Abstract
Helicobacter pylori is a pathogen related to severe diseases such as gastric cancer; because of rising antimicrobial-resistant strains, failure to eradicate H. pylori with antibiotics has increased worldwide. Multidrug-resistant H. pylori and gastric cancer is common in Mongolia; therefore, we aimed to explore alternative antimicrobial treatments and the genomes of resistant strains in this country. A total of 361 H. pylori strains isolated from patients in Mongolia were considered. Minimal inhibitory concentrations for two fluoroquinolones (ciprofloxacin and moxifloxacin), rifabutin, and furazolidone were determined via two-fold agar dilution. Genomic mutations in antibiotic-resistant strains were identified by next-generation sequencing using the Illumina Miseq platform and compared with genes from a reference H. pylori strain (26695). The resistance rate of H. pylori strains to quinolones was high (44% to ciprofloxacin and 42% to moxifloxacin), and resistance to rifabutin was low (0.5%); none were resistant to furazolidone. Most quinolone-resistant strains possessed gyrA gene mutations causing amino acid changes (e.g., N87K, A88P, and D91G/Y/N). While one rifabutin-resistant strain had amino acid-substituting mutations in rpoB (D530N and R701C), the other had three novel rpoB mutations; both rifabutin-resistant strains were sensitive to furazolidone. Overall, our findings suggest that rifabutin and/or furazolidone may be an alternative, effective H. pylori treatment in patients who have failed to respond to other treatment regimens.
Keywords: Helicobacter pylori, antibiotic resistance, ciprofloxacin, moxifloxacin, rifabutin, furazolidone, next-generation sequencing, resistant gene mutation, Mongolia
1. Introduction
Over the last 40 years, antibiotic resistance to Helicobacter pylori has increased globally, with higher rates in less-developed countries [1]. Because of its resistance rate, H. pylori has emerged as one of sixteen antibiotic-resistant pathogens that pose the most serious threat to human health, according to the World Health Organization [2,3,4]. Both the American College of Gastroenterology and the European Helicobacter and Microbiota Study group have changed their guidelines regarding H. pylori infection to offer the treatment for positive cases; in addition, H. pylori was included in the latest version of the World Health Organization’s International Classification of Diseases (ICD-11) as a cause of gastritis [5].
The most popular treatment regimens for H. pylori use a combination of two antibiotics (amoxicillin, clarithromycin, metronidazole, or tetracycline) with a proton-pump inhibitor (PPI) and/or bismuth subcitrate [6]. According to the Maastricht VI/Florence consensus report, first-line treatment efficacy is more than 90% for individuals who are being treated for the first time. However, if an individual fails to respond to the first-line treatment regime, secondary antibiotics or alternative antibiotics may be required. Currently, rifabutin- and/or furazolidone-based regimens (e.g., ciprofloxacin, moxifloxacin, levofloxacin) are used when eradication fails multiple times because of fluoroquinolone resistance [7].
Antimicrobial resistance mechanisms to fluoroquinolones, nitrofurans (e.g., furazolidone), and rifamycin derivatives (e.g., rifabutin) are well studied, especially in H. pylori. Fluoroquinolones exert their bactericidal activity by inhibiting two essential bacterial type II topoisomerases, DNA gyrase (Gyr) and topoisomerase IV; both are heterotetrametric enzymes consisting of two A subunits and two B subunits, and modulate the chromosomal supercoiling required for DNA synthesis, transcription, and cell division [8,9]. Target-mediated resistance due to mutations on the GyrA or/and GyrB subunits of gyrase is one of the main reasons for fluoroquinolone regimen failure [10], with resistance occurring because of either a single or dual mutations in the gyrA or gyrB codon regions [11,12]. We, and others, previously determined that amino acid-substituting mutations in the gyrA gene (N87K, D91N, D91G, D91Y) were frequently observed in highly resistant H. pylori strains [13,14]; however, there are some resistant strains without identified mutations. Apart from fluoroquinolones, furazolidone is increasingly used to treat H. pylori infection. Furazolidone primarily works through 2-oxoglutarate oxidoreductase subunit D and/or pyruvate–ferredoxin oxidoreductase subunit D to bind free radicals and counteract DNA replication and protein production [15,16]. Finally, rifabutin inhibits the DNA-dependent RNA polymerase of H. pylori at very low concentrations in vitro [17,18]; resistance to rifabutin is related to amino acid substitution in the β subunit of RNA polymerase (RpoB).
