Abstract
Recently, a promoter for the essential gene ftsI, which encodes penicillin-binding protein 3 of Escherichia coli, was precisely localized 1.9 kb upstream from this gene, at the beginning of the mra cluster of cell division and cell envelope biosynthesis genes (H. Hara, S. Yasuda, K. Horiuchi, and J. T. Park, J. Bacteriol. 179:5802–5811, 1997). Disruption of this promoter (Pmra) on the chromosome and its replacement by the lac promoter (Pmra::Plac) led to isopropyl-β-d-thiogalactopyranoside (IPTG)-dependent cells that lysed in the absence of inducer, a defect which was complemented only when the whole region from Pmra to ftsW, the fifth gene downstream from ftsI, was provided in trans on a plasmid. In the present work, the levels of various proteins involved in peptidoglycan synthesis and cell division were precisely determined in cells in which Pmra::Plac promoter expression was repressed or fully induced. It was confirmed that the Pmra promoter is required for expression of the first nine genes of the mra cluster: mraZ (orfC), mraW (orfB), ftsL (mraR), ftsI, murE, murF, mraY, murD, and ftsW. Interestingly, three- to sixfold-decreased levels of MurG and MurC enzymes were observed in uninduced Pmra::Plac cells. This was correlated with an accumulation of the nucleotide precursors UDP–N-acetylglucosamine and UDP–N-acetylmuramic acid, substrates of these enzymes, and with a depletion of the pool of UDP–N-acetylmuramyl pentapeptide, resulting in decreased cell wall peptidoglycan synthesis. Moreover, the expression of ftsZ, the penultimate gene from this cluster, was significantly reduced when Pmra expression was repressed. It was concluded that the transcription of the genes located downstream from ftsW in the mra cluster, from murG to ftsZ, is also mainly (but not exclusively) dependent on the Pmra promoter.
The rigid, shape-determining material in bacterial cell walls is a giant polymer of periodic structure named peptidoglycan or murein. Its biosynthesis is a complex process involving many different cytoplasmic and membrane steps (33). Conditional-lethal mutants of Escherichia coli altered at different levels of this metabolic pathway have been described previously, and most of the mutations were mapped in several regions of the chromosome (10, 27, 36). One of them, at 2 min on the E. coli map, was studied in great detail because it contained a large cluster of genes, from mraZ to envA, that code for proteins involved in cell envelope biosynthesis and cell division. It was designated either mra for murein region A (7, 19, 27) or dcw for division and cell wall (1, 9, 34, 35). Through earlier work by our and other laboratories, the complete physical map and DNA sequence of the whole 17-kb region were determined and the function of most of the genes present in this cluster were identified. However, with the exception of the cell division genes ftsQ, ftsA, and ftsZ, whose transcription had been investigated in great detail (4, 9, 11, 29, 31, 34, 35, 37), the crucial question of how the genes from this large mra cluster are transcribed was still open. A promoter for the ftsI gene (also named pbpB), which encodes penicillin-binding protein 3 (PBP3) (7, 19, 28), and the three genes upstream of it was recently identified (13). Interestingly, the inactivation of this promoter (Pmra) on the chromosome and its replacement by an inducible promoter (Plac) led to isopropyl-β-d-thiogalactopyranoside (IPTG)-dependent cells that could grow in the absence of inducer only when a plasmid carrying at least the mraZ-ftsW region was present (13). It was thus concluded that the Pmra promoter was essential for expression of the first nine genes of the mra cluster. In fact, we report here that uninduced Pmra::Plac cells were significantly depleted of the products of the different genes located downstream from ftsW, indicating that the main proportion of the transcription of these genes (from murG to ftsZ) derives from the Pmra promoter.
MATERIALS AND METHODS
Materials.
Acetyl coenzyme A (acetyl-CoA), glucosamine-1-phosphate (GlcN-1-P), UTP, and UDP–N-acetylglucosamine (UDP-GlcNAc) were bought from Sigma. Peptidoglycan nucleotide precursors and the dipeptide d-Ala–d-Ala were prepared as described previously (20–24). UDP-[14C]GlcNAc (7.4 GBq · mmol−1), [3H]UMP (0.5 TBq · mmol−1), and l-[14C]alanine (5.6 GBq · mmol−1) were purchased from Amersham; [14C]acetyl-CoA (1.9 GBq · mmol−1) was from ICN; d-[14C]glutamic acid (1.9 GBq · mmol−1) was obtained from American Radiolabeled Chemicals (St. Louis, Mo.); and [meso-14C]diaminopimelic acid ([meso-14C]A2pm; 11.5 GBq · mmol−1) was from CEA (Saclay, France).
Bacterial strains and plasmids.
JE7968 (Pmra::Plac) and JE7970 (Pmra::Plac recA1) are derivatives of W3110 and carry the Plac cassette inserted into the HindIII site within Pmra on the chromosome (13). Thus, they are dependent on a lac inducer for growth. The Plac cassette is composed of the cat gene followed by two transcriptional terminators of the rrnB operon, the lacIq gene in the orientation opposite of and Plac in the same orientation as Pmra. MC1061-5 (ΔlacX74) (17) was used as a host in the lacZ operon fusion experiment. pHR416 is a plasmid derived from pSY396 which carries the entire mra cluster (21-kb AatII fragment) (13). pHR477, pHR478, pHR479, pHR431, pHR439, pHR427, and pHR426 are mini-F plasmids carrying the chromosomal fragment from Pmra to mraY, murD, ftsW, murG, ftsQ, ftsA, and ftsZ, respectively. pHR485 carries ftsW under the control of the aadA promoter in the vector pGB2 (13). The mini-F plasmids used in the operon fusion experiments are depicted in Fig. 1. The vector was pFZY1ΔH (13), a derivative of pFZY1 (17).
