Abstract
Quorum sensing control mediated by N-acyl homoserine lactone (AHL) signaling molecules has been established as a key feature of the regulation of exoenzyme production in many gram-negative bacteria. In Chromobacterium violaceum ATCC 31532 a number of phenotypic characteristics, including production of the purple pigment violacein, hydrogen cyanide, antibiotics, and exoproteases are known to be regulated by the endogenous AHL N-hexanoyl-l-homoserine lactone (HHL). In this study we show that C. violaceum produces a set of chitinolytic enzymes whose production is regulated by HHL. The chitinolytic activity was induced in strains grown in the presence of chitin as the sole carbon source and quantitated in the secreted proteins by using p-nitrophenol analogs of disaccharide, trisaccharide, and tetrasaccharide oligomers of N-acetylglucosamine. By using 4-methylumbelliferyl analogs of the same oligomers of N-acetylglucosamine as substrates for proteins separated and renatured by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, at least six enzymes were detected: a chitobiase with high specificity to a dimeric substrate of 87 kDa, two N-acetylglucosaminidases with apparent molecular masses of 162 and 133 kDa, two endochitinases of 108 and 67 kDa, and a chitobiosidase of 56 kDa. In addition, two unidentified bands of >205 kDa were found where a tetrameric chitin derivative was used as a substrate. A pleiotropic mini-Tn5 mutant of C. violaceum (CV026) that is defective in HHL production and other quorum-sensing-regulated factors was also found to be completely deficient in chitinolytic activity. Growth of this mutant on minimal medium with chitin supplemented with culture supernatant from the C. violaceum wild-type strain or 10 μM synthetic HHL restored chitinase production to the level shown by the parental strain. These results constitute the most complete evidence so far for regulation of chitinolytic activity by AHL signaling in a gram-negative bacterium.
Many species of bacteria are known to synthesize enzymes capable of degrading chitin, an insoluble linear polymer consisting of β-1,4-linked N-acetylglucosamine (GlcNAc) units that forms the main structural component of cell walls of most fungi and arthropods (33, 43). Among gram-negative bacteria, chitinolytic activity has been described for strains from the genera Aeromonas (3, 16), Alteromonas (53), Enterobacter (5), Pseudomonas (11, 54, 57), Serratia (20, 29, 39), Ewingella (17), and Vibrio (1, 18, 60). Although studies on chitinolytic activity in Vibrio (1, 21, 46) and Pseudoalteromonas (49) spp., have shown that soluble oligosaccharides liberated by the action of extracellular chitinase on chitin elicit the induction of expression of a number of proteins, little is known at present about the genetic regulation of chitinolytic enzyme expression in gram-negative bacteria.
In strain Pseudomonas fluorescens BL915, studied by Gaffney et al. (11), expression of uncharacterized chitinolytic activity is regulated by a two-component system consisting of a transmembrane environmental sensor protein (LemA) and a cytoplasmic response regulator protein (GacA) (7, 25, 38). Cloning of the gacA regulatory region from strain BL915 in certain heterologous soil isolates of P. fluorescens was found to stimulate expression of otherwise latent chitinase genes (11), indicating that global regulation by two-component regulators may be a common feature of the regulation of chitinase expression.
It has emerged over the last few years that expression of many phenotypic characteristics in late-growth-phase bacterial cultures, including cell differentiation and the production of secondary metabolites and exoenzymes, is a cell density-dependent phenomenon mediated by intercellular communication in a process known as quorum sensing (10). In gram-negative bacteria, quorum sensing control is typically regulated by N-acyl homoserine lactone molecules (AHLs). These signal molecules have been reported to control a multitude of characteristics (reviewed in references 9 and 48), including extracellular enzyme production in Pseudomonas aeruginosa (19, 24, 32, 57), Erwinia carotovora (19, 35), and Chromobacterium violaceum (50, 51). At the core of the majority of AHL-based quorum sensing systems so far described are genes encoding protein products with homology to LuxI and LuxR from Vibrio fischeri (8, 10). Proteins with homology to LuxI are responsible for AHL biosynthesis, whereas LuxR homologs act as transcriptional activators, interacting with the accumulated AHL small molecule signals to stimulate gene expression (for reviews see references 9, 28, and 48). There is good evidence to suggest that in Pseudomonas AHL-mediated regulation may in turn be controlled by a global GacA-LemA regulation system (37). Thus far, more than 16 genera of gram-negative bacteria have been reported to utilize AHL regulation in the control of a variety of metabolic traits (9, 48). However, despite the widespread nature of this means of communication in gram-negative bacteria, the involvement of AHL signaling in the regulation of chitinase(s) has only been demonstrated in Pseudomonas aeruginosa PAO1, where butanoyl-l-homoserine lactone synthesized by the VsmI (RhlI) protein was shown to restore uncharacterized extracellular chitinolytic activity in the P. aeruginosa pleiotropic mutant PAN067 (57).
A strain of C. violaceum, ATCC 12472, selected from a variety of chitin-utilizing bacterial species as the most active in chitin degradation, has previously been shown to grow on crystalline or colloidal chitin as its sole carbon and nitrogen source (45). More recently, in C. violaceum ATCC 31532 (a strain originally isolated as a monobactam antibiotic producer [55]), the production of a variety of factors, including violacein pigment, antibiotics, hydrogen cyanide, and proteases, has been shown to be controlled by the endogenous AHL inducer molecule N-hexanoyl-l-homoserine lactone (HHL) (26, 50, 56). We describe here the existence of a number of chitinolytic enzymes in C. violaceum ATCC 31532 and demonstrate that chitinolytic activity is controlled by quorum sensing regulation mediated by the endogenous AHL HHL.
MATERIALS AND METHODS
Cultures and growth media.
