG1363 grown exponentially in SA glucose medium to an OD450 of approximately 0.8. Total RNA from a 20-ml culture was isolated according to the method of Arnau and coworkers (3).
Primer extension.
A synthetic oligonucleotide, 5′-TTTCCTGTTCACAACCTTGC-3′, complementary to the sense strand covering nucleotides 726 to 745 was radioactively labelled at its 5′ end with [γ-32P]ATP and T4 polynucleotide kinase and used for primer extensions on 20 μg of total RNA isolated from L. lactis MG1363 as previously described (16). The elongation was performed at 41°C with SuperScript II reverse transcriptase (Gibco BRL).
Nucleotide sequence accession number.
The nucleotide sequence reported in this paper has been submitted to the EMBL data library and assigned the accession no. AJ000109.
RESULTS
Cloning and sequencing of the carB region of the chromosome.
A part of the carB gene of L. lactis encoding the large subunit of CPSase (CPSase B) was obtained by chance. During the sequencing of clones obtained from a partial Sau3A library in pBR322, plasmid pKS2, which contains two Sau3A fragments from the L. lactis chromosome (3), turned out to include a 4-kb Sau3A fragment which was shown to harbor part of a putative open reading frame showing a high degree of similarity to those encoding the C-terminal parts of the large subunits of CPSases from different organisms. A 1,700-bp HindIII-Sau3A fragment was subcloned in the E. coli vector pRC1 (28), thus creating pSJ24. The fragment harbors an internal part of the carB open reading frame (Fig. 2). This plasmid was allowed to integrate into the chromosome of MG1363 by homologous recombination, resulting in strain MB35, which carries an insertion in carB (Fig. 2).
FIG. 2.
Genetic maps the carB regions of plasmids and strains used in this work. The physical map and the positions of selected restriction endonuclease sites are shown. The carB DNAs contained in the different plasmids are shown with lines. The dotted line in pKS2 indicates that the cloned lactococcal DNA extends to a Sau3A site at 6.1 kb. A P indicates the position of the carB promoter. The maps of the chromosomal DNA in the carB regions of the wild type (WT), MB35, MB36, MB37, and MB38 are shown. The broken lines represent the E. coli plasmid DNA, which is not drawn to scale. The erythromycin resistance genes of MB36, MB37, and MB38 are not shown. The carB terminator and the carB attenuator are shown by omega- and double-omega-like structures, respectively.
In order to clone the N-terminal-encoding part of the CPSase B gene by marker rescue, chromosomal DNA was extracted from MB35, digested with different restriction endonucleases, ligated, and transformed to E. coli to select for erythromycin resistance. A rescue plasmid (pSJ50) obtained from the SpeI digest was subjected to further analysis. By performing Southern blotting experiments on chromosomal DNA isolated from L. lactis MG1363 with probes derived from pKS2 and pSJ50, it was shown that the lactococcal DNAs present on the two plasmids overlap (not shown). By combining the sequencing data obtained from pKS2 and pSJ50, the sequence of the carB region was determined. Two open reading frames encoding 157 and 1,064 amino acids that are transcribed in the same direction were found by computer analysis of the DNA sequence. The 1,064-amino-acid product of an open reading frame encoding a protein with a theoretical size of 117 kDa showed a high degree of identity (70%) to the CPSase B from Lactobacillus plantarum (12) and 66% identity to the same enzyme from B. subtilis encoded by the pyrAB gene (40). Upstream of the putative carB gene, one expects to find the carA gene encoding the small CPSase subunit. Surprisingly, the 157-amino-acid product of the open reading frame showed no homology whatsoever to the small CPSase subunits from other organisms. Instead, this open reading frame product showed high degrees of identity to glutathione peroxidases from various organisms: 54% identity to B. subtilis (46), 52% identity to S. cerevisiae (5), 48% identity to Synechocystis spp. (47), and 49% identity to Chlamydia reinhardtii (29). Upstream of both reading frames, translational initiation signals can be identified (Table 2).
TABLE 2.