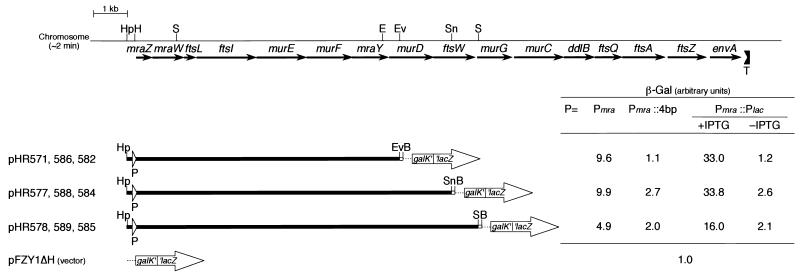
FIG. 1.
lacZ operon fusion experiments on the Pmra-murG region of the mra cluster. The orientation and approximate sizes of the mra cluster genes (black arrows) and a rho-independent terminator (a bracket with a T beneath it) are shown at the top. Below, filled bars denote the chromosomal fragments cloned into the promoter assay mini-F vector pFZY1ΔH. Their left ends, cut by HpaI, were joined to the filled EcoRI site of the vector. Short open bars denote an SmaI-BamHI part of the Ω interposon polylinker (8). Open arrowheads with P’s beneath them represent either Pmra, disrupted Pmra (Pmra::4 bp), or Pmra replaced by Plac (Pmra::Plac). The plasmid numbers at the left are for those with Pmra, Pmra::4 bp, and Pmra::Plac, respectively. β-Gal activities are averages of measurements from three transformants of MC1061-5 grown in buffered L broth-glucose-thymine medium (13) containing ampicillin, with (+) or without (−) IPTG at a concentration of 1 mM. The figure is drawn to scale except for the galK′-′lacZ fusion gene (large white arrows) and its short upstream region, including the polylinker (dotted line) of the vector (13, 17). Abbreviations for restriction sites (only relevant sites are shown): B, BamHI; E, EcoRI; Ev, EcoRV; H, HindIII; S, SmaI; Sn, SnaBI.
Recombinant DNA procedures.
These methods were essentially based on those of Sambrook et al. (30). The β-galactosidase (β-Gal) assay and the unit definition used for it were as described by Koop et al. (17).
Growth conditions.
Unless otherwise noted, 2YT medium (26) was used for growing cells. Cell growth was monitored at 600 nm with a spectrophotometer (model 240; Gilford Instrument Laboratories, Inc., Oberlin, Ohio). For strains carrying drug resistance genes, antibiotics were used at the following concentrations (in micrograms per milliliter): ampicillin, 100; kanamycin, 30; spectinomycin, 30; tetracycline, 12.5; and chloramphenicol, 10.
Preparation of crude enzyme.
Exponential-phase cells (0.5-liter cultures) of the different strains listed in Table 1 were grown at 37°C in 2YT medium, in the presence or absence of 1 mM IPTG. Strains requiring IPTG for growth were first grown in its presence, and the cultures were then diluted about 100-fold into prewarmed medium lacking the inducer. The first effects of the depletion of IPTG on cell morphology (loss of rod shape) and cell growth (arrest of growth followed by the onset of cell lysis) were observed after a time period that depended on the strain being used: 2 h for JE7968 and JE7970 and more than 3 h for JE7970(pHR477/pHR485). The 1-h delay observed with the latter strain was due to the fact that only one biosynthetic activity (MurD) was depleted in that strain on IPTG deprivation. At that time (the final optical density of the culture was approximately 0.7), the cells were harvested and washed with 40 ml of cold 20 mM potassium phosphate buffer (pH 7) containing 0.3 mM MgCl2 and 1 mM β-mercaptoethanol. The wet cell pellet was suspended in 5 ml of the same buffer and disrupted by sonication (Sonicator 150; T. S. Ultrasons, Annemasse, France) for 5 min with cooling. The resulting suspension was centrifuged at 4°C for 30 min at 200,000 × g with a Beckman TL100 centrifuge. The supernatant was dialyzed overnight at 4°C against 100 volumes of the same phosphate buffer, and the resulting solution (40 to 50 mg of protein per 5 ml), designated as crude soluble enzyme, was stored at −20°C. The pellet, consisting of membrane proteins, was resuspended in 1 ml of the same buffer. Protein concentrations were determined by the method of Lowry et al. (18), using bovine serum albumin as a standard.
TABLE 1.
Specific activities of peptidoglycan-synthesizing enzymes in E. coli strainsa
| Strain | IPTG | Relative amt
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| MurE | MurF | MraY | MurD | MurG | MurC | Ddl | GlmU | ||
| W3110 | − | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| JE7968 | − | 0.25 | 0.2 | 0.35 | 0.25 | 0.4 | 0.3 | 0.7 | 1.0 |
| + | 3.0 | 2.0 | 1.8 | 2.1 | 2.0 | 3.4 | 1.3 | 1.0 | |
| JE7970 | − | 0.20 | 0.3 | 0.4 | 0.3 | 0.4 | 0.4 | 0.7 | 1.0 |
| + | 2.5 | 2.1 | 2.2 | 2.1 | 2.2 | 3.5 | 1.2 | 1.0 | |
| JE7970(pHR479) | − | 1.0 | 1.1 | 1.0 | 1.2 | 0.3 | 0.15 | 0.8 | 1.0 |
| + | 2.5 | 2.2 | 1.5 | 2.7 | 1.5 | 3.2 | 1.2 | 1.0 | |
| JE7970(pHR477/pHR485) | − | 1.0 | 1.2 | 0.9 | 0.05 | 0.4 | 0.4 | 0.8 | 1.0 |
| + | 2.5 | 2.2 | 1.5 | 2.0 | 2.0 | 3.2 | 1.3 | 1.0 | |
| JE7970(pHR439) | − | 1.0 | 1.1 | 1.1 | 1.0 | 1.1 | 1.1 | 1.1 | 1.0 |
Cells were grown exponentially at 37°C in 2YT medium supplemented (+) or not supplemented (−) with 1 mM IPTG. IPTG-requiring strains were first grown in the presence of the inducer and then diluted into prewarmed medium without IPTG. Growth was then continued until the first effects on cell growth were observed, from 2 to 3 h later, depending on the strain used (see the text). Crude extracts (soluble and membrane fractions) were prepared and analyzed for the different enzyme activities. A value of 1 for the wild-type strain W3110 corresponds (in nanomoles · minute−1 · milligram of protein−1) to 1.5 for MurE, 3 for MurF, 0.6 for MurD, 0.04 for MraY, 0.02 for MurG, 0.06 for MurC, 0.8 for Ddl, and 103 for GlmU, respectively. The specific activity of GlmU, an enzyme involved in early steps of peptidoglycan synthesis but encoded by a gene from a separate chromosomal region (25), was used here as an internal standard to control the absence of variability among the extracts.