Three related strains of C. violaceum were used in this work: the wild-type C. violaceum ATCC 31532 (CVWT) (HHL producer) and two mutants affected in quorum sensing regulation obtained after mini-Tn5 mutagenesis of a spontaneous streptomycin-resistant mutant of CVWT. CV017 (Smr mini-Tn5 Hgr) produces HHL and carries a genetically uncharacterized mutation causing derepression of the HHL-inducible violacein pigment production at 30°C; CV026 (Smr mini-Tn5 Hgr cviI::Tn5xylE Kmr) is a non-HHL producer derived from CV017 as a result of mini-Tn5 insertion in the cviI gene encoding the LuxI homolog, CviI. This mutant is nonpigmented unless provided with exogenous AHL and thus acts as a biosensor (26, 56). The Enterobacter agglomerans strains IC1270 and 40b described previously (5, 47) were used as controls. For bacterial growth, liquid or solid (1.5% [wt/vol] agar) Luria broth (LB) and agar or liquid synthetic medium (SM) (31) with 0.4% (wt/vol) glucose or sucrose and 10% (vol/vol) LB were used. To induce chitinolytic activity, bacteria were grown in SM with 0.2% (wt/vol) colloidal chitin and 10% (vol/vol) LB. The colloidal chitin was prepared by the method of Rodriguez-Kabana et al. (41) by partial hydrolysis with 10 N HCl followed by repeated washings with water to give a final pH of 6.0 to 6.5. Where indicated, strain CV026 was grown in SM with colloidal chitin supplemented with HHL at a final concentration of 10 μM or with cell-free culture medium of the strain CVWT (10% [vol/vol]). The conditioned culture medium was obtained by growing the CVWT strain in LB for 24 h at 28°C with aeration. The cells were centrifuged (4,000 rpm at 4°C, 15 min), and then the supernatant was filtered through 0.45-μm (pore size) filters (Schleicher and Schuell) and stored at −12°C.
Detection of chitinolytic activity on agar plates and in liquid medium.
Cells were seeded onto plates with semiminimal medium consisting of a mixture of SM and LB (10% [vol/vol]) supplemented with colloidal chitin (0.2% [wt/vol]) and solidified with 1.5% agar or were placed in tubes containing the same medium but without agar. Where indicated, HHL (10 μM) or cell-free supernatant (10% [vol/vol]) of the CVWT strain (instead of LB) was added. The plates and tubes were incubated at 30°C for 72 to 96 h until zones of clearing of the chitin could be seen around the colonies or until the degradation of the chitin particles was observed in the liquid growth medium.
Preparation of extracellular and intracellular proteins.
Cells were grown in SM with 0.2% colloidal chitin as the sole carbon source and LB (10% [vol/vol]), or in SM with glucose (0.4% [vol/vol]) and LB (10% [vol/vol]) for 72 h at 28°C with aeration. Either HHL (10 μM) or cell-free supernatants (10% [vol/vol]) of the strain CVWT (instead of LB) were added as appropriate. The cells were centrifuged, and the supernatant was filtered through 0.45-μm filters (Schleicher and Schuell). Intracellular proteins were extracted from cells with a French pressure cell press (Aminco) at 1,500 lb/in2. Debris was separated by centrifugation, and the extracts were filtered as described above. Phenylmethylsulfonyl fluoride (final concentration, 0.2 mM) (Sigma) was added to the filtrates as a protease inhibitor. Filtrates containing extracellular or intracellular proteins were used for solution assays of chitinolytic enzymes. For analysis by gel electrophoresis, the filtrates were first dialyzed and concentrated in Micro-ProDiCon membranes (molecular weight cutoff, 25 kDa) against distilled water at 4°C in a Micro-ProDiCon negative-pressure microprotein dialysis-concentrator (Bio-Molecular Dynamics).
Assays of chitinolytic activity in solutions.
Initial experiments used carboxymethyl-chitin-remazol brilliant violet (CM-chitin-RBV) (42) to detect endochitinase activity in C. violaceum culture supernatants on agar plates and after gel electrophoresis of extracellular proteins. Although this method is a useful indicator of the presence of endochitinase activity against a polymeric chitin substrate, the use of a range of labeled GlcNAc oligomers as assay substrates is more instructive for determining specific chitinase activities. In order to measure chitinolytic activity in solutions, a chromogenic assay procedure with p-nitrophenyl-labeled substrates was performed according to the method of Roberts and Selitrennikoff (40) with minor modifications (14). The following chromogenic oligomers of GlcNAc were used as substrates: p-nitrophenyl-N-acetyl-β-d-glu- cosaminide (pNP-GlcNAc), p-nitrophenyl-β-d-N,N′-diacetylchitobiose [pNP- (GlcNAc)2], and p-nitrophenyl-β-d-N,N′,N"-triacetylchitotriose [pNP-(GlcNAc)3] (Sigma). The standard reaction mixture contained ca. 10 μg of the proteins tested in 0.1 M phosphate buffer (pH 6.5) and 10 μl of stock solution (1 to 2 mg ml−1) of one of the three above-mentioned substrates. The reaction mixture was incubated at 40°C in a water bath until a slight yellow-green color appeared. The reaction was terminated by adding an equal volume of 0.2 M Na2CO3. The release of the chromophore p-nitrophenol (pNP) from the substrates was measured at 410 nm, and 1 U of enzymatic activity was defined as 1 μmol of pNP/μg of protein/h. Protein content was determined with the Bio-Rad protein assay reagent and bovine serum albumin as a standard.
Detection of chitinolytic enzymes after SDS-PAGE.
Proteins concentrated in the Micro-ProDiCon system were prepared in sample buffer (22) without 2-mercaptoethanol (except when specifically indicated) and incubated for 10 min at room temperature prior to loading. Where sample buffer containing 2-mercaptoethanol was used, the samples were boiled for 4 min prior to loading. The proteins were separated by sodium dodecyl sulfate (SDS)–7.5% polyacrylamide gel electrophoresis (PAGE). Enzymes were reactivated in the gels by removing SDS by the casein-EDTA procedure (27) as modified by Haran et al. (13). Enzyme activity was detected on gels by using fluorescent substrates as described by Tronsmo and Harman (52). The chitinolytic enzymes appeared as fluorescent bands under UV light because of enzymatic hydrolysis of fluorescent 4-methylumbelliferone from the GlcNAc mono- and oligosaccharides. The following substrates were used: 4-methylumbelliferyl-N-acetyl-β-d-glucosaminide (4-MU-GlcNAc); 4-methylumbelliferyl-β-d-N,N′-diacetylchitobioside [4-MU-(GlcNAc)2]; and 4-methylumbelliferyl-β-d-N,N′,N"-triacetylchitotriose [4-MU-(GlcNAc)3] (Sigma). These compounds served as analogs of disaccharide, trisaccharide, and tetrasaccharide chitin derivatives, respectively, with the 4-methylumbelliferyl group linked by β-1,4 linkage to the GlcNAc monosaccharide (in the case of 4-MU-GlcNAc) or oligosaccharides [in the case of 4-MU-(GlcNAc)2 and 4-MU-(GlcNAc)3]. Each lane contained about 120, 20, and 20 μg of protein for detection with tetrameric, trimeric, and dimeric chitin derivatives, respectively. The molecular weights of the renaturated chitinases were estimated by using high-range prestained standards (Bio-Rad Laboratories). Proteins separated by SDS-PAGE were stained with Coomassie brilliant blue G-250 prepared as described by Neuhoff et al. (30). Each lane contained about 30 μg of protein.
