Abstract
Drosophila melanogaster was first used for research in the early 1900’s by scientists located in the northeastern corridor of the United States, gaining prominence with the establishment of the famous “fly room” by Thomas Hunt Morgan at Columbia University circa1908. Several reasons for using D. melanogaster in research are well known; easy and inexpensive to breed, short lifespan, amongst others. But why was this insect species flourishing in a temperate northeast region of the New World during the late 1800’s when they originated in the tropical forests of sub-Saharan Africa millions of years ago? The purpose of this review is to provide an overview of the experimental underpinnings for a temperature sensitive mechanism that likely contributed to the rather unique ability of Drosophila melanogaster to successfully colonize temperate regions on a global scale. It also furnishes an interesting historical insight into how ancestral genetics serendipitously held the keys to the journey of D. melanogaster becoming such a popular research organism. While numerous papers have been published detailing different aspects of the work, this is the first comprehensive review. Herein, I discuss the discovery of a small thermosensitive intron in D. melanogaster (termed dmpi8) that controls midday siesta levels. Like many day-active animals, Drosophila exhibits a robust genetically based midday siesta that is protective in warm climates. Yet long bouts of daytime inactivity might be counterproductive in temperate climates, especially since daylength in these regions is shorter during the cooler months. Evidence discussed in this review strongly indicates that targeting of dmpi8 splicing efficiency by natural selection enhanced the ability of D. melanogaster to scale daytime sleep levels commensurate with a wide range of local climates. Surprisingly, dmpi8 splicing regulates midday siesta levels in trans by controlling the expression of a nearby anti-siesta gene called daywake. The “fortuitous” genetic arrangement of a thermosensitive intron in proximity to an anti-siesta gene might have contributed to the cosmopolitan nature of D. melanogaster and its historical journey in becoming a popular research organism.
Introduction
The genus Drosophila contains around 1,600 described species that vary in many attributes and are found all over the world from tropical and desert, to temperate and alpine regions1. Of all these species, D. melanogaster has a unique position in the scientific literature, a prima-donna subject of human inquiry. A recent search of Pubmed using “Drosophila melanogaster” as the query returns over 60,000 citations, whereas the next highest, Drosophila simulans barely cracks 1,500. Five Nobel Prizes have been awarded to nine scientists for work primarily done using Drosophila melanogaster as a model organism. Studies using D. melanogaster, have been foundational in understanding evolution, genetics, development and neurobiology (e.g.,2,3). By combining cutting-edge computing and imaging, it is now possible to visualize 3-D interactive maps of every single neuron and synapse in the adult brain of D. melanogaster. How did D. melanogaster come to be such a favorite organism for probing minds, from high school students to the world’s most prestigious research institutes?
The answer to this question begins in the United States towards the start of the 20th century. Serendipity, exchange of ideas, re-discovery of Mendelian genetics, interest in a nagging question and constraints imposed by the size of a room all played major parts. Charles Woodworth, an entomologist, is the first scientist credited with breeding Drosophila melanogaster, introducing it to William Castle at Harvard whose work inspired Thomas Morgan, then at Columbia University (for historical reviews, see3–6). This series of events led to the first “Fly Room”. Morgan was attracted to the easy and cheap breeding of D. melanogaster that could be maintained in large numbers with the help of students even in a small room. Fast breeding times, large numbers and easily recognized physical phenotypes were ideal for experimental probing into the basis of heredity. Morgan and colleagues isolated a white-eyed mutant in 1910, following that up with the discovery of many more mutants and established chromosomes as key units of inheritance—for which he won a Nobel Prize in 1933. Perhaps appropriately, the white mutatant is the most widely used laboratory strain for Drosophilist, serving as host for generating tens of thousands of genetically altered strains by simply following the co-rescue of the wildtype red eye color. Indeed, with the demonstration that transgenic D. melanogaster could be generated by P-element transformation in the early 1980’s7,8, this solidified its place as a workhorse animal model system, which continues unabated with the development of ever-more innovative genetic tool kits and realms of data to be mined.
Yet a much different story might have occurred if D. melanogaster was not already endemic to parts of the United States where these early pioneers set up shop. Sure, nowadays wherever you might be, these flies have likely pestered your kitchens and buffet tables, especially those with ripening fruit. But that was not always the case, at least not in the United States. D. melanogaster was first described by the German entomologist Johann Wilhem Meigen in 1830. The first report of D. melanogaster in the United States was in the northeast in 1875, quickly radiating west over the next few decades4. Irrespective of when D. melanogaster first arrived in the New World, it was already a cosmopolitan strain that closely associated with humans.
It is thought that the genus Drosophila originated in the lowlands of sub-Sahara tropical Africa approximately 40–50 million years ago9–11 (Fig. 1). More recent findings have fine-tuned this hypothesis and suggest D. melanogaster originated in the tropical forests of current day Southwestern Africa (Zimbabwe and Zambia)12,13. Expansion of D. melanogaster into Europe and Asia is thought to have occurred via a single “out-of-Africa” bottleneck some 12–19,000 years ago, around the time of the last ice age, followed by a quick population expansion throughout Africa and Eurasia14,15. The America’s, Australia, Japan and other places separated by large bodies of water that experienced more recent human migration or commerce from Africa and Eurasia, have several 100 years at most in providing a home for naturally-breeding populations of D. melanogaster. This tropical species is now endemic to temperate regions and high altitudes around the world.
Fig 1.
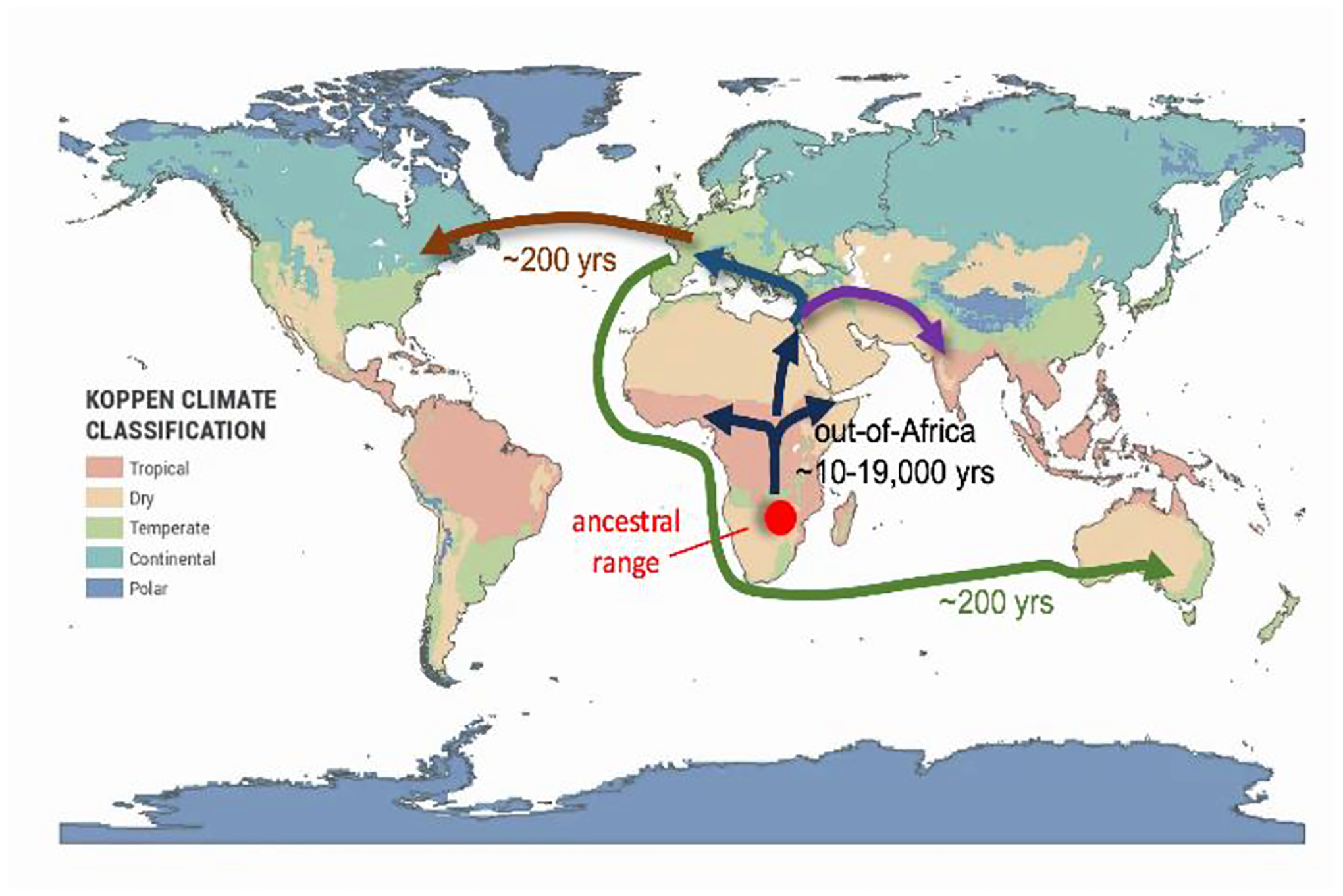
Proposed historical expansion of D. melanogaster beyond its ancestral range in Africa (red circle), with emphasis on the more recent colonization of North America and Australia. Map based on EarthHow.com; Drosophila melanogaster migration routes based on refns.13,15
Given the diversity of Drosophila it is not surprising that these species adapted to survival in geographical locations that differ greatly in temperature, humidity, diets, etc. However, even for the closely related nine species in the D. melanogaster species subgroup (erecta, mauritiana, melanogaster, orena, santomea, sechellia, simulans, teissieri and yakuba), only D. melanogaster and D. simulans are cosmopolitan and apparently opportunistic in human commensalism16. Of the two, D. melanogaster is easily more widespread around the globe. How and when D. melanogaster became a human commensal is not clear.
