Abstract
Changes in intracellular 3′,5′ cyclic AMP (cAMP) concentration regulate the development of natural competence in Haemophilus influenzae. In Escherichia coli, cAMP levels are modulated by a cAMP phosphodiesterase encoded by the cpdA gene. We have used several approaches to demonstrate that the homologous icc gene of H. influenzae encodes a functional cAMP phosphodiesterase and that this gene limits intracellular cAMP and thereby influences competence and other cAMP-dependent processes. In E. coli, expression of cloned icc reduced both cAMP-dependent sugar fermentation and β-galactosidase expression, as has been shown for cpdA. In H. influenzae, an icc null mutation increased cAMP-dependent sugar fermentation and competence development in strains where these processes are limited by mutations reducing cAMP synthesis. When endogenous production of cAMP was eliminated by a cya mutation, an icc strain was 10,000-fold more sensitive to exogenous cAMP than an icc+ strain. The icc strain showed moderately elevated competence under noninducing conditions, as expected, but had subnormal competence increases at onset of stationary phase in rich medium, and on transfer to a nutrient-limited medium, suggesting that excessive cAMP may interfere with induction. Consistent with this finding, a cya strain cultured in 1 mM cAMP failed to develop maximal competence on transfer to inducing conditions. Thus, by limiting cAMP levels, the H. influenzae cAMP phosphodiesterase may coordinate its responses to nutritional stress, ensuring optimal competence development.
The cyclic nucleotide 3′,5′ cyclic AMP (cAMP) is a central mediator of the responses of enteric bacteria to changes in the nutritional environment. Expression of a large number of cAMP-controlled genes is elevated when intracellular cAMP levels peak at the onset of starvation (24) or stationary phase (38).
In enteric bacteria, intracellular levels of cAMP are determined by the relative rates of its synthesis, excretion, and degradation (4). Adenylate cyclase, which synthesizes cAMP, has only basal activity during exponential growth on preferred sugars and is stimulated by the phosphoenolpyruvate:sugar phosphotransferase system (PTS) when these sugars are depleted from the medium. cAMP is excreted when preferred sugars are restored (7), but the regulatory significance of excretion is unclear and awaits isolation of excretion-defective mutants. Finally, cAMP levels can be reduced by cAMP-specific phosphodiesterases, which catalyze cleavage of 3′,5′-cAMP to 5′-cAMP. Although the regulatory role of cAMP phosphodiesterases has been unclear (9), Imamura et al. recently demonstrated that the cAMP phosphodiesterase produced by the Escherichia coli cpdA gene modulates intracellular cAMP levels and proposed that such phosphodiesterases may regulate expression of cAMP-dependent genes (20).
In Haemophilus influenzae, cAMP regulates development of natural competence, and cells mutant in the adenylate cyclase gene (cya) or the cAMP receptor protein (CRP) gene (crp) cannot become competent (11, 14). Although direct measurement of cAMP concentrations in H. influenzae has not been achieved (14), competence is undetectable during exponential growth, when cAMP levels are expected to be low. It develops spontaneously when exogenous cAMP is added or when endogenous cAMP levels are expected to be high, for example, at the onset of stationary phase in rich medium. The highest levels of competence are seen when exponential-phase cells are transferred to the nutrient-limited medium MIV (18), when intracellular cAMP levels are expected to peak.
Competent cells can take up several hundred kilobases of DNA from their environment. Some of this DNA may be homologously recombined into the chromosome; nonrecombined DNA is degraded, and the nucleotides are reused (35). The requirement of cAMP for competence induction suggests that H. influenzae’s ability to take up DNA from the environment may be a response to nutritional stress, with the degraded DNA serving as a source of nucleotides. To explore this possibility, we are investigating the regulation of intracellular cAMP levels in H. influenzae and their relationship to competence.