RESULTS
Chitinolytic activity.
C. violaceum CVWT and CV017 hydrolyzed colloidal chitin after 72 to 96 h of growth on semiminimal agar (SM plus LB at a 10:1 ratio) supplemented with colloidal chitin as the sole carbon source. Large zones of clearing around the growing bacteria were observed (Fig. 1). Two additional strains were tested on the same plate as the controls. Enterobacter agglomerans IC1270 was previously shown to produce a set of chitinolytic enzymes (5) and was used as a positive control; E. agglomerans 40b (47) exhibits no detectable chitinolytic activity and was used as the negative control. The HHL-deficient mutant (CV026) derived from CV017 was unable to form zones of chitin clearing on the solid medium or to degrade colloidal chitin added to the liquid SM medium (Fig. 2, left). However, supplementation of the growth medium with HHL (10 μM) restored chitinolytic activity (Fig. 2, right). The same effect was observed if cell-free supernatant of CVWT (10% [vol/vol]) was added instead of HHL (data not shown).
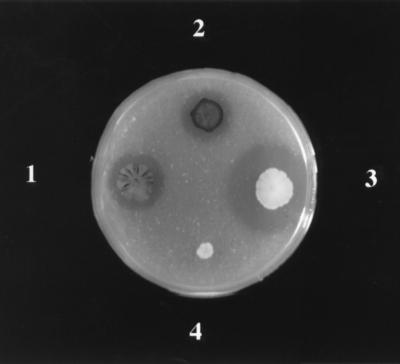
FIG. 1.
Assay of chitinolytic activity on plates with SM agar medium supplemented with colloidal chitin (0.2% [wt/vol]). Clearing zones of colloidal chitin formed around the colonies of C. violaceum strains CVWT (1) and CV017 (2). Strains E. agglomerans IC1270 (3) and 40b (4) were seeded on the plate as positive and negative controls, respectively. The plate was incubated at 28°C for 4 days.
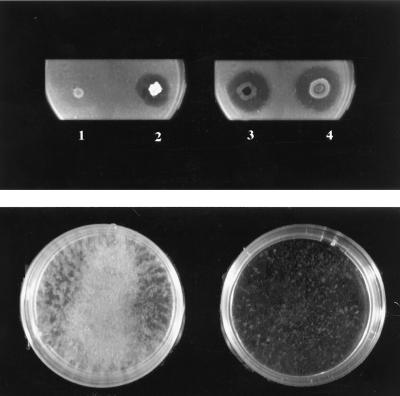
FIG. 2.
(Top) Assay of chitinolytic activity on a plate with SM agar supplemented with colloidal chitin (0.2% [wt/vol]) (left) and on a plate with SM agar supplemented with colloidal chitin (0.2 [wt/vol]) plus HHL (10 μM) (right): 1 and 3, growth of mutant CV026; 2 and 4, growth of E. agglomerans IC1270 strain (positive control). Plates were incubated at 28°C for 4 days. (Bottom) Assay of chitinolytic activity of mutant CV026 grown in liquid SM medium with colloidal chitin (0.2% [wt/vol]) (left) and in the same medium supplemented with HHL (10 μM) (right). The colloidal chitin was almost completely hydrolyzed in the HHL-supplemented medium, whereas particles of nonhydrolyzed chitin are clearly visible in the unsupplemented medium. Bacteria were grown on the medium indicated at 28°C for 72 h with agitation (200 rpm).
These results suggested that the chitinolytic activity secreted into the culture medium by C. violaceum is subject to quorum sensing regulation stimulated by the HHL signal molecule. To investigate this chitinolytic activity further with a quantitative assay, supernatants from 72-h cultures of the C. violaceum wild-type, CV017, and CV026 strains grown in liquid SM with chitin and of the CV026 strain grown in the same medium supplemented with HHL (10 μM) were assayed. The chitinolytic activity of extracellular proteins was examined by assessing the release of the chromophore pNP in reaction mixtures with chromogenic analogs of di- and trioligomers of GlcNAc. The activity was observed with both of these substrates (Fig. 3), but the level was found to be strain specific; in the extracellular proteins of strain CVWT it was higher in the reaction with the pNP monosaccharide derivative (pNP-GlcNAc) and lower with pNP-(GlcNAc)2 (Fig. 3, columns 1 and 2), while proteins secreted by strain CV017 exhibited almost the same level of activity with both substrates (Fig. 3, columns 3 and 4). Almost no activity was found in secreted proteins of the mutant CV026 (Fig. 3, columns 5 and 6) grown on SM with chitin. However, when CV026 was grown on SM with chitin supplemented with HHL, the chitinolytic activity was restored to a level comparable to that of strain CV017 (Fig. 3, columns 7 and 8). The same restoration of chitinolytic activity was observed in a culture of CV026 when a filtrate of conditioned culture medium of strain CVWT was added to SM plus chitin medium (data not shown). These results indicate that when grown in chitin-containing medium, C. violaceum CVWT and CV017 produce chitinolytic enzyme(s) in their culture supernatants which exhibit activity against di- and trioligomers of GlcNAc. Only very weak chitinolytic activity was detected in fractions containing intracellular proteins. The chitinolytic activity was restored in culture supernatants of the HHL-deficient and chitinase-deficient mutant CV026 by growth in the presence of either synthetic HHL or culture supernatant from CVWT. None of these strains showed constitutive chitinolytic activity when the chitin in the medium was replaced with glucose or sucrose, independent of HHL supplementation (data not shown).