It’s presumed ancestral range around Zimbabwe (Fig. 1) is associated with the marula fruit, a resource that even today’s cosmopolitan strains prefer12. It is proposed that the harvesting of large quantities of marula fruit by indigenous peoples of Southwestern Africa some ~10,000 years ago might explain the commensalism. Intriguingly, large supplies of marula fruit were kept in caves, which may have seeded the current ability of D. melanogaster to share indoor living with humans.
While adapting to cohabitating with humans and becoming a generalist or opportunistic feeder certainly helps explains its worldwide distribution, there is more to the story. Most notably, natural populations of D. melanogaster show extensive genetic differentiation as a function of latitudinal and altitudinal clines that show parallel changes, patterns consistent with temperature as the driving selection pressure (reviewed in,17–19). Additionally, there is genome-wide correlation between clinal and seasonal variation in natural D. melanogaster populations suggesting common and dynamic mechanisms in establishing fluctuations in allele frequencies that are selected for real-time climate adaptation20. Indeed, the direction of allele frequency change at seasonally variable polymorphisms can be predicted by weather conditions in the weeks prior to sampling21.
These studies not only indicate that adaptation to local temperatures increases the fitness of even cosmopolitan D. melanogaster strains, but that it can occur rapidly, suggesting a rich source of historically sustained genomic variants within these populations that can be readily selected in distinct combinations to optimize organismal function to local temperature ranges. In this regard it is important to note that the presumed ancestral range of D. melanogaster, while tropical, sits close to the edge of the Tropic of Capricorn. Seasonal temperatures can fluctuate between lows of 9–18° to highs of 24–30°C depending on altitude. Thus, it is likely that even in its ancestral range, D. melanogaster as an ectotherm had to find flexible strategies to adapt to long periods of cooler and warmer temperatures. Therefore, although the world-wide expansion of D. melanogaster to temperate regions beyond its ancestral range is due to its association with humans, adapting to local temperatures plays a key role in enhancing the fitness of natural populations. One of those traits, and the subject of this review, is sleep. Specifically, midday sleep, or more commonly referred to as “siesta” (meaning sixth hour or midday).
Temperature, daily activity patterns and midday siesta.
Virtually all animals exhibit some form of sleep or quiescent state that has a characteristic timing during a daily cycle and length. In general, invertebrate and vertebrate animals are mainly active at particular times of day, most notably during daylight hours (diurnal), the night (nocturnal), dawn/dusk (crepuscular) or cathemeral (both day and night)22,23. Patterns of daily activity are mainly constrained by duration of illumination and thermal limits suitable for organismic function. Daily wake-sleep cycles and their timing are governed by internal cell-based circadian (~24 hr) clocks24–27. Sleep is also regulated by homeostatic pathways, where sleep drive builds with length of wake28–30. In addition, environmental cues, biotic factors and internal physiological state (e.g., hunger) have major influences on wake-sleep behavior (e.g., 31).
Ambient temperature features prominently in sleep behavior (32–35). Ever try to sleep at night when it’s hot? Early studies analyzing garter snakes showed that ambient temperature has strong effects on the daily activity patterns of animals36,37. On cool days snakes exhibit peak activity during the middle of the day which is suppressed at higher temperatures where they are mainly active in late night-early morning. Many day-active animals whether they can actively thermoregulate their body temperature (homeotherms) or not (poikilotherms) avoid excess activity during the middle of hot days. The increase in midday quiescence on hot days is commonly referred to as siesta. Although largely abandoned in the modern world, studies of several preindustrial societies showed that midafternoon napping increased in frequency and duration during the summer compared to winter38. Like humans, nighttime sleep in D. melanogaster is more fragmented and less intense at warmer temperatures, whereas daytime sleep intensity increases with temperature39–42 (further discussed below).
Midday siesta aligns well with the timing of post-lunch drowsiness or postprandial sleep/fatigue, which is widely observed irrespective of temperature43–45. This early afternoon drowsiness is essentially a feature of the dynamics underlying the circadian timing system and sleep homeostatic pathways46,47. Depending on the time-of-day, the balance between sleep drive and wake drive fluctuates. Your circadian clock strongly pushes wake from post afternoon to early nighttime hours. Thus, midday is a more vulnerable state for sleepiness, which can be enhanced by heat, feeding and possibly other factors. While midday siesta is most associated with avoiding heat, there are other environmental hazards, most notably exposure to noxious levels of radiation from UV and/or blue light. Moreover, the dangers of heat/UV exposure are generally more severe for smaller animals since they have a larger surface-to-volume ratio making them at increased risk for water-loss and desiccation.
Many studies have shown the benefits of short daytime napping (“power-napping”), such as improving cognitive functions and lowering blood pressure48. However, excessive daytime napping or sickness behavior is linked to poor prognosis for many medical disorders, such as Parkinson’s, Alzheimer’s and diabetes49,50. Intriguingly, daytime skin temperature for individuals with several of these disorders are higher than usual, reinforcing the notion that elevated daytime body temperature is sleep promoting51,52. Large scale analysis of genetic variation in human sleep behavior reveals that nighttime and daytime sleep are governed by different and/or non-overlapping pathways, serving different functions53–55. Indeed, studies in Drosophila have clearly established different mechanisms that underlie daytime and nighttime sleep56.
Discovery of the thermosensitive dmpi8 intron regulating midday siesta in D. melanogaster.
Some 25 years ago we sought to understand how seasonal changes in temperature affect the distribution of daily activity patterns in D. melanogaster, studies inspired by earlier work analyzing garter snakes (see above). We hoped to use the genetic tools available in Drosophila to gain mechanistic insights; essentially, are the changes in activity patterns an acute reaction to ambient temperature or involve an underlying thermosensing mechanism?
The standard laboratory conditions for analyzing daily wake-sleep behavior in Drosophila is based on measuring fly locomotor activity levels. The standard lab conditions are maintaining flies for several days at 25°C while exposed to 12hr light: 12hr dark cycles [LD; where Zeitgeber time 0 (ZTO) is defined as lights-on]57. Subjecting Drosophila to days of 12 hr light followed by 12 hr dark is reasonable considering they originated around equatorial Africa. In the classic experimental design, individual flies are placed in small glass tubes whereby locomotor activity movement can be tracked by counting “beam brakes” using an infra-red source positioned across a photomultiplier tube. This system was refined and marketed by a small company (at that time) called Trikinetics that was situated a short distance from the labs of Drs. Hall and Rosbash at Brandies University in Waltham, Massachusetts, USA. Working together in the early 1980’s, they optimized the “Drosophila Activity Monitor” (or DAM) system. Since then, other systems have been developed58,59 but the Trikinetics DAM system is still the most popular due to its ease of use and scalability. Indeed, numerous central clock genes were identified using the Trikinetics system in large-scale mutant screens, playing an assisting role in the 2017 Nobel Prize for Medicine and Physiology awarded to Drs. Hall, Rosbash and Young (Rockefeller University) for ground-breaking discoveries on the mechanisms underlying circadian rhythms60.
We found that the daily distribution of activity in D. melanogaster is heavily influenced by ambient temperature41 (Fig. 2). At the standard temperature of 25°C, D. melanogaster exhibit two prominent clock-controlled activity peaks, a “morning” peak (M) centered around the lights-on transition and an “evening” peak (E) centered around the light-off transition61 (Fig. 2, bottom) At cooler temperatures, flies show increased daytime activity whereby the morning and evening peaks are closer together with elevated activity levels during the midday41. As temperatures increase, the morning activity peak shifts into the late night, whereas the evening peak is delayed into the early night with a concomitant large dip in midday activity41,62.
Fig. 2.
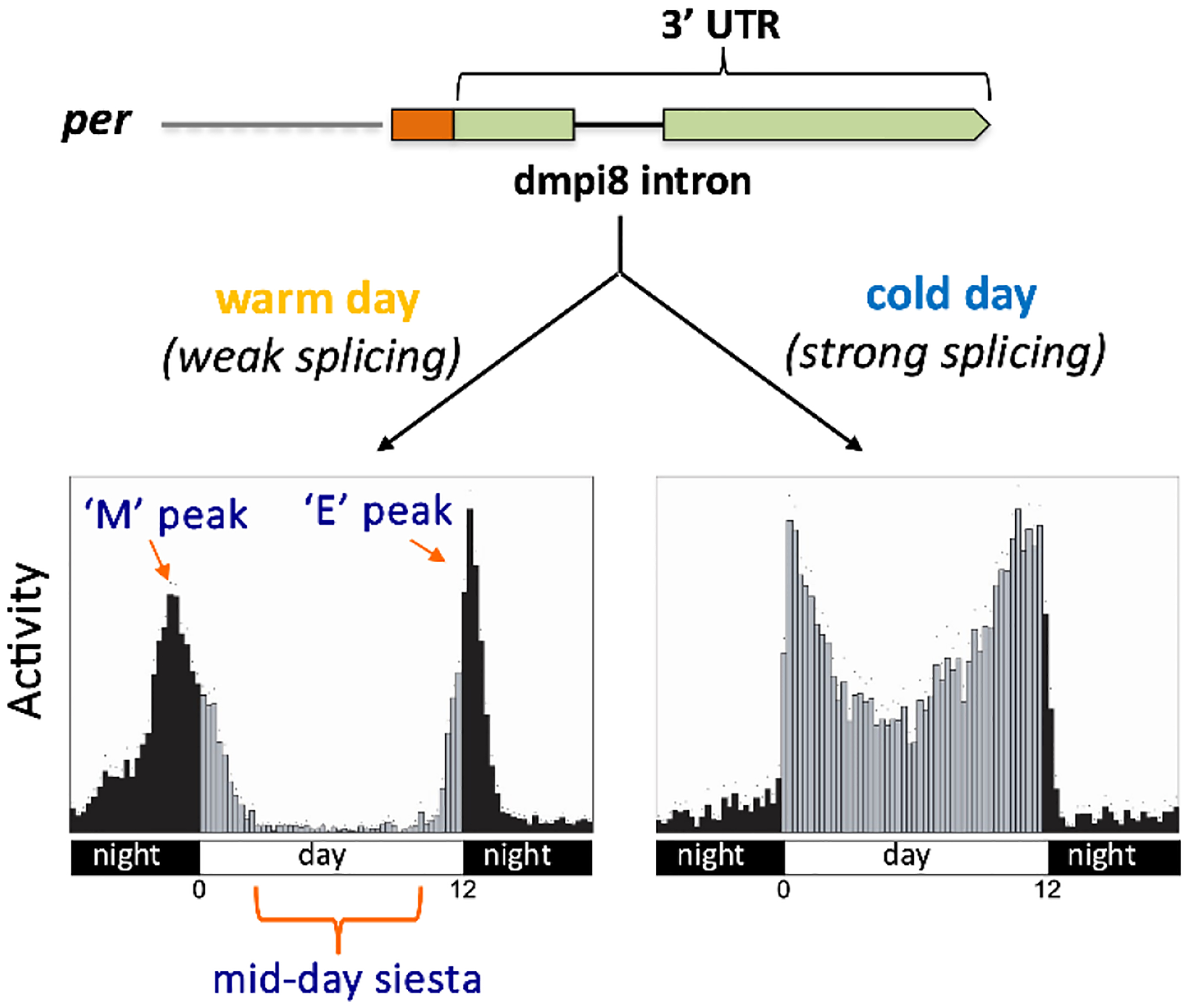
Thermal adaptation of midday siesta levels in D. melanogaster is regulated by thermosensitive splicing of the dmpi8 intron found in the per 3’ UTR. M = morning peak of activity; E = evening peak of activity.