Sequence properties of the gpo-carB region
| Section of gpo-carB region | Nucleotide coordinatesa | Sequenceb or no. of codons |
|---|---|---|
| gpo translation initiation | 150–165 | CCAGGAGGTAAACAAATGAA |
| gpo open reading frame | 163–636 | 157 codons |
| carB promoter | 658–694 | TGAGACTTGTAAAAGGTCAATTATTTTGCTAAACTAATAAACAATAA |
| carB translation initiation | CTACAAAGGAGAAAAACAAATGCC | |
| carB open reading frame | 1,064 codons | |
| carB terminator | ACTGTCCCAAATGGGGCTTTTTTTTTTTTT |
The nucleotide numbers refer to the sequence submitted to the database.
The putative start codons are shown in boldface letters, and the nucleotides of the mRNA complementary to the 3′ end of the 16S rRNA from L. lactis (3′-UCUUUCCUCCA-5′) are indicated by underscoring. The −35 and −10 sequences are shown by underscoring, and the first nucleotide to be transcribed is shown in boldface type. The inverted repeats in the putative terminator are shown by double underlining.
The carB gene is transcribed as a monocistronic message.
Since the carB gene is preceded by the functionally unrelated gene gpo, which encodes glutathione peroxidase, it is tempting to believe that the carB gene is transcribed by a promoter present in the intercistronic region between gpo and carB. In order to assay promoter activity, a PCR fragment covering the entire intercistronic region and parts of the gpo and carB open reading frames was amplified and cloned into the promoter probe vector pAK80 (22), thus generating pSJ60 (Fig. 2). After transformation into MG1363, the specific β-galactosidase activity was measured in exponentially growing cells and determined to be 0.3, thus demonstrating the presence of a promoter in the intercistronic region just upstream of carB, since the specific activity of cells harboring the pAK80 vector alone is less than 0.001 (Table 3).
TABLE 3.
β-Galactosidase activities of strains carrying different carB::lacLM fusions
| Strain | Plasmid | Chromosomal lacLM fusion | Relevant genotype | β-Galactosidase activitya
|
|
|---|---|---|---|---|---|
| No supplement | Uracil added | ||||
| MB36 | pSJ23 carB | carB::lacLM erm carB | 0.26 | 0.025 | |
| MB37 | pSJ61 carB | carB::lacLM erm | 0.084 | 0.033 | |
| MB38 | pSJ66 carB terminator | erm | <0.001 | NDb | |
| MG1363 | pSJ60 | None | carB::lacLM erm | 0.30 | ND |
| MG1363 | pAK80 | None | erm | <0.001 | ND |
Cells were grown in BIV minimal medium supplied with arginine. The β-galactosidase activity was determined with the formula OD420/(OD450 × min × ml of culture).
ND, activity not determined.
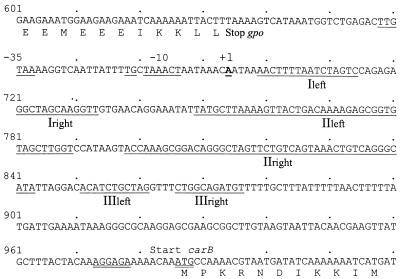
In order to map the precise location of the promoter, the 5′ end of the transcript was determined by primer extension on RNA isolated from MG1363. The result is presented in Fig. 3. This experiment mapped the first nucleotide to be transcribed (+1) to position 695 (Fig. 4). This finding is supported by sequence analysis, as an extended −10 sequence (TGCTAAACT) can be identified. In lactococcal promoters an extended −10 sequence is characterized by a TGN sequence immediately in front of the −10 sequence (TATAAT) (50). Furthermore, by 17 nucleotides upstream of the −10 sequence there is a −35 sequence (TTGTAA) (Fig. 4).
FIG. 3.
Primer extension mapping of the 5′ end of the carB mRNA. Sequencing ladders generated with the oligonucleotide used for the primer extension were loaded next to the reaction mixture. The DNA sequence of the sense strand around the first nucleotide in the transcript (designated +1) is presented, and the −10 sequence is boxed.
FIG. 4.
Sequence of the gpo-carB intercistronic region. The numbers refer to the sequences submitted to the database. The amino acids of the glutathione peroxidase and CPSase large subunit derived from the DNA sequence are shown. The translational start site of carB is indicated with double underscoring. The −10 and −35 sequences of the carB promoter are indicated with underscoring, and the first nucleotide to be transcribed is marked +1. The right and left stems of loops I, II, and III (Fig. 5) are shown.