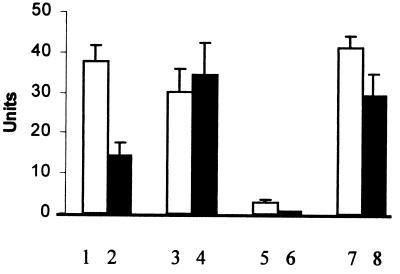
FIG. 3.
Chitinolytic activity in extracellular proteins of C. violaceum strains. Assays were performed with pNP-GlcNAc (columns 1, 3, 5, and 7) or pNP-(GlcNAc)2 (columns 2, 4, 6, and 8). Activity shown was found in extracellular proteins of strains CVWT (columns 1 and 2), CV017 (columns 3 and 4), and CV026 (columns 5 and 6) grown in SM with chitin. Activity shown in columns 7 and 8 was found in extracellular proteins of strain CV026 grown in SM containing chitin and supplemented with HHL (10 μM). Values (mean ± standard error) were determined from 11 independently obtained preparations.
Identification of chitinolytic enzymes.
Preliminary experiments with CM-chitin-RBV to detect endochitinase activity directly in C. violaceum wild-type and mutant culture supernatants and after SDS-PAGE separation of extracellular proteins showed endochitinase activity in CV017 (data not shown). To determine the specific chitinolytic activities of extracellular proteins which had been renatured following their separation by SDS-PAGE, a set of three fluorescent chitin derivatives were used. Chitinolytic enzymes appear as fluorescent bands under UV light as a result of hydrolysis of the fluorescent substrate 4-methylumbelliferone from the GlcNAc mono- and oligosaccharides. Although no fluorescent bands were seen with extracellular proteins of mutant CV026 growing on medium containing chitin (Fig. 4, lanes 1), supplementation of the same culture medium with 10 μM HHL resulted in a band pattern similar to that obtained for CV017 (Fig. 4, lanes 2 and 3). Extracellular proteins exhibiting chitinolytic activity were designated according to their apparent molecular masses. Bands of Chit133 and Chit162 were detected with all three substrates used; bands of Chit67 and Chit108 were detected with analogs of trimeric [4-MU-(GlcNAc)2] and tetrameric [4-MU-(GlcNAc)3] chitooligosaccharides, and a band of Chit87 appeared only with the analog of dimeric chitin [4-MU-(GlcNAc)]. Chit56 activity was revealed only with the trimeric analog, although the activity was lower than for Chit67.
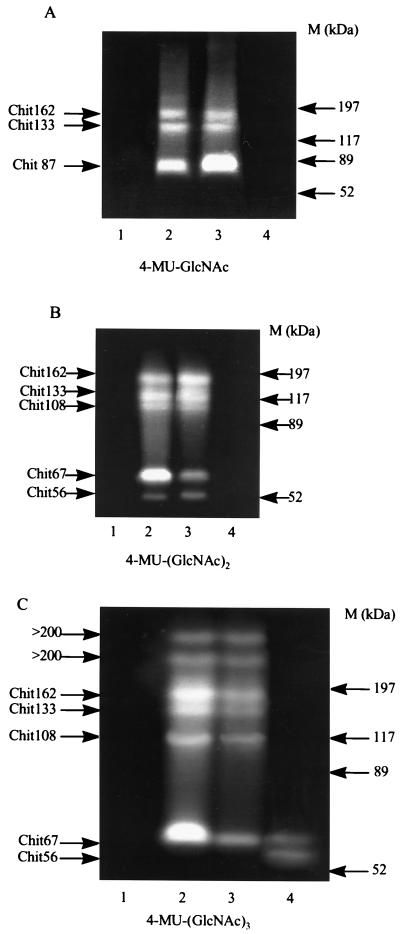
FIG. 4.
Detection of chitinolytic enzymes in extracellular proteins after separation by SDS-PAGE. Chitinolytic activity was detected with the substrates 4-MU- GlcNAc (A), 4-MU-(GlcNAc)2 (B), and 4-MU-(GlcNAc)3 (C). Lanes 1 and 2, extracellular proteins from strains CV026 and CV017 grown on SM with chitin; lane 3, extracellular proteins from strain CV026 grown on SM with chitin supplemented with HHL (10 μM); lane 4, extracellular proteins of strain CV017 boiled for 4 min in sample buffer containing 2-mercaptoethanol prior to loading. High-range prestained SDS-PAGE standards (Bio-Rad) were used as size markers (M). The positions of protein size markers are shown on the right.
Most of these bands of chitinolytic activity were found to be similarly nonrenatured by casein-EDTA after being boiled in sample buffer with 2-mercaptoethanol (Fig. 4, lanes 4). However, bands of Chit56 and Chit67 were just visible with 4-MU-(GlcNAc)3 after this treatment (Fig. 4C, lane 4), and only the band of 67 kDa could be observed with the 4-MU-(GlcNAc)2 after extended (up to 1 h instead of a few minutes) incubation of the gel with the substrate. Two additional bands of chitinolytic activity, each with a molecular mass higher than 200 kDa, were observed in secreted proteins when the tetrameric, but not the dimeric or trimeric, chitin derivatives were used as substrates. However, these bands were absent from preparations boiled in the presence of 2-mercaptoethanol (Fig. 4C, lane 4) or heated at 55°C for 3 min (data not shown).
Bands corresponding to the chitinolytic enzymes described above could be observed by SDS-PAGE after Coomassie blue staining of extracellular proteins produced by strains CV017 and CVWT grown in the presence of colloidal chitin or mutant CV026 grown on SM plus chitin supplemented with HHL. These proteins were not visible or were very weakly visible when strains CV017 and CVWT were grown on glucose (or sucrose) instead of chitin or when the mutant CV026 was grown on SM plus chitin without the addition of HHL (data not shown).