Since clock mechanisms not only control the pace of daily rhythms but also its phasing, we initially assumed temperature-induced changes in the oscillatory dynamics of central clock proteins was the basis for the thermosensitivity in daily activity patterns. A key mechanistic logic of circadian clocks is that some attribute in the expression of one or more core clock genes (e.g., levels, phosphorylated state) oscillates with an approximately 24 hr period that is inextricably linked to clock progression and rhythm generation63,64. Thus, we focused on circadian genes, especially period (per), critical to setting the pace and phase of the clock65.
The most relevant observation we made was that the splicing efficiency of a small intron (86–89 bp, depending on fly strain) in the 3’ untranslated region of per was modulated by temperature, being more efficient at colder temperatures41 (Fig. 2, top). We eventually called this intron dmpi8 (Drosophila melanogaster per intron 8). Transgenic flies where the small 3’ UTR intron was either missing or could not be spliced both led to a similar delay in evening activity and longer midday siesta41. This suggests that active splicing per se and not just the presence or absence of the dmpi8 intron is the key molecular signal modulating daily activity patterns. Based on our findings that splicing of dmpi8 advanced the phase of per mRNA and protein levels41 we proposed a “clock-centric” explanation for the function of this intron in the thermal adaptation of daily activity patterns. Essentially, we postulated that the cold-enhanced splicing of dmpi8 advances daily cycles in per mRNA and protein levels leading to an earlier ‘evening’ activity which therefore decreases the duration of midday dip in activity41. As reviewed below, our initial per-based model turned out to be wrong because we had not anticipated the discovery of nearby gene that was indirectly regulated by dmpi8 splicing.
Multiple weak splicing signals and thermosensitivity.
Comparative biology has been instrumental in the study of adaptation. Using this approach, we asked if thermal plasticity in siesta is also present in closely related Drosophila species that are only indigenous to their ancestral range in tropical Africa. As an initial test case we examined D. yakuba, which diverged from D. melanogaster about 5 million years ago66. Unlike D. melanogaster, midday siesta in D. yakuba is always prominent even at cool temperatures67. Intriguingly, a small intron is also present in the 3’ UTR of the per gene in D. yakuba (termed dyp3’), however its splicing efficiency is not modulated by temperature, consistent with the lack of thermo-responsiveness in siesta. This thermal difference in splicing efficiency between dmpi8 and dyp3’ introns was recapitulated in a simplified tissue culture system indicating that all the signals required for thermosensitive splicing are contained within the intron and nearby flanking regions67.
Comparison of dmpi8 and dyp3’ showed that the 5’ and 3’ splicing signals for dmpi8 are extremely weak67 (Fig. 3). The first and rate-limiting step in pre-mRNA splicing is the recognition of the 5’ss by U1 snRNA and associated protein factors, which generally involves 5–8 base-pair interactions between the −3 and +6 nucleotides of the 5’ss68–70. The consensus 5’ss in D. melanogaster is (−3)MAGGTAAGT(+6) (where M=any nucleotide; GT= 5’splice site, where G=position +1)71,72. Optimizing the 5’ss of dmpi8, especially in conjunction with a stronger 3’ splice site, increased dmpi8 splicing efficiency and abolished its thermal sensitivity67. Importantly, transgenic flies wherein dimpi8 was replaced by the same intron with optimized 5 and 3’ splice sites (termed dmpi8UP) manifest reduced siesta compared to wildtype controls (termed dmpi8WT) at all temperatures tested. Intriguingly, the dmpi8 5’ss has the potential for 5 bp interactions with U1 snRNA, whereas the per 3’ intron from D. yakuba has an extra base pairing at the critical +6 position67 (Fig. 3). Presumably, the 5 base-pairing interactions between U1 snRNA and dmpi8 is sufficiently destabilized at higher but physiologically relevant temperatures to decrease spliceosome binding and overall splicing rate below that occurring at cooler temperatures. In contrast, the 6 base-pairing interactions between U1 snRNA and dyp3’ are not rate-limiting within the physiologically relevant temperature range for this species. Thus, it is possible that the absence of one critical base-pairing interaction between the 5’ss of dmpi8 and U1 snRNA facilitated the world-wide expansion of D. melanogaster to temperate regions by providing a thermally sensitive molecular throttle operating within physiologically relevant temperature ranges.
Fig. 3.
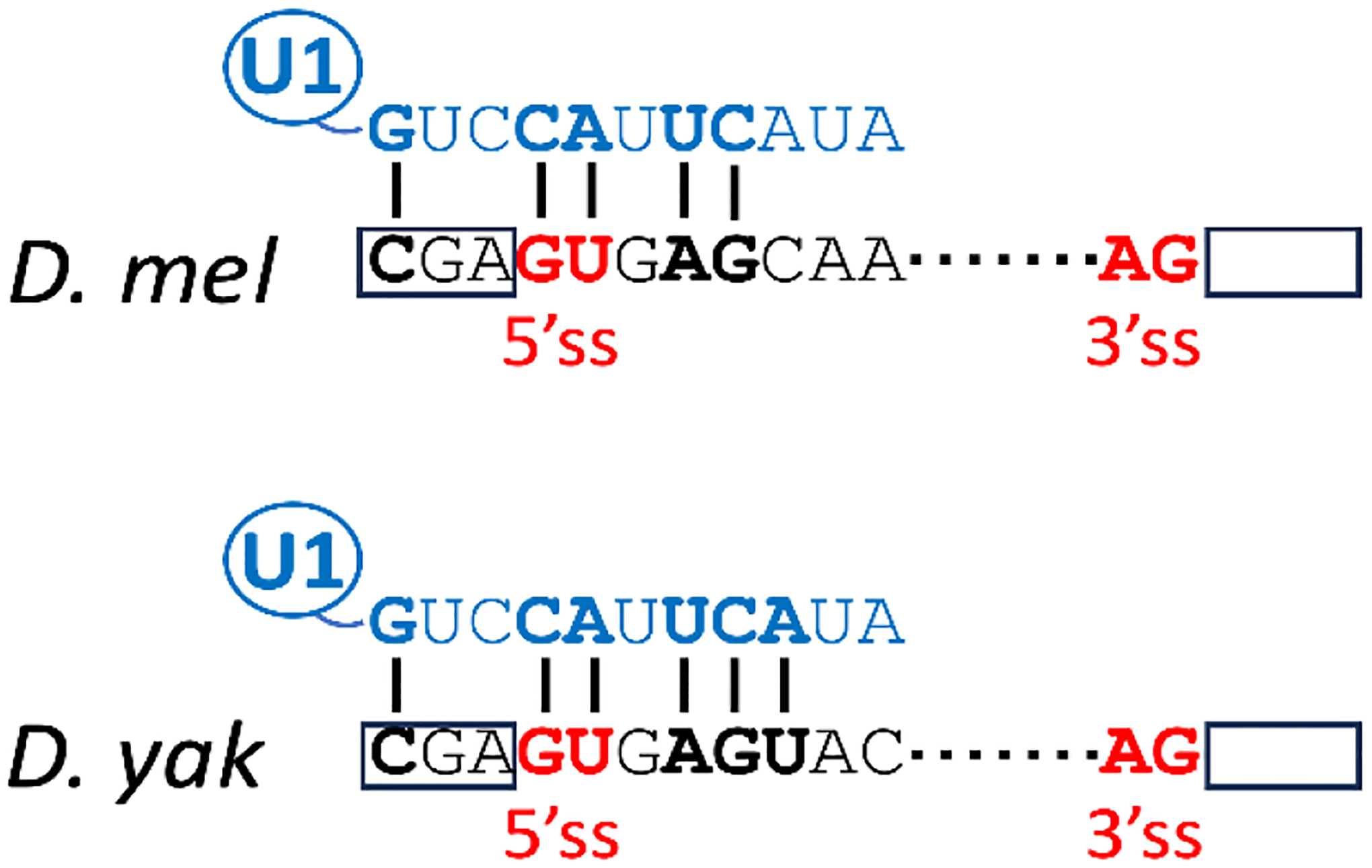
Weak 5’ss based on only 5 bp interactions with U1 contributes to thermal sensitivity in dmpi8 splicing efficiency, whereas the 6 bp for the D. yakuba intron is not thermosensitive for U1 binding within physiological temperatures.
