Abstract
Cyclic di-GMP (c-di-GMP) is the specific nucleotide regulator of β-1,4-glucan (cellulose) synthase in Acetobacter xylinum. The enzymes controlling turnover of c-di-GMP are diguanylate cyclase (DGC), which catalyzes its formation, and phosphodiesterase A (PDEA), which catalyzes its degradation. Following biochemical purification of DGC and PDEA, genes encoding isoforms of these enzymes have been isolated and found to be located on three distinct yet highly homologous operons for cyclic diguanylate, cdg1, cdg2, and cdg3. Within each cdg operon, a pdeA gene lies upstream of a dgc gene. cdg1 contains two additional flanking genes, cdg1a and cdg1d. cdg1a encodes a putative transcriptional activator, similar to AadR of Rhodopseudomonas palustris and FixK proteins of rhizobia. The deduced DGC and PDEA proteins have an identical motif structure of two lengthy domains in their C-terminal regions. These domains are also present in numerous bacterial proteins of undefined function. The N termini of the DGC and PDEA deduced proteins contain putative oxygen-sensing domains, based on similarity to domains on bacterial NifL and FixL proteins, respectively. Genetic disruption analyses demonstrated a physiological hierarchy among the cdg operons, such that cdg1 contributes 80% of cellular DGC and PDEA activities and cdg2 and cdg3 contribute 15 and 5%, respectively. Disruption of dgc genes markedly reduced in vivo cellulose production, demonstrating that c-di-GMP controls this process.
Cyclic di-GMP (c-di-GMP) acts as a reversible and highly specific positive effector of cellulose (1,4-β-d-glucan) synthase in the cellulose-producing bacterium Acetobacter xylinum (34, 35; for a review, see reference 36). This unique nucleotide stimulates the enzyme reaction rate up to 200-fold (activation constant [Kact] = 0.35 μM), such that the in vitro rate of cellulose synthesis may reach 50% of that in the intact cell (2, 32). c-di-GMP activation of purified cellulose synthase is kinetically independent of the concentration of the substrate UDP-glucose (UDP-glc), and its effect is exerted by binding directly to the enzyme at a regulatory site distinct from the catalytic site (35).
The intracellular concentration of c-di-GMP is maintained by the opposing action of the enzymes diguanylate cyclase (DGC), which catalyzes its formation from two molecules of GTP, and Ca2+-sensitive phosphodiesterase A (PDEA), which in conjunction with phosphodiesterase B (PDEB) catalyzes its degradation to 5′-GMP (34). The pathway to c-di-GMP synthesis occurs via the linear dinucleotide pppGpG in two distinct PPi-releasing steps; the degradative pathway is initiated by PDEA, which cleaves a single phosphodiester bond in the cyclic structure, yielding the inactive linear dimer pGpG, which is converted to 5′-GMP by PDEB.
Within the cell, c-di-GMP is tightly associated with the c-di-GMP binding protein (CDGBP), a membrane protein which exhibits saturable and reversible c-di-GMP binding with high affinity (46). The equilibrium of the reaction is markedly and specifically shifted towards the binding direction by K+, such that the intracellular concentration of free c-di-GMP is only 10% of the overall intracellular c-di-GMP concentration (5 to 10 μM), consistent with the high K+ level in the cell. CDGBP is apparently structurally associated with the cellulose synthase, since both activities reside within highly purified cellulose synthase preparations and comigrate in gel filtration of solubilized membrane preparations. This structural association and implied physical proximity within the membrane may enable CDGBP to functionally regulate synthase activity in vivo by modulating the intracellular level of free c-di-GMP.
Four proteins essential for bacterial cellulose synthesis are encoded by the bcs operon in A. xylinum (47). The first gene in the operon, bcsA, encodes the subunit of the cellulose synthase which binds the substrate UDP-glc and presumably catalyzes the polymerization of 1,4-β-d-glucan from glucose units (23). The second gene, bcsB, encodes the subunit which binds the activator c-di-GMP (25). Examination of a series of synthetic cyclic dimer and trimer analogs of c-di-GMP for the ability to interact with cellulose synthase and PDEA revealed that although the two enzymes have a similar high degree of specificity for the c-di-GMP structure, their cyclic dinucleotide sites are not identical (35).
c-di-GMP may also be produced and active in other cellulose-producing systems. In Agrobacterium tumefaciens, the cellular occurrence of c-di-GMP and DGC and PDEA activities have been demonstrated, and addition of exogenous c-di-GMP to cell extracts enhances cellulose synthesis (4, 42). Cotton fiber extracts contain peptides which specifically bind c-di-GMP with high affinity in photolabeling studies (5). Labeling is most pronounced in fibers harvested at the developmental phase of maximal cellulose production, suggesting a physiological role for c-di-GMP in this process. Additional evidence that the plant kingdom utilizes c-di-GMP-dependent pathways for cellulose synthesis has been provided by the discovery of a unique saponin in both Pisum sativum and A. xylinum which acts as a specific inhibitor of the DGC reaction (29, 30). The intracellular abundance of this compound in numerous plant systems may be a significant factor which has thus far prevented in vitro detection of DGC activity in plants.
In another significant finding, plant homologs of A. xylinum bcsA genes (39, 47) and of A. tumefaciens celA genes (24) encoding catalytic subunits of cellulose synthase have been isolated from cotton and rice cDNA libraries (31). These plant celA genes encode proteins containing three regions of conserved sequence with respect to the bacterial gene products, within which are found highly conserved subdomains proposed to be critical for catalysis and/or UDP-glc binding.
Here we report the isolation of the cdg1, cdg2, and cdg3 operons, which encode homologous isoforms of DGC and PDEA. Each of these operons is organized with a pdeA gene upstream of a dgc gene, yet genetic disruption analyses indicate that they contribute differentially to cellular PDEA and DGC enzymatic activities. cdg1 is responsible for 80% of each activity and contains two flanking genes of as-yet-unknown function, cdg1a and cdg1d. The cdg1a gene product is similar in sequence to known prokaryotic transcriptional activators, suggesting that it has a regulatory role. The proteins encoded by the dgc and pdeA genes display a high degree of identity within each isoenzyme set, and significant structural conservation is also apparent between the two isoenzyme sets. In their N termini, all six isoenzymes contain domains similar to those found in various oxygen-sensing proteins, suggesting an oxygen-mediated mechanism of signal transduction for c-di-GMP metabolism and, ultimately, cellulose production. Further downstream, the DGC and PDEA sequences share a lengthy consensus motif, consisting of two adjacent domains termed GGDEF (16) and EAL.
The organization of the cdg operons is distinguished by the juxtaposition of genes encoding enzymes of opposing action on the same genetic unit and by its multiplicity. The coordinate expression of pdeA and dgc provides a requisite balance of PDEA and DGC for achieving the optimal concentration of c-di-GMP, which is essential for a rate of cellulose synthesis in tune with environmental conditions. The presence of homologs in noncellulose-producing bacteria raises the possibility that c-di-GMP may be involved in additional cellular functions.
MATERIALS AND METHODS
Bacterial and phage strains and plasmids.
The Escherichia coli strains used in this work were MM294 (thi-1 hsdR17 endA1 supE44), DG101 (thi-1 hsdR17 endA1 supE44 lacIq lacZΔM15), DG98 [thi-1 hsdR17 endA1 supE44 lacIq lacZM15 proC::Tn10 (F′ lacIq lacZM15 proC+)], and K802 (recA lacY1 supE44 galK galT22 rtbD1 metB1 hsdR2). The A. xylinum strains used were the following: 1306-3, an isolate from strain B42 (North Regional Research Laboratories, Peoria, Ill.); 1306-11 and 1306-21, glucose dehydrogenase mutants of strain 1306-3 (47); and 1499, a strain from M. Benziman’s collection. In addition, the recombinant strains listed in Table 1 were derived in the course of this work as described below. Bacteriophages M13mp18 and M13mp19 and plasmids pUC18, pUC19 (49), pBR322 (9), and pACYC184 (10) were used for DNA manipulations. Derivation of the bacteriophage pKT230cos5, shuttle vectors pUC18-824 and pUC19-824, and R-20 medium have been described previously (47).
TABLE 1.