DISCUSSION
A number of phenotypic traits of C. violaceum are known to be controlled by quorum sensing regulation mediated by an AHL molecule. The results presented here extend this plethora of factors to include the most complete example of quorum-sensing-regulated control of chitinolytic activity enabling a gram-negative bacterium to utilize chitin as a growth source. In this system, quorum sensing regulation is mediated by HHL synthesized by C. violaceum. Two of the test strains (CVWT and its derivative CV017) produce HHL molecules and release them into the culture medium, while CV026, a derivative of CV017 with a mini-Tn5 insertion in the autoinducer synthase gene, is deficient in HHL production. In the presence of HHL, either produced by the CVWT or CV017 strain or supplied exogenously for CV026, all of the strains exhibited strong chitinolytic activity, as determined by the formation of clearing zones on chitin agar, by degradation of colloidal chitin fine particles in liquid medium, and by the release of pNP from the chromogenic chitooligosaccharide analogs. No activity or only very weak activity was found for intracellular protein of the strains, suggesting that the chitinolytic enzymes are almost completely extracellular.
We used a set of fluorescent 4-MU glucosides of GlcNAc mono- and oligosaccharides as substrates to identify the chitinolytic activity of extracellular proteins renatured after their separation by SDS-PAGE. The same pattern of bands showing chitinolytic activity was observed in the preparation of proteins secreted by the mutant strains CV017 and CV026 when chitinolytic activity in the latter was restored by induction with HHL in the growth medium. Chitinolytic activity was detected only when the bacteria were grown in an inducing medium containing colloidal chitin. The enzymes detected differed in their substrate specificities. Those designated according to their apparent molecular masses as Chit162 and Chit133 released fluorescent 4-MU from all three substrates. Fluorescent bands corresponding to Chit108 and Chit67 appeared as a result of the release of 4-MU from trimeric [4-MU-(GlcNAc)2] and tetrameric [4-MU-(GlcNAc)3] chitin analogs, but not from the dimeric analog 4-MU-GlcNAc. The band of Chit56 was detected only when 4-MU-(GlcNAc)2 was used, and its activity with this substrate was apparently weaker than that of Chit67. Finally, the band of Chit87 was detected only with the dimeric chitin derivative 4-MU-GlcNAc. All of these protein bands could also be detected by Coomassie staining.
According to the nomenclature suggested by Sahai and Manocha (43), the chitinolytic enzymes are divided into three principal types. Endochitinases (EC 3.2.1.14) are defined as enzymes catalyzing the random hydrolysis of 1,4-β linkages of GlcNAc at internal sites over the entire length of the chitin microfibrils. The products of the reaction are soluble, low-molecular-mass multimers of GlcNAc, such as chitotetraose, chitotriose, and diacetylchitobiose. Exochitinases, also termed “chitobiosidases” or chitin-1,4-β-chitobiosidases (14), catalyze the progressive release of diacetylchitobiose units in a stepwise fashion as the sole product from the chitin chains, so that no monosaccharides or oligosaccharides are formed. The third is N-acetyl-β-1,4-d-glucosaminidase (EC 3.2.1.30), a chitinolytic enzyme which also acts in exo-splitting mode on diacetylchitobiose and higher analogs of chitin, including chitotriose and chitotetraose, resulting in GlcNAc monomers. This definition inherently includes the chitobiase activity which specifically hydrolyzes diacetylchitobiose, forming GlcNAc monomers. Based on this nomenclature, the protein band of Chit87, which appears to be due to hydrolysis of a fluorescent product from 4-MU-GlcNAc, but not from trimeric [4-MU-(GlcNAc)2] or tetrameric [4-MU-(GlcNAc)3], analogs of chitin, can be regarded as a chitobiase. This type of activity, specifically hydrolyzing the dimeric chitin analog, has been observed previously in some acaropathogenic fungi (4).
The enzymes of 108 and 67 kDa can be classified as chitinases with an endo-mode of chitin splitting. They produced a fluorescent product from 4-MU-(GlcNAc)2 and 4-MU-(GlcNAc)3 (indicating random cleavage at internal sites along the entire length of the chitin analog), but they did not hydrolyze 4-MU from the dimeric analog. Chit56 produced 4-MU only from the trisaccharide analog 4-MU-(GlcNAc)2, and consequently we defined this enzyme to be a chitobiosidase. Chitobiosidase activity has been reported for several bacteria, including E. agglomerans (5), Serratia marcescens (2), and Bacillus cereus (36). Unlike the other chitinases produced by the C. violaceum strains studied, the 56- and 67-kDa proteins retained chitinolytic activity after renaturation irrespective of prior treatment with 2-mercaptoethanol. Similar characteristics have been reported for E. agglomerans enzymes of 50 and 58 kDa (5, 6). However, at present it is unclear why Chit56 renatured after treatment with 2-mercaptoethanol should be active against the tetrameric substrate when it exhibits only weak activity for a trimeric substrate known to be specific for chitobiosidase.
Experiments with C. violaceum grown in SM medium containing glucose, sucrose, or 10% LB instead of colloidal chitin as the carbon source indicated that the chitinolytic activity was not induced under these growth conditions either in the presence or absence of HHL. These results support the assumption that colloidal chitin, or more specifically low-molecular-weight breakdown products of chitin, are required for the induction of chitinolytic enzyme production in C. violaceum. The colloidal chitin used in these experiments was prepared by partial hydrolysis with 10 N HCl and sterilized by autoclaving. Under these conditions d-glucosamine and GlcNAc, known inducers for the synthesis of chitinase (44), are released. In previous studies colloidal chitin prepared by this technique has been shown to induce a wide spectrum of chitinolytic enzymes, including N-acetylglucosaminidase, endochitinase, and chitobiosidase in the bacterial species Aeromonas caviae (16), E. agglomerans (5, 6), and Bacillus cereus (36) and in the fungal species Trichoderma harzianum (13) and Hirsutella sp. (4). Moreover, the results for growth of the wild-type and mutant strains of C. violaceum on colloidal chitin clearly indicate that induction with an AHL (HHL) is likewise an absolute requirement for the expression of chitinolytic activity, even in the presence of the substrate-level inducers. It is significant that induction with HHL stimulates the coordinate expression of a combination of chitinolytic enzymes with different specificities for polymeric chitin and degradation products, as this enables C. violaceum to use the resultant GlcNAc subunits as a sole source of carbon and nitrogen for growth. A similar situation has previously been observed in a mutant of E. agglomerans, in which a single Tn5 insertion was found to downregulate the coordinate expression of several chitinases (5).