D santomea and D. simulans flies were also analyzed in the same study67. D. santomea diverged from D. yakuba about 400,000 years ago and is endemic to São Tomé, one of the Gulf of Guinea islands in west-equatorial Africa73. Whereas D. santomea flies exhibit little to no changes in siesta as a function of temperature, D. simulans does. Consistent with our model, the per 3’ intron in D. santomea is flanked by strong splice sites and shows constant high splicing efficiency over a range of temperatures, in sharp contrast to the per3’ intron in D. simulans. Thus, at least for the D. melanogaster subgroup, there is a strong correlation between multiple weak splice sites flanking a small intron in the 3’ UTR of per, thermosensitive splicing and adaptability of siesta levels to changes in ambient temperature. In temperate climates, cooler seasons are associated with shorter days. Therefore, the cold-enhanced splicing efficiency of dmpi8 might allow D. melanogaster to fulfill its daytime activities despite short days, giving it an advantage over flies that are hard-wired to sleep during the midday despite favorable thermal conditions67.
Importantly, just because D. yakuba and D. santomea appear limited in decreasing midday siesta on cool days, they do show thermal adaptation in other behaviors. For example, D. yakuba prefers warmer temperatures (mean = 25.47 °C) compared to D. santomea (22.56 °C), consistent with their distribution to either low altitudes or higher elevation, respectively74,75. Indeed, this climate-based spatial segregation may have contributed to the reproductive isolation and speciation of D. yakuba and D. samtomea75. Studies in D. melanogaster have uncovered the molecular basis for thermal preference, which is based on temperature sensitive neural circuits39,76, a mechanism distinct from how dmpi8 splicing modulates midday siesta. Clearly, different mechanisms underlie a multitude of behavioral adaptations to temperature that differ in selective pressures depending on variant life history traits even in closely related species. Thus, a species like D. santomea shows evidence of adaptation to cooler temperatures but this apparently did not extend to strong plasticity in midday siesta. Might it be that for closely related species to D. melanogaster, the ‘only’ adaptive route to reducing midday siesta on cold days is dependent on having a small intron in the 3’-terminal of per with multiple weak splice sites as a basis for thermosensitive splicing efficiency?
Although not a focus of this review, that multiple weak splicing signals might underlie thermosensitive splicing was strongly supported by earlier work from Murphy and co-workers using the Moloney murine sarcoma virus ts110 mutant that shows temperature sensitivity in growth77–79. It was suggested that weakened RNA:RNA interactions between key splicing signals and snRNPs might diminish spliceosome binding at higher temperatures. An estimated ~15% of all disease-associated mutations in humans affect splice sites80–83, with positions −1 and +5 likely to be most critical to U1 binding68,84. Disease-causing mutations that weaken the 5’ss can lead to temperature sensitive splicing, as shown for an allele of Ehlers-Danlos syndrome Type VII85. In comparing the 5’ss of the per 3’-terminal intron from D. melanogaster and D. yakuba, the former is missing a base-pairing interaction at +6 with U1 snRNA. Mutations at intronic position +6 or position +3 of U1 snRNA (which base-pairs with the intronic +6 site) lead to weakened splice sites that are casually linked to several human diseases, such as familial dysautonomia86, Ehlers-Danlos syndrome87 and medulloblastoma88. Perhaps many of the splicing mutations that weaken strong 5’ss have thermal sensitive phenotypes since these sites might lack other compensatory elements to ensure robust recognition. Intriguingly, the interaction between microRNAs and mRNAs are also based on a similar number of base-pair interactions as that of U1 snRNA and the 5’ss89. This suggests that the RNA:RNA interactions key to pre-mRNA splicing and miRNA regulation are ideally suited for thermal responsiveness to temperature ranges widely observed on Earth, either on a daily or seasonal scale.
Single nucleotide polymorphisms, dmpi8 splicing and midday siesta.
As discussed above, midday siesta makes biological sense on hot days. However, on cool days sleeping during the middle of the day for a visual day-active organism might prove counter-productive. We reasoned that D. melanogaster adapted to cooler regions would exhibit smaller mid-day siestas compared to their warm adapted counterparts. Drosophila is one of the best studied species in clinal studies. These flies have been captured by many different researchers over the years and usually kept in private laboratories. The different developmental, phenotypic and behavioral traits measured as a function of geographical location are stable despite years of laboratory rearing19,90. In general, latitudinal and altitudinal clines are ascribed to differences in temperature (although other factors, such as oxygen tension is also relevant, especially for altitude).
As an initial test case we evaluated the sleep patterns of natural populations of D. melanogaster from the eastern coast of the United States, spanning from Vermont to Florida91. Differences in the daily distribution of activity as a function of latitude were not observed. However, two major haplotypes of the per 3’ UTR, termed VT1.1 and VT1.2, that include four single nucleotide polymorphisms were identified (herein termed SNP1, SNP2, SNP3 and SNP4) (Fig. 4). SNP3, which is either an A or G, had the most significant effect on dmpi8 splicing and midday siesta, whereby SNP3G has higher dmpi8 splicing efficiency and diminished midday siesta compared to SNP3A. Since all dmpi8 introns are flanked by the same suboptimal 5’ and 3’ss (Fig. 3), dmpi8 splicing remains temperature sensitive irrespective of which SNP3 is present. Thus, the main thing SNP3G does is to increase the basal and peak splicing efficiency of dmpi8 but still within a physiologically relevant thermal sensitive range.
Fig. 4.
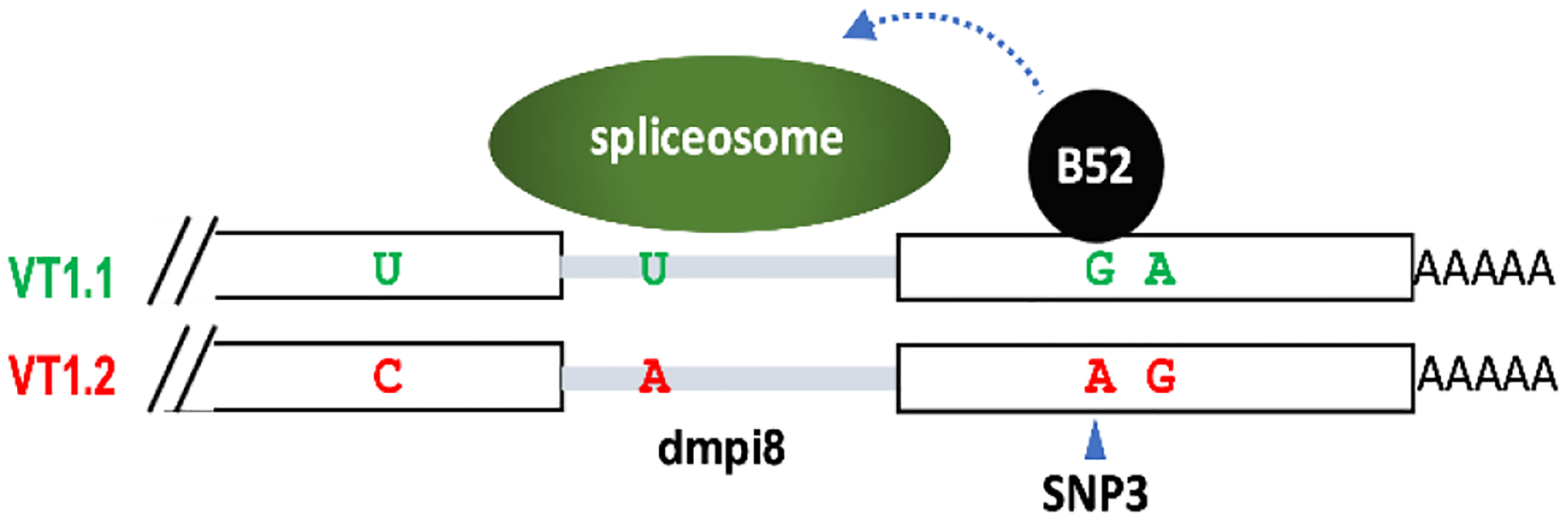
Two major per 3’ UTR haplotyes for natural populations of flies along the eastern coast of the United States. SNP3 is critical to dmpi8 splicing efficiency; when a G is present, it enhances binding of the B52 splicing factor, which stimulates overall dmpi8 splicing.
Further work showed that SNP3 is part of a binding site for B52/SRp5592, a member of the highly conserved serine/arginine (SR) family of splicing factor proteins. Binding of B52/SRp55 to the per 3’UTR is stronger when SNP3 is a G compared to A, explaining why SNP3G enhances dmpi8 splicing efficiency and reduces midday activity. Whether other SR proteins also bind the per 3’ UTR and modulate dmpi8 splicing efficiency is not clear. Although a distinct mechanism from that described for B52 binding to the per 3’ UTR in Drosophila, body temperature cycles in mammals can drive rhythmic phosphorylation of SR proteins that underlie thermosensitive regulation of global alternative splicing programs93. Thus, temperature can regulate pre-mRNA splicing in a myriad of ways, including modulating RNA secondary structure, miRNA levels and gene expression changes in splicing regulatory factors.
D. melanogaster from Africa and Australia show parallel clinal changes in dmpi8 splicing and midday siesta levels.
Analysis of daily wake-sleep patterns in natural fly populations from Africa and Australia revealed parallel decreases in midday siesta levels in cool adapted flies94,95 (Fig. 5). In the case of Africa, fly populations ranged in altitude from approximately 78 to over 3,000m above sea level. For Australia, flies were analyzed from a well-studied tropical and temperate latitudinal cline along the eastern coast96,97. The cool adapted flies still show increased midday siesta at warmer daily temperatures, but their overall baseline levels are lower at each temperature tested compared to the warm-adapted populations (Fig. 5). Thus, there is remarkable inter-continental congruence between altitudinal and latitudinal effects on midday siesta for flies from equatorial Africa and the eastern coast of Australia. Surprisingly however, although dmpi8 splicing efficiency figures prominently in the thermal adaptation of midday siesta as a function of altitude and latitude, the underlying mechanisms are distinct, as reviewed below.
Fig. 5.
