Abstract
A Pseudomonas aeruginosa gene homologous to the fabG gene, which encodes the NADPH-dependent β-ketoacyl-acyl carrier protein (ACP) reductase required for fatty acid synthesis, was identified. The insertional mutation of this fabG homolog (herein called rhlG) produced no apparent effect on the growth rate and total lipid content of P. aeruginosa cells, but the production of rhamnolipids was completely abrogated. These results suggest that the synthetic pathway for the fatty acid moiety of rhamnolipids is separate from the general fatty acid synthetic pathway, starting with a specific ketoacyl reduction step catalyzed by the RhlG protein. In addition, the synthesis of poly-β-hydroxyalkanoate (PHA) is delayed in this mutant, suggesting that RhlG participates in PHA synthesis, although it is not the only reductase involved in this pathway. Traits regulated by the quorum-sensing response, other than rhamnolipid production, including production of proteases, pyocyanine, and the autoinducer butanoyl-homoserine lactone (PAI-2), were not affected by the rhlG mutation. We conclude that the P. aeruginosa rhlG gene encodes an NADPH-dependent β-ketoacyl reductase absolutely required for the synthesis of the β-hydroxy acid moiety of rhamnolipids and that it has a minor role in PHA production. Expression of rhlG mRNA under different culture conditions is consistent with this conclusion.
Pseudomonas aeruginosa is a bacterium that can be isolated from many different habitats, including water, soil, and plants (5). P. aeruginosa is also an opportunistic human pathogen that causes serious nosocomial infections (8). The secretion of numerous toxic compounds and hydrolytic enzymes has been correlated with its pathogenicity (19). These exoproducts include different proteases, such as elastase, LasA protease, and alkaline protease, as well as phospholipase C, exotoxin A, pyocyanine, and rhamnolipids. The production of these compounds is considered to be a virulence-associated trait and is coordinately regulated by a mechanism called “quorum sensing” (11), which depends on the production of N-acylated homoserine lactones harboring acyl substituents of two different lengths; PAI-1 contains a 12-carbon chain (22), while PAI-2 contains a butanoyl moiety (23). These small diffusible signaling molecules activate gene expression at high bacterial densities through interaction with specific transcriptional activators, LasR (22) and RhlR (20), respectively.
The role of these exoproducts in soil or aquatic habitats has not been determined, but it is clear that environmental and clinical P. aeruginosa isolates do not represent different populations, since it has been shown that there is a major clone common to pathogenic and environmental isolates of this bacterium (26).
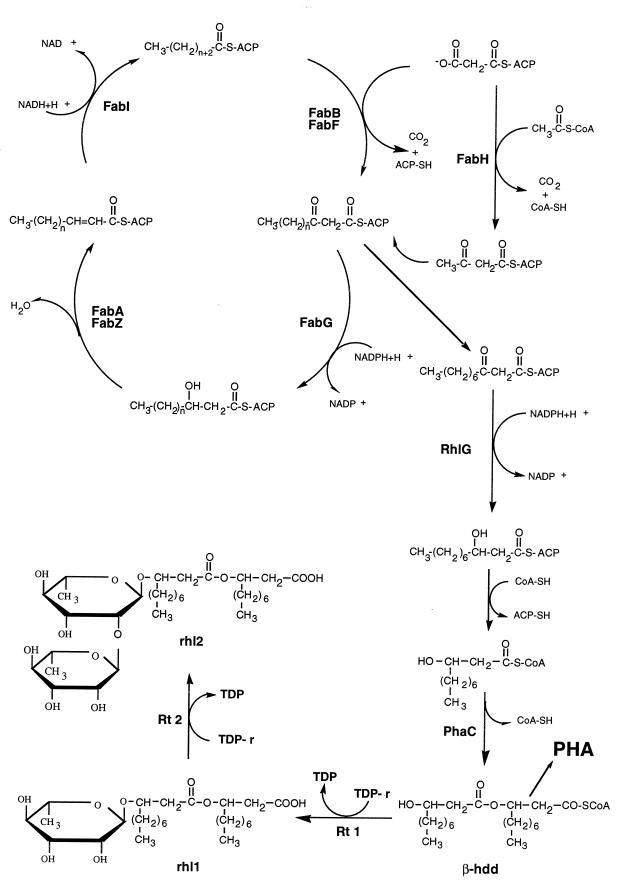
Rhamnolipids are glycolipids produced by P. aeruginosa which reduce water surface tension and emulsify oil. These compounds are biodegradable and have potential industrial and environmental applications (14, 17). Recently, rhamnolipids have been found to have antagonistic effects on economically important zoosporic plant pathogens, thus opening up their use as biocontrol agents (29). The rhamnolipids produced by P. aeruginosa in liquid cultures (Fig. 1) are mainly rhamnosyl-β-hydroxydecanoyl-β-hydroxydecanoate (monorhamnolipid) and rhamnosyl-rhamnosyl-β-hydroxydecanoyl-β-hydroxydecanoate (dirhamnolipid). Rhamnolipid biosynthesis proceeds through two rhamnose transfers from TDP–l-rhamnose (3). For the synthesis of monorhamnolipid, the enzyme rhamnosyltransferase 1 (Rt 1) catalyzes the rhamnose transfer to β-hydroxydecanoyl-β-hydroxydecanoate, while Rt 2 synthesizes dirhamnolipid from TDP–l-rhamnose and monorhamnolipid. Genes coding for biosynthesis, regulation, and induction of Rt 1 enzyme are organized in tandem in the rhlABRI gene cluster around min 38 of the P. aeruginosa chromosome (20). The genes encoding Rt 2 have yet to be described.
FIG. 1.
Schematic representation of the fatty acid biosynthetic pathway showing the deduced role of the RhlG protein in the production of rhamnolipids and PHAs. Initiation of the fatty acid biosynthetic cycle, catalyzed by FabH, requires acetyl-CoA and malonyl-ACP to form aceto-acetyl-ACP. Subsequent cycles are initiated by condensation of malonyl-ACP with acyl-ACP, catalyzed by FabB and FabF. In the second step, the resulting β-ketoester is reduced to a β-hydroxyacyl-ACP by FabG. The third step in the cycle is catalyzed by either FabA or FabZ. The fourth and final step is the conversion of trans-2-enoyl-ACP to acyl-ACP, a reaction catalyzed by FabI. TDP-r, thymidine-diphospho-l-rhamnose; PhaC, PHA synthase; rhl 1, monorhamnolipid; rhl 2, dirhamnolipid; β-hdd, β-hydroxydecanoyl-β-hydroxydecanoate.
Polyhydroxyalkanoates (PHAs) are bacterial storage compounds, which are synthesized by the polymerization of β-hydroxyacids by the PHA synthases (PhaC), with the coenzyme A (CoA)-linked fatty acids as substrates (Fig. 1) (31). The NADPH-dependent β-ketoacyl-CoA reductase (PhaB) is responsible for the reduction step in the production of the β-hydroxyacids. These storage compounds are intracellularly deposited as granules in many species. P. aeruginosa mainly produces PHAs consisting of medium-chain-length polymers, mainly poly-β-hydroxydecanoate (30).