We have previously shown that the synthesis of cAMP by adenylate cyclase is regulated by a simple fructose-specific PTS and that this in turn regulates competence development (22). Efflux of cAMP has not been studied. H. influenzae does, however, possess a cpdA homolog, icc (15, 20). Below, we show that icc encodes a cAMP phosphodiesterase which functions together with adenylate cyclase to fine-tune intracellular cAMP levels and to modulate competence development (and other cAMP-dependent processes) according to growth rate and/or nutrient availability.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Strains and plasmids used in this study are listed in Tables 1 and 2. Strain RR801 was derived from a ptsI::kan crr::cat strain obtained from M. Gwinn (17). Double-mutant strains RR819, RR820, and RR821 were constructed by transforming strains RR668, RR745, and RR801 to spectinomycin resistance with limiting RR812 chromosomal DNA.
TABLE 1.
Bacterial strains used in this study
| Strain | Genotype | Source and/or reference |
|---|---|---|
| H. influenzae Rd | ||
| KW20 | Wild type | 1 |
| RR812 | icc::spc | This study |
| RR668 | cya::mini-Tn10kan | 14 |
| RR745 | ptsI::mini-Tn10kan | 22 |
| RR801 | crr::cat | This study |
| RR819 | ptsI::mini-Tn10kan icc::spc | This study |
| RR820 | crr::cat icc::spc | This study |
| RR821 | cya::mini-Tn10kan icc::spc | This study |
| MAP7 | str kan nov nal spc vio stv | J. Setlow (6) |
| E. coli | ||
| W3110 | Wild type | R. Redfield |
| SH8150 | cpdA::kan | H. Niki (20) |
| GM2163 | dam | New England BioLabs |
TABLE 2.
Plasmids used in this study
| Plasmid | Insert/vector | Source (reference) |
|---|---|---|
| pKRP13 | spc cassette | G. J. Phillips |
| pCAT19 | cat cassette | C. Fuqua |
| pGEMsxy | sxy/pGEM-T | L. Bannister |
| pGEM+ | Control insert/pGEM-T | This study |
| pAX923 | cpdA/pACYC184 | H. Niki (20) |
| pCMP | icc/pGEM-T | This study |
| pCMP::spc | icc::spc/pGEM-T | This study |
Culture conditions.
H. influenzae cells were grown aerobically at 37°C in brain heart infusion broth (BHI; Difco) supplemented with hemin and NAD (sBHI), with the recommended concentrations of antibiotics (6). Additional hemin was applied to sBHI plates greater than 24 h old. E. coli strains were routinely grown aerobically in Luria-Bertani broth at 37°C, with the recommended concentrations of antibiotics (5).
Competence and transformation.
Spontaneous competence development by H. influenzae was followed during growth in sBHI, while maximal competence was induced by transfer of exponential-phase cells to MIV starvation medium (18). Competence was assessed as transformation frequency. E. coli cells were made competent by transfer to cold CaCl2 and transformed by standard procedures (5).
Sugar fermentation assays.
Sugar fermentation by H. influenzae was assayed in Difco phenol red broth (PRB) supplemented with NAD, hemin, 10% BHI, and 1% sugar to be tested. After overnight growth in loosely capped tubes, results were scored by measurement of culture pH. Sugar fermentation by E. coli was scored on Difco MacConkey agar plates containing 1% lactose or maltose.
Cloning and mutagenesis of icc.
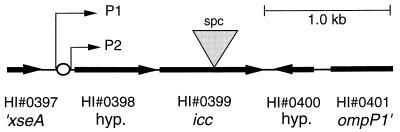
The availability of the full H. influenzae genome sequence (36) has simplified the construction of directed mutations in genes of interest. A 3-kb region of KW20 chromosomal DNA containing the icc operon (genes HI 0398 and HI 0399) (15) was amplified by PCR using the following forward and reverse primers (H. influenzae genome positions are given in parentheses): 5′-GCGTAAATCGCCAAGTGACGG-3′ (bp 419090 to 419111) and 5′-GCATTCCGTTCAACTTGGGC-3′ (bp 422142 to 422122). The resulting fragment was cloned into the vector pGEM-T (Promega) to generate pCMP. A gel-purified spectinomycin resistance gene (spc) cassette from BamHI-cut plasmid pKRP13 was ligated into BclI-cut pCMP (prepared from the dam E. coli strain GM2123) (Fig. 1), giving pCMP::spc. MIV-competent KW20 cells were transformed to spectinomycin resistance with ApaI-linearized pCMP::spc. Southern blotting of genomic DNA from the new icc::spc strain RR812 with spc and icc PCR product probes confirmed that it contained a single cassette insertion in the icc gene.