The experimental conditions used here, with colloidal chitin as the sole carbon source and in the presence of chitin breakdown products and synthetic or endogenous HHL, were designed to be optimal for expression of AHL-regulated chitinolytic activity. However, although we used up-to-date and sensitive methods for the determination of chitinolytic enzymes (2, 52), the possibility remains that some chitinase activity may go undetected because of denaturation by treatment with SDS. In this analysis we have therefore restricted our interpretation to the enzymes which could be observed through their functional activity against the particular substrates used.
There is a wealth of data supporting the important role of chitinolytic enzymes in microbial antagonism, including antagonism between different bacteria and between bacteria and fungi in the rhizosphere (for reviews see references 12 and 23). Although C. violaceum usually constitutes only a minor component of the total microflora found in soil and water, some strains isolated from rhizosphere soil of maize and used for the inoculation of maize seeds were found to significantly increase plant yield (15). This could be the result of antagonism towards other soilborne bacterial and fungal plant pathogens. It has been suggested that quorum sensing signaling by AHLs may contribute to the success of a bacterium in competition with other rhizosphere inhabitants (34, 59). As C. violaceum also produces antibiotics and hydrogen cyanide under AHL-mediated control (56), a combination of these factors may be important. Results from preliminary in vitro experiments show that C. violaceum is able to suppress the growth of the fungal phytopathogens Pythium aphanidermatum and Rhizoctonia solani and that in the HHL-deficient mutant this correlates with supplementation of HHL to the growth medium (58). Future studies will focus on how other bacterial regulatory systems interact with quorum sensing signaling to modulate the expression of a number of factors, including chitinolytic enzyme production, which may contribute to successful biocontrol of plant pathogens.
ACKNOWLEDGMENTS
This work was supported in part by grant no. T06045 from the Biotechnology and Biological Sciences Research Council (Swindon, Wiltshire, United Kingdom) (to P.W., G.S.A.B.S, and B.W.B.) and a grant from the U.S. Agency for International Development (Washington, D.C.), U.S.-Israel CDR-CAR program, to I.C. and L.S.C. (grant no. TA-MOU-97-CA16-012).
REFERENCES
- 1.Bassler B L, Yu C, Lee Y C, Roseman S. Chitin utilization by marine bacteria. Degradation and catabolism of chitin oligosaccharides by Vibrio furnissii. J Biol Chem. 1991;266:24276–24286. [PubMed] [Google Scholar]
- 2.Brurberg M B, Nes I F, Eijsink V G H. Comparative studies of chitinases A and B from Serratia marcescens. Microbiology. 1996;142:1581–1589. doi: 10.1099/13500872-142-7-1581. [DOI] [PubMed] [Google Scholar]
- 3.Chen J P, Nagayama F, Chang M C. Cloning and expression of a chitinase gene from Aeromonas hydrophila in Escherichia coli. Appl Environ Microbiol. 1991;57:2426–2428. doi: 10.1128/aem.57.8.2426-2428.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chernin L, Gafni A, Moses-Koch R, Gerson U, Szteinberg A. Chitinolytic activity of the acaropathogenic fungi Hirsutella thompsonii and Hirsutella necatrix. Can J Microbiol. 1997;43:440–446. [Google Scholar]
- 5.Chernin L, Ismailov Z, Haran S, Chet I. Chitinolytic Enterobacter agglomerans antagonistic to fungal plant pathogens. Appl Environ Microbiol. 1995;61:1720–1726. doi: 10.1128/aem.61.5.1720-1726.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chernin L S, de la Fuenta L, Sobolev V, Haran S, Vorgias C E, Oppenheim A B, Chet I. Molecular cloning, structural analysis, and expression in Escherichia coli of a chitinase gene from Enterobacter agglomerans. Appl Environ Microbiol. 1997;63:834–839. doi: 10.1128/aem.63.3.834-839.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corbell N, Loper J E. A global regulator of secondary metabolite production in Pseudomonas fluorescens Pf-5. J Bacteriol. 1995;177:6230–6236. doi: 10.1128/jb.177.21.6230-6236.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engebrecht J, Silverman M. Nucleotide sequence of the regulatory locus controlling expression of bacterial genes for bioluminescence. Nucleic Acids Res. 1987;15:10455–10467. doi: 10.1093/nar/15.24.10455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuqua C, Winans S C, Greenberg E P. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 10.Fuqua W C, Winans S C, Greenberg E P. Quorum sensing in bacteria: the LuxR-LuxI family of cell-density responsive transcriptional regulators. J Bacteriol. 1994;176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaffney T D, Lam S T, Ligon J, Gates K, Frazelle A, Di Maio J, Hill S, Goodwin S, Torkewitz N, Allshouse A M, Kempt H-J, Becker J O. Global regulation of expression of antifungal factors by a Pseudomonas fluorescens biological-control strain. Mol Plant-Microbe Interact. 1994;7:455–463. doi: 10.1094/mpmi-7-0455. [DOI] [PubMed] [Google Scholar]
- 12.Haran S, Schikler H, Chet I. Molecular mechanisms of lytic enzymes involved in the biocontrol activity of Trichoderma harzianum. Microbiology. 1996;142:2321–2331. [Google Scholar]
- 13.Haran S, Schikler H, Oppenheim A, Chet I. New components of the chitinolytic system of Trichoderma harzianum. Mycol Res. 1995;99:441–446. [Google Scholar]
- 14.Harman G E, Hayes C K, Lorito M, Broadway R M, Di Pietro A, Deterbauer C, Tronsmo A. Chitinolytic enzymes of Trichoderma harzianum: purification of chitobiosidase and endochitinase. Phytopathology. 1993;83:313–318. [Google Scholar]