The fatty acid synthetase system of Escherichia coli as well as that of most bacteria and plants is a dissociated fatty acid type of system (i.e., different reactions are catalyzed by separate proteins encoded by separate genes) (7). This biosynthetic pathway has been widely studied at the molecular level in E. coli and is encoded by a cluster of genes called fab genes which have been cloned and sequenced. As shown in Fig. 1, each round of elongation requires four chemical reactions. Initiation requires acetyl-CoA and malonyl-acyl carrier protein (ACP) to form aceto-acetyl-ACP. The first cycle is initiated by Kas III (FabH). Subsequent cycles are initiated by condensation of malonyl-ACP with acyl-ACP, catalyzed by Kas I (FabB) and Kas II (FabF). In the second step, the resulting β-ketoester is reduced to a β-hydroxyacyl-ACP by a single, NADPH-dependent β-ketoacyl-ACP reductase (FabG). The third step in the cycle is catalyzed by either the fabA- or fabZ-encoded β-hydroxyacyl-ACP dehydratases. The fourth and final step is the conversion of trans-2-enoyl-ACP to acyl-ACP, a reaction catalyzed by a single NADH-dependent enoyl-ACP reductase (FabI).
Recently the complete P. aeruginosa fab gene cluster sequence was deposited in the GenBank database (accession no. U91631). In addition, the P. aeruginosa fabA and fabB genes have been characterized (15). The objective of this work is to present evidence for the existence of a P. aeruginosa gene (rhlG) encoding a FabG homolog which is specifically involved in the synthesis of the β-hydroxyacid moiety of rhamnolipids. There have been no previous reports on the nature of the enzymes involved in the synthesis of the β-hydroxyacid moiety of rhamnolipids, and it has been assumed that they are the same proteins involved in fatty acid synthesis. Evidence is also presented suggesting that RhlG has a role in PHA synthesis.
MATERIALS AND METHODS
Microbiological procedures.
The bacterial strains and plasmids used in this work are shown in Table 1. P. aeruginosa strains were routinely grown on Luria- Bertani medium (LB), Pseudomonas isolation agar (PIA [Difco]), or PPGAS (the phosphate-limited medium designed for rhamnolipid production) (34) at 29°C. M9 minimal medium supplemented with 0.05% NH4Cl and gluconate 0.2% (MM + gluconate) was used to induce the production of PHAs. The antibiotic concentrations used for P. aeruginosa PAO1 and W51D, respectively, were as follows: carbenicillin, 250 and 50 μg/ml; gentamicin, 250 and 30 μg/ml; and tetracycline, 150 and 50 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Strains | ||
| P. aeruginosa | ||
| W51D | Strain able to degrade surfactants | 28 |
| W51D-10 | W51D rhlG::Tc mutant | This work |
| PAO1 | Wild-type strain | B. H. Hollowayb |
| ACP5 | PAO1 rhlG::Tc mutant | This work |
| PAO R1 | PAO1 lasR::Tc mutant | 12 |
| C. violaceum | ||
| ATCC 31532 | Wild-type strain | 16 |
| CV026 | ATCC 31532 nonpigmented mutant | 16 |
| Plasmids | ||
| pJQ200mp18 | Cloning vector Gmr, unable to replicate in Pseudomonas | 25 |
| pJC1 | pJQ200mp18 with a 600-bp rhlG internal fragment | This work |
| pJC2 | pJC1 with a Tcr cassette cloned into the single SmaI site of rhlG | This work |
| pUCP20 | pUC19-derived E. coli-Pseudomonas shuttle vector, Cbr | 33 |
| pJC3 | pUCP20 with the PAO1 rhlG gene obtained by PCR | This work |
| pJC4 | pUCP20 with 7 kb of W51D DNA, including the rhlG gene | This work |
The abbreviations used represent resistance to carbenicillin (Cbr), gentamicin (Gmr), and tetracycline (Tcr).
Monash University, Clayton, Victoria, Australia.
Exoproducts and PHA determination.
Pyocyanine was extracted with chloroform from the culture supernatant and determined by A690 as described previously (6). Protease production was measured by halo formation in LB plates containing 1% skim milk and inoculated with 20 μl of a saturated liquid culture. Total rhamnolipid concentration was determined from culture supernatants of cells grown on PPGAS medium at 29°C for 48 h by measuring the rhamnose concentration after acid hydrolysis by the orcinol method (4). The production of butanoyl-homoserine lactone (PAI-2) by different P. aeruginosa strains was determined by using the biosensor developed for the detection of small-chain N-acyl-homoserine lactones based on violacein production by Chromobacterium violaceum mutant strain CV026 (16). The wild-type C. violaceum strain ATCC 31532 produces violacein induced by the autoinducer N-hexanoyl-l-homoserine lactone, while mutant CV026 only produces this pigment when given medium supplemented with this autoinducer or related compounds, such as the P. aeruginosa PAI-2 autoinducer. PHA was determined after 24 h of growth under nitrogen-deprived conditions (30). Cells were harvested by centrifugation and washed with 100 mM Tris–100 mM NaCl buffer (pH 7). Cells were ruptured by sonication, and the extract was digested with 1.8% sodium hypochlorite for 1 h. After centrifugation, the pellet was washed twice with ethanol and once with acetone. The PHA concentration is expressed as milligrams of PHA per milligram of protein.
Fatty acid analysis.
Total cell lipids were extracted by the method of Folch et al. (10). Briefly, 1 ml of the culture was washed twice and then brought back to the original volume. The following reagents were added with vortexing after each addition: 2 ml of 2:1 methanol-chloroform, 1 ml of 1 N KCl acidified with 0.1 N HCl, and 1 ml of chloroform. In some samples, a white emulsion phase formed between the aqueous and organic phases. In this case, the sample was placed in the refrigerator overnight to allow the emulsion phase to settle. The lower phase (chloroform) was removed and evaporated at 45°C under a nitrogen stream. The fatty acids were analyzed by gas chromatography after methyl esterification (18). Chloroform (0.5 ml) was added, and the sample was vortexed. Two milliliters of BF3-methanol was added, and the mixture was heated at 80°C for 1 h in an airtight Teflon sealed screw-cap tube (18). The resulting fatty acid methyl esters (FAMEs) were extracted three times with 1 ml of hexane, and the three fractions were combined. Finally, the hexane was evaporated at 45°C under a nitrogen stream, and the FAMEs were brought to a concentration of 200 μl with chloroform.
Electron microscopy.
PHA production by different P. aeruginosa strains was visualized by electron microscopy. Cells were treated for electron microscopic observation as follows. They were washed three times with phosphate buffer at pH 7.2, fixed with 2% glutaraldehyde for 2 h, and washed with phosphate buffer. Further fixation with 2% osmium tetroxide for 2 h was done; all of these procedures were carried out at 4°C. Fixed cells were washed and then dehydrated by passage through a graded ethanol series. After exposure to propylene oxide, samples were placed in L. R. White resin as recommended by the manufacturer. Ultrathin sections were incubated with uranyl acetate, washed with distilled water, treated with lead citrate, washed again, and observed.