In Mongolia, in recent studies, it was reported that multidrug resistance (MDR) in H. pylori strains is high: 51% for amoxicillin, clarithromycin, metronidazole, levofloxacin, and minocycline [14] and 60.8% for amoxicillin, clarithromycin, metronidazole, erythromycin, and nitrofuran [13]. Because of the high rate of MDR, alternative drugs to treat H. pylori are needed. In this study, we aimed to investigate the efficacy of new antimicrobials against H. pylori strains with MDR; specifically, we studied ciprofloxacin (CIP), moxifloxacin (MOXI), furazolidone (FUR), and rifabutin (RFB). We also aimed to define genomic mutations underlying antibiotic resistance using next-generation sequencing analysis of H. pylori. Overall, we found that furazolidone and rifabutin may be key to treating H. pylori strains with MDR in Mongolia.
2. Results
2.1. Sample and Antibiotic Resistance Distribution
All 361 H. pylori strains previously reported by our group [14] were identified in this study. The average age of the participants was 44.3 ± 13.4 (mean ± SD) years, and 26.9% (97/361) were male. The resistance status of all strains to seven antibiotics is shown in Table 1.
Table 1.
Demographics and antibiotic resistance rate from a previous study and the present study.
| Sex | Total Strains Tested | Antibiotic-Resistant Strains, n (%) | ||||||
|---|---|---|---|---|---|---|---|---|
| n | AMX * | CLR * | MNZ * | LVF * | MINO * | RFB | FUR | |
| Total | 361 | 43 (11.9) |
108 (29.9) | 284 (78.7) | 149 (41.3) | 1 (0.3) |
2 (0.6) |
0 (0) |
| Female | 259 | 31 (11.7) |
85 (32.2) |
218 (82.5) |
113 (42.8) |
0 (0) |
1 (0.4) |
0 (0) |
| Male | 94 | 12 (12.4) |
23 (23.7) |
66 (68.0) |
36 (37.1) |
1 (1.0) |
1 (1.0) |
0 (0) |
| Unknown gender | 8 | 1 (12.5) |
2 (25) |
7 (87.5) |
4 (50) |
0 (0) |
0 (0) |
0 (0) |
AMX, amoxicillin; CLR, clarithromycin; MNZ, metronidazole; LVF, levofloxacin; MINO, minocycline; RFB, rifabutin; FUR, furazolidone. * Data from our previous study [8].
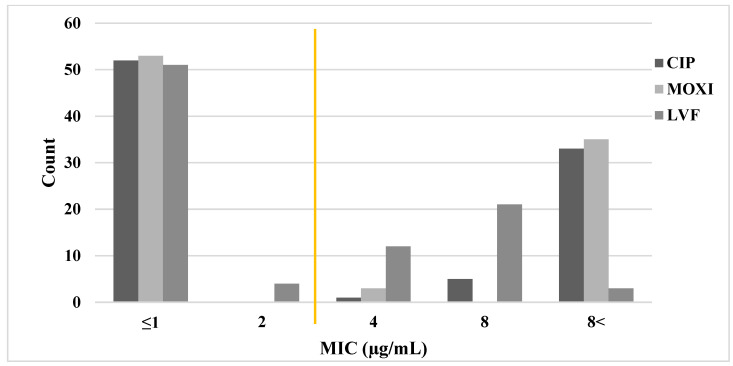
Initially, we selected 91 representative H. pylori strains from a total of 361 strains (25% of all strains), which were selected based on five antimicrobial resistance statuses identified previously [14]: 20 strains sensitive to all antibiotics, 21 strains resistant to a single antibiotic, 20 strains resistant to two antibiotics, 20 strains resistant to three antibiotics, and 10 strains resistant to five antibiotics. These 91 strains were used to determine MICs for the quinolone group antibiotics CIP and MOXI, which were compared with a previously reported MIC for levofloxacin (LVF). We found that H. pylori strains exhibited a similar level of resistance to all three quinolone group antibiotics (LVF, 44%; CIP, 43%; and MOXI, 42%), which was consistent across sexes (Table 2 and Figure 1).