FIG. 1.
The 3-kb H. influenzae icc operon fragment cloned in pCMP. xseA, exonuclease VII, large subunit; ompP1, outer membrane protein; hyp., hypothetical protein; P1 and P2, putative promoter sites; open circle, putative CRP-binding site; inverted triangle, location of spc cassette insertion at bp 1573 (a BclI site) of pCMP::spc.
β-Galactosidase assay.
Cells were grown in M9 minimal glucose medium (5) supplemented with 0.5% Casamino Acids and 1 mM isopropyl-β-d-thiogalactopyranoside. β-Galactosidase activity was assayed by using a modification (33) of the method described by Miller (25).
RESULTS
Analysis of the icc gene.
The gene now known to encode the E. coli cAMP-specific phosphodiesterase was originally characterized as affecting only the expression of the lacZ gene (19), and the gene sequence was submitted to GenBank under the name icc. Imamura et al. subsequently demonstrated that this gene encodes a 3′,5′-cAMP-specific phosphodiesterase (20) and renamed it cpdA. The existence of an H. influenzae icc homolog was noted (15, 20).
The predicted 274-amino-acid Icc protein shows 53.3% identity and 71.3% similarity to its E. coli homolog, CpdA (36). Gapped TBLASTN (3) searching of the GenBank databases and available bacterial genome sequences detected putative Icc homologs with extensive amino acid sequence identities (E [expect value] < 1e-30) in only three other bacterial species, all members of the gamma subdivision of the proteobacteria (31) (Table 3). In addition, cAMP-specific phosphodiesterases have been purified from the proteobacteria Serratia marcescens (30), Salmonella typhimurium (2), and Klebsiella aerogenes (10). As expected, the icc-containing organisms with complete genome sequences available also contain adenylate cyclase (cya) homologs: H. influenzae, E. coli, Pseudomonas aeruginosa, and S. typhimurium. These findings also suggest that 3′,5′-cAMP phosphodiesterases may be limited to the proteobacteria.
TABLE 3.
Eubacterial species whose genomes contain Icc homologs
| Organism | Icc homologa
|
No. of aa | E valueb | Source/accession no. (reference) | |
|---|---|---|---|---|---|
| % Identical | % Similar | ||||
| Actinobacillus actinomycetemcomitansc | 64.2 | 78.2 | 271 | 1e-106 | QUAGCT (37) |
| Escherichia coli | 53.3 | 71.3 | 231 | 4e-77 | GenBank/D16557 (16) |
| Pseudomonas aeruginosac | 27.7 | 46 | 260 | 1e-34 | PAGP (32) |
Gapped TBLASTN (3) searches of the completely sequenced genomes of the following bacterial species found no sequences with significant identity to Icc: Borrelia burgdorferi (36), Chlamydia trachomatis (12), Helicobacter pylori (36), Neisseria meningitidis (34, 36), Synechocystis strain PCC 6803 (21), Treponema pallidum (36), Bacillus subtilis (8), Mycoplasma genitalium (36), and Streptococcus pyogenes (37).
Likelihood that an identified sequence match could have occurred by change.
Sequencing and/or analysis of the genome is incomplete.
The icc gene may belong to a conserved nucleotide-processing operon. The upstream gene HI 0398 is separated from icc (HI 0399) by only 13 bp (Fig. 1). Similarly, an HI 0398 homolog is found immediately upstream of the icc homologs of P. aeruginosa and Actinobacillus actinomycetemcomitans and is separated by one gene from the E. coli icc homolog cpdA (36). HI 0398 and its homologs all contain a signature sequence common to members of the MutT family of proteins, whose members hydrolyze nucleoside diphosphate linkages in various substrates (29). Although we found no putative icc-specific promoters (by sequence comparison with the E. coli promoter consensus), we identified two potential promoters upstream of the HI 0398 initiation codon: P1 at bp 419321 to 419351 and P2 at bp 419417 to 419445.
We identified a putative binding site for CRP immediately 5′ to P2 (5′-TTTTGTGACTCACTTCAAACTC-3′ at bp 419394 to 419416) by comparison with the E. coli CRP-binding consensus (7) (Fig. 1). This finding suggests that expression of icc is not constitutive but may be induced as cAMP levels rise.