Regarding the 3′ end of the carB transcript, a potential transcriptional terminator can be identified at positions 4191 to 4218 (Table 2). In order to show that transcription does not extend past the putative terminator, a promoterless lacLM gene was integrated into the chromosome immediately after the stretch of thymine residues following the stem-loop structure (strain MB38) (Fig. 2). The rationale for this was twofold. First, if another pyrimidine- or arginine-biosynthetic gene is expressed from the carB promoter, the integration of the plasmid will disrupt transcription, thus resulting in a polar mutation, and the strain will acquire a pyrimidine or arginine requirement. Second, the amount of β-galactosidase produced by this strain will reflect the amount of transcription extending past the putative terminator structure. The phenotype of the resulting MB38 strain was determined by growth on minimal medium in the absence and presence of arginine and/or uracil in the BIV glucose minimal medium. Strain MB38 required neither uracil nor arginine, thus demonstrating that no biosynthetic gene involved in these pathways is located downstream of carB and transcribed from the carB promoter. As shown in Table 3, no detectable β-galactosidase activity could be identified in BM38, thus showing that transcription from the carB promoter is terminated no later than after the stem-loop structure ending at position 4205.
In conclusion, the data presented here unambiguously demonstrate that the identified carB gene of L. lactis is transcribed as a monocistronic mRNA.
The physiological effect of a carB mutation.
In order to elucidate the role of the carB gene product, the insertion mutant MB35 was subjected to phenotypic analysis. As previously mentioned, MB35 carries a truncated carB gene, lacking one third of its coding region for the C terminus (Fig. 2). Carbamoylphosphate is required for the biosynthesis of pyrimidines and arginine. To test whether the carB mutation would confer a pyrimidine requirement on the cell, the abilities of the strain to grow in the absence and presence of uracil were tested with the SA glucose medium, which includes arginine. An effect of uracil on growth was observed, since addition of uracil to the SA glucose medium resulted in a slight increase in the growth rate of MB35.
L. lactis has the ability to degrade arginine by the arginine deiminase pathway (8), thus forming carbamoylphosphate as an intermediate. To test whether arginine could serve as carbamoylphosphate donor, which subsequently could be utilized in pyrimidine biosynthesis, MB35 (carB) was propagated in the BIV glucose medium in the absence and presence of arginine and/or uracil. Arginine was clearly required for growth, whereas uracil alone was unable to facilitate growth. The addition of a surplus of uracil in addition to arginine resulted in a 60% increase in growth rate. Addition of arginine to the BIV medium resulted in a slight increase in the growth rate of the wild-type strain MG1363. To further test the effect of the carB mutation on growth, strain MB35 was analyzed for its ability to grow on different precursors in the arginine biosynthetic pathway. Citrulline, but not ornithine, was able to support growth of the mutant. Since carbamoylphosphate is required for the conversion of ornithine into citrulline (Fig. 1), the data show that the carB mutation results in a shortage of carbamoylphosphate in the cell. This finding suggests that L. lactis harbors only one gene encoding the large CPSase subunit. Furthermore, the results imply that L. lactis MG1363 has the ability to degrade arginine to carbamoylphosphate, which can subsequently be exploited as a precursor in pyrimidine biosynthesis. An alternative explanation may account for the observations made. If the biosynthesis of arginine and UMP is compartmentalized, meaning that two different CPSases are parts of larger complexes that include either arginine- or pyrimidine-specific biosynthetic enzymes, and carbamoylphosphate is bound to these complexes at all times, then the carB gene described in this paper encodes only the arginine-specific CPSase, since strain MB35 carrying the carB mutation requires arginine but not uracil.
The expression of carB is regulated by pyrimidines.