- 15.Hussain A, Vancura V. Formation of biologically active substances by rhizosphere bacteria and their effect on plant growth. Folia Microbiol. 1970;15:468–478. doi: 10.1007/BF02880191. [DOI] [PubMed] [Google Scholar]
- 16.Inbar J, Chet I. Evidence that chitinase produced by Aeromonas caviae is involved in the biological control of soil-borne plant pathogens by this bacterium. Soil Biol Biochem. 1991;23:973–978. [Google Scholar]
- 17.Inglis P W, Peberdy J F. Production and purification of a chitinase from Ewingella americana, a recently described pathogen of the mushroom, Agaricus bisporus. FEMS Microbiol Lett. 1997;157:189–194. [Google Scholar]
- 18.Jannatipour M, Soto-Gil R W, Childers L C, Zyskind J W. Translocation of Vibrio harveyi N,N′-diacetychitobiase to the outer membrane of Escherichia coli. J Bacteriol. 1987;169:3785–3791. doi: 10.1128/jb.169.8.3785-3791.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones S, Yu B, Bainton N J, Birdsall M, Bycroft B W, Chhabra S R, Cox A J R, Golby P, Reeves P J, Stephens S, Winson M K, Salmond G P C, Stewart G S A B, Williams P. The lux autoinducer regulates the production of exoenzyme virulence determinants in Erwinia carotovora and Pseudomonas aeruginosa. EMBO J. 1993;12:2477–2482. doi: 10.1002/j.1460-2075.1993.tb05902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joshi S, Kozlowski M, Selvaraj G, Iyer V N, Davies R W. Cloning of the genes of the chitin utilization regulon of Serratia liquefaciens. J Bacteriol. 1988;170:2984–2988. doi: 10.1128/jb.170.7.2984-2988.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keyhani N O, Wang L-X, Lee Y C, Roseman S. The chitin catabolic cascade in the marine bacterium Vibrio furnissii. Characterization of an N,N′-diacetyl-chitobiose transport system. J Biol Chem. 1996;271:33409–33413. doi: 10.1074/jbc.271.52.33409. [DOI] [PubMed] [Google Scholar]
- 22.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Lam S T, Gaffney T D. Biological activities of bacteria used in plant pathogen control. In: Chet I, editor. Biotechnology in plant disease control. New York, N.Y: Wiley-Liss, Inc.; 1993. pp. 291–320. [Google Scholar]
- 24.Latifi A, Winson M K, Foglino M, Bycroft B W, Stewart G S A B, Lazdunski A, Williams P. Multiple homologues of LuxR and LuxI control expression of virulence determinants and secondary metabolites through quorum sensing in Pseudomonas aeruginosa PAO1. Mol Microbiol. 1995;17:333–343. doi: 10.1111/j.1365-2958.1995.mmi_17020333.x. [DOI] [PubMed] [Google Scholar]
- 25.Laville J, Voisard C, Keel C, Maurhofer M, Defago G, Haas D. Global control in Pseudomonas fluorescens mediating antibiotic synthesis and suppression of black root rot of tobacco. Proc Natl Acad Sci USA. 1992;89:1562–1566. doi: 10.1073/pnas.89.5.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McClean K H, Winson M K, Fish L, Taylor A, Chhabra S R, Camara M, Daykin M, Lamb J H, Swift S, Bycroft B W, Stewart G S A B, Williams P. Quorum sensing in Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology. 1997;143:3703–3711. doi: 10.1099/00221287-143-12-3703. [DOI] [PubMed] [Google Scholar]
- 27.McGrew B R, Green D M. Enhanced removal of detergent and recovery of enzymatic activity following sodium dodecyl sulfate-polyacrylamide gel electrophoresis: use of casein in gel wash buffer. Anal Biochem. 1990;189:68–74. doi: 10.1016/0003-2697(90)90045-b. [DOI] [PubMed] [Google Scholar]
- 28.Meighen E A. Genetics of bacterial bioluminescence. Annu Rev Genet. 1994;28:117–139. doi: 10.1146/annurev.ge.28.120194.001001. [DOI] [PubMed] [Google Scholar]
- 29.Monreal J, Rees E T. The chitinase of Serratia marcescens. Can J Microbiol. 1969;15:689–696. doi: 10.1139/m69-122. [DOI] [PubMed] [Google Scholar]
- 30.Neuhoff V, Arold N, Taube D, Ehrhardt W. Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis. 1988;9:255–262. doi: 10.1002/elps.1150090603. [DOI] [PubMed] [Google Scholar]
- 31.Okon Y, Chet I, Henis Y. Effect of lactose, ethanol and cycloheximide on the translocation pattern of radioactive compounds and sclerotium formation in Sclerotium rolfsii. J Gen Microbiol. 1973;74:251–255. [Google Scholar]
- 32.Passador L, Cook J M, Gambello M J, Rust L, Iglewski B H. Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science. 1993;260:1127–1130. doi: 10.1126/science.8493556. [DOI] [PubMed] [Google Scholar]
- 33.Perrakis A, Wilson K S, Chet I, Oppenheim A B, Vorgias C E. Phylogenetic relationships of chitinases. In: Muzzarelli R A A, editor. Chitin enzymology. Ancona, Italy: European Chitin Society; 1993. pp. 217–232. [Google Scholar]
- 34.Pierson III L S, Pierson E A. Phenazine antibiotic production in Pseudomonas aureofaciens: role in rhizosphere ecology and pathogen suppression. FEMS Microbiol Lett. 1996;136:101–108. [Google Scholar]
- 35.Pirhonen M, Flego D, Heikinheimo R, Palva E T. A small diffusible signal molecule is responsible for the global control of virulence and exoenzyme production in the plant pathogen Erwinia carotovora. EMBO J. 1993;12:2467–2476. doi: 10.1002/j.1460-2075.1993.tb05901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pleban S, Chernin L, Chet I. Chitinolytic activity of an endophytic strain of Bacillus cereus. Lett Appl Microbiol. 1997;25:284–288. doi: 10.1046/j.1472-765x.1997.00224.x. [DOI] [PubMed] [Google Scholar]
- 37.Reimmann C, Beyeler M, Latifi A, Winteler H, Foglino M, Lazdunski A, Haas D. The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol Microbiol. 1997;24:309–319. doi: 10.1046/j.1365-2958.1997.3291701.x. [DOI] [PubMed] [Google Scholar]
- 38.Rich J J, Kinscherf T G, Kitten T, Willis D K. Genetic evidence that the gacA gene encodes the cognate response regulator for the lemA sensor in Pseudomonas syringae. J Bacteriol. 1994;176:7468–7475. doi: 10.1128/jb.176.24.7468-7475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roberts R L, Cabib E. Serratia marcescens chitinase: one step purification and use for the determination of chitin. Ann Chem. 1982;127:402–412. doi: 10.1016/0003-2697(82)90194-4. [DOI] [PubMed] [Google Scholar]