Demonstration of a functional icc-encoded phosphodiesterase.
To determine whether the E. coli and H. influenzae cAMP phosphodiesterase homologs have similar activities, we compared the effects of overexpression of each on cAMP-dependent processes in E. coli. Expression of the E. coli lactose and maltose utilization operons is cAMP dependent, and Imamura et al. have shown that overexpression of cpdA reduces transcription of lacZ and catabolism of lactose analogs (20). As judged by colony color on MacConkey agar plates supplemented with 1% lactose or maltose, plasmids carrying the E. coli and H. influenzae homologs of cpdA reduced fermentation of both sugars by E. coli W3110 to similar degrees. Disruption of the icc gene by gene cassette insertion in pCMP::spc abolished this effect, while vector controls pACYC184 and pGEMsxy had no effect on sugar fermentation (23).
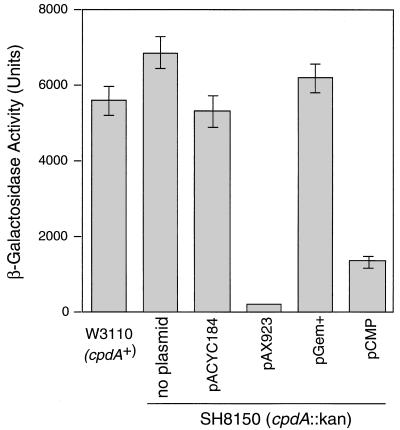
β-Galactosidase activity is a more sensitive indicator of intracellular cAMP concentration in cells expressing different phosphodiesterase homologs. We found that pCMP, like pAX923 (20), reduced β-galactosidase activity (presented as mean ± standard error of the mean [SEM]) by at least 75% (Fig. 2). These data confirm that the H. influenzae gene icc encodes a cAMP phosphodiesterase.
FIG. 2.
β-Galactosidase activity in E. coli cells carrying cpdA/icc plasmids. Activity is in Miller units (25). Mean values from three replicates are shown; error bars represent SEM. (SEM for pAX923 is too small to be visible on this scale.)
Regulation of H. influenzae cAMP levels by icc.
To determine whether the icc gene limits intracellular cAMP levels in H. influenzae, we constructed an icc mutant strain and assessed the effects on known cAMP-dependent processes: ribose fermentation and competence development.
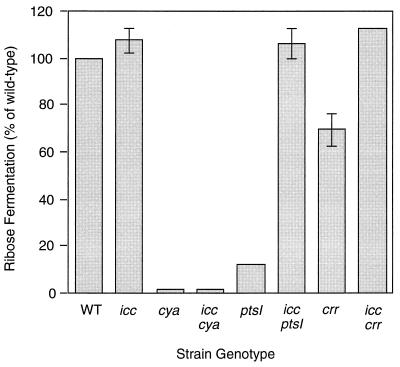
We have previously shown that utilization of ribose by H. influenzae is cAMP dependent. PRB assays demonstrated that icc disruption has little effect on ribose fermentation in a wild-type background (Fig. 3). We predicted, however, that if the icc mutation prevents degradation of cellular cAMP, it should restore ribose fermentation to strains with low cAMP (ptsI or crr) but not to a strain totally lacking cAMP (cya). As expected, an icc cya strain remained unable to ferment ribose, but icc ptsI and icc crr strains fermented ribose at almost wild-type levels, presumably because they cannot degrade the small amounts of cAMP that they synthesize (Fig. 3).
FIG. 3.
Effect of icc disruption on ribose fermentation in wild-type (WT), cya, and PTS-disrupted backgrounds. A 100-μl aliquot of exponentially growing wild-type or mutant strains was inoculated into 5 ml of supplemented PRB plus 1% ribose. After rotation of the cultures overnight, we measured the pH of each culture and calculated the pH change (ΔpH). The degree of ribose fermentation of each mutants strain was expressed as a percentage of wild-type ΔpH (wild-type H. influenzae cultures in 1% ribose showed an average pH drop from 7.47 to 5.43). Mean values from three replicates are shown. Error bars represent SEM. (SEM values for cya, icc cya, ptsI, and icc crr strains are too small to be visible on this scale.)