Table 2.
Quinolone-resistant H. pylori strains from 91 representative Mongolian strains.
| Total Strains Tested | Antibiotic-Resistant Strains, n (%) | |||
|---|---|---|---|---|
| n (%) | LVF | CIP | MOXI | |
| Total | 91 (100) | 40 (44) | 39 (43) | 38 (42) |
| Female | 62 (68) | 28 (45) | 28 (45) | 27 (43) |
| Male | 27 (30) | 11 (41) | 11 (41) | 11 (41) |
| Unknown | 2 (2) | 1 (50) | 0 | 0 |
LVF, levofloxacin; CIP, ciprofloxacin; MOXI, moxifloxacin.
Figure 1.
Minimal inhibitory concentration (MIC) distributions for moxifloxacin (MOXI), ciprofloxacin (CIP), and levofloxacin (LVF) against H. pylori. MIC (µg/mL) was determined by agar dilution assays of 91 representative Mongolian H. pylori strains. The MIC for levofloxacin was determined previously [14]. The yellow line represents the cutoff for resistance (≤2 µg/mL, sensitive; >2 µg/mL, resistant).
Among the 91 tested strains, 49 were sensitive to quinolones: 42 to all three and 7 to one or two (LVF-R, MOXI-S, CIP-S: two strains; LVF-R, MOXI-R, CIP-S: one strain; LVF-R, MOXI-S, CIP-R: two strains; LVF-S, MOXI-R, CIP-R: two strains).
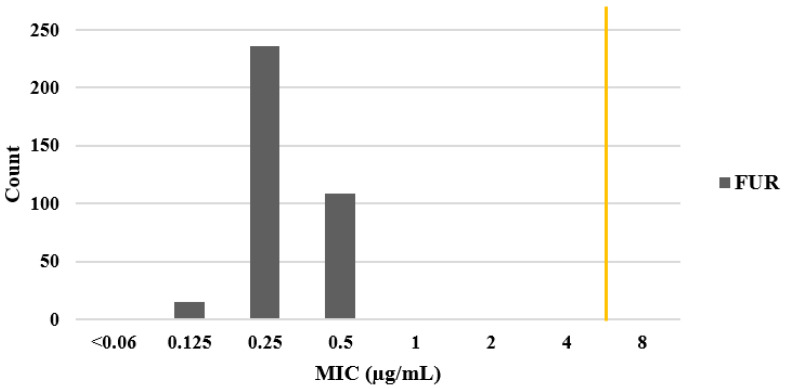
Two further antibiotics, FUR and RFB, were tested in these 91 strains. We found no resistance to FUR; however, one strain was resistant to RFB (strain UB217, MIC = 4.0 µg/mL). To find whether there was more resistance to RFB or not and to FUR against H. pylori strains, MICs for FUR and RFB were then determined for the remaining 270 strains. Tests for all 361 strains showed that resistance rates to FUR and RFB were very low: 0% (no strains) and 0.5% (2 strains), respectively. MICs for FUR were determined as <0.064 µg/mL (1 strain), 0.125 µg/mL (15 strains), 0.25 µg/mL (236 strains), and 0.5 µg/mL (109 strains). In all 361 H. pylori strains, FUR had an MIC less than the 4 µg/mL cut-off required to define antimicrobial resistance (Figure 2).
Figure 2.
Minimal inhibitory concentration (MIC) distribution for furazolidone (FUR). MIC (µg/mL) was determined by agar dilution assays of 361 Mongolian H. pylori strains. The yellow line represents the cutoff for resistance (≤4 µg/mL, sensitive; >4 µg/mL, resistant).