- 40.Roberts W K, Selitrennikoff C P. Plant and bacterial chitinases differ in antifungal activity. J Gen Microbiol. 1988;134:169–176. [Google Scholar]
- 41.Rodriguez-Kabana R, Godoy G, Morgan-Jones G, Shelby R A. The determination of soil chitinase activity: conditions for assay and ecological studies. Plant Soil. 1983;75:95–106. [Google Scholar]
- 42.Saborowski R, Buchholz F, Vetter R-A H, Wirth S J, Wolf G A. A soluble, dye-labelled chitin derivative adapted for the assay of krill chitinase. Comp Biochem Physiol. 1993;105B:673–678. [Google Scholar]
- 43.Sahai A S, Manocha M S. Chitinases of fungi and plants: their involvement in morphogenesis and host-parasite interaction. FEMS Microbiol Rev. 1993;11:317–338. [Google Scholar]
- 44.Smith R J, Grula E A. Chitinase is an inducible enzyme in Beauveria bassiana. J Invest Pathol. 1983;42:319–326. [Google Scholar]
- 45.Streichsbier F. Utilization of chitin as sole carbon and nitrogen source by Chromobacterium violaceum. FEMS Microbiol Lett. 1983;19:129–132. [Google Scholar]
- 46.Svitil A L, Ni Chadhain S M, Moore J A, Kirchman D L. Chitin degradation proteins produced by the marine bacterium Vibrio harveyi growing on different forms of chitin. Appl Environ Microbiol. 1997;63:408–413. doi: 10.1128/aem.63.2.408-413.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swift S, Winson M K, Chan P F, Bainton N J, Birdsall M, Reeves P J, Rees C E D, Chhabra S R, Hill P J, Throup J P, Bycroft B W, Salmond G P C, Williams P, Stewart G S A B. A novel strategy for the isolation of luxI homologues: evidence for the widespread distribution of a LuxR:LuxI superfamily in enteric bacteria. Mol Microbiol. 1993;10:511–520. doi: 10.1111/j.1365-2958.1993.tb00923.x. [DOI] [PubMed] [Google Scholar]
- 48.Swift S, Throup J P, Salmond G P C, Williams P, Stewart G S A B. Quorum sensing: a population-density component in the determination of bacterial phenotype. Trends Biochem Sci. 1996;21:214–219. [PubMed] [Google Scholar]
- 49.Techkarnjanaruk S, Pongpattanakitshote S, Goodman A E. Use of a promoterless lacZ gene insertion to investigate chitinase gene expression in the marine bacterium Pseudoalteromonas sp. strain S9. Appl Environ Microbiol. 1997;63:2989–2996. doi: 10.1128/aem.63.8.2989-2996.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thompson J M, Winson M K, Bycroft B W, Williams P, Stewart G S A B. Abstracts of the 133rd Ordinary Meeting of the Society for General Microbiology 1996. Reading, United Kingdom: Society for General Microbiology; 1996. Quorum sensing regulation of protease production in Chromobacterium violaceum, abstr., P-8; p. 26. [Google Scholar]
- 51.Throup J, Winson M K, Bainton N J, Bycroft B W, Williams P, Stewart G S A B. Signalling in bacteria beyond bioluminescence. In: Campbell A, editor. Bioluminescence and chemiluminescence: fundamentals and applied aspects. Chichester, United Kingdom: Wiley; 1995. pp. 89–92. [Google Scholar]
- 52.Tronsmo A, Harman G. Detection and quantification of N-acetyl-β-d-glucosaminidase, chitobiosidase, and endochitinase in solutions and on gels. Anal Biochem. 1993;208:74–79. doi: 10.1006/abio.1993.1010. [DOI] [PubMed] [Google Scholar]
- 53.Tsujibo H, Orikoshi H, Shiotani K, Hayashi M, Umeda J, Miyamoto K, Imada C, Okami Y, Inamori Y. Characterization of chitinase C from a marine bacterium, Alteromonas sp. strain O-7, and its corresponding gene and domain structure. Appl Environ Microbiol. 1998;64:472–478. doi: 10.1128/aem.64.2.472-478.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang S-L, Chang W-T. Purification and characterization of two bifunctional chitinases/lysozymes extracellularly produced by Pseudomonas aeruginosa K-187 in a shrimp and crab shell powder medium. Appl Environ Microbiol. 1997;63:380–386. doi: 10.1128/aem.63.2.380-386.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wells J S, Trejo W H, Principe P A, Bush K, Georgopapadakou N, Bonner D P, Sykes R B. SQ 26,180, a novel monobactam. I. Taxonomy, fermentation and biological properties. J Antibiot. 1982;35:184–188. doi: 10.7164/antibiotics.35.184. [DOI] [PubMed] [Google Scholar]
- 56.Winson M K, Bainton N J, Chhabra S R, Bycroft B W, Salmond G P C, Williams P, Stewart G S A B. Abstracts of the 94th Annual Meeting of the American Society for Microbiology 1994. Washington, D.C: American Society for Microbiology; 1994. Control of N-acyl-l-homoserine lactone-regulated expression of multiple phenotypes in Chromobacterium violaceum, abstr. H-71; p. 212. [Google Scholar]
- 57.Winson M K, Camara M, Latifi A, Foglino M, Chhabra S R, Daykin M, Bally M, Chapon V, Salmond G P C, Bycroft B W, Lazdunski A, Stewart G S A B, Williams P. Multiple N-acyl-l-homoserine lactone signal molecules regulate production of virulence determinants and secondary metabolites in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:9427–9431. doi: 10.1073/pnas.92.20.9427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Winson, M. K., and L. S. Chernin. Unpublished data.
- 59.Wood D W, Gong F, Daykin M M, Williams P, Pierson L S., III N-Acyl-homoserine lactone-mediated regulation of phenazine gene expression by Pseudomonas aureofaciens 30-84 in the wheat rhizosphere. J Bacteriol. 1997;179:7663–7670. doi: 10.1128/jb.179.24.7663-7670.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wortman A T, Somerville C C, Colwell R R. Chitinase determinants of Vibrio vulnificus: gene cloning and applications of a chitinase probe. Appl Environ Microbiol. 1986;52:142–145. doi: 10.1128/aem.52.1.142-145.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]