PDEA and DGC activities and in vivo cellulose production in A. xylinum recombinant strainsa
| Strain | Site of mutation | PDEA activity (%) | DGC activity (%) | Cellulose production (%) |
|---|---|---|---|---|
| 1306-21 (WT) | None | 100 | 100 | 100 |
| Dis1 | pdeA1 | 20 | 20 | 85 |
| Dis4 | dgc1 | 98 | 21 | 80 |
| ABT8 | Upstream of cdg1a | 23 | 22 | 78 |
| ABT9 | pdeA2 | 85 | 85 | 98 |
| TRT150 | dgc2 | 98 | 86 | 95 |
| ABT2 | Partial deletion (pdeA2+dgc2) | 89 | 91 | 99 |
| ABT21 | dgc1 dgc2 | 105 | 4 | 36 |
| ABT11 | pdeA1 pdeA2 | 5 | 5 | 68 |
| ABT1 | pdeA1 pdeA2 dgc2 | 5 | 4 | 70 |
Recombinant strains of A. xylinum disrupted in cdg1 and/or cdg2 were derived, and whole cells were assayed for cellulose production, as described in Material and Methods. Soluble and membrane fractions were prepared from logarithmic-phase cultures and assayed for PDEA and DGC activities.
Preparation of polyclonal antibodies and inhibition assays.
GTP-agarose binding peptides were isolated from A. xylinum 1499, as described previously (32, 33), and were designated p77, p64, p61, and p59 according to their mobilities on a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel. In A. xylinum 1306-11, which was used for most of this investigation, p59 and p61 could not be distinguished from each other. The individual peptides were excised from gels and used to generate polyclonal antisera in rabbits. Immunoglobulin G fractions were purified from each antiserum by standard procedures. For inhibition assays, crude preparations containing 0.1 mg of DGC or PDEA were incubated at 4°C for 1 h in the presence of 20 μl of immunoglobulin G (12 to 25 mg/ml) in a final volume of 80 μl in buffer containing 50 mM Tris (pH 7.5), 10 mM MgCl2, and 1 mM EDTA (TME). Protein A-Sepharose (3 mg) was added, and incubation was continued for 2 h. Following centrifugation, supernatants were assayed for DGC or PDEA activity.
Purification of PDEA.
Washed membranes (25) from A. xylinum 1306-21 were homogenized in TME, and the supernatant was incubated with 1% Brij 58 at 4°C for 1 h. Brij 58 was shown to selectively inactivate DGC and thus prevent its binding to GTP-agarose (45). Following loading onto a GTP-agarose column, washing was performed sequentially with 25 volumes of TME containing 1 mM DTT (TME-DTT), 15 volumes of TME-DTT containing 1 mM ATP, and 10 volumes of TME-DTT. PDEA was eluted with 10 volumes of buffer containing 50 mM Tris (pH 7.5), 10 mM EDTA, 0.2 M KCl, and 1 mM DTT. Fractions (0.9 ml) were collected in vials containing 40 μl of 1 M MgCl2 and 100 μl of glycerol. The PDEA obtained exhibited a specific activity of 2,300 nmol of c-di-GMP degraded/min/mg of protein, which represents a 600-fold purification with respect to the crude extract. SDS-PAGE revealed a highly purified single band with an apparent molecular mass of 77 kDa.
Purification of DGC.
DGC was precipitated with 30% saturated ammonium sulfate from the membrane-free fraction of A. xylinum 1306-21 extracts (32, 33) and was further purified by affinity chromatography on a GTP-agarose column. After loading, the column was washed first with 40 column volumes of TME-DTT, then with 20 volumes of 5 mM ATP in TME-DTT, and finally with 20 volumes of 50 mM Tris (pH 7.5) containing 1 mM DTT. DGC was eluted with 10 volumes of 50 mM Tris (pH 7.5) containing 10 mM EDTA, 0.2M KCl, and 1 mM DTT. Fractions were collected as described for PDEA purification. The bulk of the DGC activity was eluted from fraction volumes between 2 and 7 ml. The enzyme exhibited a specific activity of 650 nmol of c-di-GMP formed/min/mg of protein, which represents an overall purification of 500-fold with respect to the crude extract. SDS-PAGE revealed one major band with an apparent molecular mass of 60 kDa.
Amino acid sequence determination of PDEA and DGC.
Affinity-purified PDEA p77 was applied to a preparative SDS-PAGE gel, and the protein from the excised band was transferred to a polyvinylidene fluoride membrane. N-terminal amino acid sequencing was performed by automated Edman degradation (Applied Biosystems model 470A sequenator).
Since attempts to obtain N-terminal amino acid sequence information for the DGC p59 peptide were unsuccessful, the protein excised from SDS-PAGE gels was partially digested by oxidation with performic acid and dialyzed against 0.1% trifluoroacetic acid–25% CH3CN–H2O. The preparation was dried, resuspended in buffer (200 μl) containing 3 M guanidine chloride, 25 mM Tris (pH 9.1), and Achromobacter lysyl endopeptidase (0.005 μg/μl), and incubated at 40°C for 4 h. Digestion products were separated by high-performance liquid chromatography (HPLC) on a C4 reverse-phase column D-1 (Merck). Elution was performed with solvents A (0.1% trifluoroacetic acid in H2O) and B (0.085% trifluoroacetic acid–85% CH3CN–H2O) by using a gradient of 0 to 70% solvent B. Peptides from five different column fractions were sequenced by Edman degradation. PCR primers were designed on the basis of the amino acid sequence of fraction 5.
Chromosomal DNA isolation, library construction, and dot blot analysis.
A. xylinum cells were grown in R-20 medium (11) in the presence of 0.1% (vol/vol) cellulose as described previously (25), resuspended in 50 mM Tris (pH 8.0)–25% sucrose–lysozyme (1 mg/ml), and incubated at 30°C for 10 min. The preparation was supplemented with 75 mM EDTA, proteinase K (0.3 mg/ml), RNase A (60 μg/ml), and RNase T1 (7.5 U/ml) and incubated at 37°C for 30 min. SDS was added to 1%, and incubation was continued for 30 min. DNA was extracted repeatedly with phenol-chloroform-isoamyl alcohol (25:24:1), precipitated with ethanol, and resuspended to 1.9 mg/ml in 10 mM Tris (pH 8.0)–1 mM EDTA. Chromosomal DNA libraries were prepared in the vector pKT230cos5 as described previously (47) and in the vector pUC18-824 by using strain 1306-3 chromosomal DNA partially digested with EcoRI (3 to 6 kb).
Dot blot analysis of strain 1306-3 chromosomal DNA digested with PstI was performed by using 32P-end-labeled oligonucleotide pools as probes. Molecular biology procedures were performed in accordance with standard protocols (37).
Primer design and PCR amplification for cloning the gene encoding DGC.
On the basis of the amino acid sequence information obtained from an internal peptide of DGC p59, four forward primer pools corresponding to amino acid residues 5 to 9 (Ala-Glu-Thr-Asp-Thr) were synthesized: Cel-1 [5′-GCNGA(G/A)ACTGA(C/T)AC-3′], Cel-2 [5′-GCNGA(G/A)ACCGA(C/T)AC-3′], Cel-3 [5′- GCNGA(G/A)ACAGA(C/T)AC-3′], and Cel-4 [5′-GCNGA(G/A)ACGGA (C/T)AC-3′]. Eight reverse primer pools corresponding to amino acid residues 17 to 22 (Gly-Gly-Phe-Asn-Thr-Ala) were synthesized: Cel-5 [5′-GC(G/T)CA(A/G)TT(G/A)AA(G/T)CC(G/T)CC-3′], Cel-6 [5′-GC(G/T)CA(A/G)TT(G/A)AA(G/T)CC(C/A)CC-3′], Cel-7 [5′-GC(C/A)CA(A/G)TT(G/A)AA(G/T)CC(G/T)CC-3′], Cel-8 [5′-GC(C/A)CA(A/G)TT(G/A)AA(G/T)CC(G/T)CC-3′], Cel-9 [5′-GC(G/T)CA(A/G)TT(G/A)AA(G/T)CC(C/A)CC-3′], Cel-10 [5′-GC (C/A)CA(A/G)TT(G/A)AA(G/T)CC(C/A)CC-3′], Cel-11 [5′-GC(C/A)CA (A/G)TT(G/A)AA(G/T)CC(G/T)CC-3′], and Cel-12 [5′-GC(C/A)CA(A/G)TT(G/A)AA(G/T)CC(C/A)CC-3′].