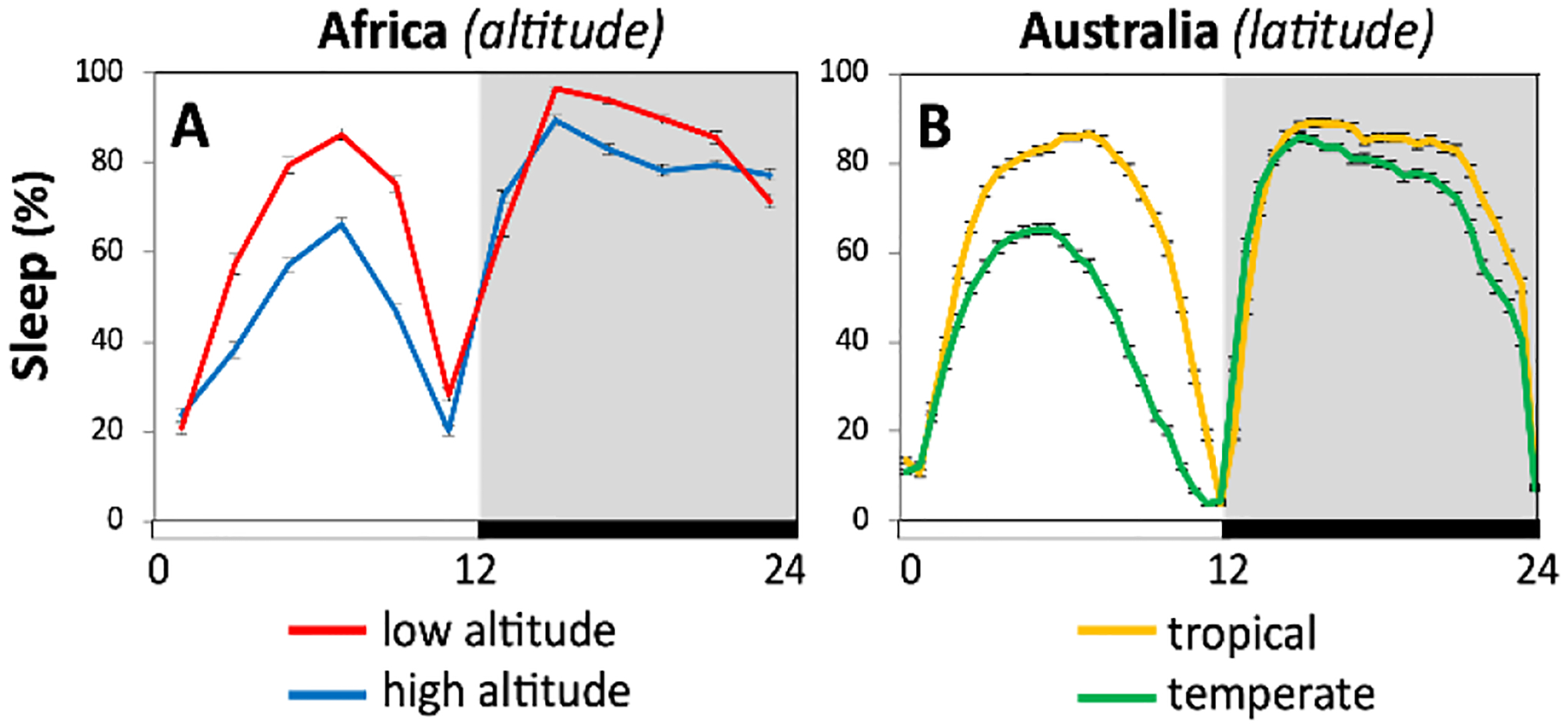
Parallel latitudinal and altitudinal clines in decreased midday siesta for natural populations of D. melanogaster from high altitudes in Africa (A), and temperate regions along the eastern coast of Australia (B). Shown are daily sleep profiles.
Natural populations of flies from high altitudes in Africa exhibit increased dmpi8 splicing efficiency compared to their low-altitude counterparts, consistent with their decreased midday siesta94. Sequencing of the per3’ UTR from flies representing different altitudes across eastern and western regions of equatorial Africa identified at least a dozen SNPs that generated some 10 different haplotypes of varying frequency (Fig 6A, top). This is consistent with the origins of D. melanogaster and the likelihood that these natural variants represent rich ancestral diversity13. Nonetheless, we could not find any evidence of altitudinal cline or clines in any of the per 3’ UTR variants either examined individually or in combination. Thus, although dmpi8 splicing efficiency was likely targeted by natural selection to adjust midday siesta as a function of altitude in equatorial Africa, this is not based on cis-acting elements in the per gene but might reside in trans-acting factors (e.g., Fig. 4).
Fig. 6.
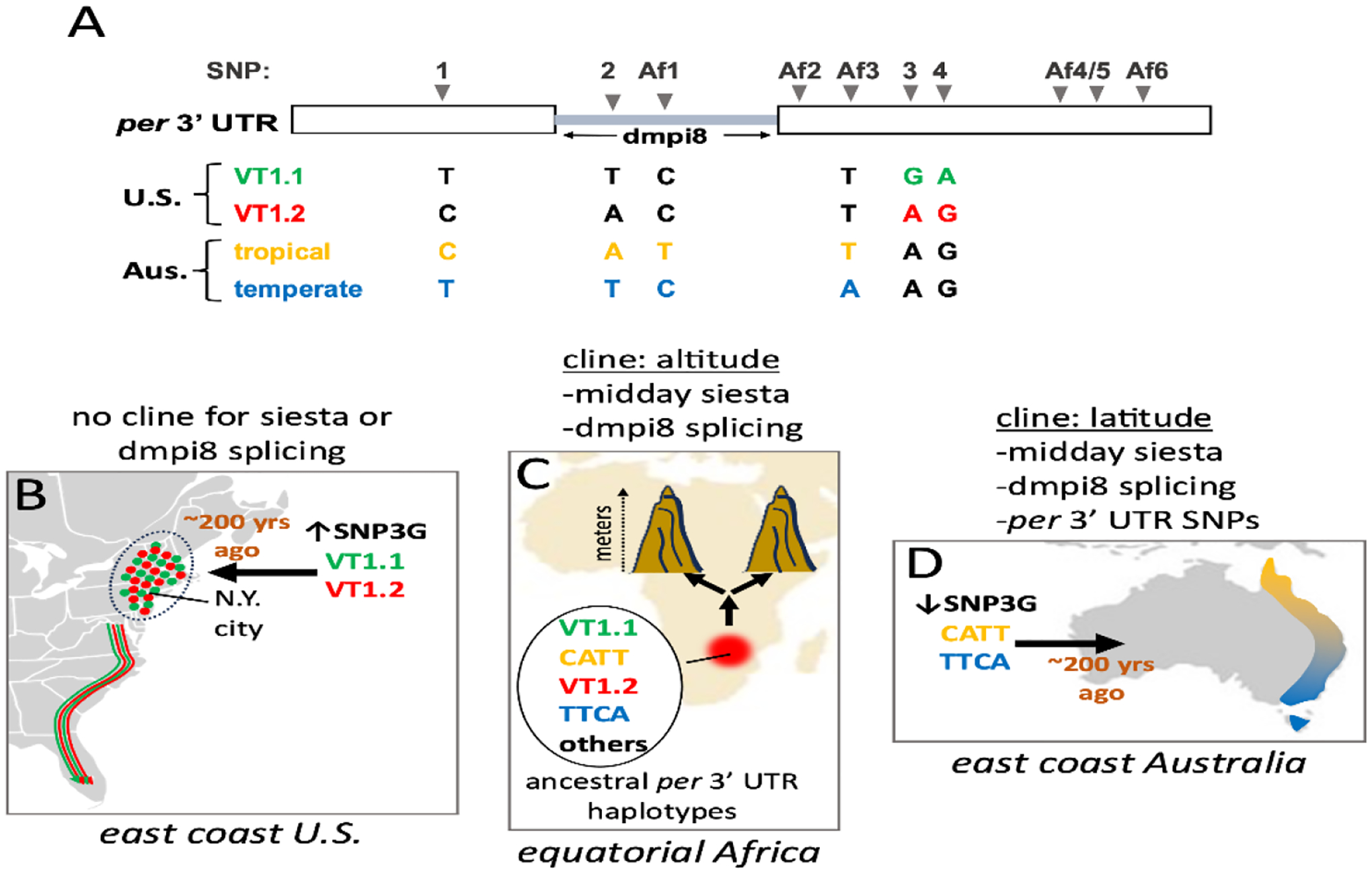
Summary of clinal relationships between midday siesta, dimpi8 splicing efficiency and per 3’ UTR SNPs. (A) Schematic of of the major SNPs in the per 3’ UTR found in natural populations of D. melanogaster that we sampled. (B, C, D) While all per3’ UTR SNPs identified to date are found in Africa, presumably also in ancestral populations (C), it is possible that the founding populations for North America (B) and Australia (D) differed in per3’ UTR haplotypes, perhaps explaining why latitudinal clines in dmpi8 splicing and midday siesta are observed in Australia but not the northeastern United States.
A different conclusion emerged from our studies of tropical and temperate D. melanogaster populations representing the ‘tips’ of a well-studied latitudinal cline from the eastern coast of Australia96,97 (Fig. 6). Monthly averages for the tropical regions range from 21–31°C, whereas forthe temperate region it is 8–16 °C. Unlike the natural populations we examined from the eastern coast of the United States and Africa where SNP3G is present in high frequency (approx. 50–60%), this variant is rare in the Australian populations (approx. 25%)95. For the SNP3A containing flies from Australia we identified two major haplotypes containing four single nucleotide polymorphisms (SNPs) in the 3’ UTR of per (Fig. 6A, top). Two of the SNPs were similar to those identified in flies from the eastern coast of the United States and also seen in African flies (i.e., SNPs 1 and 2). Two additional SNPs seen in Australian flies were not observed in flies from the United States but were found in African flies (i.e., Af1 and Af3). Similar to SNP3, each of these four SNPs (i.e., SNP1, SNP2, Af1 and Af3) were also limited to one of two nucleotide possibilities. The two most abundant of these haplotypes exhibit a reciprocal tropical-temperate distribution in relative frequency. In tropical regions, 75% of the populations evaluated have the C/A/T/T combination of SNPs (SNP1/SNP2/Af1/Af3). Conversely, 75% of the populations from temperate regions have the T/T/C/A combination. The other less prominent SNP3A haplotypes either did not show spatial variation or were too minor to draw any solid conclusions.