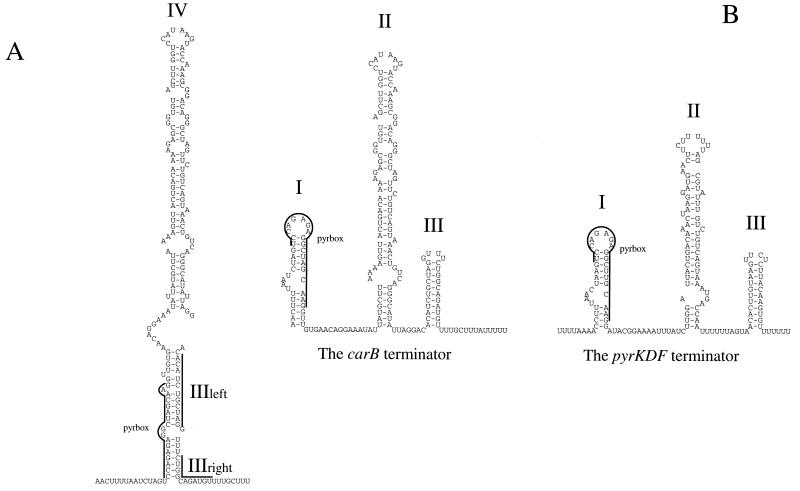
The leader of the carB mRNA has the potential to fold into two mutually exclusive structures: a putative terminator and a putative antiterminator (Fig. 5). Furthermore, sequence analysis showed that immediately after the transcriptional start site of the mRNA, the carB leader is equipped with a sequence that has extremely high levels of similarity to the three PyrR binding sites of B. subtilis, all of which are found at the same position with respect to that of the antiterminator (49). Exactly the same sequence is found in the pyrKDF operon of L. lactis (2). Moreover, pyrKDF mRNA can be folded into a structure similar to the one found in the carB leader (Fig. 5B). In order to analyze whether the expression of the carB gene is subject to regulation by pyrimidines, the β-galactosidase content of L. lactis carrying a carB::lacLM fusion was monitored. The HindIII-Sau3A fragment from pKS2 harboring an internal part of the carB open reading frame and the PCR fragment covering the carB promoter used for construction of pSJ60 were subcloned in the E. coli vector pSMA500 (35) as 1,700- and 1,250-bp fragments, respectively (Fig. 2). The resulting plasmids were designated pSJ23 and pJS61. These plasmids were allowed to integrate into the chromosome of MG1363 by homologous recombination. The strain obtained by transformation with pSJ23 (MB36) has acquired a carB mutation identical to that found in MB35 in addition to the carB::lacLM fusion (Fig. 2), whereas MB37 which was obtained by transformation with pSJ61 is like the wild type with respect to carB despite the fact that this strain carries a carB::lacLM fusion on the chromosome. MB36 and MB37 were grown in BIV minimal medium supplied with arginine in the absence and presence of uracil, and their levels of β-galactosidase synthesis were assayed. The results are presented in Table 3. The absence of uracil led to a 10-fold induction of expression of the carB gene in a carB mutant, whereas a wild-type background led to a 3-fold reduction in the induction of expression. It should be noted that since MB36 carries the same carB mutation as MB35, in the absence of uracil, strain MB36 must be starved for pyrimidines. In order to investigate whether the expression of the carB gene is regulated by arginine, the amounts of β-galactosidase produced by strains growing in BIV minimal medium in the absence and presence of arginine were assayed. This experiment was conducted only with strain MB37, since MB36 is unable to grow in the absence of arginine. The expression of the carB gene was not repressed in the presence of exogenous arginine, whereas its expression was repressed threefold by exogenous uracil in the absence of arginine.
FIG. 5.
Sequences of the carB and pyrKDbF attenuators. (A) carB antiterminator structure IV. The putative binding site of the PyrR protein is indicated with a line marked pyrbox. The stems of the terminator are shown as IIIleft and IIIright. (B) carB and pyrKDF terminator structures. The putative PyrR binding site in domain I is indicated with a line marked pyrbox. The actual terminator structure is designated III.
DISCUSSION
L. lactis harbors only one carB allele.
As previously stated, the carB strain requires arginine but not uracil for growth. Two different models may account for this observation: either only one carB allele is present, and the carbamoylphosphate requirement for pyrimidine biosynthesis is fulfilled by degradation of arginine, or the gene described in this paper is the arginine-specific carB allele used in the compartmentalized biosynthesis of arginine. It is highly unlikely that an arginine-specific CPSase is regulated by uracil. However, carB expression in L. lactis was found to be regulated by uracil. Therefore, the evidence points to the conclusion that L. lactis harbors only one carB allele.