Nucleic acid procedures.
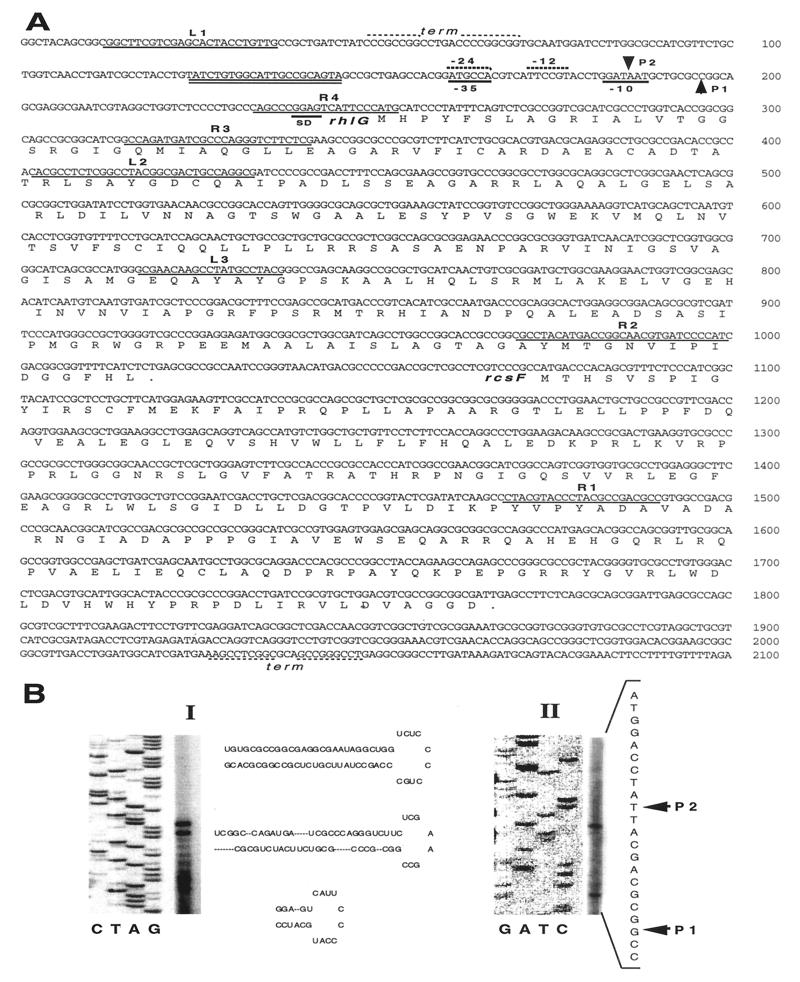
DNA isolation, cloning and sequencing, Southern and Northern blotting, and nick translation procedures were carried out as described previously (27). RNA was isolated with the RNaid PLUS kit (BIO101, Inc.). Primer extension analysis was done with two primers (R3 and R4 [Fig. 2]), both located in the 5′ region of the rhlG gene from P. aeruginosa PAO1 (Pseudomonas Genome Project contig 1780). The templates used for sequencing reactions were obtained by PCR of total DNA from P. aeruginosa PAO1 with the oligonucleotides L1 and R3 or R4 (Fig. 2). The sequencing reactions were done with the Thermo Sequenase radiolabeled terminator cycle sequencing kit (Amersham Life Science, Inc.).
FIG. 2.
Characterization of the transcription arrangement of the P. aeruginosa PAO1 rhlG and rcsF genes. (A) Nucleotide sequence of the genes and regulatory sequences. The sequence and position of the oligonucleotides used during this work are shown in the figure and identified as Ln or Rn, depending on their polarity (L oligonucleotides amplify the sequence from 5′ to 3′, and R oligonucleotides have the opposite polarity). The sequence corresponding to the lux box is double underlined. Arrows indicate the two transcription start sites detected (P1 and P2). SD (Shine-Dalgarno) indicates the ribosome binding site sequence for mRNA translation. The sequences corresponding to putative transcriptional termination sites (term) are shown. (B) Primer extension analysis of the rhlG gene with two different oligonucleotides as primers. In panel BI, the primer extension analysis was done with oligonucleotide R3 and revealed the existence of the mRNA secondary structures shown. In panel BII, the oligonucleotide R4 was used, and two transcription start points indicated as P1 and P2 were found.
Genetic manipulations.
P. aeruginosa matings (34) and transformation (21) were done as reported previously. The PAO1 and W51D rhlG::Tc mutants (ACP5 and W51D-10, respectively [Table 1]) were constructed by selection of double recombination events with plasmid pJC2. This plasmid is a derivative of plasmid pJC1, which contains a 600-bp rhlG internal fragment from P. aeruginosa W51D. A 1.4-kb tetracycline resistance gene from plasmid pBSL141Tc (1) was cloned on the unique SmaI site of the rhlG fragment contained in plasmid pJC1, rendering plasmid pJC2 (Table 1). The W51D rhlG internal fragment was obtained by PCR with the oligonucleotides L2′ (CGAACTCTGCAGGTACGGCGAGTGCATCGG) and R2′ (GATGCTGCAGATGTTGCCGGTCATGTAGGC) (corresponding to the positions in the PAO1 rhlG gene of the L2 and R2 oligonucleotides shown in Fig. 2), with the recognition site for the PstI endonuclease incorporated on the flanking ends of both of them. Plasmid pJC3 is a pUCP20 (33) derivative containing the PAO1 rhlG gene obtained by PCR with oligonucleotides L1 and R1 (Fig. 2). Plasmid pJC4 is a pUCP20 derivative containing a 7-kb EcoRI W51D DNA fragment which includes the rhlG gene. The sequence of the L1 oligonucleotide is not present in the W51D rhlG gene region.
Computer analysis of the DNA and protein sequences.
Computer analyses of the sequences were carried out by using the GENE WORKS program (IntelliGenetics, Inc.) and the University of Wisconsin Genetics Computer Group (UWGCG) programs. The sequences of different P. aeruginosa PAO1 contigs were obtained from the Pseudomonas Genome Project web site (http://www.pseudomonas.com).
Nucleotide sequence accession number.
The sequence of the W51D rhlG gene has been deposited in the GenBank database under accession no. AF052586.
RESULTS AND DISCUSSION
Identification and sequencing of the P. aeruginosa W51D rhlG gene.