The forward and reverse oligonucleotide pools were used in combinations for 32 PCR amplifications. Reaction mixtures (50 μl) contained 10 mM Tris (pH 8.3), 50 mM KCl, 3 mM MgCl2, 100 μg of gelatin per ml, 20 μM deoxynucleotide triphosphates, a 500 μM concentration of each 5′ and 3′ primer pool, 2 U of Taq polymerase (Perkin-Elmer Cetus), and 50 ng A. xylinum 1306-3 chromosomal DNA. Thirty cycles of amplification (93°C for 1 min, 35°C for 1 min, and 68°C for 30 s) were performed. Under these conditions, primer pool combinations Cel-2 and Cel-5 and Cel-2 and Cel-8 produced a product of the expected length (53 bp). When the annealing temperature was raised from 35 to 42°C, only the combination of Cel-2 and Cel-8 produced the 53-bp product. To prepare a 32P-labeled probe, 1 ng of the 53-bp DNA product was amplified under the same conditions by using [32P]dCTP (10 μl of an 800-Ci/mmol concentration; New England Nuclear Corp.).
Design of oligonucleotide probes for cloning the gene encoding PDEA.
On the basis of the N-terminal acid sequence determined for affinity-purified PDEA p77, the following oligonucleotide pools were designed based on the first five residues (Pro-Asp-Ile-Thr-Ala): Cel-117 [5′-CC(G/C)GATAT(A/T/C)ACNGC-3′], Cel-118 [5′-CC(A/T)GATAT(A/T/C)ACNGC-3′], Cel-119 [5′-CC(G/C)GACAT(A/T/C)ACNGC-3′], Cel-120 [5′-CC(A/T)GACAT(A/T/C)ACNGC-3′], Cel-121 [5′-CC(G/C)GACATCAC(G/C)GC-3′], and Cel-122 [5′-CC(A/T)GATAT(A/T)AC(A/T)GC-3′].
RNA isolation, primer extension analysis, and nucleotide sequence determination.
To map the transcription initiation site of the operon cdg2, cellular RNA was isolated from A. xylinum 1306-3 and primer extension (49) was carried out with oligonucleotide primer Cel-213 (5′-TAGCAGAAAGCAGGACATCGGCGC-3′). Nucleotide sequences of selected chromosomal library clones or subclones were determined on both strands by using specifically synthesized primers and the dideoxy chain termination method (39). DNA and amino acid sequences were analyzed with the Genetics Computer Group Wisconsin Package (version 9.0). Multiple sequence alignments were generated with the Genetics Computer Group program PileUp and edited with GeneDoc (28a).
Gene disruptions and generation of recombinant strains.
The β-lactamase gene, encoding ampicillin resistance (Ampr), and the streptomycin fusion gene, encoding streptomycin resistance (Strr), were shown to be functional in A. xylinum. The Ampr gene was isolated from pBR322 as an EcoRI-AlwNI fragment. The Strr gene was isolated from pKT230cos5 as a 1.5-kb BamHI-PvuII fragment, cloned into the BamHI-EcoRV sites of pACYC184, and reisolated as a 1.68-kb BamHI-EcoRI fragment for further manipulations. Mutated genes were constructed in the vector pACYC184, which does not replicate in A. xylinum. Linearized vector constructs were used for transformation of A. xylinum by electroporation (47), and transformants were selected on plates containing the appropriate antibiotic(s).
For insertional inactivation of pdeA1, a 4.2-kb EcoRI fragment from cosmid 3F3 (Fig. 1) was subcloned into pACYC184. The Ampr or Strr gene was inserted between the EcoRV sites, and the resultant plasmids were used to transform A. xylinum 1306-21, yielding strains Dis1 and ABT3, respectively.
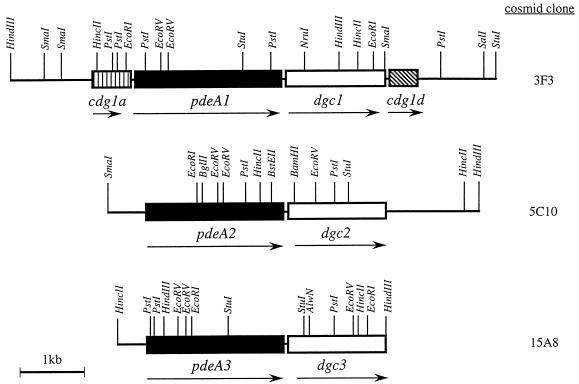
FIG. 1.
Genetic organization of the cdg1, cdg2, and cdg3 operons and their flanking regions in A. xylinum, located on cosmid clones 3F3, 5C10, and 15A8, respectively. The pdeA and dgc genes are indicated by solid and open boxes, respectively. Arrows indicate the transcriptional directions of the genes.
For insertional inactivation of dgc1, a 3.5-kb StuI fragment from cosmid 3F3 (Fig. 1) was subcloned into the EcoRV site of pACYC184. Following insertion of the Ampr gene at the NruI site and transformation of A. xylinum 1306-21, strain Dis4 was obtained.
To disrupt the 5′ upstream region of the cdg1 operon, a 5.4-HindIII fragment from cosmid 3F3 (Fig. 1) was subcloned into pACYC184. The 0.4-kb SmaI fragment upstream of cdg1a was removed, and the Ampr gene was inserted. The resultant plasmid was used to transform A. xylinum 1306-21, yielding strain ABT8.
For dgc2 insertional mutants, a 2.9-kb HindIII-BamHI fragment from cosmid 5C10 (Fig. 1) was subcloned into pACYC184. The Ampr gene was inserted into the EcoRV site, and the resultant plasmid was linearized and transformed into A. xylinum 1306-21, yielding strain TRT150. The strain TRT151 was mutated at the same site of dgc2 by insertion of the Strr gene.
For pdeA2 mutants, the 5.6-kb SmaI-HindIII fragment from cosmid 5C10 carrying the coding regions of pdeA2 and dgc2 (Fig. 1) was manipulated. A deletion mutant was obtained by subcloning into pUC18 and excision of the 1.8-kb PstI fragment. The shortened 3.8-kb fragment was subcloned into pACYC184, and the Ampr gene was inserted at the PstI site. Transformation of A. xylinum 1306-21 with this construct yielded strain ABT2. For a pdeA2 insertional mutant, the 5.6-kb SmaI-HindIII fragment was subcloned into pACYC184, followed by insertion at the BglII site of the Strr gene, yielding strain ABT9.
To generate a double mutant disrupted in pdeA1 and pdeA2, strain ABT9 was transformed with the plasmid used to obtain strain Dis1, yielding the Strr Ampr strain ABT11. Similarly, to generate a double mutant disrupted in dgc1 and dgc2, strain TRT151 was transformed with the plasmid used to obtain Dis4, yielding the Strr Ampr strain ABT21. To obtain a strain effectively disrupted in pdeA1, dgc1, pdeA2, and dgc2, the plasmid used to obtain ABT3 was used to transform ABT2, yielding the Strr Ampr strain ABT1.
Southern analyses were carried out for each mutant strain to verify that the desired chromosomal recombination event had occurred (data not shown).
Enzymatic assays.
Soluble and membrane fractions were prepared from washed membranes and assayed for activities of PDEA, DGC, and cellulose synthase as described previously (25). The latter assay was conducted in the presence of excess exogenous c-di-GMP.
In vivo cellulose production.
Cells grown in R-20 medium were analyzed for cellulose formation as described previously (47).
Nucleotide sequence accession numbers.
The DNA sequences of the cdg1, cdg2, and cdg3 operons have been submitted to the GenBank database under accession no. AF052517, AF052518, and AF052519, respectively.
RESULTS
Purification and amino acid sequence determination of DGC and PDEA.
GTP-agarose affinity chromatography has been effectively utilized to purify both DGC and PDEA in active forms from A. xylinum. The affinity of DGC for GTP appears to be via its substrate binding site, while that of PDEA is via an inhibitory site distinct from its substrate or product binding sites (36). A membrane-free preparation derived from extracts of A. xylinum 1499 was applied to a GTP-agarose column (32, 33). Elution with 5 mM GTP yielded a fraction containing four polypeptides, which were designated p77, p64, p61, and p59. Inhibition studies employing antisera raised against each polypeptide indicated that p59 and p61 are associated with DGC activity while p77 is associated with PDEA activity.
Since anti-p59 antiserum most effectively inhibited DGC activity, p59 was selected as the most likely DGC candidate. Attempts to directly determine its N-terminal amino acid sequence were unsuccessful, however, and so the protein was partially digested into peptides which were separated by HPLC. The amino acid sequence of one peptide was determined to be Leu-Ser-Glu-Leu-Ala-Glu-Thr-Asp-Thr-Leu-Thr-Ala-Leu- Leu-Asn-Arg-Gly-Gly-Phe-Asn-Thr-Ala-Leu-Ser-Ala-Ala- Leu-Gly-Xaa-Xaa-Xaa-Lys.
N-terminal amino acid sequencing of PDEA p77 yielded the sequence -Pro-Asp-Ile-Thr-Ala-Leu-Thr-Thr-Glu-Ile-Leu- Leu-Pro-Ala-Leu-Glu-Arg-Ala-COOH.