Development of competence for natural transformation is also cAMP dependent and is our most sensitive indicator of changes in intracellular cAMP levels in H. influenzae (e.g., Fig. 4). Wild-type H. influenzae strain KW20 reaches transformation frequencies (TF) as high as 10−2. The cya mutation completely abolishes competence development (TF < 10−7) (14), while ptsI strains show a ∼200-fold reduction in competence (TF = 7 × 10−5) (22). As expected, competence remained undetectable (TF < 10−7) in an icc cya strain, but disruption of the phosphodiesterase gene in a ptsI background boosted competence 10-fold (TF = 7.6 × 10−4) (23). These data confirm that H. influenzae cells lacking the icc-encoded phosphodiesterase have increased intracellular cAMP levels.
FIG. 4.
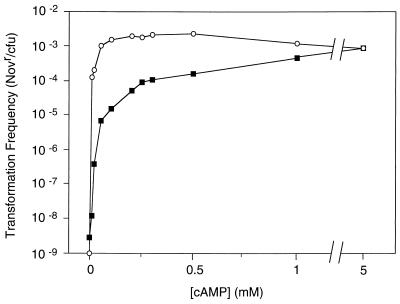
cAMP sensitivity of icc cya (○) and icc+ cya (■) H. influenzae strains. Strains were grown to mid-exponential phase in sBHI, and competence was induced by transfer of cells to MIV medium containing 0 to 5 mM cAMP, as described. Cells were incubated with MAP7 chromosomal DNA (1 μg ml−1) for 30 min; transformation frequency was calculated as the number of novobiocin-resistant (Novr) transformants divided by the total number of cells. Representative results are shown.
Sensitivity of H. influenzae icc strains to exogenous cAMP.
Alper and Ames (2) found that the presence of a cpd mutation in S. typhimurium strains lacking endogenous cAMP (cya mutants) reduced 10-fold the exogenous cAMP required for expression of catabolic operons. We examined the cAMP sensitivity of H. influenzae cya strains carrying icc+ and icc alleles by assaying the sensitivity of competence development to exogenous cAMP. As expected, the icc strain was much more sensitive to exogenous cAMP: the lowest concentration tested (10 nM) increased competence 10,000-fold in this strain but only 3-fold in the icc+ strain (Fig. 4). These data imply that the H. influenzae icc strain is unable to degrade exogenously supplied cAMP.
Regulation of competence development by a cAMP phosphodiesterase.
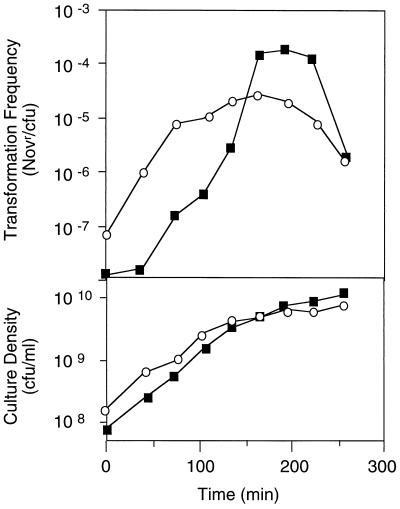
We found that RR812 developed spontaneous competence unusually early during exponential growth in rich medium (Fig. 5). This observation is consistent with a model in which Icc regulates intracellular cAMP concentrations throughout growth, not merely when cAMP levels are high.
FIG. 5.
Spontaneous competence of RR812 (icc) (○) and KW20 (■) in sBHI. Transformation frequencies were assayed as described in the legend to Fig. 4. Representative results are shown. Novr, novobiocin resistant.