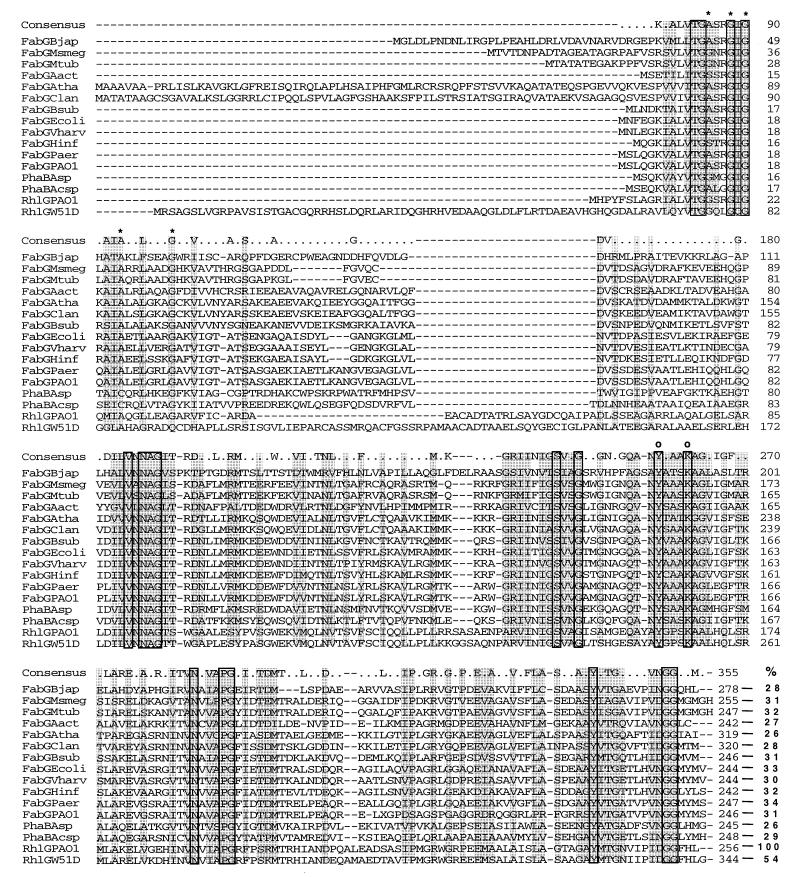
P. aeruginosa W51D is a bacterium which is able to degrade at least 70% of a branched-chain alkylbenzene sulfonate mixture and is resistant to high concentrations of this surfactant (28). In order to study the W51D surfactant catabolic pathway, we have isolated several transposon mutants affected in the degradation of presumed intermediates of the surfactant degradation (unpublished results). During the DNA sequencing of a mutant unable to degrade citronellol, we detected a linked open reading frame (ORF) which was homologous to the E. coli fabG gene, having 36% amino acid sequence identity along the entire length. This ORF, called rhlG, has the characteristic codon usage and bias of its GC composition in the third position of each codon of the Pseudomonas genes (32). The alignment of the protein deduced from the sequence of the rhlG gene with the proteins deposited in the GenBank database confirmed the presence of the characteristic signature for NADPH binding, as well as the characteristic motifs of dehydrogenases. As shown in Fig. 3, these sequences are conserved in all of the sequenced bacterial and plant FabG proteins. However, the chromosomal region surrounding this fabG homolog did not show the presence of other fab genes, as has been reported for P. aeruginosa PAO1 and E. coli.
FIG. 3.
Multiple alignment of the RhlG deduced amino acid sequence with different NADPH-dependent β-ketoacyl-ACP reductase (FabG) and NADPH-dependent ketoacyl-CoA reductase (PhaB) proteins. Residues within rectangles correspond to identical amino acids, and those shaded are conserved among most of the proteins analyzed. The percentage of identity of the different proteins with PAO1 RhlG is shown in the bottom right column of the figure. Asterisks mark the residues which form the NADPH binding signature, and circles show the amino acids conserved in dehydrogenases. FabGBjap, FabG from Bradyrhizobium japonicum; FabGMsmeg, FabG from Mycobacterium smegmatis; FabGMtub, FabG from Mycobacterium tuberculosis; FabGAact, FabG from Actinobacillus actinomycetemcomitans; FabGAtha, FabG from Arabidopsis thaliana; FabGClan, FabG from Cuphea lanceolata; FabGBsub, FabG from Bacillus subtilis; FabGEcoli, FabG from E. coli; FabGVharv, FabG from Vibrio harveyi; FabGHinf, FabG from Haemophilus influenzae; FabGPaer, FabG from P. aeruginosa (GenBank database accession no. U91631); FabGPAO1, FabG from P. aeruginosa PAO1 (contig 1761); PhaBAsp, PhaB from Alcaligenes sp. strain SH69; PhaBAcsp, PhaB from Acinetobacter sp. strain RA3849; RhlGPAO1, RhlG from P. aeruginosa PAO1 (contig 1780); RhlGW51D, RhlG from P. aeruginosa W51D (GenBank database accession no. AF052586).
Another ORF encoding a protein with a sequence 44% identical to that of E. coli RcsF, which is involved in regulation of capsular production (13), was detected downstream of rhlG. This genetic arrangement and the fact that a fabG gene has been already described in P. aeruginosa PAO1 led us to the hypothesis that this is a novel gene which encoded a second functional NADPH-dependent β-ketoacyl reductase. In order to test this hypothesis, we constructed an rhlG::Tc mutant according to the strategy shown in Fig. 4. The inactivation of the W51D rhlG gene did not produce a fatty acid auxotrophy or a decrease in growth rate (data not shown). These results showed that the RhlG protein is not responsible for the total cellular fatty acid synthesis, so a FabG protein should also exist in strain W51D. However, this evidence was not enough to determine the expression and functionality of the rhlG gene product.
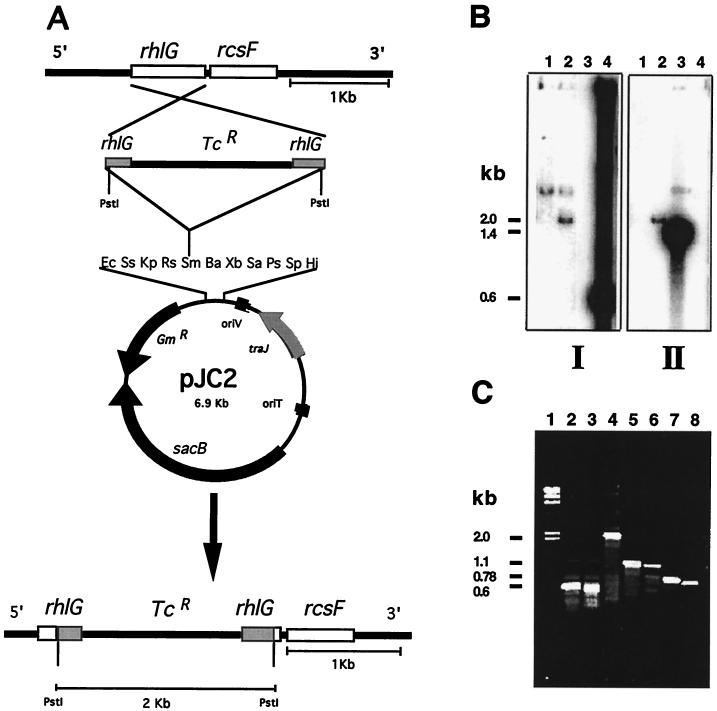
FIG. 4.