Enzymatic assays. (i) GlcN-1-P acetyltransferase (GlmU).
The two-step formation of UDP-GlcNAc from GlcN-1-P, catalyzed by the bifunctional enzyme GlcN-1-P acetyltransferase (GlmU), in a standard assay mixture containing 50 mM Tris-HCl buffer (pH 8.0), 2 mM UTP, 3 mM MgCl2, 0.5 mM [14C]acetyl-CoA (700 Bq), 2 mM GlcN-1-P, and enzyme (10 μg of protein) in a final volume of 100 μl was monitored.
(ii) l-Alanine-adding enzyme (MurC).
The formation of UDP–N-acetylmuramyl (MurNAc)–l-Ala in a standard assay mixture containing 100 mM Tris-HCl buffer (pH 8.6), 5 mM ATP, 20 mM MgCl2, 2 μM l-[14C]alanine (2 KBq), 1 mM UDP-MurNAc, and enzyme (250 μg of protein) in a final volume of 50 μl was monitored.
(iii) d-Glutamic acid-adding enzyme (MurD).
The formation of UDP– MurNAc–l-Ala–d-Glu in a standard assay mixture containing 100 mM Tris-HCl buffer (pH 8.6), 5 mM ATP, 5 mM MgCl2, 25 μM d-[14C]glutamic acid (500 Bq), 25 μM UDP–MurNAc–l-alanine, and enzyme (5 μg of protein) in a final volume of 50 μl was monitored.
(iv) meso-A2pm-adding enzyme (MurE).
The formation of UDP-MurNAc tripeptide in a standard assay mixture containing 100 mM Tris-HCl buffer (pH 8.6), 5 mM ATP, 100 mM MgCl2, 0.1 mM [meso-14C]A2pm (500 Bq), 0.2 mM UDP–MurNAc–l-Ala–d-Glu, and enzyme (50 μg of protein) in a final volume of 75 μl was monitored.
(v) d-Alanyl–d-alanine-adding enzyme (MurF).
The formation of UDP- MurNAc pentapeptide in a standard assay mixture containing 100 mM Tris-HCl buffer (pH 8.6), 5 mM ATP, 100 mM MgCl2, 70 μM d-[14C]Ala–d-Ala (500 Bq), 70 μM UDP–MurNAc–l-Ala–γ-d-Glu–meso-A2pm, and enzyme (20 μg of protein) in a final volume of 100 μl was monitored.
(vi) d-Alanine:d-alanine ligase (Ddl).
The standard assay mixture contained 50 mM Tris-HCl buffer (pH 8.6), 5 mM ATP, 20 mM MgCl2, 50 μM d-[14C]Ala (1 KBq), 0.12 mM UDP–MurNAc–l-Ala–γ-d-Glu–meso-A2pm, and enzyme (20 μg of protein) in a final volume of 50 μl. Because the reaction product, d-Ala–d-Ala, inhibits the activity of d-alanine:d-alanine ligase (Ddl) (20), it was quantitatively converted to UDP-MurNAc pentapeptide by coupling the Ddl activity to that of the MurF present in the extract.
(vii) Phospho-MurNAc pentapeptide translocase (MraY).
The reaction for the substitution of [3H]UMP for the UMP moiety of UDP-MurNAc pentapeptide was used as an assay for translocase activity in a 20-μl reaction mixture consisting of 50 mM Tris-HCl buffer (pH 7.5), 12.5 mM MgCl2, 13 μM [3H]UMP (500 Bq), 0.16 mM UDP-MurNAc pentapeptide, and membranes (60 μg of protein).
In all cases (assays i to vii), reaction mixtures were incubated at 37°C for 30 min, reactions were terminated by the addition of 10 μl of acetic acid, and reaction products were separated by high-voltage electrophoresis on Whatman 3MM filter paper in 2% formic acid (pH 1.9) for 1 to 1.5 h at 40 V/cm, using an LT36 apparatus (Savant Instruments, Hicksville, N.Y.). The radioactive spots were located by overnight autoradiography with type R2 films (3M, St. Paul, Minn.) or with a radioactivity scanner (Multi-Tracermaster model LB285; EG&G Wallac/Berthold, Evry, France). The radioactive spots were cut out, and the radioactivity in each was counted in a Betamatic IV liquid scintillation spectrophotometer (Kontron Instruments) with a solvent system consisting of 2 ml of water and 13 ml of Aqualyte mixture (J. T. Baker Chemicals, Deventer, The Netherlands).
(viii) N-Acetylglucosaminyltransferase (MurG).
The standard reaction mixture contained, in a final volume of 25 μl, 100 mM Tris-HCl buffer (pH 7.5), 40 mM MgCl2, 30 mM ATP, 0.7 mM UDP-MurNAc pentapeptide, 2 μM UDP-[14C]GlcNAc (1 KBq), and membranes (150 μg of protein). Membranes were incubated first with UDP-MurNAc pentapeptide for 10 min at 35°C to generate undecaprenyl-pyrophosphoryl MurNAc pentapeptide (via MraY) before addition at time zero of the radioactive substrate and a 10-min incubation at 35°C. The reaction was stopped by placing tubes in a boiling-water bath for 2 min, and the reaction mixtures were analyzed by descending chromatography for 16 h on Whatman 1 filter paper in isobutyric acid–1 M NH4OH (5:3, vol/vol). Spots corresponding to products (peptidoglycan and lipid intermediate) and remaining UDP-GlcNAc substrate were detected and their radioactivity was counted as described above.
Pool levels of peptidoglycan precursors.
Cells of W3110 and JE7970 derivatives (1-liter cultures) were grown exponentially at 37°C in 2YT medium in the absence or presence of IPTG. When the optical density of the cultures reached 0.7, the cells were rapidly chilled to 0°C and harvested in the cold. The extraction of peptidoglycan nucleotide precursors with boiling water and cold trichloroacetic acid and the analytical procedure used for their quantitation were as previously described (20, 21).