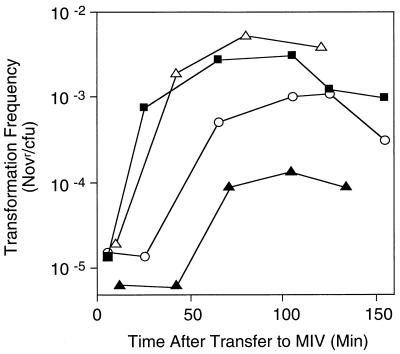
Our simplest model predicted that cAMP phosphodiesterase inactivation should increase intracellular cAMP concentrations and that an icc strain should achieve at least wild-type levels of competence. It was therefore surprising to discover that the icc strain failed to reach wild-type levels of competence in both rich and nutrient-limited media (Fig. 5 and 6). We subsequently determined that this was not due to a simple toxic effect of high intracellular cAMP, since up to 10 mM exogenous cAMP did not significantly affect H. influenzae viability. Moreover, icc disruption itself does not reduce viability of this strain—it grows with a normal doubling time in batch culture. Instead, we reasoned that the timing of an increase in intracellular cAMP relative to growth rate or nutrient availability might be significant in the induction of maximal competence. To investigate this, we assessed competence of a cya strain provided with 1 mM exogenous cAMP either continuously or only after transfer of cells to starvation medium. We found that cya cells that encounter a dramatic increase in cAMP coincident with nutrient limitation develop wild-type competence levels. However, cells cultured in 1 mM cAMP prior to transfer to MIV show delayed and reduced competence, similarly to the icc strain (Fig. 6). These data suggest that the timing of a cAMP increase is critical in competence induction and that Icc regulates this timing in H. influenzae.
FIG. 6.
MIV-induced competence of RR812 (icc) and KW20 compared with RR668 (cya) precultured with or without 1 mM cAMP. Transformation frequencies were assayed as described in the legend to Fig. 4. Filled squares, KW20; open circles, RR812 (icc); open triangles, RR668 transferred to MIV plus 1 mM cAMP; filled triangles, RR668 (cya) cultured in sBHI plus 1 mM cAMP before transfer to MIV plus 1 mM cAMP. Representative results are shown. Novr, novobiocin resistant.
DISCUSSION
Why bacteria limit increases in cellular cAMP.
To optimize transcription of cAMP-dependent genes involved in nutrient transport and catabolism, bacterial cells must closely regulate cAMP levels throughout growth. Because cAMP is a very stable molecule, active mechanisms for cAMP elimination are needed to quickly reduce cAMP concentrations whenever conditions improve. It has also been suggested that cAMP phosphodiesterases may protect cells from the transcriptional repression that can result from excessive cAMP (2). Such repression may be caused by the cAMP-CRP complex directly: although this complex is better known as a transcriptional activator, it can also function as a transcriptional repressor (7). In addition, excessive cAMP may prevent efficient transcription of cAMP-dependent genes by overloading the CRP complex: the CRP protein has two identical subunits, each of which binds one molecule of cAMP, and the CRP-cAMP2 complex has much lower affinity for CRP-binding sites than the CRP-cAMP1 complex (7).
Uncontrolled cAMP increases may repress transcription of competence genes.
It was puzzling that disruption of the H. influenzae phosphodiesterase gene reduced maximal competence levels in both stationary phase and MIV (Fig. 5 and 6), even though ribose fermentation data (Fig. 3) implied that intracellular cAMP concentrations were raised. However, competence is also thought to require at least one cAMP-independent regulatory event (14), and the uncontrolled increase in intracellular cAMP in the icc strain may interfere with necessary coordination between the cAMP-dependent and cAMP-independent signals. Thus, one role of the cAMP phosphodiesterase may be to regulate the timing of increases in intracellular cAMP.
The biological function of competence.
In H. influenzae, competence is induced by conditions of nutrient limitation and absolutely requires an increase in cAMP, an established indicator of nutritional stress. Ferenci and colleagues have demonstrated that E. coli cAMP levels peak during the transition to starvation, allowing transient hunger-response expression of cAMP-dependent genes (13, 26–28). The pattern of competence development by H. influenzae suggests that it may also be a hunger response. We have previously shown that adenylate cyclase activity is influenced by the PTS, which responds to changes in carbon source availability. We have now shown that an H. influenzae cAMP phosphodiesterase participates in regulation of cAMP levels and maximal competence development.
ACKNOWLEDGMENTS
This work was supported by an operating grant to R.J.R. from the Medical Research Council of Canada. L.P.M. was supported by a studentship from the Canadian Cystic Fibrosis Foundation.
We thank H. Niki for his gift of E. coli strains and plasmids, and we thank the members of the Redfield lab for helpful discussions.