Molecular characterization of the PAO1 rhlG mutant ACP5. (A) Schematic representation of the strategy to construct the rhlG mutants (ACP5 and W51D-10). (B) Southern blotting hybridization with the 600-bp insert of plasmid pJC1 (I) and the 1.4-kb Tcr resistance cassette (II) used as probes. Lanes correspond to DNA samples digested with PstI endonuclease from the PAO1 genome (lane 1), the ACP5 genome (lane 2), the Tcr cassette (lane 3), and the PCR product of the amplification of the W51D genome with oligonucleotides L2′ and R2′ (lane 4). (C) Amplification by PCR with different oligonucleotides specific for the rhlG and rcsF genes. Lanes correspond to the following DNA samples: 1, λ phage genome digested with HindIII; 2, PCR product with W51D DNA as a template and the L2′ and R2 oligonucleotides as primers; 3, PCR product with PAO1 DNA as a template and the L2 and R2 oligonucleotides as primers; 4, PCR product with ACP5 DNA as a template and the L2 and R2 oligonucleotides as primers; 5, PCR product with W51D DNA as a template and the L2′ and R1 oligonucleotides as primers; 6, PCR product with PAO1 DNA as a template and the L2 and R1 oligonucleotides as primers; 7, PCR product with W51D DNA as a template and the L3 and R1 oligonucleotides as primers; and 8, PCR product with PAO1 DNA as a template and the L3 and R1 oligonucleotides as primers.
Identification of the rhlG gene in the P. aeruginosa PAO1 genome.
We decided to study the functionality of the RhlG protein in the P. aeruginosa PAO1 strain for two reasons: approximately 95% of its genome has been sequenced (http://www.pseudomonas.com), and the existence of the fabG gene had already been reported in this strain (GenBank database accession no. U91631). We identified the PAO1 rhlG gene in contig 1780 of the Pseudomonas Genome Project, showing the characteristic codon usage and bias of GC composition in the third position of each codon of the Pseudomonas genes (32). The genetic arrangement of PAO1 is similar to that of strain W51D, in which the rcsF gene is downstream (Fig. 2). The deduced PAO1 RhlG protein consists of 256 amino acids, with a predicted molecular mass of 26,813 Da and has amino acids 54% identical to those of the W51D RhlG protein (Fig. 3). The great divergence between both rhlG genes is mainly due to differences in the sequences at their 5′ ends. If the sequences are compared after deletion of the first 52 amino acids of the PAO1 RhlG protein and 112 amino acids of the corresponding protein in W51D, they have 91% identical amino acid sequences. Furthermore, both proteins contain the motifs important for their putative catalytic capabilities (Fig. 3). The difference between the amino-terminal sequences of PAO1 and W51D RhlG proteins is striking, considering that both strains belong to the same species. The significance of this variability is not clear to us. The DNA sequence of the first 300 nucleotides of the PAO1 rhlG 5′ region (Fig. 2) was confirmed by us, and we found only three differences. This result rules out the possibility that the divergence between the rhlG genes is due to major inaccuracies in the reported sequence in contig 1780.
We confirmed that the rhlG genes were conserved and that rcsF was present downstream in PAO1 and W51D strains by PCR amplification of total DNA (Fig. 4C). The following oligonucleotides were used as primers: L2 or L2′ (the oligonucleotide corresponding to the W51D rhlG gene sequence in the same region) and R2, L2 or L2′ and R1, and L3 and R1 (Fig. 2A). The amplified product was a DNA band of the same size from either strain (Fig. 4C), thus validating the high degree of homology between rhlG genes and the conservation of the genetic arrangement inferred from the analysis of the sequence obtained from the Pseudomonas Genome Project in contig 1780.
The sequences of the PAO1 fabG gene from contig 1761 of the Pseudomonas Genome Project and GenBank (accession no. U91631) were compared. The two PAO1 fabG gene DNA sequences are not identical. This inconsistency may result from inaccuracies in the sequence of the Pseudomonas Genome Project. We compared both PAO1 FabG protein sequences to the deduced protein sequences of the PAO1 RhlG protein and found the amino acids were 33 and 34% identical, respectively (Fig. 3). This is further evidence that RhlG is an NADPH-dependent β-ketoacyl reductase. The PAO1 RhlG protein also aligned with FabG proteins of different origin, as well as with PhaB proteins from Alcaligenes sp. strain SH-69 and Acinetobacter sp. strain RA3849 (Fig. 3). This result is not surprising, since the PhaB proteins are NADPH-dependent acetoacetyl reductases which participate in PHA synthesis. The significance of the RhlG homology with PhaB proteins is discussed below.
Expression of the rhlG gene in P. aeruginosa PAO1.
We carried out primer extension experiments to determine whether the rhlG was expressed in strain PAO1 grown for 48 h on PPGAS, a medium designed to increase rhamnolipid production (34). Two oligonucleotides derived from the DNA sequence reported in contig 1780 corresponding to the 5′ end of the rhlG gene were used as primers (Fig. 2). These experiments revealed the presence of a specific rhlG mRNA, confirming that the gene is expressed under these culture conditions (Fig. 2BI and BII). When the R3 oligonucleotide was used, the extension was aborted very near the putative RhlG protein start codon, suggesting the existence of an mRNA region with a secondary structure that prevented DNA polymerization by reverse transcriptase beyond this point (Fig. 2BI). The DNA sequence within this region predicted the formation of several loops in the mRNA (Fig. 2BI), which could play a role in the regulation of the rhlG gene expression at the posttranscriptional level.
Two mRNA start sites were observed when the oligonucleotide R4 was used as a primer in extension experiments (Fig. 2BII). R4 is complementary to the mRNA sequence in which the extension of the primer was aborted with oligonucleotide R3 (Fig. 2A). The most frequent mRNA start site seems to be transcribed from a putative ς54 type of promoter, although the −12/−24 regions do not present all the elements which have been claimed to be important in these promoters. A similar situation has been found in the rhlAB ς54 promoter (24). The second, less abundant mRNA start site is a typical ς70 type of promoter. These two promoters overlap at their respective −24 and −35 regions. Between nucleotides −43 and −63 (with respect to the putative ς54 promoter), the sequence ATCTGTGGCATTGCCGCAGTA corresponding to a “lux box” is present (Fig. 2A). The presence of this regulatory sequence strongly suggests that the rhlG gene is regulated at the transcriptional level by one of the two LuxR homologs forming part of the quorum-sensing type of response in P. aeruginosa, LasR or RhlR. The characteristics of the rhlG promoter region (two promoters, one of which is a noncanonical ς54 type of promoter, and the presence of a lux box) are very similar to those present in the promoter region of the rhlAB operon, which encodes the Rt 1 enzyme (24). In the case of this key enzyme for rhamnolipid biosynthesis, RhlR positively regulates its transcription (20), and the alternative sigma factor ς54 is involved in its expression (24). As will be shown later, RhlG protein is involved in the synthesis of one of the rhamnolipid precursors, so it is likely that the structural similarity between the promoter regions of the rhlG gene and the rhlAB operon reflects that they are subject to similar genetic regulation. This possibility was examined further (see below).