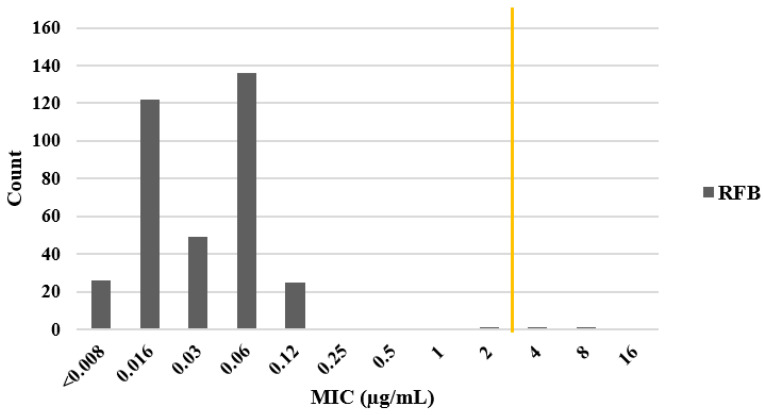
For all 361 H. pylori strains, RFB had a MIC of 0.03 µg/mL against 136 strains (37.6%) and an MIC of 0.008 µg/mL against 122 strains (33.7%). Two strains showed RFB resistance (UB217, MIC 4.0 µg/mL and Ke9, MIC 8 µg/mL) (Figure 3).
Figure 3.
Minimal inhibitory concentration (MIC) distribution for rifabutin (RFB) against H. pylori. MIC (µg/mL) was determined by agar dilution assays of 361 Mongolian H. pylori strains. The yellow line represents the cutoff for resistance (≤2 µg/mL, sensitive; >2 µg/mL, resistant).
Next, we focused on whether these two RFB-resistant strains were multiple-antibiotic-resistant. Strain Ke9 was resistant to four other antibiotics, MNZ and the three quinolones, but was sensitive to AMX, CLA, MINO, and FUR. Strain UB217 was resistant to six other antibiotics, but sensitive to MINO and FUR (Table 3).
Table 3.
Resistance status of the two RFB-resistant strains to eight other antibiotics and their minimal inhibitory concentration (µg/mL).
| No. | Strain Name | RFB | FUR | AMX | CLR | MNZ | MINO | LVF | MOXI | CIP |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | KE-9 | 8 | 0.25 | 0.06 | 0.03 | 32 | 0.06 | 8 | 4≤ | 4 |
| 2 | UB-217 | 4 | 0.5 | 0.24 | 2 | 32 | 0.25 | 4 | 8≤ | 8 |
RFB, rifabutin; FUR, furazolidone; AMX, amoxicillin; CLR, clarithromycin; MNZ, metronidazole; MINO, minocycline; LVF, levofloxacin; MOXI, moxifloxacin; CIP, ciprofloxacin. Green box indicates resistance.
2.2. Genotypic Determination of Antibiotic Resistance
Separately from the agar dilution tests in this study, we performed full-genome sequencing of 229 H. pylori strains (63%), including 77 (85%) sequences from the representative 91 strains. Protein sequence alignment was performed using these 77 strains for genes related to quinolone resistance and all 229 strains for genes related to rifabutin resistance. A total of 10 genomes from the 10 strains sensitive to all nine antibiotics and a partial genome from reference strain H. pylori 26695 were used as reference gene sequences. The potential genotypes of quinolone- and rifamycin-resistant H. pylori strains are shown in Supplementary Table S1.