A PCR oligonucleotide product was used to isolate a DGC gene, dgc.
The amino acid sequence of the DGC p59 internal peptide was used to design a series of degenerate oligonucleotide primers for isolating the coding region of the DGC gene via PCR. A single PCR product of the expected length (53 bp) was generated from primer pool combination Cel-2 and Cel-8 by using A. xylinum 1306-11 chromosomal DNA as the template. The nucleotide sequence of the product was determined to be GCCGAGACCGATACCCTGACCGGCCTGCTCAACCGCGGAGGCTTCAACACGGC. Conceptual translation of the PCR product sequence yields an amino acid sequence identical to that determined for the p59 internal peptide (residues 5 to 21), except for substitution of glycine for alanine at the 12th residue, possibly reflecting sequence diversity in strains 1499 and 1306-11.
The 53-bp PCR product, representing a portion of the DGC gene coding region, was used to screen an A. xylinum 1306-11 chromosomal DNA library prepared in pKT230cos5. Twenty positive clones were obtained, which upon restriction mapping grouped into three distinct classes, represented by cosmids 5C10, 3F3, and 15A8 (Fig. 1). Of these clones, only 5C10 DNA was capable of generating the 53-bp PCR product when used as a template in combination with primers Cel-2 and Cel-8, and it was thus selected for nucleotide sequence analysis.
dgc and pdeA reside on the cdg operon.
Nucleotide sequence analysis of cosmid 5C10 indicated two open reading frames (ORFs) over 4,798 bp separated by a stretch of 108 bp (Fig. 1). A possible ribosome binding site precedes each of these ORFs at −1 and −8 bp upstream from the initiating ATG codons. The downstream ORF, encoding a peptide of 574 amino acids with a calculated molecular mass of 64.6 kDa, was identified as the coding region of a dgc gene, based on the fact that its deduced sequence contains a stretch matching the amino acid sequence determined for the DGC p59 internal peptide.
The upstream ORF encodes a peptide of 752 amino acids with a calculated molecular mass of 83.8 kDa. Although the N terminus of the deduced protein does not match the N-terminal sequence determined for purified PDEA p77, this ORF was designated as a gene for PDEA, since genetic disruption experiments described below demonstrated that this coding region is associated with PDEA activity. The discrepancy between the deduced N terminus and the N-terminal sequence determined for purified PDEA p77 was later accounted for upon discovery of three distinct pdeA genes. Their deduced amino acid sequences are about 65% identical, but that encoded by the pdeA gene described here contains an N-terminal tail of 23 amino acids that is not present in the other two PDEA sequences.
The genes encoding PDEA and DGC which were discovered on cosmid clone 5C10 were later designated pdeA2 and dgc2, respectively, on the basis of their relative contribution of enzymatic activities within the cell, as described below (Table 1). The genes pdeA2 and dgc2 are likely organized as an operon, since primer extension experiments indicated a transcriptional start site upstream of pdeA2 (Fig. 2) but none was identified upstream of dgc2 (data not shown). Also, insertional mutation of pdeA2 in strain ABT9 decreased both PDEA and DGC enzymatic activities (Table 1), suggesting a polar effect on the translation of the downstream dgc2 gene. On the basis of these observations, we conclude that pdeA2 and dgc2 are cotranscribed as a polycistronic mRNA and designate this genetic unit as the operon for cyclic diguanylate (cdg2).
FIG. 2.

Primer extension analysis of operon cdg2. Total RNA extracted from A. xylinum 1306-3 was hybridized with 32P-end-labeled primer Cel-213. cDNA was synthesized with avian myeloblastosis virus reverse transcriptase and the four deoxyribonucleotide triphosphates. The length of the extension product (1) was measured against the terminal products produced in DNA sequencing reactions by using the same primer. G, A, T, and C indicate the individual sequencing reactions.
Recombinant strains disrupted in dgc2 and pdeA2 exhibit some reduction in DGC and PDEA activities.
Recombinant A. xylinum strains TRT150, ABT9, and ABT2 were examined for DGC and PDEA enzymatic activities (Table 1). Surprisingly, the reduction in DGC and PDEA activities as a result of disruption in dgc2, pdeA2, or both was no greater than 15% with respect to that of the wild-type (WT) strain. However, reduction in PDEA2 activity as a result of pdeA2 inactivation was accompanied by a decrease in DGC activity, indicating physical linkage of pdeA2 and dgc2. Thus, we concluded that dgc2 and pdeA2 are not the sole genes responsible for the DGC and PDEA activities in A. xylinum and that there must be additional dgc and pdeA genes which either compensated for the deficiencies in the mutant strains or are responsible for the bulk of the activities.
An additional cdg operon contains four ORFs.
Utilizing the N-terminal amino acid sequence determined for PDEA p77, degenerate oligonucleotide probes were synthesized and used for dot blot hybridization of A. xylinum 1306-3 chromosomal DNA. Oligonucleotide pool Cel-121 yielded the strongest signal and was thus used to screen a 1306-3 chromosomal library constructed in the shuttle vector pUC18-824. Six identical clones were identified, and the nucleotide sequence of an 0.8-kb PstI fragment from one clone, PDEA-7A, was determined. That sequence was subsequently found to be carried on cosmid clone 3F3; thus, the complete nucleotide sequence of the 3F3 insert was determined.
Analysis of a 5,910-bp region from cosmid 3F3 revealed the presence of four ORFs, encoding proteins with calculated molecular masses of 23, 84.5, 64.6, and 15.2 kDa (Fig. 1). Small intergenic regions of 23, 30, and 6 bp separate the ORFs, suggesting that they are arranged as a single transcriptional unit, and an inverted repeat sequence (CCTGCATGAGCCT…AGGCTCATGCAGG) that could serve as a bacterial transcriptional terminator is present at the 3′ end of the fourth ORF. Putative ribosome binding sites immediately precede the initiating ATG codons of the second, third, and fourth ORFs. The second ORF encodes a protein of 757 amino acid residues which exhibits 58% overall identity with the product of pdeA2, and its N-terminal region matches the sequence determined for purified PDEA p77. Therefore, we conclude that this ORF carries an additional pdeA gene.
The third ORF encodes a protein of 575 amino acid residues which exhibits 54% overall identity with the product of dgc2. The N-terminal region of this protein (Ser-Leu-Lys-His- Asp-Asp-Arg-Leu-Arg-Ala-Leu-Thr-His-Gln-Asp-Ser-Asp) matches the N-terminal amino acid sequence determined for DGC purified from the mutant TRT150, which is disrupted in dgc2. Thus, this ORF appears to encompass an additional dgc gene, specifically the isoenzyme remaining intact in TRT150 which appears to be responsible for the bulk of the cellular DGC activity.
Based on the similarity of the second and third ORFs to pdeA2 and dgc2, respectively, we conclude that the genetic unit on which they reside represents an additional and distinct operon for cyclic diguanylate, designated cdg1 in accordance with its relative contribution of enzymatic activities as presented below. The four potential genes are designated sequentially as cdg1a, pdeA1, dgc1, and cdg1d.
CDG1A is similar to prokaryotic transcriptional regulators.
cdg1a encodes a protein similar in size (211 amino acid residues) and structure to members of the Crp-Fnr family of transcriptional regulators which are involved in oxygen control of various processes. Thus, CDG1A is 38% identical to AadR of Rhodopseudomonas palustris, which functions in anaerobic growth (13), 37% identical to FixK proteins of Bradyrhizobium japonicum (7) and Azorhizobium caulinodans (20), which are activators in the nitrogen fixation cascade, and 31% identical to FnrN2 of Rhizobium leguminosarum (17), which can activate genes involved in anaerobic respiration (40). The most striking similarity is within a helix-turn-helix motif found in the C-terminal region (positions 156 to 170), a structural feature shown to be involved in DNA binding in Fnr (41). Also partially conserved in CDG1A is a series of glycine residues in the N-terminal region, which in Crp has been shown to form a β-roll structure thought to be important for nucleotide binding (44).
Similarity of CDG1D to other proteins.
The protein encoded by cdg1d is similar in length (160 amino acid residues) and sequence (44 and 40% identity, respectively) to hypothetical proteins of Bacillus subtilis (GenBank accession no. Y14082) and E. coli (GenBank accession no. U14003).
Genetic disruptions reveal a hierarchy of enzymatic activities between operons cdg1 and cdg2.