Construction of a P. aeruginosa PAO1 rhlG::Tc mutant.
The high degree of similarity of the PAO1 and W51D rhlG genes, excluding their 5′ ends (Fig. 3), enabled us to construct a PAO1 rhlG::Tc mutant (ACP5 [Table 1]). Plasmid pJC2, which contains an rhlG internal fragment from strain W51D with a Tcr cassette insertion, was transferred by transformation to strain PAO1, and Tcr Gms transformants which were putative double recombinants carrying an interrupted rhlG gene were selected (Fig. 4). One of these transformants is the ACP5 mutant (Table 1), which indeed seems to be the product of a double recombination event in which the rhlG gene is interrupted by the Tcr cassette; this conclusion is drawn from the analysis by Southern blot hybridization and PCR amplification as shown in Fig. 4B and C, respectively. The Southern blot hybridization analysis (Fig. 4B) shows that mutant ACP5 contains, as expected, a 2-kb PstI fragment with homology with both the rhlG gene and the Tcr cassette (lanes 2 in Fig. 4BI and BII) and that this fragment is not present in the PAO1 genome (lanes 1, Fig. 4BI and BII). Unexpectedly, however, the ACP5 DNA retained hybridization with the 3.2-kb PstI rhlG homologous band. This result can be explained by the presence of heterogeneity in the chromosomes of strain ACP5, in which not all of the rhlG copies contain a Tcr cassette, or by the presence in the PAO1 chromosome of an rhlG homolog (probably fabG), which gives a hybridization signal of the same size when DNA is digested with PstI. In order to distinguish between these possibilities, PCR was performed in which rhlG-specific oligonucleotides (L2 and R2 in Fig. 2) were used to amplify the PAO1 and ACP5 genomes. We found that the expected 600-bp DNA fragment is amplified from PAO1, while a single 2-kb band is amplified from the ACP5 genome (Fig. 4C, lanes 3 and 4), these results clearly show that all of the rhlG gene copies in ACP5 contain a 1.4-kb insert (the Tcr cassette), so the most likely explanation is that we are detecting an rhlG homologous gene by Southern blot hybridization, probably fabG.
Effect of the rhlG inactivation in P. aeruginosa PAO1.
Mutant ACP5 does not have a fatty acid auxotrophy, grows at the same rate as its PAO1 parental strain, and does not show any significant change in its total lipid profile. Furthermore, the total lipid profiles of the parent and mutant strains were identical (data not shown). This suggests that there must be a functional FabG protein that is responsible for the synthesis of total cellular lipids and other essential products which contain a fatty acid moiety, such as the lipid A molecule (9).
P. aeruginosa produces different secondary metabolites which contain a lipid moiety, such as the autoinducers PAI-1 and PAI-2, as well as rhamnolipids and PHAs. Therefore, we investigated whether the production of some of these compounds was affected by the cassette insertion in the rhlG gene (mutant ACP5). Mutants affected in the production of any of the autoinducers are defective in total protease production (2, 22). We used this phenotype as a criterion to evaluate autoinducer production. It was found that mutant ACP5 has the same proteolytic activity as the PAO1 parental strain (data not shown), suggesting that autoinducer production is not affected. Rhamnolipid production in mutant ACP5 is completely abrogated (Table 2), suggesting that the RhlG protein is involved in the reaction leading to the production of the β-hydroxydecanoyl precursor of rhamnolipids (Fig. 1). In order to obtain direct evidence of the involvement of RhlG protein in rhamnolipid production and to rule out that the phenotype of mutant ACP5 was due to a polar effect of the Tcr cassette insertion in rhlG (and not to an inactivation of this gene), we complemented in trans the ACP5 mutant with plasmid pJC3, which contains the PAO1 rhlG gene (Table 1). The results obtained (Table 2) clearly show that the presence in trans of the rhlG gene is sufficient to restore the ACP5 capability to produce rhamnolipids.
TABLE 2.
Production of rhamnolipids and pyocyanine by mutant ACP5 and its parental strain, PAO1
| Strain | Concn (%) ofa:
|
||
|---|---|---|---|
| Rhamnolipid | Pyocyanine | PHA | |
| PAO1 | 150 ± 15 (100) | 0.59 (100) | 311 ± 13 (100) |
| ACP5 | <2 | 0.24 (40.6) | 87 ± 4 (27.9) |
| ACP5/pJC3 | 125 ± 25 (83.3) | 0.60 (101.6) | 243 ± 30 (78) |
| ACP5/pJC4 | 146 ± 10 (97.3) | 0.64 (108.4) | 305 ± 5 (98) |
| PAO R1 | <2 | <0.05 | NDb |
Rhamnolipid concentration is expressed as micrograms of rhamnose in rhamnolipids per milliliter of culture. The concentration of pyocyanine is expressed as the A690 of the chloroform-extracted culture supernatant. PHA was measured after 24 h of growth on MM + gluconate and is expressed as milligrams of PHA per milligram of protein.
ND, not determined.
It was apparent that mutant ACP5 produces lower levels of pigment than strain PAO1 (Table 2). It has been reported that production of both rhamnolipids and pyocyanine is induced by PAI-2-mediated activation (20), so we measured PAI-2 production by using the C. violaceum CV026 biosensor (16). Mutant ACP5 produced PAI-2 autoinducer at levels similar to those produced by PAO1 (data not shown). At present, we do not have a clear explanation for the reduction in pigment formation by mutant ACP5, but both rhamnolipid production and pyocyanine production are restored upon introduction of a functional rhlG gene in plasmid pJC3 (Table 2).
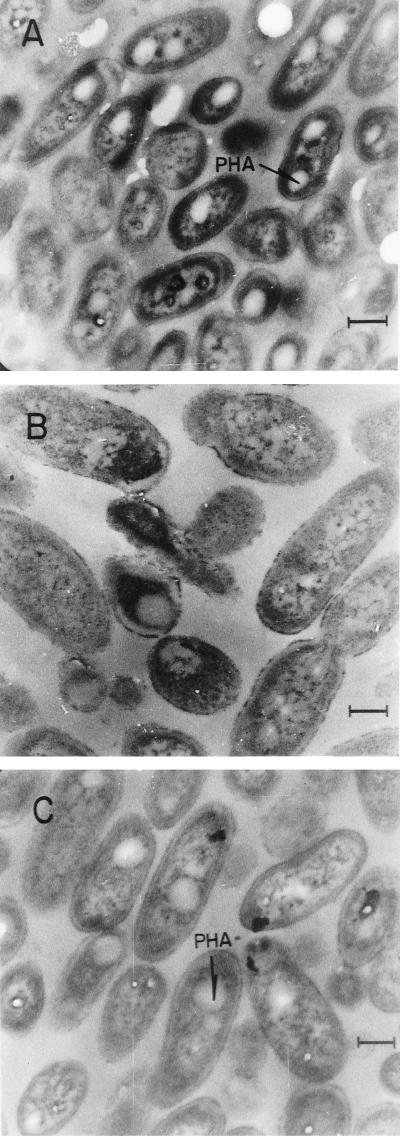
P. aeruginosa is known to produce PHA by using fatty acids from the de novo synthesis as precursors (30). The production of total PHA is reduced in mutant ACP5 at 24 h of growth (Table 2), but reaches the same level as PAO1 after 96 h of growth (data not shown). This defect in PHA synthesis can be observed in the electron micrographs taken after 24 h of growth as a decrease in the number and size of granules in mutant ACP5 (Fig. 5). This deficiency is due to rhlG inactivation, since plasmid pJC3 restores PHA production (Table 2). These results suggest that RhlG plays a role in biosynthesis of fatty acids used as substrates for PHA production, but that it is not an absolute requirement.