Isolation of sacculi and quantitation of peptidoglycan.
Exponential-phase cells (0.5-liter cultures) of W3110 or JE7970 derivatives were grown as described above, in the absence or presence of IPTG. Harvested cells were washed with a cold 0.85% NaCl solution and centrifuged again. Bacteria were then rapidly resuspended with vigorous stirring in 20 ml of a hot (95 to 100°C) aqueous 4% sodium dodecyl sulfate solution for 30 min. After standing overnight at room temperature, the suspensions were centrifuged for 30 min at 200,000 × g and the pellets were washed several times with water. The final suspensions, made in 2-ml volumes of water, were homogenized by brief sonication. Aliquots were hydrolyzed and analyzed as previously described (24), and the peptidoglycan content of the sacculi was expressed in terms of its muramic acid content.
RESULTS
Levels of peptidoglycan-synthesizing enzymes in Pmra::Plac cells.
To examine how far into the mra cluster the function of the Pmra promoter was required, we recently disrupted this promoter on the chromosome and replaced it with the inducible lac promoter (13). An IPTG-dependent strain, JE7968, resulted from this construction; this strain was shown to lyse when deprived of lac inducer during exponential growth, most probably because expression of one or more of the genes involved in peptidoglycan synthesis was repressed. To control this point, the levels of the enzymes encoded by the different genes of the mra cluster were determined in cells of strain JE7968 that were either grown continuously in the presence of 1 mM IPTG or depleted of lac inducer for approximately 2 h (until the first effects on cell growth were observed). In most cases (for MurE, MurF, MraY, MurD, MurG, and MurC), the levels of enzymes observed in induced JE7968 cells were two- to threefold higher than those of the parental W3110 cells (Table 1). One exception concerned Ddl, whose activity was increased by only 20 to 30%. As expected, inverse variations were observed in IPTG-depleted cells, which contained three- to fivefold less of the different enzymes mentioned above than the wild-type strain W3110, with the same exception of Ddl, which was decreased by only 30% (Table 1). The concomitant and similar variations of all these enzymes clearly indicated that transcription of the corresponding genes was mainly dependent on the Pmra promoter.
As reported previously, the IPTG requirement of strain JE7968 was complemented only if the chromosomal fragment extending from Pmra to at least ftsW (inclusive) was provided in trans on a plasmid (13). Some other constructs were made in which expression of only one of these genes was impaired when IPTG was absent—for instance, ftsW in strain JE7970(pHR478) and murD in strain JE7970(pHR477/pHR485). Each of these strains required IPTG for growth and filamented or lysed when deprived of lac inducer, depending on the function of the gene product in either cell division or peptidoglycan synthesis. For instance, here we observed that cells of JE7970(pHR477/pHR485) stopped growing and lysed after about 3 h of exponential growth in the absence of IPTG. An analysis of the cell content at that time was consistent with a defective expression of d-glutamic acid-adding enzyme (MurD): a 20-fold-reduced level of this enzyme was detected (Table 1), large amounts of UDP–MurNAc–l-Ala (the nucleotide substrate of MurD) were accumulated, and all precursors located downstream in the pathway were depleted. This resulted in a 40% lower peptidoglycan content, which was probably the minimum value compatible with cell viability (data not shown).
Cells of JE7968 or JE7970 (a recA derivative of the former) became IPTG independent for growth when transformed with a plasmid (pHR479) carrying as a minimal complementing fragment the Pmra-ftsW region. However, the growth rate of this strain in the absence of IPTG was relatively low compared to that of the wild-type strain or to its own rate in the presence of IPTG (generation times at 37°C were 70, 35, and 39 min, respectively). As expected, JE7970(pHR479) cells contained wild-type levels of MurE, MurF, MraY, and MurD enzymes but three- and sixfold-reduced levels of MurG and MurC activities, respectively (Table 1). Since the cells were grown continuously in the absence of IPTG, these values represented the production of these enzymes under conditions in which expression from the Pmra promoter was maximally repressed. The cells were still viable under these conditions, implying that MurG and MurC activities were somewhat in excess in a wild-type strain and that this residual activity was enough to sustain peptidoglycan synthesis, at least at a rate sufficient for cell integrity. Further analyses showed that these cells accumulated UDP-GlcNAc and UDP-MurNAc, the nucleotide substrates of the MurG and MurC enzymes, respectively (Table 2). This finding confirmed the partial depletion of these enzymes from the cell content and indicated a significant reduction of the flow of metabolites in the pathway for peptidoglycan synthesis at both enzymatic steps. Consequently, the pool of UDP–N-acetylmuramyl pentapeptide, the end product of the cytoplasmic steps, was decreased twofold and the cell peptidoglycan content was decreased by 30% (Table 2). It was previously established that the peptidoglycan content of E. coli cells could be reduced by up to 40 to 50% without loss of cell integrity (reference 25 and references therein). Table 2 shows that the pools of precursors and the peptidoglycan content of strain JE7970(pHR439), which carries murG and murC in addition on the plasmid, were normal. The same results were observed when strain JE7970(pHR479) was grown in the presence of IPTG. This finding confirmed that the variations described above were clearly correlated with the partial depletion of the MurG and MurC enzymes from the cell content. At first approximation, the reduced growth rate could be attributed to the depletion of both enzymes, since the same strain carrying in addition murG and murC on the plasmid, JE7970(pHR439), grew faster, with a generation time of 50 min.
TABLE 2.
Abnormal peptidoglycan content and pools of peptidoglycan precursors in uninduced JE7970(pHR479) cellsa
| Strain | IPTG | Amt (nmol/liter of culture) of:
|
|||
|---|---|---|---|---|---|
| UDP-GlcNAc | UDP-MurNAc | UDP-MurNAc pentapeptide | Peptido- glycan | ||
| W3110 | − | 70 | 25 | 230 | 2,600 |
| JE7970(pHR479) | − | 240 | 150 | 90 | 1,900 |
| + | 75 | 30 | 190 | 3,000 | |
| JE7970(pHR439) | − | 75 | 30 | 230 | 2,500 |
Cells were grown exponentially at 37°C in 2YT medium in the absence (−) or presence (+) of 1 mM IPTG. In all cases, cells were harvested at an optical density of 0.7, corresponding to approximately 230 mg of bacterial cell (dry weight) per liter of culture. Cell peptidoglycan was extracted and quantitated in terms of its muramic acid content (20, 24). Nucleotide precursors were extracted, purified, and quantitated as previously described (20, 21).