A series of recombinant A. xylinum strains differentially disrupted in cdg1 and cdg2 were assayed for DGC and PDEA enzymatic activities (Table 1). Dis1, lacking a functional pdeA1, displayed only 20% of WT PDEA activity, whereas ABT9, disrupted in pdeA2, displayed 85% of WT activity. Likewise, a mutation in dgc1 (Dis4) reduced DGC activity to 21% of that of WT, while a mutation in dgc2 (TRT150) caused only a 14% reduction in activity. These results clearly indicate that the operon cdg1 is the source of approximately 80% of both DGC and PDEA activities. This was further supported by the results provided by strains ABT8 and ABT2. The former, insertionally inactivated upstream of cdg1, displayed residual activities for both enzymes of 25%, whereas the latter, a deletion mutant lacking major portions of both pdeA2 and dgc2, displayed 90% of WT activities.
The polar effect of disruption in each pdeA gene on downstream dgc expression was shown by the reduction in DGC activity in the single pdeA mutants, Dis1 and ABT9, and in the double pdeA mutant, ABT11. Conversely, in the single dgc mutants, Dis4 and TRT150, and in the double dgc mutant, ABT21, reduction in DGC activity was not accompanied by a depression of PDEA activity.
Identification of operon cdg3.
The observation that recombinant strains doubly disrupted in either pdeA (ABT11), dgc (ABT21), or both (ABT1) displayed residual enzymatic activities of about 5% (Table 1) suggested that there might be other functional pdeA and dgc genes in A. xylinum. Since we had demonstrated that two of the three cosmid clones isolated on the basis of hybridization with the DGC-derived PCR probe carried cdg operons, we suspected that the remaining cosmid (15A8) carried another cdg operon, which was indeed shown by nucleotide sequence analysis.
Nucleotide sequence analysis of a 4,675-bp region of cosmid 15A8 indicated two ORFs, encoding proteins of calculated molecular masses of 82.5 and 55.1 kDa (Fig. 1). The deduced amino acid sequence of the upstream ORF (740 residues) displays 71 and 63% identity with the deduced PDEA1 and PDEA2 sequences, respectively, while that of the downstream ORF (493 residues) displays 49% identity with both the deduced DGC1 and DGC2 sequences. The intragenic region is 50 bp in length, and each ORF is preceded by the sequence CGAGGTAAC 3 bp upstream of each initiating ATG codon. This sequence is highly homologous to the postulated ribosomal binding sites of cdg1 and cdg2. Based on this information, we designated this operon cdg3 and the two potential genes pdeA3 and dgc3.
Intracellular c-di-GMP concentration governs in vivo cellulose synthesis.
In vitro cellulose synthase activity did not significantly differ from that of WT in any of the A. xylinum recombinant strains (data not shown). Likewise, in vivo cellulose production was not appreciably decreased in the single cdg1 or cdg2 mutants (Table 1). Although cdg1 clearly provides the bulk of the DGC activity, the level of cellulose production achieved in the absence of a functional dgc1 gene was about 80% of the WT level in each of the cdg1 mutants. More significant reduction of cellulose production in vivo occurred in the double mutants, with the greatest decrease displayed by ABT21 (36% of the cellulose production by WT). In this case, c-di-GMP formation was severely compromised, while enzymatic breakdown of the nucleotide activator was unhampered, apparently resulting in a decrease of cellular c-di-GMP to a level insufficient for optimal synthase activation and, in turn, cellulose synthesis. In the pdeA double mutant, ABT11, cellulose production was inhibited to a lesser extent, perhaps since the decrease in c-di-GMP production which occurred as a result of the polar effect noted above was also accompanied by a 95% reduction in PDEA activity.
These results suggest that c-di-GMP production supplied by DGC isoenzymes distinct from DGC1 is sufficient for near-optimal activation of the cellulose synthase and, thus, cellulose biogenesis. The observation that the double mutants, with 5% of DGC activity, still maintain respectable levels (36 to 70%) of cellulose production is consistent with the low Kact value (0.35 μM) calculated for c-di-GMP with respect to the cellulose synthase (36) and the modulation of the intracellular level of free c-di-GMP by the cellulose synthase-associated c-di-GMP binding protein (46).
The DGC and PDEA proteins share a conserved domain structure occurring in various bacterial proteins.
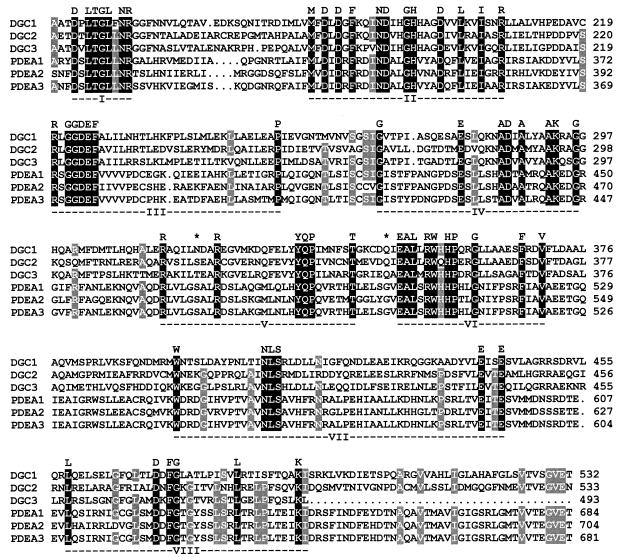
Comparative alignment of the DGC and PDEA protein sequences reveals that all six isoenzymes share significant structural conservation over a stretch of 390 amino acid residues, encompassing one-half to two-thirds the length of each protein (Fig. 3). Common to all the proteins is a consensus sequence of 62 invariant amino acid residues which may be arranged in eight regions. Regions I through IV were previously recognized as a domain structure by alignment of PleD, a motility-associated response regulator of Caulobacter crescentus, with various bacterial proteins of unknown function, and was designated GGDEF according to the conserved sequence within region III (16).
FIG. 3.
Multiple alignment of the deduced amino acid sequences of the pdeA and dgc genes of A. xylinum showing the conserved GGDEF (regions I to IV) and EAL (regions V to VIII) domains. Residues identical in all six sequences are shaded in black and indicated in the consensus line; residues identical in five of six sequences are shaded in gray. In the region shown, the DGC and PDEA isoenzyme sets have 23 to 29% identity, compared to 77 to 89% identity among the PDEA sequences and 47% identity among the DGC sequences. Putative Q-linker regions are delimited by asterisks.
Putative Q-linkers are found in the vicinity of regions V and VI (Fig. 3). These flexible segments tether functionally distinct domains in bacterial regulatory proteins and contain relatively high concentrations of glutamine, arginine, glutamic acid, serine, and proline residues in regions of “coil” secondary structure (48). All the isoenzymes meet these criteria according to their primary structure and predictions for secondary structure (22), except for PDEA3, which lacks the coil characteristic. The putative Q-linkers appear to separate the upstream GGDEF domain (regions I through IV) from a downstream domain (regions VI through VIII), which we designate EAL (see Fig. 6) according to a conserved sequence within region VI.
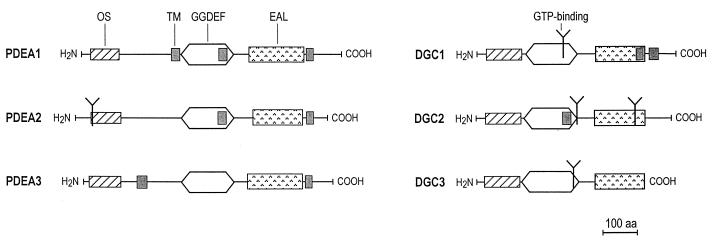
FIG. 6.
Proposed domain structure of the PDEA and DGC isoenzymes of A. xylinum. The GGDEF and EAL domains form a motif conserved among all six isoenzymes. N-terminal regions putatively function in oxygen sensing (OS). Predicted sites of GTP binding may indicate catalytic sites of DGC proteins. TM, putative transmembrane helices; aa, amino acids.