The car genes are usually members of operons.
The results presented in this work demonstrate that the carB gene from L. lactis is transcribed as a monocistronic message. This finding is in contrast to the observation made for most other organisms, namely, that the carB gene is part of an operon consisting of either the carA and carB genes alone or the carA and carB genes as members of a larger operon that includes other pyrimidine-biosynthetic genes. However, exceptions to this paradigm have been reported; in P. aeruginosa the existence of a 216-amino-acid-encoding open reading frame with unknown function between carA and carB has been demonstrated, although the three genes are part of the same operon transcribed from a promoter upstream of carA (26). Neisseria seems to be a true exception. Sequences between carA and carB that vary in size from 2.2 to 3.7 kb were found among different Neisseria species. Furthermore, putative transcription terminators in the intergenic DNA were identified by sequence analysis of one species. Whether these structures were of physiological relevance was not indicated by experimental data (27). Recently, the carA gene from L. lactis has been cloned in our lab, and both Southern blot analysis and PCR failed to demonstrate linkage between carA and carB in L. lactis (45).
Arginine is degraded in L. lactis.
Originally, L. lactis subsp. lactis was distinguished from L. lactis subsp. cremoris by the ability of L. lactis subsp. lactis to degrade arginine by means of the arginine deiminase pathway (8). The first step in the pathway is the deamination of arginine to citrulline, which subsequently is phosphorolytically cleaved into ornithine and carbamoylphosphate. The latter energy-rich derivative can either be used for pyrimidine biosynthesis as described in this work or be degraded to carbon dioxide and ammonia with formation of ATP, thus generating one energy-rich bond per molecule of arginine. It has been shown that arginine uptake in lactococci is mediated by an energy-independent arginine-ornithine antiporter (11). Therefore, arginine can be used as an energy source. Strain MG1363 has been classified as L. lactis subsp. cremoris based on genetic evidence (19, 25, 41). Originally MG1363 was considered a strain of L. lactis subsp. lactis based on its physiological traits, including its capability to degrade arginine. The results presented here further confirm that L. lactis MG1363 degrades arginine through the arginine deiminase pathway.
Glutathione peroxidase encoded by the gpo gene protects against oxidizing elements.
Glutathione is a tripeptide (l-γ-glutamyl-l-cysteinylglycine) and is present in relatively large amounts in L. lactis (13). It is important as a scavenger of free radicals and in the control of the redox potential in the cell. In addition, glutathione is involved in transpeptidation and reduction of thiol groups in proteins and it acts as a cofactor in the reduction of ribonucleotides (39). In addition to completely reduced oxygen as found in water, partially reduced forms, such as singlet oxygen, superoxide anions, hydrogen peroxide, and hydroxyl radicals, are present in organisms growing in an aerobic environment. All these compounds are highly reactive, and they can oxidize proteins and damage DNA, and may oxidize membrane fatty acids, leading to peroxidation of the lipids. Aerobic organisms produce these compounds as metabolic by-products, but all oxygen-tolerant organisms, like L. lactis, are exposed to these powerful agents and must protect themselves against cell damage (14). In E. coli three different activities that protect against the reactive oxygen species noted above have been identified: superoxide dismutase, catalase, and peroxidase (20). In L. lactis, a gene encoding superoxide dismutase (sodA) has been identified (43). Based on the finding that an L. lactis mutant lacking superoxide dismutase is viable in an aerobic environment, Sanders and coworkers (43) concluded that an additional oxygen-protecting mechanism must be present in L. lactis. The glutathione peroxidase found in this work may fulfill this role.
The expression of the carB gene is regulated by pyrimidines.
In this work we have been able to demonstrate that the carB gene is regulated by the presence of uracil in the growth medium. By analyzing the sequence of the carB leader, a structure including a pyrR binding site similar to the one found in the pyrKDF leader can be identified (2). The mechanism by which B. subtilis regulates its expression of the pyr operon by transcriptional attenuation through the PyrR regulatory protein has been studied in great detail (32–34). The structures that may be formed by the RNA transcribed from the carB and pyrKDF (2) operons in L. lactis are similar to the structures found in the RNA transcribed from the B. subtilis pyr operon (33). Figure 5B shows the structures believed to result in termination at the attenuator. Stem-loop structure I, including the PyrR binding site, is highly homologous to a similar structure found in the B. subtilis pyr operon designated the anti-antiterminator by Lu and coworkers (34). These findings strongly suggest the presence of a PyrR homologue in L. lactis.