All of these results taken together suggested, first, that transcription of each gene located within the Pmra-ftsW region was under the control of the sole Pmra promoter and, second, that transcription of genes distal to ftsW originated not only from Pmra but also from another promoter(s). The finding that 70 to 80% of the MurG and MurC activities was lost on repression of Plac in Pmra::Plac cells further indicated that Pmra was not absolutely required for, but governed the main part of, the transcription of the corresponding genes.
A promoter other than Pmra contributes to the expression of the murG and murC genes.
To examine the contribution of Pmra to the expression of genes in the mra cluster, we also made fusions between the proximal region of the mra cluster and the promoterless galK′-′lacZ gene on a promoter assay mini-F vector, pFZY1ΔH (Fig. 1). The cloned DNA was from the HpaI site just upstream of Pmra, to the EcoRV site toward the 5′ end of murD (pHR571), to the SnaBI site around the middle of ftsW (pHR577), and to the SmaI site toward the 5′ end of murG (pHR578). Pmra was then disrupted at the HindIII site within it by the filling-in reaction of T4 DNA polymerase (Pmra::4bp; pHR586, pHR588, and pHR589 respectively), which has been shown to completely abolish the Pmra activity (13), or displaced by insertion of the Plac cassette into the HindIII site (Pmra::Plac; pHR582, pHR584, and pHR585 respectively).
β-Gal assays, in the absence of functional Pmra (Pmra::4bp) or when the Pmra::Plac activity was repressed, indicated that the level of transcription detectable at the EcoRV site was very low compared to the background β-Gal activity measured for the pFZY1ΔH vector (Fig. 1). Interestingly, Boyle et al. recently localized a promoter for the ftsW gene between the EcoRI and BglII sites in the mraY-murD intergenic region (3). Since this region is just upstream from the EcoRV site (Fig. 1), a contribution of this promoter to the low-level transcription detected here is likely. At the SnaBI and SmaI sites, on the other hand, a small but significant amount of transcription was detected. There is likely to be a promoter(s), somewhere between the EcoRV site within murD and the SnaBI site within ftsW, which is responsible for the residual expression of murG and downstream genes observed in uninduced Pmra::Plac cells.
In the presence of a functional Pmra on the operon fusion plasmids, a higher β-Gal activity was detected at the SmaI site within murG (Fig. 1). The transcription originating from Pmra seems to proceed to murG and probably beyond, although it is not necessarily required for the growth and division of the cell. This is consistent with the increase in MurG, MurC, Ddl, and other enzymatic activities observed in induced JE7970 cells. The activity of Pmra was not as high as that of Plac fully induced with 1 mM IPTG.
The finding that the β-Gal activity detected at the SmaI site, in the presence of functional Pmra or when the Pmra::Plac activity was induced, was about half of that at the EcoRV and SnaBI sites was intriguing. Transcriptional attenuation or differential degradation of mRNA may be occurring.
Relative amounts of MraW, PBP3, and FtsZ in E. coli mutant strains.
We also characterized the expression of the genes encoding MraW, PBP3, and FtsZ in strain JE7970 after depletion of IPTG inducer for 2 h, just before lysis of the cells, by measuring the relative amounts of the proteins detected with specific antibodies by an immunoblotting assay. Increases in the levels of the three proteins, compared with their levels in the parental strain, W3110, were found under induction conditions, and decreases were observed after depletion of the inducer (Table 3). Note that in this case the amount of protein, and not the enzymatic activity, was measured. About a twofold increase was found for MraW and a sevenfold increase was evident for PBP3, suggesting some specific regulation of these two genes at the level of transcription and/or translation. FtsZ also showed a small (1.5-fold) increase in the presence of inducer. Levels of the three gene products under depletion conditions and after induction with IPTG were in agreement with the results obtained with the mur gene products and suggested that transcription from Pmra can proceed up to ftsZ.
TABLE 3.
Relative amounts of MraW, PBP3, and FtsZ in E. coli mutant strainsa
| Strain | IPTG | Relative amt ± SD of:
|
||
|---|---|---|---|---|
| MraW | PBP3 | FtsZ | ||
| W3110 | − | 100 | 100 | 100 |
| + | 106 ± 13 | 88 ± 9 | 109 ± 16 | |
| JE7970 | − | 71 ± 11 | 49 ± 3 | 70 ± 12 |
| + | 207 ± 23 | 660 ± 192 | 148 ± 11 | |
| JE7970(pHR431) | − | 114 ± 20 | 334 ± 36 | 72 ± 8 |
| JE7970(pHR439) | − | 88 ± 2 | 361 ± 28 | 59 ± 10 |
| JE7970(pHR427) | − | 141 ± 22 | 75 ± 3 | 209 ± 13 |
| JE7970(pHR426) | − | 132 ± 15 | 70 ± 5 | 217 ± 16 |
| JE7970(pHR416) | − | 117 ± 19 | 71 ± 13 | 208 ± 31 |
Cells were grown exponentially at 37°C in 2YT medium supplemented (+) or not supplemented (−) with 1 mM IPTG. Crude extracts (total for MraW and FtsZ and membrane fractions for PBP3) were prepared by differential centrifugation after disrupting cells with a French press. Proteins were fractionated in a Tricine–sodium dodecyl sulfate–8% acrylamide gel and detected by immunoblotting with specific antibodies and by the chemiluminiscence method. The absorbances of the corresponding bands in the film from at least three different experiments were measured and are expressed here relative to the amount of protein detected in parental strain W3110.
Two revertants of strains JE7967 and JE7970 that were still chloramphenicol resistant but became IPTG independent were isolated by simply growing cells in the absence of IPTG until stationary phase and plating them on 2YT-chloramphenicol medium. Expression of the three genes in these strains also became IPTG independent, and the levels of the proteins were quite similar in the absence or presence of the inducer (data not shown). This result indicated that the defect in the revertant strains could be some modification of the lac promoter and/or operator that did not allow the correct binding or expression of LacI. Whatever the changes in these strains, they confirmed that expression from the Pmra::Plac promoter can proceed up to ftsZ.