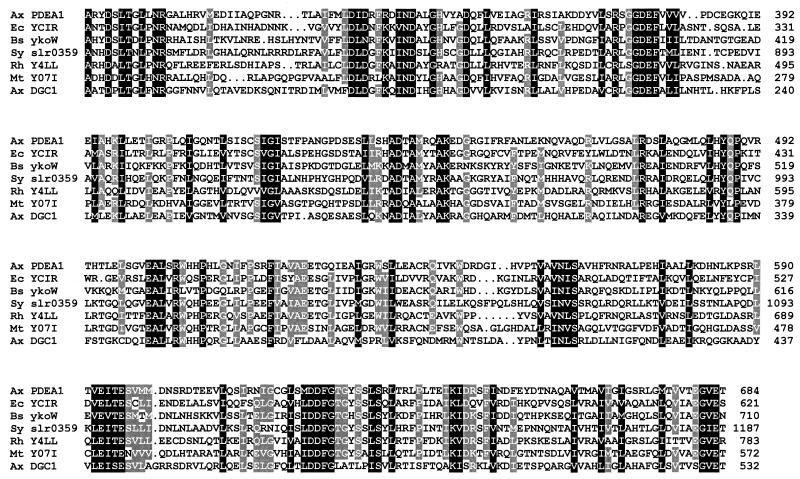
Database searches (3) conducted with the GGDEF and EAL domain sequences revealed that a large number of bacterial proteins of unconfirmed function significantly resemble the PDEA (up to 55% identical) and DGC (up to 33% identical) proteins. The most similar sequences are presented in the alignment shown in Fig. 4. Notably, most of the domain consensus residues derived in Fig. 3 are identical or conservatively replaced among all these proteins, and the greatest degree of conservation is seen in the final 90 residues of the alignment, where the similarity extends significantly beyond the consensus residues.
FIG. 4.
Alignment of bacterial predicted protein sequences showing conservation of GGDEF and EAL domains. Residues with identity or conservative substitution in 100 and ≥80% of the sequences are shaded in black and gray, respectively. Identities displayed between each sequence and the A. xylinum (Ax) PDEA1 and DGC1 sequences, respectively, are shown in parentheses below. The amino acid sequences shown are from E. coli (Ec) YCIR, GenBank accession no. D90766 (50 and 28%); Bacillus subtilis (Bs) ykoW, GenBank Z99110 (50 and 26%); Synechocystis strain PCC6803 (Sy) slr0359, GenBank D63999 (49 and 30%); Rhizobium sp. strain NGR234 (Rh) Y4LL, GenBank AE000083 (48 and 32%); Mycobacterium tuberculosis (Mt) Y07I, GenBank Z75555 (31 and 32%).
The N-terminal regions of the DGC and PDEA proteins are similar to oxygen-sensing domains on bacterial proteins.
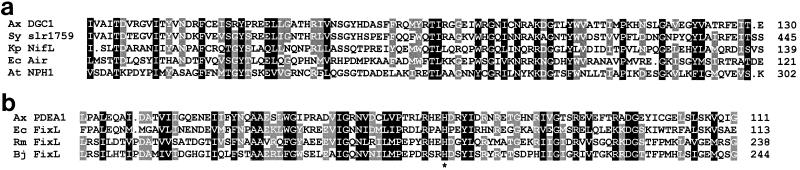
Database searches (3) revealed that the N-terminal regions of the DGC deduced amino acid sequences (corresponding to residues 29 to 130 on DGC1) (Fig. 5a) display 58% identity with the sequence of a putative sensory transduction histidine kinase from Synechocytis sp. strain PCC6803 and 33 to 30% identity with the sequences of NifL proteins from nitrogen-fixing diazotroph bacteria, as exemplified by that of Klebsiella pneumoniae (14). NifL acts as an oxygen-responsive negative regulator of the transcriptional activator NifA (26). The same DGC sequence also displays 34 and 25% identity, respectively, with the Aer sequence of E. coli, implicated in aerotactic responses (8), and the NPH1 sequence of Arabidopsis thaliana, implicated in phototropism (19). This region (Fig. 5a) has been proposed to contain a motif for a sensory domain shared among diverse prokaryotic and eukaryotic proteins which are regulated by environmental signals of light, oxygen, or voltage and have thus been termed LOV domains (19).
FIG. 5.
Multiple alignment of the N-terminal regions of the DGC1 and PDEA1 sequences showing conservation with domains on other proteins. Residues with identity or conservative substitution in 100 and ≥75% of the sequences are shaded in black and gray, respectively. (a) The amino acid sequences shown are from A. xylinum (Ax) DGC1, Synechocystis strain PCC6803 (Sy) slr1759 (sensory transduction histidine kinase, GenBank accession no. D90903), Klebsiella pneumoniae (Kp) NifL (GenBank X13303), E. coli (Ec) Aer (GenBank U28379), and Arabidopsis thaliana (At) NPH1 (GenBank AF030864). (b) Amino acid sequences shown are A. xylinum (Ax) PDEA1, E. coli (Ec) FixL (GenBank D90790), Rhizobium meliloti (Rm) FixL (GenBank Z70305), and Bradyrhizobium japonicum (Bj) FixL (GenBank P23222). An asterisk indicates a His residue conserved among FixL and PDEA proteins.
An N-terminal region of the PDEA protein sequence (corresponding to residues 13 to 111 of the PDEA1 sequence) displays 44 to 34% identity with mid-regions of the rhizobial FixL protein sequences and 47% identity with the sequence of a putative FixL protein from E. coli (Fig. 5b). FixL acts as an oxygen-responsive positive activator in the nitrogen fixation cascade (6, 12). That region of FixL from Rhizobium meliloti (12) shown in Fig. 5b is essential for oxygen-binding hemoprotein activities (27), and His-194 is implicated in the heme binding function (28). This conserved residue among FixL proteins corresponds to an invariant His residue among the PDEA proteins (His-67 on PDEA1), which could thus play an analogous regulatory role in c-di-GMP turnover.
Predicted transmembrane regions and GTP binding sites of the DGC and PDEA proteins.
The DGC and PDEA protein sequences were analyzed for putative models of transmembrane topology (18). According to these predictions, DGC1 has two possible transmembrane helices and DGC2 has one (Fig. 6). PDEA1 has three such possible regions, while PDEA2 and PDEA3 each have two. Analyses for Prosite motifs indicate potential GTP binding sites (44) on DGC1 (Ala-257 to Thr-264), DGC2 (Ala-298 to Gln-306 and Ala-470 to Gly-483), DGC3 (Ala-297 to Gln-304), and PDEA2 (Ala-61 to Ser-68), as shown in Fig. 6.
DISCUSSION
We previously established that the enzymes DGC and PDEA catalyze the synthesis and degradation, respectively, of c-di-GMP in A. xylinum and thus most probably play key regulatory roles for cellulose biosynthesis (34). We now report characterization of the dgc and pdeA genes controlling cellular turnover of c-di-GMP and their organization on three unlinked operons, cdg1, cdg2, and cdg3. Each of the three DGC and three PDEA protein sequences encoded by these genes have a homologous domain structure. Genetic disruption experiments provide conclusive evidence that the intracellular concentration of c-di-GMP, representing the net result of its synthesis and degradation, governs cellulose biogenesis in vivo.
The discovery of the three cdg operons reveals an unusual genetic organization in bacteria. Juxtaposition of genes encoding enzymes of opposing action on the same genetic unit is rare. To our knowledge, the only other case is that of the E. coli polyphosphate operon which contains the gene ppk, encoding polyphosphate kinase, upstream of the gene ppx, encoding exopolyphosphatase (1). The multiplicity of the cdg operons is also noteworthy, since bacterial isoenzymes are uncommon. In E. coli, three different phospho-2-keto-3-deoxyheptonate aldolase isoenzymes catalyze the first step in the biosynthesis of aromatic amino acids (15), and each is subject to feedback inhibition. Anabaena has five different genes encoding adenylate cyclases which have similar C-terminal catalytic domains and differ in their N-terminal regulatory domains (21).
The cdg operons display a functional hierarchy, as indicated by analyses of recombinant strains disrupted at various regions in the cdg operons. Thus, cdg1 is the source of 80% of PDEA and DGC enzymatic activities, while cdg2 and cdg3 contribute 15 and 5%, respectively. If the operons contributed equally to enzymatic activities, the drastic differences in loss of function observed between the strains disrupted in cdg1 and those disrupted in cdg2 (Table 1) could not be expected. An alternate explanation is that mutation in operon cdg1 could exert a vectorial effect on the activities controlled by the other operon, if they were physically or functionally linked. This possibility may be dismissed, however, since disruption in dgc1 resulted in reduction of only DGC and not PDEA activity in Dis4. Vectorial effects are seen only within each operon, consistent with the physical linkage of one pdeA gene with one dgc gene.