Importantly, natural populations and transgenic flies with the major tropical isoform (C/A/T/T) manifest increased daytime sleep and reduced dmpi8 splicing compared to those carrying the temperate variant (T/T/C/A)95. Two of the SNPs (SNP2 and Af1) fall within the dmpi8 intron, whereas SNP1 and Af3 are positioned 5’ and 3’ to the intron, respectively (Fig. 6A, top). It is currently unclear why the C/A/T/T combination leads to reduced dmpi8 splicing efficiency compared to T/T/C/A. Perhaps similar to the mode-of-action at SNP3, the C/A/T/T and T/T/C/A variants differentially affect the binding efficiencies of trans-acting splicing factors such as B52. While we cannot rule out demography as a contributing factor for the inverse geographical distribution of the T/T/C/A and C/A/T/T haplotypes, gene flow between D. melanogaster populations has been shown to be extensive and quite symmetrical along the Australian latitudinal cline97. Irrespective, the maintenance of an inverse temperate-tropical distribution for the T/T/C/A and C/A/T/T haplotypes suggests active selection.
All the per 3’ UTR SNPs we observed in flies from the United States and Australia are present in African populations, suggesting that the different SNPs and SNP combinations in the dper3’ UTRs currently observed in cosmopolitan strains reflect those originating in the ancestral African populations. The C/A/T/T and T/T/C/A variants are not prominent along the eastern coast of the United States. Likewise, the major two haplotypes we noted in the United States, VT1.1 and VT1.2, are not observed in the Australian populations we studied. Although analysis of more populations is required, it is possible that the founding D. melanogaster strains that swept through the United States and Australia were different (Fig. 6B, D). In this regard, it is possible that early invasion of flies with high frequency of SNP3A-containing C/A/T/T and T/T/C/A strains allowed for a more subtle thermal adaptation of midday siesta via varying dmpi8 splicing efficiency. Perhaps a high prevalence of SNP3G-containing flies with much higher baseline dmpi8 splicing efficiency creates too shallow of a thermosensitive response to create a robust cline in dmpi8 splicing efficiency and hence mid-day siesta94. Our findings suggest that that for a large portion of D. melanogaster from Australia, thermal adaptation of midday siesta along the eastern coast also involved spatial selection at the level of ancestrally derived SNPs in the dper3’ UTR that differentially set dmpi8 splicing efficiency. Interestingly, two other studies surveying natural populations that included the America’s, Europe, East Asia but only one population from Australia did not find a connection between latitude (in the aggregate) and daytime sleep, but they both observed that nighttime sleep behavior changes as a function of latitude98,99. While limits in sampling size might have contributed to a lack of correlation between daytime sleep and latitude in those studies, the combined results suggest that clines in midday siesta might be limited to certain latitude-longitude combinations and/or specific regions varying in altitude. An analysis of dmpi8 splicing efficiency and per 3’ UTR haplotypes might provide clues as to why clines in D. melanogaster midday siesta are not always observed despite other traits that clearly show thermal adaptation.
In summary, there is a remarkable parallel co-evolution in D. melanogaster mid-day siesta as a function of altitude and latitude from two continents. In both cases, clinal changes in dmpi8 splicing efficiency are observed, indicating this molecular mechanism is a key target of natural selection. However, in flies from Australia a major mechanism is cis-acting SNPs that regulate daily splicing efficiency of the dmpi8 intron, which is not apparent in African flies. This suggest that even in multi-continent parallel adaptation of a behavioral trait that involves a similar molecular step (i.e., dmpi8 splicing efficiency), there is regional variety in the solutions ‘found’ by natural selection despite shared genetic variation. Additional studies are needed to show how clinal differences in midday siesta provide enhanced fitness in the wild.
Dmpi8 splicing modulates midday siesta in-trans by regulating the expression of the nearby daywake gene.
Despite the overwhelming evidence that splicing of the dmpi8 intron in the 3’ UTR of per regulates midday siesta in a manner consistent with a real-world role in thermal adaptation, the possible connection of dmpi8 to per function was a mystery. As noted above, flies carrying the dmpi8UP version have higher dmpi8 splicing efficiency and reduced midday siesta compared to those with dmpi8WT40. Strangely, the reduced daytime sleep observed in dmpi8UP flies during LD cycles continues when they are exposed for several more days in constant light (LL). Without discussing too many details, prior work showed that PER protein levels are extremely low and do not accumulate in constant light conditions; in addition, behavioral and molecular circadian rhythms are abolished in constant light100. Thus, changes in PER protein levels are unlikely to explain how higherdmpi8 splicing efficiency connects to lower daytime sleep. Perhaps the effects of dmpi8 splicing efficiency on daytime sleep levels is independent of per function?
Indeed, we showed that dmpi8 splicing efficiency regulates daytime sleep in-trans by modulating the levels of a small slightly overlapping reverse-oriented gene that we called daywake (dyw, originally termed the ‘0.9’ gene due to its size in kb)101. By a mechanism that is still not understood, splicing of the dmpi8 intron somehow increases the levels of dyw mRNA. We suspect that spliceosome binding at the per dmpi8 intron is somehow co-opted to stimulate mRNA levels of the reverse transcribed dyw. Irrespective, numerous lines of evidence indicate that increases in the levels of dyw lead to decreases in daytime sleep levels by lowering arousal thresholds to sensory modalities (i.e., visible light)101. The ‘fortuitous’ alignment of an intron whose splicing is enhanced by cold (dmpi8) and can act in-trans to stimulate expression of a nearby “anti-siesta” gene (dyw) might have provided D. melanogaster with the ability to reduce midday sleep when conditions are favorable (i.e., cool), giving it a competitive advantage in adapting to temperate climates. DYW is a member of the takeout-family of juvenile hormone binding proteins (JHBPs) that are part of a larger lipid carrying superfamily102. How it functions to control daytime sleep-wake balance is not clear. Nonetheless, pertinent to this review we have not observed natural variations in dyw gene sequences that might suggest a role in clinal adaptation (unpublished observations). Intriguingly, the effects of dmpi8 splicing on circadian rhythms are very small41,103. Thus, the overriding physiological contribution of dmpi8 splicing thermosensitivity is reflected via its ability to modulate dyw expression, hence generating a robust mechanism for the thermal adaptation of daytime wake-sleep balance in D. melanogaster.
Conclusion
It is intriguing to ponder that the serendipitous genomic alignment of a small intron with multiple weak splice sites ‘precisely’ oriented next to an anti-siesta gene allowed D. melanogaster to widely colonize temperate regions of the New World, setting the stage for northeasterners such as Morgan and other early pioneers to adopt it as a cheap, easily bred, naturally available species that could be exploited for genetic analysis. The rest is history, as they say.
Acknowledgement Statement:
I would like to especially thank all the people in my laboratory that over the course of more than two decades contributed to the research reviewed in this article. Special thanks go to John Majercak, Wen-Feng Chen, Kwang Low, Weihuan Cao, Yong Yang and Zhichao Zhang.
Footnotes
Conflict of Interest Statement:
None
Dedication Statement:
This article is dedicated to the memory of Professor Jerry Pelletier of McGill University in Montreal, a close friend and wonderful scientist who recently passed.