FIG. 5.
Electron micrographs of the P. aeruginosa strains PAO1 (A), ACP5 (B), and ACP5/pJC4 (C) grown for 24 h on MM + gluconate. Some of the PHA granules are pointed out. Micrographs were taken at a ×20,000 magnification.
These findings suggested the existence in PAO1 of other reductases involved in PHA production. As mentioned above, RhlG is homologous to PhaB proteins (Fig. 3), so we decided to search in the Pseudomonas Genome Project for PhaB homologs. We found that contig 983 contains an ORF coding for a protein with amino acids 30% identical to those of PhaB from Acinetobacter sp. strain RA3849 and 26% identical to those of RhlG from P. aeruginosa PAO1. It is very likely that the detected PHA synthesis in mutant ACP5 is due to the presence of an alternative pathway in which the reduction step is catalyzed by the putative acetoacetyl-CoA reductase encoded by the PAO1 phbB gene. Since this enzyme is expected to be used in polyhydroxybutanoyl synthesis, it would be interesting to determine whether the lengths of the fatty acid moiety of PHAs produced by mutant ACP5 are different from those of the PHAs produced by the wild-type strain, PAO1.
Plasmid pJC4, which contains 7 kb of the W51D chromosome, including the rhlG gene (Table 1), complemented in trans mutant ACP5 for rhamnolipid and pigment production and PHA synthesis (Table 2 and Fig. 5), suggesting that this gene has the same function in rhamnolipid and PHA synthesis in both P. aeruginosa strains.
Regulation of rhlG expression in P. aeruginosa PAO1.
To obtain additional evidence in support of the involvement of the RhlG protein in rhamnolipid and PHA synthesis, the concentration of rhlG mRNA was quantified under different culture conditions. The maximum rhlG mRNA concentration is found under conditions in which rhamnolipid production is maximum (that is, the stationary phase of growth on PPGAS medium) (Table 3), but there is also considerable expression when bacteria are grown for 48 h on LB or MM + gluconate medium (Table 3). It is important to point out that in the latter medium, PAO1 also produced rhamnolipids (37 μg/ml after 24 h of growth). The level of expression of the rhlG gene in the exponential phase of growth was low under all culture conditions studied (Table 3). These results provide additional evidence of the involvement of RhlG in the production of secondary metabolites, such as rhamnolipids and PHA.
TABLE 3.
Relative concentration of rhlG mRNA on different media and time of growtha
| Strain | mRNA concn with growth time givena
|
|||||
|---|---|---|---|---|---|---|
| PPGAS
|
LB
|
MM + gluconate
|
||||
| 6 h | 48 h | 6 h | 48 h | 6 h | 48 h | |
| PAO1 | 45 | 417 | 35 | 209 | 24 | 248 |
| PAO R1 | —b | 365 | NDc | ND | ND | ND |
mRNA concentration is expressed as the ratio of the RNA hybridization detected by autoradiography scanning to total RNA concentration used in the experiment.
—, not detected.
ND, not determined.
The DNA sequence of the rhlG promoter region suggested that the rhlG gene was regulated at the transcriptional level by one of the two LuxR homologs forming part of the quorum-sensing type of response in P. aeruginosa, LasR or RhlR. To obtain additional evidence in this respect, we determined the rhlG mRNA concentration of the PAO R1 strain (a PAO1 lasR mutant) grown on PPGAS medium. We used this mutant because it has been reported to be defective in both quorum-sensing regulatory circuits present in P. aeruginosa (12, 24). Table 2 shows that in agreement with these observations, PAO R1 lacks rhamnolipid and pyocyanine production when grown on PPGAS medium for 48 h. Unexpectedly, the level of PAO R1 rhlG mRNA concentration after 48 h of growth on PPGAS is only slightly lower than that of the wild-type PAO1 strain (Table 3), thus ruling out the direct involvement of LasR as the transcriptional activator of the rhlG gene. It is still possible that the RhlR protein activates rhlG transcription, since it has been shown that rhlR mRNA is expressed at a significant level in the PAO R1 mutant (24).
This is the first report of the existence in P. aeruginosa of a ramification of the fatty acid biosynthetic pathway specifically involved in rhamnolipid production. Figure 1 shows the proposed role of RhlG protein in the rhamnolipid biosynthesis pathway. At present, we do not know whether the RhlG substrate is β-ketoacyl linked to ACP or to CoA. Our model (Fig. 1) shows the substrate to be β-ketoacyl-ACP, because most of the RhlG homologs are FabG-like enzymes (Fig. 3). We propose that CoA-β-hydroxyacids are the precursors of rhamnolipids, since the PHA synthases only use as a substrate the CoA-linked fatty acids (31), and the lipid moiety of rhamnolipids (β-hydroxydecanoyl-β-hydroxydecanoate) seems to be the product of these enzymes.
In summary, a new gene, rhlG, involved in rhamnolipid biosynthesis has been identified. The deduced RhlG protein shows significant sequence homology with numerous NADPH-dependent ketoacyl reductases. Complementation studies and measurement of the rhlG mRNA suggest that the RhlG protein is required for rhamnolipid biosynthesis and can be used in PHA production, but is not necessary for fatty acid synthesis.
ACKNOWLEDGMENTS
We thank Paul Gaytán, Eugenio López, and Filiberto Sánchez for technical support.
This research was founded in part by the National Institute of Environmental Health Sciences (grant P42 ES04940). Jesús Campos held a CONACyT scholarship during the development of this work.