Comparative examination of the predicted DGC and the PDEA protein sequences reveals a high degree of identity within each isoenzyme set, as well as significant structural conservation between the two sets (Fig. 6). All six isoenzymes contain GGDEF and EAL motif patterns in their C-terminal regions and putative sensory domains in their N-terminal regions. Putative GTP binding sites, likely corresponding to catalytic sites, are found on the DGC isoenzyme sequences within the GGDEF domain. Although it is difficult to postulate the molecular basis of the functional hierarchy displayed by the isoenzymes, differences in the microenvironments of the GTP sites could account for variation in catalytic rates and thus specific activities of the DGC isoenzymes. That the DGC isoenzymes do indeed vary with respect to specific activity has been shown by purification and characterization of DGC activities from strains Dis4, TRT150, and ABT21 (11), although relative expression patterns of the corresponding genes have yet to be examined.
Considering that A. xylinum is an obligate aerobe, c-di-GMP may act as a metabolic-state signal of oxygen tension, in addition to functioning as an activator of cellulose synthesis. Since cellulose production enables formation of a buoyant pellicle (36) through which contact with the atmosphere may be achieved, a regulatory system based on response to oxygen tension may be operative. The presence of putative sensory domains on the DGC and PDEA proteins, most similar to oxygen-sensing domains of the NifL and FixL proteins, respectively, could provide the capability for an oxygen-induced response. In their respective cascades, NifL is a negative repressor activated by aerobic conditions (26), whereas FixL is a positive activator induced by anaerobic conditions (27). By analogy, DGC and PDEA may each be activated by opposite extremes of oxygen tension, consistent with their other mutually exclusive modes of regulation, which are discussed below. Thus, under stressed physiological conditions requiring enhanced cellulose production, induction of dgc1 would enable the most efficient production of c-di-GMP for rapid cellulose synthase activation. At the same time, the product of cdg1a may act as a transcriptional activator specific for the c-di-GMP binding protein (46), which would also be required at a higher concentration to ensure targeting of c-di-GMP for cellulose synthase activation. We have indeed shown that plasmid-mediated overexpression of CDG1A in A. xylinum results in increased production of c-di-GMP binding protein (45). The similarity of CDG1A to the oxygen-sensitive transcriptional regulators AadR (13) and FixK (7) suggests that its expression may also be influenced by oxygen tension. On the other hand, under conditions that are unstressed with respect to oxygen supply, the cdg2 and cdg3 operons might operate constitutively to ensure a lower level of cellulose synthesis.
The genetic disruption experiments presented here indicate that c-di-GMP synthesis is probably essential for cellulose synthesis, although conclusive proof awaits disruption of the third DGC allele. The proposed essential role of c-di-GMP for cellulose synthesis is consistent with our findings at the physiological level, which show that excessive in vivo depletion of cellular c-di-GMP following replacement of cellular K+ with Na+ results in almost complete inhibition of cellulose synthesis (46). Such cation replacement enhances release of c-di-GMP from its bound form in association with the c-di-GMP binding protein, leaving it vulnerable to degradation by PDEA. Although c-di-GMP synthesis is essential for cellulose synthase activation, its degradation is probably not essential for the process, as suggested by the observation that c-di-GMP can activate cellulose synthase in the complete absence of PDEA (25). Thus, DGC and PDEA enzymatic activities within the cell could be coordinated such that when one is operative, the other is negatively affected, a mechanism distinct from the classical G-protein model whereby a GTP-to-GDP exchange reaction is required for target activation. A number of factors that may facilitate this regulation exist: (i) GTP, the substrate of the DGC reaction, acts as an inhibitor of PDEA activity at a site distinct from its catalytic site (36); (ii) Ca2+ inhibits PDEA but not DGC activity (36); and (iii) a glycosidic triterpenoid saponin (GTS) isolated from A. xylinum acts as a noncompetitive inhibitor of the DGC reaction, possibly by preventing release of the c-di-GMP product, and does not affect the PDEA reaction (30). A terminal glucosyl residue essential for GTS activity could be a locus of regulation, since the intracellular level of glucosidase activity may influence the level of biologically active GTS.
The identification of the GGDEF and EAL domains shared by all the DGC and PDEA isoenzymes suggests that either or both domains are involved with conserved regulatory mechanisms, or with guanyl-nucleotide binding, a function common to both enzymatic activities. Since these domains are also found in numerous bacterial proteins of undefined function, they could coordinate distinct regulatory activities required among diverse proteins. Alternatively, if these regions are specifically associated with c-di-GMP metabolism, the possibility arises that c-di-GMP has wider significance as a regulatory molecule for processes other than cellulose synthesis.
ACKNOWLEDGMENT
This work was supported by the Weyerhaeuser Co., Tacoma, Wash.
REFERENCES
- 1.Akiyama M, Crooke E, Kornberg A. An exopolyphosphatase of Escherichia coli. J Biol Chem. 1993;268:633–639. [PubMed] [Google Scholar]
- 2.Aloni Y, Cohen Y, Benziman M, Delmer D P. Solubilization of UDP-glucose: 1,4-β-d-glucan 4-β-d-glucosyl transferase (cellulose synthase) from Acetobacter xylinum. J Biol Chem. 1983;258:4419–4423. [PubMed] [Google Scholar]
- 3.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Amikam D, Benziman M. Cyclic diguanylic acid and cellulose synthesis in Agrobacterium tumefaciens. J Bacteriol. 1989;171:6649–6655. doi: 10.1128/jb.171.12.6649-6655.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amor Y, Mayer R, Benziman M, Delmer D P. Evidence for a cyclic diguanylic acid-dependent cellulose synthase in plants. Plant Cell. 1991;3:989–995. doi: 10.1105/tpc.3.9.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anthamatten D, Hennecke H. The regulatory status of the fixL- and fixJ-like genes in Bradyrhizobium japonicum may be different from that in Rhizobium meliloti. Mol Gen Genet. 1991;225:38–48. doi: 10.1007/BF00282640. [DOI] [PubMed] [Google Scholar]
- 7.Anthamatten D, Scherb B, Hennecke H. Characterization of a fixLJ-regulated Bradyrhizobium japonicum gene sharing similarity with the Escherichia coli fnr and Rhizobium meliloti fixK genes. J Bacteriol. 1992;174:2111–2120. doi: 10.1128/jb.174.7.2111-2120.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bibikov S I, Biran R, Rudd K E, Parkinson J S. A signal transducer for aerotaxis in Escherichia coli. J Bacteriol. 1997;179:4075–4079. doi: 10.1128/jb.179.12.4075-4079.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bolivar F, Rodrigues R L, Greene P J, Betlach M C, Heynecker H L, Boyer H W, Crosa J H, Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2:95–113. [PubMed] [Google Scholar]
- 10.Chang A C Y, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen, A., and M. Benziman. Unpublished data.