References:
- 1.O’Grady PM & DeSalle R Phylogeny of the Genus Drosophila. Genetics 209, 1–25 (2018). 10.1534/genetics.117.300583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bellen HJ, Tong C & Tsuda H 100 years of Drosophila research and its impact on vertebrate neuroscience: a history lesson for the future. Nat Rev Neurosci 11, 514–22 (2010). 10.1038/nrn2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Letsou A & Bohmann D Small flies--big discoveries: nearly a century of Drosophila genetics and development. Dev Dyn 232, 526–8 (2005). 10.1002/dvdy.20307 [DOI] [PubMed] [Google Scholar]
- 4.Keller A Drosophila melanogaster’s history as a human commensal. Current Biology 17, R77–R81 (2007). DOI 10.1016/j.cub.2006.12.031 [DOI] [PubMed] [Google Scholar]
- 5.Markow TA The secret lives of Drosophila flies. Elife 4(2015). 10.7554/eLife.06793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stephenson R & Metcalfe NH Drosophila melanogaster: a fly through its history and current use. J R Coll Physicians Edinb 43, 70–5 (2013). 10.4997/JRCPE.2013.116 [DOI] [PubMed] [Google Scholar]
- 7.Rubin GM & Spradling AC Genetic transformation of Drosophila with transposable element vectors. Science 218, 348–53 (1982). 10.1126/science.6289436 [DOI] [PubMed] [Google Scholar]
- 8.Spradling AC & Rubin GM Transposition of cloned P elements into Drosophila germ line chromosomes. Science 218, 341–7 (1982). 10.1126/science.6289435 [DOI] [PubMed] [Google Scholar]
- 9.Lachaise D et al. Historical Biogeography of the Drosophila-Melanogaster Species Subgroup. Evolutionary Biology-New York 22, 159–225 (1988). [Google Scholar]
- 10.Izumitani HF, Kusaka Y, Koshikawa S, Toda MJ & Katoh T Phylogeography of the Subgenus Drosophila (Diptera: Drosophilidae): Evolutionary History of Faunal Divergence between the Old and the New Worlds. PLoS One 11, e0160051 (2016). 10.1371/journal.pone.0160051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Russo CA, Takezaki N & Nei M Molecular phylogeny and divergence times of drosophilid species. Mol Biol Evol 12, 391–404 (1995). 10.1093/oxfordjournals.molbev.a040214 [DOI] [PubMed] [Google Scholar]
- 12.Mansourian S et al. Wild African Drosophila melanogaster Are Seasonal Specialists on Marula Fruit. Curr Biol 28, 3960–3968 e3 (2018). 10.1016/j.cub.2018.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pool JE et al. Population Genomics of sub-saharan Drosophila melanogaster: African diversity and non-African admixture. PLoS Genet 8, e1003080 (2012). 10.1371/journal.pgen.1003080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arguello JR, Laurent S & Clark AG Demographic History of the Human Commensal Drosophila melanogaster. Genome Biol Evol 11, 844–854 (2019). 10.1093/gbe/evz022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haudry A, Laurent S & Kapun M Population Genomics on the Fly: Recent Advances in Drosophila. Methods Mol Biol 2090, 357–396 (2020). 10.1007/978-1-0716-0199-0J5 [DOI] [PubMed] [Google Scholar]
- 16.Lachaise D & Silvain JF How two Afrotropical endemics made two cosmopolitan human commensals: the Drosophila melanogaster-D. simulans palaeogeographic riddle. Genetica 120, 17–39 (2004). 10.1023/b:gene.0000017627.27537.ef [DOI] [PubMed] [Google Scholar]
- 17.Adrion JR, Hahn MW & Cooper BS Revisiting classic clines in Drosophila melanogaster in the age of genomics. Trends Genet 31, 434–44 (2015). 10.1016/j.tig.2015.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fabian DK et al. Spatially varying selection shapes life history clines among populations of Drosophila melanogaster from sub-Saharan Africa. J Evol Biol 28, 826–40 (2015). 10.1111/jeb.12607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mayekar HV et al. Clinal variation as a tool to understand climate change. Front Physiol 13, 880728 (2022). 10.3389/fphys.2022.880728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodrigues MF, Vibranovski MD & Cogni R Clinal and seasonal changes are correlated in Drosophila melanogaster natural populations. Evolution 75, 2042–2054 (2021). 10.1111/evo.14300 [DOI] [PubMed] [Google Scholar]
- 21.Machado HE et al. Broad geographic sampling reveals the shared basis and environmental correlates of seasonal adaptation in Drosophila. Elife 10(2021). 10.7554/eLife.67577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bennie JJ, Duffy JP, Inger R & Gaston KJ Biogeography of time partitioning in mammals. Proc Natl Acad Sci U S A 111, 13727–32 (2014). 10.1073/pnas.1216063110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Daan S Adaptive daily strategies in behavior, in Biological rhythms 275–298 (Springer, 1981). [Google Scholar]
- 24.Cederroth CR et al. Medicine in the Fourth Dimension. Cell Metab 30, 238–250 (2019). 10.1016/j.cmet.2019.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koronowski KB & Sassone-Corsi P Communicating clocks shape circadian homeostasis. Science 371 (2021). 10.1126/science.abd0951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takahashi JS Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet 18, 164–179 (2017). 10.1038/nrg.2016.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Young MW & Kay SA Time zones: a comparative genetics of circadian clocks. Nat Rev Genet 2, 702–15 (2001). 10.1038/35088576 [DOI] [PubMed] [Google Scholar]
- 28.Keene AC & Duboue ER The origins and evolution of sleep. J Exp Biol 221 (2018). 10.1242/jeb.159533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sehgal A & Mignot E Genetics of sleep and sleep disorders. Cell 146, 194–207 (2011). 10.1016/j.cell.2011.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shafer OT & Keene AC The Regulation of Drosophila Sleep. Curr Biol 31, R38–R49 (2021). 10.1016/j.cub.2020.10.082 [DOI] [PubMed] [Google Scholar]
- 31.Beckwith EJ & French AS Sleep in Drosophila and Its Context. Front Physiol 10, 1167 (2019). 10.3389/fphys.2019.01167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harding EC, Franks NP & Wisden W The Temperature Dependence of Sleep. Front Neurosci 13, 336 (2019). 10.3389/fnins.2019.00336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mattingly SM et al. The effects of seasons and weather on sleep patterns measured through longitudinal multimodal sensing. NPJ Digit Med 4, 76 (2021). 10.1038/s41746-021-00435-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Obradovich N, Migliorini R, Mednick SC & Fowler JH Nighttime temperature and human sleep loss in a changing climate. Sci Adv 3, e1601555 (2017). 10.1126/sciadv.1601555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okamoto-Mizuno K & Mizuno K Effects of thermal environment on sleep and circadian rhythm. J Physiol Anthropol 31, 14 (2012). 10.1186/1880-6805-31-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heckrotte C The effect of the environmental factors in the locomotory activity of the plains garter snake (Thamnophis radix radix). Animal Behaviour 10, 193–207 (1962). [Google Scholar]
- 37.Sweeney BM & Hastings JW Effects of temperature upon diurnal rhythms. Cold Spring Harb Symp Quant Biol 25, 87–104 (1960). 10.1101/sqb.1960.025.01.009 [DOI] [PubMed] [Google Scholar]
- 38.Yetish G et al. Natural sleep and its seasonal variations in three pre-industrial societies. Curr Biol 25, 2862–2868 (2015). 10.1016/j.cub.2015.09.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alpert MH, Gil H, Para A & Gallio M A thermometer circuit for hot temperature adjusts Drosophila behavior to persistent heat. Curr Biol 32, 4079–4087 e4 (2022). 10.1016/j.cub.2022.07.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cao W & Edery I A novel pathway for sensory-mediated arousal involves splicing of an intron in the period clock gene. Sleep 38, 41–51 (2015). 10.5665/sleep.4322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Majercak J, Sidote D, Hardin PE & Edery I How a circadian clock adapts to seasonal decreases in temperature and day length. Neuron 24, 219–30 (1999). 10.1016/s0896-6273(00)80834-x [DOI] [PubMed] [Google Scholar]
- 42.Parisky KM, Agosto Rivera JL, Donelson NC, Kotecha S & Griffith LC Reorganization of Sleep by Temperature in Drosophila Requires Light, the Homeostat, and the Circadian Clock. Curr Biol 26, 882–92 (2016). 10.1016/j.cub.2016.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lehrskov LL et al. The role of IL-1 in postprandial fatigue. Mol Metab 12, 107–112 (2018). 10.1016/j.molmet.2018.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Monk TH The post-lunch dip in performance. Clin Sports Med 24, e15–23, xi–xii (2005). 10.1016/j.csm.2004.12.002 [DOI] [PubMed] [Google Scholar]
- 45.Murphy KR et al. Postprandial sleep mechanics in Drosophila. Elife 5(2016). 10.7554/eLife.19334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Borbely AA A two process model of sleep regulation. Hum Neurobiol 1, 195–204 (1982). [PubMed] [Google Scholar]
- 47.Deboer T Sleep homeostasis and the circadian clock: Do the circadian pacemaker and the sleep homeostat influence each other’s functioning? Neurobiol Sleep Circadian Rhythms 5, 68–77 (2018). 10.1016/j.nbscr.2018.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mantua J & Spencer RMC Exploring the nap paradox: are mid-day sleep bouts a friend or foe? Sleep Med 37, 88–97 (2017). 10.1016/j.sleep.2017.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Panossian LA & Veasey SC Daytime sleepiness in obesity: mechanisms beyond obstructive sleep apnea-a review. Sleep 35, 605–15 (2012). 10.5665/sleep.1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schapira AH Excessive daytime sleepiness in Parkinson’s disease. Neurology 63, S24–7 (2004). 10.1212/wnl.63.8_suppl_3.s24 [DOI] [PubMed] [Google Scholar]
- 51.Most EI, Scheltens P & Van Someren EJ Increased skin temperature in Alzheimer’s disease is associated with sleepiness. J Neural Transm (Vienna) 119, 1185–94 (2012). 10.1007/s00702-012-0864-1 [DOI] [PubMed] [Google Scholar]
- 52.Te Lindert BHW & Van Someren EJW Skin temperature, sleep, and vigilance. Handb Clin Neurol 156, 353–365 (2018). 10.1016/B978-0-444-63912-7.00021-7 [DOI] [PubMed] [Google Scholar]
- 53.Lane JM et al. Genome-wide association analyses of sleep disturbance traits identify new loci and highlight shared genetics with neuropsychiatric and metabolic traits. Nat Genet 49, 274–281 (2017). 10.1038/ng.3749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lane JM et al. Genetics of circadian rhythms and sleep in human health and disease. Nat Rev Genet 24, 4–20 (2023). 10.1038/s41576-022-00519-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lopez-Minguez J, Morosoli JJ, Madrid JA, Garaulet M & Ordonana JR Heritability of siesta and night-time sleep as continuously assessed by a circadian-related integrated measure. Sci Rep 7, 12340 (2017). 10.1038/s41598-017-12460-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ishimoto H, Lark A & Kitamoto T Factors that Differentially Affect Daytime and Nighttime Sleep in Drosophila melanogaster. Front Neurol 3, 24 (2012). 10.3389/fneur.2012.00024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chiu JC, Low KH, Pike DH, Yildirim E & Edery I Assaying locomotor activity to study circadian rhythms and sleep parameters in Drosophila. J Vis Exp (2010). 10.3791/2157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Garbe DS et al. Context-specific comparison of sleep acquisition systems in Drosophila. Biol Open 4, 1558–68 (2015). 10.1242/bio.013011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gilestro GF Video tracking and analysis of sleep in Drosophila melanogaster. Nat Protoc 7, 995–1007 (2012). 10.1038/nprot.2012.041 [DOI] [PubMed] [Google Scholar]