Expression of the three gene products was also studied in the Pmra::Plac strain carrying plasmids with different fragments of the mra cluster. The results are presented in Table 3. MraW was expressed at more or less the same level in all strains. Only a small increase in expression (15 to 30%) was found in extracts of JE7970(pHR431), JE7970(pHR427), JE7970(pHR426), and JE7970(pHR416) cells, and a small decrease in expression (less than 10%) was evident in JE7970(pHR439) that was difficult to correlate with the presence of plasmids. However, the expression of PBP3 and FtsZ showed opposite variations in these strains. In strains JE7970(pHR431) and JE7970(pHR439), PBP3 was increased by three- to fourfold and FtsZ was decreased by 30 to 40%. Inversely, in strains JE7970(pHR427), JE7970(pHR426), and JE7970(pHR416), FtsZ was increased twofold and the level of PBP3 was reduced by 25 to 30%. The main difference among the plasmids is the presence or absence of an ftsA-encoding fragment in which the main ftsZ promoters were previously identified (4, 9, 29, 37).
DISCUSSION
Previous data on the transcription of genes from the mra cluster mainly concerned the distal part, including cell division genes ftsQ, ftsA, and ftsZ as well as envA (2, 4, 9, 11, 29, 32, 37). How the mur genes from the proximal region are expressed was not known until the recent demonstration that the promoter Pmra, originally identified as being required for the expression of ftsI, also directs expression of the five downstream genes (13). Earlier we suggested the existence of several promoters in the ftsI-murC region (22), based on the observation that multicopy plasmids (pUC18 derivatives) with individual genes cloned in the orientation opposite that of the vector promoter fully complemented the specific defects of thermosensitive mutant strains and allowed significant overproduction of the corresponding enzymes. This was observed in particular with the murE, murD, murG, and murC genes. One exception concerned murF, whose expression apparently required the whole sequence of the upstream gene murE, suggesting that cotranscription may occur at least in that case (22). In the other cases, it was conceivable that a cumulative effect of very weak promoters at high copy numbers or transcriptional readthrough from another promoter on the plasmid vector resulted in sufficient expression and functional complementation. In the present work, expression of all of these genes was shown to be mainly (or completely) dependent on the Pmra promoter. In particular, we here provided evidence that repression of the Pmra promoter resulted in a dramatic depletion of the murD gene product, as well as the arrest of peptidoglycan synthesis and attendant cell lysis. This suggested that expression of the chromosomal murD gene was exclusively dependent on this promoter and, consequently, that putative promoters from the region 5′ to murD were not functional in vivo when present in only one copy. Strains defective in the product of only the ftsI or the ftsW gene were also previously constructed (13). In each case, depletion of the lac inducer resulted in a cell division defect, which was corrected by the addition of a plasmid carrying the defective gene. Strains with conditionally defective expression of the murE, murF, or mraY gene have not been constructed, but more likely the phenomena observed with these strains had confirmed that these genes are essential and are exclusively expressed from the Pmra promoter.
We confirmed that the Pmra promoter was not absolutely required for expression of genes distal to ftsW in the mra cluster. However, it was clear that a major proportion of the transcription of these genes also originated from this promoter, at least for the murG and murC genes, whose expression was reduced by three- and sixfold, respectively, on repression of Pmra. The residual expression of these gene products suggested the presence of another efficient promoter. As shown by the lacZ fusion experiments described in this work, this promoter might be somewhere between the EcoRV site lying within murD and the SnaBI site within ftsW. Since the ftsW gene is essentially dependent on the Pmra promoter for expression, this other promoter most probably resides within the sequence of ftsW, and future work will be devoted to its identification. It was noteworthy that all of the genes of the mra cluster are tightly packed together and that almost 100% of the DNA in this chromosomal region (EMBL entry EC2MIN) is coding. In particular, the murG gene, coding for the N-acetylglucosaminyltransferase, was shown to overlap the preceding ftsW gene by 4 bases (15, 23). Since the next gene in the cluster, murC, was found to be separated from murG by 100 bases (14), it was tempting to consider that the murG gene is also cotranscribed exclusively from Pmra, together with the nine preceding genes. In that case, it should be assumed that the residual transcription which occurred on IPTG deprivation in the Pmra::Plac strain was sufficient to provide enough MurG molecules to support normal cell growth. However, as discussed previously (13), the basal level of expression of genes depending on the Pmra promoter was unlikely to be sufficient in the absence of IPTG, considering the presence of lacIq in the strain and the addition of glucose in the growth medium used, both of which lead to a maximal repression of this promoter. The fact that JE7970(pHR479) cells contained three- to sixfold-lower levels of MurG and MurC enzymes was more consistent with another promoter contributing to their expression.
Under conditions of IPTG induction from the Pmra::Plac promoter in strain JE7970, levels of MraW and PBP3 proteins increased by largely different factors, two- and sevenfold, respectively. This effect was observed only with these two proteins. The simple explanation that a greater stability of PBP3 accounts for this differential expression is unlikely since the turnover of PBP3 had been shown to be very high (unpublished observation). Then, these data suggest some specific regulation of the expression of the two genes at the level of transcription and/or translation. In this sense, weak promoters were identified for each of the four first genes of the mra cluster (12), but their involvement in the differential expression of these two proteins remains to be established.
We here observed that the specific activity of Ddl was decreased by 30% in Pmra::Plac cells deprived of IPTG and, inversely, was increased by 30% in induced cells. These levels of variation were very low compared to those of enzymes encoded by other genes of the mra cluster. This showed that expression of ddlB was, at least in part, dependent on the Pmra promoter, as demonstrated for genes murG and murC. However, it should be noted that two genes, ddlA and ddlB, for two ligases with almost identical kinetic properties were identified in E. coli (38), both theoretically contributing to the Ddl activity determined in cell extracts. Since the ddlA gene belongs to a completely separate chromosomal region, the variations observed here more likely represented the specific contribution of the DdlB enzyme to the total activity. Since a null mutation in either ddlA or ddlB had no apparent effect on cell growth or morphology, at least under laboratory growth conditions, the reason for this duplication remains unclear.