REFERENCES
- 1.Alexeyev M F, Shokolenko I N, Croughan T P. Improved antibiotic resistance gene cassettes and omega elements for Escherichia coli vector construction and in vitro deletion/insertion mutagenesis. Gene. 1995;160:63–67. doi: 10.1016/0378-1119(95)00108-i. [DOI] [PubMed] [Google Scholar]
- 2.Brint J M, Ohman D E. Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family. J Bacteriol. 1995;177:7155–7163. doi: 10.1128/jb.177.24.7155-7163.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burger M M, Glaser L, Burton R M. The enzymatic synthesis of rhamnose-containing glycolipids by extracts of Pseudomonas aeruginosa. J Biol Chem. 1963;238:2595–2602. [PubMed] [Google Scholar]
- 4.Chandrasekaran E V, Bemiller J N. Constituent analyses of glycosaminoglycans. Methods Carbohydr Chem. 1980;8:89–96. [Google Scholar]
- 5.Costerton J W. Pseudomonas aeruginosa in nature and disease. In: Sabath C D, editor. Pseudomonas aeruginosa: the organism, diseases it causes and their treatment. Bern, Switzerland: Hans Huber Publishers; 1980. pp. 15–24. [Google Scholar]
- 6.Cox C D. Role of pyocyanin in the acquisition of iron from transferrin. Infect Immun. 1986;52:263–270. doi: 10.1128/iai.52.1.263-270.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cronan J E, Jr, Rock C O. Biosynthesis of membrane lipids. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 612–636. [Google Scholar]
- 8.Döring G, Maier M, Müller E, Zoubair B, Tümmler B, Kharazmi A. Virulence factors of Pseudomonas aeruginosa. Antibiot Chemother. 1987;39:136–148. doi: 10.1159/000414341. [DOI] [PubMed] [Google Scholar]
- 9.Dotson G D, Kaltashov I A, Cotter R J, Raetz C R H. Expression cloning of a Pseudomonas gene encoding a hydroxydecanoyl-acyl carrier protein-dependent UDP-GlcNAc acyltransferase. J Bacteriol. 1998;180:330–337. doi: 10.1128/jb.180.2.330-337.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Folch J, Lees M, Sloane Stanley G H. A simple method for the isolation and purification of total lipids from animal tissue. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- 11.Fuqua W C, Winans S C, Greenberg E P. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum sensing transcriptional regulators. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 12.Gambello M J, Iglewski B H. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J Bacteriol. 1991;173:3000–3009. doi: 10.1128/jb.173.9.3000-3009.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gervais F G, Drapeau G R. Identification, cloning, and characterization of rcsF, a new regulator gene for exopolysaccharide synthesis that suppresses the division mutation ftsZ84 in Escherichia coli K-12. J Bacteriol. 1992;174:8016–8022. doi: 10.1128/jb.174.24.8016-8022.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herman D C, Zhang Y, Miller R M. Rhamnolipid (biosurfactant) effects on cell aggregation and biodegradation of residual hexadecane under saturated flow conditions. Appl Environ Microbiol. 1997;63:3622–3627. doi: 10.1128/aem.63.9.3622-3627.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoang T T, Schweizer H P. Fatty acid biosynthesis in Pseudomonas aeruginosa: cloning and characterization of the fabAB operon encoding β-hydroxyacyl-acyl carrier protein dehydratase (FabA) and β-ketoacyl-acyl carrier protein synthase I (FabB) J Bacteriol. 1997;179:5326–5332. doi: 10.1128/jb.179.17.5326-5332.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McClean K H, Winson M K, Fish L, Taylor A, Chhabra S R, Camara M, Daykin M, Lamb J H, Swift S, Bycroft B W, Stewart G S A B, Williams P. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology. 1997;143:3703–3711. doi: 10.1099/00221287-143-12-3703. [DOI] [PubMed] [Google Scholar]
- 17.Miller, R. M. 1995. Biosurfactant-facilitated remediation of metal-contaminated soils. Environ. Health Perspect. 103(Suppl.):59–62. [DOI] [PMC free article] [PubMed]
- 18.Morrison W R, Smith L M. Preparation of fatty acid methyl esters and dimethylacetals from lipids with boron fluoride-methanol. J Lipid Res. 1964;5:600–608. [PubMed] [Google Scholar]
- 19.Nicas T I, Iglewski B H. The contribution of exoproducts to virulence of Pseudomonas aeruginosa. Can J Microbiol. 1985;31:387–392. doi: 10.1139/m85-074. [DOI] [PubMed] [Google Scholar]
- 20.Ochsner U A, Reiser J. Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:6424–6428. doi: 10.1073/pnas.92.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olsen R H, DeBusscher G, McCombie W R. Development of broad-host-range vectors and gene banks: self-cloning of the Pseudomonas aeruginosa PAO chromosome. J Bacteriol. 1982;150:60–69. doi: 10.1128/jb.150.1.60-69.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pearson J P, Gray K M, Passador L, Tucker K D, Eberhard A, Iglewski B H, Greenberg P. Structure of the autoinducer required for the expression of Pseudomonas aeruginosa virulence genes. Proc Natl Acad Sci USA. 1994;91:197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pearson J P, Passador L, Iglewski B H, Greenberg P. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:1490–1494. doi: 10.1073/pnas.92.5.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pearson J P, Pesci E C, Iglewski B H. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J Bacteriol. 1997;179:5756–5767. doi: 10.1128/jb.179.18.5756-5767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quandt J, Hynes M F. Versatile suicide vectors which allow direct selection for gene replacement in Gram-negative bacteria. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 26.Römling U, Wingender J, Müller H, Tümmler B. A major Pseudomonas aeruginosa clone common to patients and aquatic habitats. Appl Environ Microbiol. 1994;60:1734–1738. doi: 10.1128/aem.60.6.1734-1738.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 28.Soberón-Chávez G, Haïdour A, Ramos J L, Campos J, Ortigoza J. Selection and preliminary characterization of a Pseudomonas aeruginosa strain mineralizing some isomers in a branched-chain dodecylbenzene sulfonate mixture. World J Microbiol Biotechnol. 1996;12:367–362. doi: 10.1007/BF00340213. [DOI] [PubMed] [Google Scholar]
- 29.Stanghellini M E, Miller R M. Biosurfactants: their identity and potential efficiency in the biological control of zoosporic plant pathogens. Plant Dis. 1997;81:4–12. doi: 10.1094/PDIS.1997.81.1.4. [DOI] [PubMed] [Google Scholar]
- 30.Timm A, Steinbüchel A. Formation of polyesters consisting of medium-chain-length 3-hydroxyalkanoates acids from gluconate by Pseudomonas aeruginosa and other fluorescent pseudomonads. Appl Environ Microbiol. 1990;56:3360–3367. doi: 10.1128/aem.56.11.3360-3367.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Timm A, Steinbüchel A. Cloning and molecular analysis of the poly(3-hydroxyalkanoates acid) gene locus of Pseudomonas aeruginosa PAO1. Eur J Biochem. 1992;209:15–30. doi: 10.1111/j.1432-1033.1992.tb17256.x. [DOI] [PubMed] [Google Scholar]
- 32.West S E H, Iglewski B H. Codon usage in Pseudomonas aeruginosa. Nucleic Acids Res. 1988;16:9323–9329. doi: 10.1093/nar/16.19.9323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.West S E H, Schweizer H P, Dall C, Sample A K, Runyen-Janecky L J. Construction of improved Escherichia coli-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene. 1994;128:81–86. doi: 10.1016/0378-1119(94)90237-2. [DOI] [PubMed] [Google Scholar]
- 34.Wild M, Caro A D, Miller R M, Soberón-Chavez G. Selection and partial characterization of a Pseudomonas aeruginosa mono-rhamnolipid deficient mutant. FEMS Microbiol Lett. 1997;153:279–285. doi: 10.1111/j.1574-6968.1997.tb12586.x. [DOI] [PubMed] [Google Scholar]