- 12.David M, Daveran M L, Batut J, Dedieu A, Domergue O, Ghai J, Hertig C, Boistard P, Kahn D. Cascade regulation of nif gene expression in Rhizobium meliloti. Cell. 1988;54:671–683. doi: 10.1016/s0092-8674(88)80012-6. [DOI] [PubMed] [Google Scholar]
- 13.Dispensa M, Thomas C T, Kim M K, Perrotta J A, Gibson J, Harwood C S. Anaerobic growth of Rhodopseudomonas palustris on 4-hydroxybenzoate is dependent on AadR, a member of the cyclic AMP receptor protein family of transcriptional activators. J Bacteriol. 1992;174:5803–5813. doi: 10.1128/jb.174.18.5803-5813.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drummond M H, Wootton J C. Sequence of nifL from Klebsiella pneumoniae: mode of action and relationship to two families of regulatory proteins. Mol Microbiol. 1987;1:37–44. doi: 10.1111/j.1365-2958.1987.tb00524.x. [DOI] [PubMed] [Google Scholar]
- 15.Ger Y M, Chen S L, Chiang H J, Shiuan D. A single ser-180 mutation desensitizes feedback inhibition of the phenylalanine-sensitive 3-deoxy-d-arabino-hepulosonate-7-phosphate (DAHP) synthetase in Escherichia coli. J Biochem (Tokyo) 1994;116:986–990. doi: 10.1093/oxfordjournals.jbchem.a124657. [DOI] [PubMed] [Google Scholar]
- 16.Hecht G B, Newton A. Identification of a novel response regulator for the swarmer-to-stalked cell transition in Caulobacter crescentus. J Bacteriol. 1995;177:6223–6229. doi: 10.1128/jb.177.21.6223-6229.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hernando Y, Palacios J M, Imperial J, Ruiz-Argueso T. The hypBFCDE operon from Rhizobium leguminosarum biovar viciae is expressed from an Fnr-type promoter that escapes mutagenesis of the fnrN gene. J Bacteriol. 1995;177:5661–5669. doi: 10.1128/jb.177.19.5661-5669.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hofmann K, Stoffel W. Tmbase—a database of membrane spanning protein segments. Biol Chem Hoppe-Seyler. 1993;347:166–171. [Google Scholar]
- 19.Huala E, Oeller P W, Liscum E, Han I-S, Larsen E, Briggs W R. Arabidopsis NPH1: a protein kinase with a putative redox-sensing domain. Science. 1997;278:2120–2123. doi: 10.1126/science.278.5346.2120. [DOI] [PubMed] [Google Scholar]
- 20.Kaminski P A, Mandon K, Arigoni F, Desnoues N, Elmerich C. Regulation of nitrogen fixation in Azorhizobium caulinodans: identification of a fixK-like gene, a positive regulator of nifA. Mol Microbiol. 1991;5:1983–1991. doi: 10.1111/j.1365-2958.1991.tb00820.x. [DOI] [PubMed] [Google Scholar]
- 21.Katayama M, Ohmori M. Isolation and characterization of multiple adenylate cyclase genes from the cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol. 1997;179:3588–3593. doi: 10.1128/jb.179.11.3588-3593.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kneller D G, Cohen F E, Langridge R. Improvements in protein secondary structure prediction by an enhanced neural network. J Mol Biol. 1990;214:171–182. doi: 10.1016/0022-2836(90)90154-E. [DOI] [PubMed] [Google Scholar]
- 23.Lin F C, Brown R M, Jr, Drake R R, Haley B E. Identification of the uridine 5′-diphosphoglucose (UDP-glc) binding subunit of cellulose synthase in Acetobacter xylinum using the photoaffinity probe 5-azido-UDP-glc. J Biol Chem. 1990;254:4782–4784. [PubMed] [Google Scholar]
- 24.Matthysse A G, White S, Lightfoot R. Genes required for cellulose synthesis in Agrobacterium tumefaciens. J Bacteriol. 1995;177:1069–1075. doi: 10.1128/jb.177.4.1069-1075.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mayer R, Ross P, Weinhouse H, Amikam D, Volman G, Ohana P, Calhoon R D, Wong H C, Emerick A W, Benziman M. Polypeptide composition of bacterial cyclic diguanylic acid-dependent cellulose synthase and the occurrence of immunologically crossreacting proteins in higher plants. Proc Natl Acad Sci USA. 1991;88:5472–5476. doi: 10.1073/pnas.88.12.5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merrick M, Hill S, Hennecke H, Hahn M, Dixon R, Kennedy C. Repressor properties of the nifL gene product of Klebsiella pneumoniae. Mol Gen Genet. 1982;185:75–81. [Google Scholar]
- 27.Monson E K, Weinstein M, Ditta G S, Helinski D R. The FixL protein of Rhizobium meliloti can be separated into a heme-binding oxygen-sensing domain and a functional C-terminal kinase domain. Proc Natl Acad Sci USA. 1992;89:4280–4284. doi: 10.1073/pnas.89.10.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Monson E K, Ditta G S, Helsinki D R. The oxygen sensor protein, FixL, of Rhizobium meliloti. Role of histidine residues in heme binding, phosphorylation, and signal transduction. J Biol Chem. 1995;270:5243–5250. doi: 10.1074/jbc.270.10.5243. [DOI] [PubMed] [Google Scholar]
- 28a.Nicholas K B, Nicholas H B, Jr, Deerfield D W., II GeneDoc analysis and visualization of genetic variation. EMBNEW NEWS. 1997;4:14. [Google Scholar]
- 29.Ohana P, Delmer D P, Carlson R W, Glushka J, Azadi P, Bacic T, Benziman M. Identification of a novel triperpenoid saponin from Pisum sativum as a specific inhibitor of the diguanylate cyclase of Acetobacter xylinum. Plant Cell Physiol. 1998;39:144–152. doi: 10.1093/oxfordjournals.pcp.a029351. [DOI] [PubMed] [Google Scholar]
- 30.Ohana P, Delmer D P, Volman G, Benziman M. Glycosylated triterpenoid saponin: a specific inhibitor of diguanlyate cyclase from Acetobacter xylinum. Biological activity and distribution. Plant Cell Physiol. 1998;39:153–159. doi: 10.1093/oxfordjournals.pcp.a029351. [DOI] [PubMed] [Google Scholar]
- 31.Pear J R, Kawagoe Y, Schreckengost W E, Delmer D P, Stalker D M. Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc Natl Acad Sci USA. 1996;93:12637–12642. doi: 10.1073/pnas.93.22.12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ross P, Aloni Y, Weinhouse C, Michaeli D, Weinberger-Ohana P, Meyer R, Benziman M. An unusual guanyl oligonucleotide regulates cellulose synthesis in Acetobacter xylinum. FEBS Lett. 1985;186:191–196. doi: 10.1016/0014-5793(85)80706-7. [DOI] [PubMed] [Google Scholar]
- 33.Ross P, Aloni Y, Weinhouse H, Michaeli D, Weinberger-Ohana P, Mayer R, Benziman M. Control of cellulose synthesis in A. xylinum. A unique guanyl oligonucleotide is the immediate activator of cellulose synthase. Carbohydr Res. 1986;149:101–117. [Google Scholar]
- 34.Ross P, Weinhouse H, Aloni Y, Michaeli D, Ohana P, Mayer R, Braun S, de Vroom E, van der Marel G A, van Boom J H, Benziman M. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature (London) 1987;325:279–281. doi: 10.1038/325279a0. [DOI] [PubMed] [Google Scholar]
- 35.Ross P, Mayer R, Amikam D, Hugirat Y, Weinhouse H, Benziman M, de Vroom E, Fiddler A, de Paus P, Sliedregt L A J M, van der Marel G A, van Boom J H. The cyclic diguanylate acid regulatory system of cellulose synthesis in Acetobacter xylinum: chemical synthesis and biological activity of cyclic nucleotide dimer trimer and phosphothioate derivatives. J Biol Chem. 1990;265:18933–18973. [PubMed] [Google Scholar]
- 36.Ross P, Mayer R, Benziman M. Cellulose biosynthesis and function in bacteria. Microbiol Rev. 1991;55:35–58. doi: 10.1128/mr.55.1.35-58.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saxena I M, Kudlicka K, Okuda K, Brown R M., Jr Characterization of genes in the cellulose-synthesizing operon (acs operon) of Acetobacter xylinum: implication for cellulose crystallization. J Bacteriol. 1994;176:5735–5752. doi: 10.1128/jb.176.18.5735-5752.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spiro S, Guest J R. FNR and its role in oxygen-regulated gene expression in Escherichia coli. FEMS Microbiol Rev. 1990;75:399–428. doi: 10.1111/j.1574-6968.1990.tb04109.x. [DOI] [PubMed] [Google Scholar]
- 41.Spiro S, Gaston K L, Bell A I, Roberts R E, Busby S J W, Guest J R. Interconversion of the DNA-binding specificities of two related transcriptional regulators, CRP and FNR. Mol Microbiol. 1990;4:1831–1838. doi: 10.1111/j.1365-2958.1990.tb02031.x. [DOI] [PubMed] [Google Scholar]
- 42.Thelen M P, Delmer D P. Gel-electrophoretic separation, detection, and characterization of plant and bacterial UDP-glucose glucosyltransferases. Plant Physiol. 1986;81:913–918. doi: 10.1104/pp.81.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weber I T, Steitz T A. The structure of a complex catabolite gene activator and cyclic AMP refined at 2.5 Å resolution. J Mol Biol. 1987;198:311–326. doi: 10.1016/0022-2836(87)90315-9. [DOI] [PubMed] [Google Scholar]
- 45.Weinhouse, H., and M. Benziman. Unpublished data.
- 46.Weinhouse H, Sapir S, Amikam D, Shilo Y, Volman G, Ohana P, Benziman M. c-di-GMP-binding protein, a new factor regulating cellulose synthesis in Acetobacter xylinum. FEBS Lett. 1997;416:207–211. doi: 10.1016/s0014-5793(97)01202-7. [DOI] [PubMed] [Google Scholar]
- 47.Wong H C, Fear A L, Calhoon R D, Eichinger G H, Mayer R, Amikam D, Benziman M, Gelfand D H, Meade J H, Emerick A W, Bruner R, Ben-Bassat A, Tal R. Genetic organization of the cellulose synthase operon in Acetobacter xylinum. Proc Natl Acad Sci USA. 1990;87:8130–8134. doi: 10.1073/pnas.87.20.8130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wootton J C, Drummond M H. The Q-linker: a class of interdomain sequences found in bacterial multidomain regulatory proteins. Protein Eng. 1989;2:535–543. doi: 10.1093/protein/2.7.535. [DOI] [PubMed] [Google Scholar]
- 49.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequence of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]