REFERENCES
- 1.Alexander H, Leidy G. Determination of inherited traits of H. influenzae by desoxyribonucleic acid fractions isolated from type-specific cells. J Exp Med. 1951;93:345–359. doi: 10.1084/jem.93.4.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alper M D, Ames B N. Cyclic 3′,5′-adenosine monophosphate phosphodiesterase mutants of Salmonella typhimurium. J Bacteriol. 1975;122:1081–1090. doi: 10.1128/jb.122.3.1081-1090.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amin N, Peterkofsky A. A dual mechanism for regulating cAMP levels in Escherichia coli. J Biol Chem. 1995;270:11803–11805. doi: 10.1074/jbc.270.20.11803. [DOI] [PubMed] [Google Scholar]
- 5.Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene/Wiley; 1994. [Google Scholar]
- 6.Barcak G J, Chandler M S, Redfield R J, Tomb J-F. Genetic systems in Haemophilus influenzae. Methods Enzymol. 1991;204:321–342. doi: 10.1016/0076-6879(91)04016-h. [DOI] [PubMed] [Google Scholar]
- 7.Botsford J L, Harman J G. Cyclic AMP in prokaryotes. Microbiol Rev. 1992;56:100–122. doi: 10.1128/mr.56.1.100-122.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bacillus subtilis Genome Project.http://www.pasteur.fr/Bio/SubtiList.html.
- 9.Buettner M J, Spitz E, Rickenberg H V. Cyclic adenosine 3′,5′-monophosphate in Escherichia coli. J Bacteriol. 1973;14:1068–1073. doi: 10.1128/jb.114.3.1068-1073.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calcott P, Calvert T. Characterization of 3′,5′-cyclic AMP phosphodiesterase in Klebsiella aerogenes and its role in substrate-accelerated death. J Gen Microbiol. 1981;122:313–321. doi: 10.1099/00221287-122-2-313. [DOI] [PubMed] [Google Scholar]
- 11.Chandler M S. The gene encoding cyclic AMP receptor protein is required for competence development in Haemophilus influenzae Rd. Proc Natl Acad Sci USA. 1992;89:1626–1630. doi: 10.1073/pnas.89.5.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chlamydia Genome Project.http://chlamydia-www.berkeley.edu:4231/.
- 13.Death A, Ferenci T. Between feast and famine: endogenous inducer synthesis in the adaptation of Escherichia coli to growth with limiting carbohydrates. J Bacteriol. 1994;176:5101–5107. doi: 10.1128/jb.176.16.5101-5107.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dorocicz I, Williams P, Redfield R J. The Haemophilus influenzae adenylate cyclase gene: cloning, sequence, and essential role in competence. J Bacteriol. 1993;175:7142–7149. doi: 10.1128/jb.175.22.7142-7149.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J-F, Dougherty B A, Merrick J M, McKenney K, Sutton G, FitzHugh W, Fields C, Gocayne J D, Scott J, Shirley R, Liu L-I, Glodek A, Kelley J M, Weidman J F, Phillips C A, Spriggs T, Hedblom E, Cotton M D, Utterback T R, Hanna M C, Nguyen D T, Saudek D M, Brandon R C, Fine L D, Fritchman J L, Fuhrmann J L, Geoghagen N S M, Gnehm C, McDonald L A, Small K V, Fraser C M, Smith H O, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 16.GenBank.http://www.ncbi.nlm.nih.gov/.
- 17.Gwinn M L, Yi D, Smith H O, Tomb J-F. Role of two-component signal transduction and the phosphoenolpyruvate:carbohydrate phosphotransferase systems in competence development of Haemophilus influenzae Rd. J Bacteriol. 1996;178:6366–6368. doi: 10.1128/jb.178.21.6366-6368.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herriott R M, Meyer E M, Vogt M. Defined nongrowth media for stage II development of competence in Haemophilus influenzae. J Bacteriol. 1970;101:517–524. doi: 10.1128/jb.101.2.517-524.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imamura, R. 1993. Notes with GenBank submission, accession no. P36650.