ACKNOWLEDGMENTS
This work was supported by grants from the Danish government program for food science and technology (FØTEK) through the Center for Advanced Food Studies.
We sincerely appreciate the expert technical assistance of Susan Outzen Jørgensen.
REFERENCES
- 1.Andersen P S, Jansen P J, Hammer K. Two different dihydroorotate dehydrogenases in Lactococcus lactis. J Bacteriol. 1994;176:3975–3982. doi: 10.1128/jb.176.13.3975-3982.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen P S, Martinussen J, Hammer K. Sequence analysis and identification of the pyrKDF operon from Lactococcus lactis including a novel gene, pyrK, involved in pyrimidine biosynthesis. J Bacteriol. 1996;178:5005–5012. doi: 10.1128/jb.178.16.5005-5012.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnau J, Sørensen K, Appel K, Vogensen F K, Hammer K. Analysis of heat shock gene expression in Lactococcus lactis MG1363. Microbiology. 1996;142:1685–1691. doi: 10.1099/13500872-142-7-1685. [DOI] [PubMed] [Google Scholar]
- 4.Bardowski J, Ehrlich S D, Chopin A. Tryptophan biosynthesis genes in Lactococcus lactis subsp. lactis. J Bacteriol. 1992;174:6563–6570. doi: 10.1128/jb.174.20.6563-6570.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Budde, E., and U. Stahl. 1995. Direct submission, EMBL accession no. P40581.
- 6.Cancilla M R, Davidson B E, Hillier A L, Nguyen N Y, Thompson J. The Lactococcus lactis triosephosphate isomerase gene, tpi, is monocistronic. Microbiology. 1995;141:229–238. doi: 10.1099/00221287-141-1-229. [DOI] [PubMed] [Google Scholar]
- 7.Cancilla M R, Hillier A L, Davidson B E. Lactococcus lactis glyceraldehyde-3-phosphate dehydrogenase gene, gap: further evidence for strongly biased codon usage in glycolytic pathway genes. Microbiology. 1995;141:1027–1036. doi: 10.1099/13500872-141-4-1027. [DOI] [PubMed] [Google Scholar]
- 8.Crow V L, Thomas T D. Arginine metabolism in lactic streptococci. J Bacteriol. 1982;150:1024–1032. doi: 10.1128/jb.150.3.1024-1032.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cunin R, Glansdorff N, Piérard A, Stalon V. Biosynthesis and metabolism of arginine in bacteria. Microbiol Rev. 1986;50:314–352. doi: 10.1128/mr.50.3.314-352.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delorme C, Ehrlich S D, Renault P. Histidine biosynthesis genes in Lactococcus lactis subsp. lactis. J Bacteriol. 1992;174:6571–6579. doi: 10.1128/jb.174.20.6571-6579.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Driessen A J, Poolman B, Kiewiet R, Konings W N. Arginine transport in Streptococcus lactis is catalyzed by a cation exchanger. Proc Natl Acad Sci USA. 1987;84:6093–6097. doi: 10.1073/pnas.84.17.6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elagöz A, Abdi A, Hubert J C, Kammerer B. Structure and organisation of the pyrimidine biosynthesis pathway genes in Lactobacillus plantarum: a PCR strategy for sequencing without cloning. Gene. 1996;183:37–43. doi: 10.1016/s0378-1119(96)00461-1. [DOI] [PubMed] [Google Scholar]
- 13.Fahey R C, Brown W C, Adams W B, Worsham M B. Occurrence of glutathione in bacteria. J Bacteriol. 1978;133:1126–1129. doi: 10.1128/jb.133.3.1126-1129.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fridovich I. The biology of oxygen radicals. Science. 1978;201:875–880. doi: 10.1126/science.210504. [DOI] [PubMed] [Google Scholar]