The levels of FtsZ, the essential cell division protein encoded by the penultimate gene of this cluster, also varied on repression or induction of the Pmra promoter. These variations paralleled those of the mur gene products, but their levels were comparatively lower (Table 3). This suggested that a portion of the transcription of the last genes in the cluster also derived from the Pmra promoter. Dai and Lutkenhaus (4) previously reported that a strain with a null allele of ftsZ on the chromosome could not be complemented by a lambda phage (λ16-2) carrying 6 kb of DNA upstream of ftsZ, including the ftsA and ddlB genes, in which some of the promoters of ftsZ have been identified (9, 29, 37). This suggested the involvement of a far-upstream promoter, whose contribution to ftsZ expression was estimated at about 30 to 40% (4). Pmra participates in but is apparently not essential for ftsZ expression, since the Pmra::Plac strain carrying only the region up to ftsW on a plasmid grew well in the absence of IPTG. The 5′ end of the insert present in λ16-2 that contained ftsZ but failed to complement a null allele of ftsZ was in the middle of the ftsW gene (3, 15). When considering data obtained with the lacZ fusions, it could be assumed that such a promoter, responsible for at least a portion of the expression of genes from murG to ftsZ, was present upstream from the SnaBI site in the sequence of ftsW. As shown above, the levels of PBP3 and FtsZ proteins showed opposite variations in the Pmra::Plac strain carrying plasmids with different fragments of the mra cluster. The main difference among the plasmids was the presence or absence of a fragment carrying ftsA and the main promoters of ftsZ. Then, we propose that expression of FtsA in the absence of the Pmra promoter elicits a regulatory mechanism that involves repression of the specific promoter for PBP3 by FtsA. In this regard, it was previously shown that amplification of a fragment corresponding to the first genes of the cluster modified the plating capacity of a thermosensitive ftsA mutant at the restrictive temperature (16) and also that elevated levels of FtsA protein blocked cell division at some early stage (5, 6). An imbalance between FtsA and FtsZ may similarly affect the expression of the first genes of the cluster, including ftsI.
ACKNOWLEDGMENTS
This work was supported by a grant from the Centre National de la Recherche Scientifique (URA 1131) and a grant “Biotechnologies” from the Ministère de l’Education Nationale de la Recherche et de la Technologie (no. 97.C.0177), France. Work done in J.A.’s laboratory was supported by grant BI094-0789 from the Comision Interministerial de Ciencia y Tecnologia, Spain. J.A. acknowledges the institutional help of the Fundación Ramón Areces to the Centro de Biología Molecular Severo Ochoa.
We thank Kensuke Horiuchi for encouragement and discussions.
REFERENCES
- 1.Ayala J A, Garrido T, de Pedro M A, Vicente M. Molecular biology of bacterial septation. New Compr Biochem. 1994;27:73–101. [Google Scholar]
- 2.Beall B, Lutkenhaus J. Sequence analysis, transcriptional organization, and insertional mutagenesis of the envA gene of Escherichia coli. J Bacteriol. 1987;169:5408–5415. doi: 10.1128/jb.169.12.5408-5415.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyle D V, Khattar M M, Addinall S G, Lutkenhaus J, Donachie W D. ftsW is an essential cell-division gene in Escherichia coli. Mol Microbiol. 1997;24:1263–1273. doi: 10.1046/j.1365-2958.1997.4091773.x. [DOI] [PubMed] [Google Scholar]
- 4.Dai K, Lutkenhaus J. ftsZ is an essential cell division gene in Escherichia coli. J Bacteriol. 1991;173:3500–3506. doi: 10.1128/jb.173.11.3500-3506.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dai K, Lutkenhaus J. The proper ratio of FtsZ to FtsA is required for cell division to occur in Escherichia coli. J Bacteriol. 1992;174:6145–6151. doi: 10.1128/jb.174.19.6145-6151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dewar S J, Begg K J, Donachie W D. Inhibition of cell division initiation by an imbalance in the ratio of FtsA to FtsZ. J Bacteriol. 1992;174:6314–6316. doi: 10.1128/jb.174.19.6314-6316.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donachie W D. The cell cycle of Escherichia coli. Annu Rev Microbiol. 1993;47:199–230. doi: 10.1146/annurev.mi.47.100193.001215. [DOI] [PubMed] [Google Scholar]
- 8.Fellay R, Fray J, Krisch H. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of Gram-negative bacteria. Gene. 1987;52:147–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- 9.Flardh K, Garrido T, Vicente M. Contribution of individual promoters in the ddlB-ftsZ region to the transcription of the essential cell-division gene ftsZ in Escherichia coli. Mol Microbiol. 1997;24:927–936. doi: 10.1046/j.1365-2958.1997.4001762.x. [DOI] [PubMed] [Google Scholar]
- 10.Fletcher G, Irwin C A, Henson J M, Fillingim C, Malone M M, Walker J R. Identification of the Escherichia coli cell division gene sep and organization of the cell division-cell envelope genes in the sep-mur-ftsA-envA cluster as determined with specialized transducing lambda bacteriophages. J Bacteriol. 1978;133:91–100. doi: 10.1128/jb.133.1.91-100.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garrido T, Sanchez M, Palacios P, Aldea M, Vicente M. Transcription of ftsZ oscillates during the cell cycle of Escherichia coli. EMBO J. 1993;12:3957–3965. doi: 10.1002/j.1460-2075.1993.tb06073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gómez M J. Doctoral thesis. Madrid, Spain: Universidad Autónoma de Madrid; 1991. [Google Scholar]