- 20.Imamura R, Yamanaka K, Ogura T, Hiraga S, Fujita N, Ishihama A, Niki H. Identification of the cpdA gene encoding cyclic 3′,5′-adenosine monophosphate phosphodiesterase in Escherichia coli. J Biol Chem. 1996;271:25423–25429. doi: 10.1074/jbc.271.41.25423. [DOI] [PubMed] [Google Scholar]
- 21.Kazusa DNA Research Institute.http://www.kazusa.or.jp/KDRI/kazusa_home-e.html.
- 22.Macfadyen L P, Dorocicz I R, Reizer J, Saier M H J, Redfield R J. Regulation of competence development and sugar utilization in Haemophilus influenzae Rd by a phosphoenolpyruvate:fructose phosphotransferase system. Mol Microbiol. 1996;21:941–952. doi: 10.1046/j.1365-2958.1996.441420.x. [DOI] [PubMed] [Google Scholar]
- 23.Macfadyen, L. P., C. Ma, and R. J. Redfield. Unpublished data.
- 24.Matin A, Auger E A, Blum P H, Schultz J E. The genetic basis of starvation survival in non-differentiating bacteria. Annu Rev Microbiol. 1989;43:293–316. doi: 10.1146/annurev.mi.43.100189.001453. [DOI] [PubMed] [Google Scholar]
- 25.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- 26.Notley L, Ferenci T. Differential expression of mal genes under cAMP and endogenous inducer control in nutrient-stressed Escherichia coli. Mol Microbiol. 1995;16:121–129. doi: 10.1111/j.1365-2958.1995.tb02397.x. [DOI] [PubMed] [Google Scholar]
- 27.Notley L, Ferenci T. Induction of RpoS-dependent functions in glucose-limited continuous culture: what level of nutrient limitation induces the stationary phase of Escherichia coli? J Bacteriol. 1996;178:1465–1468. doi: 10.1128/jb.178.5.1465-1468.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Notley-McRobb L, Death A, Ferenci T. The relationship between external glucose concentration and cAMP levels inside Escherichia coli: implications for models of phosphotransferase-mediated regulation of adenylate cyclase. Microbiology. 1997;143:1909–1918. doi: 10.1099/00221287-143-6-1909. [DOI] [PubMed] [Google Scholar]
- 29.O’Handley S F, Frick D N, Bullions L C, Mildvan A S, Bessman M J. Escherichia coli orf17 codes for a nucleoside triphosphate pyrophosphohydrolase member of the MutT family of proteins. Cloning, purification, and characterization of the enzyme. J Biol Chem. 1996;271:24649–24654. doi: 10.1074/jbc.271.40.24649. [DOI] [PubMed] [Google Scholar]
- 30.Okabayashi T, Ide M. Cyclic 3′,5′-nucleotide phosphodiesterase of Serratia marcescens. Biochim Biophys Acta. 1970;220:116–123. doi: 10.1016/0005-2744(70)90235-4. [DOI] [PubMed] [Google Scholar]
- 31.Olsen G J, Woese C R, Overbeek R. The winds of (evolutionary) change: breathing new life into microbiology. J Bacteriol. 1994;176:1–6. doi: 10.1128/jb.176.1.1-6.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pseudomonas aeruginosa Genome Project.http://www.pseudo.com/.
- 33.Rothstein D M, Pahel G, Tyler B, Magasanik B. Regulation of expression from the glnA promoter of Escherichia coli in the absence of glutamine synthetase. Proc Natl Acad Sci USA. 1980;77:7372–7376. doi: 10.1073/pnas.77.12.7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanger Centre/John Innes Centre.http://www.sanger.ac.uk; http://www.uea.ac.uk/urp/jic/.
- 35.Stewart G J, Carlson C A. The biology of natural transformation. Annu Rev Microbiol. 1986;40:211–235. doi: 10.1146/annurev.mi.40.100186.001235. [DOI] [PubMed] [Google Scholar]
- 36.The Institute for Genome Research.http://www.tigr.org.
- 37.University of Oklahoma Advanced Center for Genome Technology.http://www.genome.ou.edu. [DOI] [PubMed]
- 38.Weichart D, Lange D, Henneberg N, Hengge-Aronis R. Identification and characterization of stationary-phase inducible genes in Escherichia coli. Mol Microbiol. 1993;10:407–420. [PubMed] [Google Scholar]