- 15.Gasson M J. Plasmid complements of Streptococcus lactis NCDO 712 and other streptococci after protoplast-induced curing. J Bacteriol. 1983;154:1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerdes K, Thisted T, Martinussen J. Mechanism of post-segregational killing by the hok/sok system of plasmid R1: sok antisense RNA regulates formation of a hok mRNA species correlated with killing of plasmid-free cells. Mol Microbiol. 1990;4:1807–1818. doi: 10.1111/j.1365-2958.1990.tb02029.x. [DOI] [PubMed] [Google Scholar]
- 17.Ghim S-Y, Neuhard J. The pyrimidine biosynthesis operon of the thermophile Bacillus caldolyticus includes genes for uracil phosphoribosyltransferase and uracil permease. J Bacteriol. 1994;176:3698–3707. doi: 10.1128/jb.176.12.3698-3707.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Godon J-J, Chopin M-C, Ehrlich S D. Branched-chain amino acid biosynthesis genes in Lactococcus lactis subsp. lactis. J Bacteriol. 1992;174:6580–6589. doi: 10.1128/jb.174.20.6580-6589.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Godon J J, Delorme C, Ehrlich S D, Renault P. Divergence of genomic sequences between Lactococcus lactis subsp. lactis and Lactococcus lactis subsp. cremoris. Appl Environ Microbiol. 1992;58:4045–4047. doi: 10.1128/aem.58.12.4045-4047.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hassan H F, Fridovich I. Enzymatic defenses against the toxicity of oxygen and of streptonigrin in Escherichia coli. J Bacteriol. 1977;129:1574–1583. doi: 10.1128/jb.129.3.1574-1583.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holo H, Nes I F. High-frequency transformation by electroporation of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl Environ Microbiol. 1989;55:3119–3123. doi: 10.1128/aem.55.12.3119-3123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Israelsen H, Madsen S M, Vrang A, Hansen E B, Johansen E. Cloning and partial characterization of regulated promoters from Lactococcus lactis Tn917-lacZ integrants with the new promoter probe vector pAK80. Appl Environ Microbiol. 1995;61:2540–2547. doi: 10.1128/aem.61.7.2540-2547.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jensen P R, Hammer K. Minimal requirements for exponential growth of Lactococcus lactis. Appl Environ Microbiol. 1993;59:4363–4366. doi: 10.1128/aem.59.12.4363-4366.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johansen E, Kibenich A. Characterization of leuconostoc isolates from commercial mixed strain mesophilic starter cultures. J Dairy Sci. 1992;75:1186–1191. [Google Scholar]
- 25.Klijn N, Weerkamp A H, de Vos W M. Identification of mesophilic lactic acid bacteria by using polymerase chain reaction-amplified variable regions of 16S rRNA and specific DNA probes. Appl Environ Microbiol. 1991;57:3390–3393. doi: 10.1128/aem.57.11.3390-3393.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwon D H, Lu C D, Walthall D A, Brown T M, Houghton J E, Abdelal A T. Structure and regulation of the carAB operon in Pseudomonas aeruginosa and Pseudomonas stutzeri: no untranslated region exists. J Bacteriol. 1994;176:2532–2542. doi: 10.1128/jb.176.9.2532-2542.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lawson F S, Billowes F M, Dillon J A. Organization of carbamoyl-phosphate synthase genes in Neisseria gonorrhoeae includes a large, variable intergenic sequence which is also present in other Neisseria species. Microbiology. 1995;141:1183–1191. doi: 10.1099/13500872-141-5-1183. [DOI] [PubMed] [Google Scholar]
- 28.Le Bourgeois P, Lautier M, Mata M, Ritzenthaler P. New tools for the physical and genetic mapping of Lactococcus strains. Gene. 1992;111:109–114. doi: 10.1016/0378-1119(92)90610-2. [DOI] [PubMed] [Google Scholar]
- 29.Leisinger, U., A. J. B. Zehnder, and R. I. F. Eggen. 1997. Direct submission, GenBank accession no. AF014927.