- 13.Hara H, Yasuda S, Horiuchi K, Park J T. A promoter for the first nine genes of the Escherichia coli mra cluster of cell division and cell envelope biosynthesis genes, including ftsI and ftsW. J Bacteriol. 1997;179:5802–5811. doi: 10.1128/jb.179.18.5802-5811.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ikeda M, Wachi M, Jung H K, Ishino F, Matsuhashi M. Nucleotide sequence involving murG and murC in the mra gene cluster of E. coli. Nucleic Acids Res. 1990;18:4014. doi: 10.1093/nar/18.13.4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishino F, Jung H K, Ikeda M, Doi M, Wachi M, Matsuhashi M. New mutations fts-36, lts-33, and ftsW clustered in the mra region of the Escherichia coli chromosome induce thermosensitive cell growth and division. J Bacteriol. 1989;171:5523–5530. doi: 10.1128/jb.171.10.5523-5530.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jung H K, Ishino F, Matsuhashi M. Inhibition of growth of ftsQ, ftsA, and ftsZ mutant cells of Escherichia coli by amplification of a chromosomal region encompassing closely aligned cell division and cell growth genes. J Bacteriol. 1989;171:6379–6382. doi: 10.1128/jb.171.11.6379-6382.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koop A H, Hartley M E, Bourgeois S. A low-copy-number vector utilizing β-galactosidase for the analysis of gene control elements. Gene. 1987;52:245–256. doi: 10.1016/0378-1119(87)90051-5. [DOI] [PubMed] [Google Scholar]
- 18.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 19.Matsuhashi M, Wachi M, Ishino F. Machinery for cell growth and division: penicillin-binding proteins and other proteins. Res Microbiol. 1990;141:89–103. doi: 10.1016/0923-2508(90)90101-u. [DOI] [PubMed] [Google Scholar]
- 20.Mengin-Lecreulx D, Flouret B, van Heijenoort J. Cytoplasmic steps of peptidoglycan synthesis in Escherichia coli. J Bacteriol. 1982;151:1109–1117. doi: 10.1128/jb.151.3.1109-1117.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mengin-Lecreulx D, Flouret B, van Heijenoort J. Pool levels of UDP-N-acetylglucosamine and UDP-N-acetylglucosamine-enolpyruvate in Escherichia coli and correlation with peptidoglycan synthesis. J Bacteriol. 1983;154:1284–1290. doi: 10.1128/jb.154.3.1284-1290.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mengin-Lecreulx D, Parquet C, Desviat L R, Plá J, Flouret B, Ayala J A, van Heijenoort J. Organization of the murE-murG region of Escherichia coli: identification of the murD gene encoding the d-glutamic-acid-adding enzyme. J Bacteriol. 1989;171:6126–6134. doi: 10.1128/jb.171.11.6126-6134.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mengin-Lecreulx D, Texier L, Rousseau M, van Heijenoort J. The murG gene of Escherichia coli codes for the UDP-N-acetylglucosamine:N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase involved in the membrane steps of peptidoglycan synthesis. J Bacteriol. 1991;173:4625–4636. doi: 10.1128/jb.173.15.4625-4636.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mengin-Lecreulx D, van Heijenoort J. Effect of growth conditions on peptidoglycan content and cytoplasmic steps of its biosynthesis in Escherichia coli. J Bacteriol. 1985;163:208–212. doi: 10.1128/jb.163.1.208-212.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mengin-Lecreulx D, van Heijenoort J. Identification of the glmU gene encoding N-acetylglucosamine-1-phosphate uridyltransferase in Escherichia coli. J Bacteriol. 1993;175:6150–6157. doi: 10.1128/jb.175.19.6150-6157.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller J H. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 27.Miyakawa T, Matsuzawa H, Matsuhashi M, Sugino Y. Cell wall peptidoglycan mutants of Escherichia coli K-12: existence of two clusters of genes, mra and mrb, for cell wall peptidoglycan biosynthesis. J Bacteriol. 1972;112:950–958. doi: 10.1128/jb.112.2.950-958.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakamura M, Maruyama I N, Soma M, Kato J, Suzuki H, Hirota Y. On the process of cellular division in Escherichia coli: nucleotide sequence of the gene for penicillin-binding protein 3. Mol Gen Genet. 1983;191:1–9. doi: 10.1007/BF00330881. [DOI] [PubMed] [Google Scholar]
- 29.Robin A, Joseleau-Petit D, D’Ari R. Transcription of the ftsZ gene and cell division in Escherichia coli. J Bacteriol. 1990;172:1392–1399. doi: 10.1128/jb.172.3.1392-1399.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 31.Sitnikov D M, Schineller J B, Baldwin T O. Control of cell division in Escherichia coli: regulation of transcription of ftsQA involves both rpoS and SdiA-mediated autoinduction. Proc Natl Acad Sci USA. 1996;93:336–341. doi: 10.1073/pnas.93.1.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sullivan N F, Donachie W D. Overlapping functional units in a cell division gene cluster in Escherichia coli. J Bacteriol. 1984;158:1198–1201. doi: 10.1128/jb.158.3.1198-1201.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Heijenoort J. Murein synthesis. In: Neidhardt F C, Curtis III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1025–1034. [Google Scholar]
- 34.Vicente M, Errington J. Structure, function and controls in microbial division. Mol Microbiol. 1996;20:1–7. doi: 10.1111/j.1365-2958.1996.tb02482.x. [DOI] [PubMed] [Google Scholar]
- 35.Vicente M, Gómez M J, Ayala J A. Regulation of transcription of cell division genes in the Escherichia coli dcw cluster. Cell Mol Life Sci. 1998;54:317–324. doi: 10.1007/s000180050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wijsman H J W. A genetic map of several mutations affecting the mucopeptide layer of Escherichia coli. Genet Res. 1972;20:65–74. doi: 10.1017/s0016672300013598. [DOI] [PubMed] [Google Scholar]
- 37.Yi Q-M, Rockenbach S, Ward J E, Lutkenhaus J. Structure and expression of the cell division genes ftsQ, ftsA, and ftsZ. J Mol Biol. 1991;184:399–412. doi: 10.1016/0022-2836(85)90290-6. [DOI] [PubMed] [Google Scholar]
- 38.Zawadzke L E, Bugg T D H, Walsh C T. Existence of two d-alanine:d-alanine ligases in Escherichia coli: cloning and sequencing of the ddlA gene and purification and characterization of the DdlA and DdlB enzymes. Biochemistry. 1991;30:1673–1682. doi: 10.1021/bi00220a033. [DOI] [PubMed] [Google Scholar]