- 60.Siwicki KK, Hardin PE & Price JL Reflections on contributing to “big discoveries” about the fly clock: Our fortunate paths as post-docs with 2017 Nobel laureates Jeff Hall, Michael Rosbash, and Mike Young. Neurobiol Sleep Circadian Rhythms 5, 58–67 (2018). 10.1016/j.nbscr.2018.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wheeler DA, Hamblen-Coyle MJ, Dushay MS & Hall JC Behavior in light-dark cycles of Drosophila mutants that are arrhythmic, blind, or both. J Biol Rhythms 8, 67–94 (1993). 10.1177/074873049300800106 [DOI] [PubMed] [Google Scholar]
- 62.Matsumoto A, Matsumoto N, Harui Y, Sakamoto M & Tomioka K Light and temperature cooperate to regulate the circadian locomotor rhythm of wild type and period mutants of Drosophila melanogaster. J Insect Physiol 44, 587–596 (1998). 10.1016/s0022-1910(98)00046-8 [DOI] [PubMed] [Google Scholar]
- 63.Edery I, Zwiebel LJ, Dembinska ME & Rosbash M Temporal phosphorylation of the Drosophila period protein. Proc Natl Acad Sci USA 91, 2260–4 (1994). 10.1073/pnas.91.6.2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hardin PE, Hall JC & Rosbash M Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature 343, 536–40 (1990). 10.1038/343536a0 [DOI] [PubMed] [Google Scholar]
- 65.Konopka RJ & Benzer S Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci USA 68, 2112–6 (1971). 10.1073/pnas.68.9.2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hahn MW, Han MV & Han SG Gene family evolution across 12 Drosophila genomes. PLoS Genet 3, e197 (2007). 10.1371/journal.pgen.0030197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Low KH, Lim C, Ko HW & Edery I Natural variation in the splice site strength of a clock gene and species-specific thermal adaptation. Neuron 60, 1054–67 (2008). 10.1016/j.neuron.2008.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Malard F, Mackereth CD & Campagne S Principles and correction of 5’-splice site selection. RNA Biol 19, 943–960 (2022). 10.1080/15476286.2022.2100971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Roca X, Krainer AR & Eperon IC Pick one, but be quick: 5’ splice sites and the problems of too many choices. Genes Dev 27, 129–44 (2013). 10.1101/gad.209759.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rosbash M & Seraphin B Who’s on first? The U1 snRNP-5’ splice site interaction and splicing. Trends Biochem Sci 16, 187–90 (1991). 10.1016/0968-0004(91)90073-5 [DOI] [PubMed] [Google Scholar]
- 71.Lim LP & Burge CB A computational analysis of sequence features involved in recognition of short introns. Proc Natl Acad Sci USA 98, 11193–8 (2001). 10.1073/pnas.201407298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mount SM et al. Splicing signals in Drosophila: intron size, information content, and consensus sequences. Nucleic Acids Res 20, 4255–62 (1992). 10.1093/nar/20.16.4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lachaise D et al. Evolutionary novelties in islands: Drosophila santomea, a new melanogaster sister species from Sao Tome. Proc Biol Sci 267, 1487–95 (2000). 10.1098/rspb.2000.1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Matute DR, Novak CJ & Coyne JA Temperature-based extrinsic reproductive isolation in two species of Drosophila. Evolution 63, 595–612 (2009). 10.1111/j.1558-5646.2008.00588.x [DOI] [PubMed] [Google Scholar]
- 75.Turissini DA, Liu G, David JR & Matute DR The evolution of reproductive isolation in the Drosophila yakuba complex of species. J Evol Biol 28, 557–75 (2015). 10.1111/jeb.12588 [DOI] [PubMed] [Google Scholar]
- 76.Hamada FN et al. An internal thermal sensor controlling temperature preference in Drosophila. Nature 454, 217–20 (2008). 10.1038/nature07001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ainsworth JR, Rossi LM & Murphy EC Jr. The Moloney murine sarcoma virus ts110 5’ splice site signal contributes to the regulation of splicing efficiency and thermosensitivity. J Virol 70, 6474–8 (1996). 10.1128/JVI.70.9.6474-6478.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cizdziel PE, de Mars M & Murphy EC Jr. Exploitation of a thermosensitive splicing event to study pre-mRNA splicing in vivo. Mol Cell Biol 8, 1558–69 (1988). 10.1128/mcb.8.4.1558-1569.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Touchman JW et al. Branchpoint and polypyrimidine tract mutations mediating the loss and partial recovery of the Moloney murine sarcoma virus MuSVts110 thermosensitive splicing phenotype. J Virol 69, 7724–33 (1995). 10.1128/JVI.69.12.7724-7733.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Anna A & Monika G Splicing mutations in human genetic disorders: examples, detection, and confirmation. J Appl Genet 59, 253–268 (2018). 10.1007/s13353-018-0444-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Daguenet E, Dujardin G & Valcarcel J The pathogenicity of splicing defects: mechanistic insights into pre-mRNA processing inform novel therapeutic approaches. EMBO Rep 16, 1640–55 (2015). 10.15252/embr.201541116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jung H, Lee KS & Choi JK Comprehensive characterisation of intronic mis-splicing mutations in human cancers. Oncogene 40, 1347–1361 (2021). 10.1038/s41388-020-01614-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Scotti MM & Swanson MS RNA mis-splicing in disease. Nat Rev Genet 17, 19–32 (2016). 10.1038/nrg.2015.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Carmel I, Tal S, Vig I & Ast G Comparative analysis detects dependencies among the 5’ splice-site positions. RNA 10, 828–40 (2004). 10.1261/rna.5196404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Weil D et al. Temperature-dependent expression of a collagen splicing defect in the fibroblasts of a patient with Ehlers-Danlos syndrome type VII. J Biol Chem 264,16804–9 (1989). [PubMed] [Google Scholar]
- 86.Ibrahim EC et al. Weak definition of IKBKAP exon 20 leads to aberrant splicing in familial dysautonomia. Hum Mutat 28, 41–53 (2007). 10.1002/humu.20401 [DOI] [PubMed] [Google Scholar]
- 87.Lloyd J, Narcisi P, Richards A & Pope FM A T+6 to C+6 mutation in the donor splice site of COL3A1 IVS7 causes exon skipping and results in Ehlers-Danlos syndrome type IV. J Med Genet 30, 376–80 (1993). 10.1136/jmg.30.5.376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Suzuki H et al. Recurrent noncoding U1 snRNA mutations drive cryptic splicing in SHH medulloblastoma. Nature 574, 707–711 (2019). 10.1038/s41586-019-1650-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hibio N, Hino K, Shimizu E, Nagata Y & Ui-Tei K Stability of miRNA 5’terminal and seed regions is correlated with experimentally observed miRNA-mediated silencing efficacy. Sci Rep 2, 996 (2012). 10.1038/srep00996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rajpurohit S, Zhao X & Schmidt PS A resource on latitudinal and altitudinal clines of ecologically relevant phenotypes of the Indian Drosophila. Sci Data 4, 170066 (2017). 10.1038/sdata.2017.66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Low KH, Chen WF, Yildirim E & Edery I Natural variation in the Drosophila melanogaster clock gene period modulates splicing of its 3’-terminal intron and mid-day siesta. PLoS One 7, e49536 (2012). 10.1371/journal.pone.0049536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang Z, Cao W & Edery I The SR protein B52/SRp55 regulates splicing of the period thermosensitive intron and mid-day siesta in Drosophila. Sci Rep 8, 1872 (2018). 10.1038/s41598-017-18167-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Preussner M et al. Body Temperature Cycles Control Rhythmic Alternative Splicing in Mammals. Mol Cell 67, 433–446 e4 (2017). 10.1016/j.molcel.2017.06.006 [DOI] [PubMed] [Google Scholar]
- 94.Cao W & Edery I Mid-day siesta in natural populations of D. melanogaster from Africa exhibits an altitudinal cline and is regulated by splicing of a thermosensitive intron in the period clock gene. BMC Evol Biol 17, 32 (2017). 10.1186/s12862-017-0880-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yang Y & Edery I Parallel clinal variation in the mid-day siesta of Drosophila melanogaster implicates continent-specific targets of natural selection. PLoS Genet 14, e1007612 (2018). 10.1371/journal.pgen.1007612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hoffmann AA & Weeks AR Climatic selection on genes and traits after a 100 year-old invasion: a critical look at the temperate-tropical clines in Drosophila melanogaster from eastern Australia. Genetica 129, 133–47 (2007). 10.1007/s10709-006-9010-z [DOI] [PubMed] [Google Scholar]
- 97.Kolaczkowski B, Kern AD, Holloway AK & Begun DJ Genomic differentiation between temperate and tropical Australian populations of Drosophila melanogaster. Genetics 187, 245–60 (2011). 10.1534/genetics.110.123059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Brown EB et al. Variation in sleep and metabolic function is associated with latitude and average temperature in Drosophila melanogaster. Ecol Evol 8, 4084–4097 (2018). 10.1002/ece3.3963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Svetec N, Zhao L, Saelao P, Chiu JC & Begun DJ Evidence that natural selection maintains genetic variation for sleep in Drosophila melanogaster. BMC Evol Biol 15, 41 (2015). 10.1186/s12862-015-0316-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Marrus SB, Zeng H & Rosbash M Effect of constant light and circadian entrainment of perS flies: evidence for light-mediated delay of the negative feedback loop in Drosophila. EMBO J 15, 6877–86 (1996). [PMC free article] [PubMed] [Google Scholar]
- 101.Yang Y & Edery I Daywake, an Anti-siesta Gene Linked to a Splicing-Based Thermostat from an Adjoining Clock Gene. Curr Biol 29, 1728–1734 e4 (2019). 10.1016/j.cub.2019.04.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Alva V & Lupas AN The TULIP superfamily of eukaryotic lipid-binding proteins as a mediator of lipid sensing and transport. Biochim Biophys Acta 1861, 913–923 (2016). 10.1016/j.bbalip.2016.01.016 [DOI] [PubMed] [Google Scholar]
- 103.Cheng Y, Gvakharia B & Hardin PE Two alternatively spliced transcripts from the Drosophila period gene rescue rhythms having different molecular and behavioral characteristics. Mol Cell Biol 18, 6505–14 (1998). 10.1128/MCB.18.11.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]