- 30.Li X, Weinstock G M, Murray B E. Generation of auxotrophic mutants of Enterococcus faecalis. J Bacteriol. 1995;177:6866–6873. doi: 10.1128/jb.177.23.6866-6873.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Llanos R, Harris C J, Hillier A J, Davidson B E. Identification of a novel operon in Lactococcus lactis encoding three enzymes for lactic acid synthesis: phosphofructokinase, pyruvate kinase, and lactate dehydrogenase. J Bacteriol. 1993;175:2541–2551. doi: 10.1128/jb.175.9.2541-2551.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu Y, Switzer R L. Evidence that the Bacillus subtilis pyrimidine regulatory protein PyrR acts by binding to pyr mRNA at three sites in vivo. J Bacteriol. 1996;178:5806–5809. doi: 10.1128/jb.178.19.5806-5809.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu Y, Switzer R L. Transcriptional attenuation of the Bacillus subtilis pyr operon by the PyrR regulatory protein and uridine nucleotides in vitro. J Bacteriol. 1996;178:7206–7211. doi: 10.1128/jb.178.24.7206-7211.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu Y, Turner R J, Switzer R L. Function of RNA secondary structures in transcriptional attenuation of the Bacillus subtilis pyr operon. Proc Natl Acad Sci USA. 1996;93:14462–14467. doi: 10.1073/pnas.93.25.14462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Madsen S M, Albrechtsen B, Hansen E B, Israelsen H. Cloning and transcriptional analysis of two threonine biosynthetic genes from Lactococcus lactis MG1614. J Bacteriol. 1996;178:3689–3694. doi: 10.1128/jb.178.13.3689-3694.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinussen J, Andersen P S, Hammer K. Nucleotide metabolism in Lactococcus lactis. Salvage pathways of exogenous pyrimidines. J Bacteriol. 1994;176:1514–1516. doi: 10.1128/jb.176.5.1514-1516.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinussen J, Hammer K. Cloning and characterization of upp, a gene encoding uracil phosphoribosyltransferase from Lactococcus lactis. J Bacteriol. 1994;176:6457–6463. doi: 10.1128/jb.176.21.6457-6463.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martinussen J, Hammer K. Powerful methods to establish chromosomal markers in Lactococcus lactis: an analysis of pyrimidine salvage pathway mutants obtained by positive selections. Microbiology. 1995;141:1883–1890. doi: 10.1099/13500872-141-8-1883. [DOI] [PubMed] [Google Scholar]
- 39.Meister A, Anderson M E. Glutathione. Annu Rev Biochem. 1983;52:711–760. doi: 10.1146/annurev.bi.52.070183.003431. [DOI] [PubMed] [Google Scholar]
- 40.Quinn C L, Stephenson B T, Switzer R L. Functional organization, nucleotide sequence and regulation of the Bacillus subtilis pyrimidine biosynthetic operon. J Biol Chem. 1991;266:9113–9127. [PubMed] [Google Scholar]
- 41.Salama M, Sandine W, Giovannoni S. Development and application of oligonucleotide probes for identification of Lactococcus lactis subsp. cremoris. Appl Environ Microbiol. 1991;57:1313–1318. doi: 10.1128/aem.57.5.1313-1318.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 43.Sanders J W, Leenhouts K J, Haandrikman A J, Venema G, Kok J. Stress response in Lactococcus lactis: cloning, expression analysis, and mutation of the lactococcal superoxide dismutase gene. J Bacteriol. 1995;177:5254–5260. doi: 10.1128/jb.177.18.5254-5260.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5467–5473. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schallert, J., and J. Martinusssen. 1997. Unpublished data.
- 46.Sorokin, A. V., V. Azevedo, E. Zumstein, N. Galleron, S. D. Ehrlich, and P. Serror. 1996. Direct submission, Swiss-Prot accession no. P52035.
- 47.Tabata, S. 1996. Direct submission, DDBJ accession no. D90913.
- 48.Terzaghi B E, Sandine W E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975;29:807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Turner R J, Lu Y, Switzer R L. Regulation of the Bacillus subtilis pyrimidine biosynthetic gene cluster by an autogenous transcriptional attenuation mechanism. J Bacteriol. 1994;176:3708–3722. doi: 10.1128/jb.176.12.3708-3722.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van de Guchte M, Kok J, Venema G. Gene expression in Lactococcus lactis. FEMS Microbiol Rev. 1992;88:73–92. doi: 10.1111/j.1574-6968.1992.tb04958.x. [DOI] [PubMed] [Google Scholar]