2.3. Quinolone Resistance
Within the gyrA gene of the 77 sequences assessed for quinolone resistance, previously reported genetic variations, such as N87K/I and D91G/Y/N substitutions, were frequently observed, although the A88P substitution was found in only one strain (Table 4). The amino acid substitutions N87K/I (p < 0.0001) and D91N/Y/G (p < 0.0001) were significantly associated with LVF, MOXI, and CIP resistance, according to Fisher’s exact test (Supplementary Table S1). We also found N87K and D91G mutations in some strains that had mixed quinolone-resistant status. Of the 39 quinolone-resistant strains, gyrase subunit gene analysis showed that 39 and 30 hypothetical mutations related to quinolone-resistant phenotypes existed in gyrA and gyrB, respectively (Supplementary Table S2). Of these 39 hypothetical gyrA mutations, V199I (p < 0.012) and N660D (p < 0.005) were significantly associated with resistance in the strains Ke86, Kh95, Ub72, Uvs113, and Ub72. However, for eight resistant strain sequences, there were no significant mutations in the quinolone resistance-determining region of gyrBE415-S454 and gyrAA71-Q110.
Table 4.
gyrA gene amino acid substitutions associated with quinolone (LVF, MOXI, and CIP)-resistant H. pylori strains.
| No. | Strain Name |
MIC (µg/mL) | gyrA Mutation | |||||
|---|---|---|---|---|---|---|---|---|
| LVF | MOXI | CIP | N87 | A88 | D91 | |||
| 1 | Uvs | 103 | 32 | 8≤ | 16≤ | I | ||
| 2 | Uvs | 61 | 16 | 8≤ | 16≤ | I | ||
| 3 | UB | 97 | 8 | 8≤ | 16≤ | I | ||
| 4 | UB | 129 | 8 | 8≤ | 16≤ | K | ||
| 5 | UB | 168 | 8 | 8≤ | 16≤ | K | ||
| 6 | UB | 189 | 8 | 8≤ | 16≤ | K | ||
| 7 | Uvs | 131 | 8 | 8≤ | 16≤ | K | ||
| 8 | Uvs | 136 | 8 | 8≤ | 16≤ | K | ||
| 9 | Kh | 160 | 8 | 8≤ | 16≤ | K | ||
| 10 | Go | 150 | 8 | 8≤ | 16≤ | K | ||
| 11 | Ke | 3 | 8 | 8≤ | 16≤ | K | ||
| 12 | Ke | 47 | 8 | 8≤ | 16≤ | K | ||
| 13 | Ke | 59 | 8 | 8≤ | 16≤ | K | ||
| 14 | Ke | 160 | 4 | 8≤ | 16≤ | K | ||
| 15 | Ke | 2 | 2 | 4 | 16≤ | K | ||
| 16 | UB | 221 | 8 | 8≤ | 16≤ | P | ||
| 17 | UB | 178 | 8 | 8≤ | 16≤ | N | ||
| 18 | UB | 198 | 8 | 8≤ | 16≤ | N | ||
| 19 | Go | 129 | 8 | 8≤ | 16≤ | N | ||
| 20 | Uvs | 97 | 4 | 8≤ | 16≤ | N | ||
| 21 | UB | 29 | 8 | 8≤ | 16≤ | G | ||
| 22 | UB | 217 | 4 | 8≤ | 8 | G | ||
| 23 | Kh | 93 | 2 | 8≤ | 16≤ | G | ||
| 24 | UB | 219 | 8 | 8≤ | 16≤ | Y | ||
| 25 | Go | 1 | 4 | 8≤ | 16≤ | Y | ||
| 26 | Ke | 1 | 4 | 8≤ | 8 | Y | ||
| 27 | Uvs | 23 | 4 | 4 | 8 | Y | ||
| 28 | Kh | 111 | 4 | 8≤ | 16≤ | Y | ||
| 29 | Ke | 87 | 16 | 1 | 16≤ | K | ||
| 30 | Ke | 119 | 8 | 1 | 16≤ | K | ||
| 31 | Ke | 4 | 0.5 | 8≤ | 16≤ | G | ||
| 32 | Ke | 133 | 8 | 8≤ | 4 | |||
| 33 | Uvs | 113 | 4 | 8≤ | 16≤ | |||
| 34 | Kh | 95 | 4 | 8≤ | 16≤ | |||
| 35 | Kh | 173 | 2 | 4 | 8 | |||
| 36 | UB | 30 | 8 | 0.5 | 0.5 | |||
| 37 | Ke | 86 | 4 | 8≤ | 0.5 | |||
| 38 | Go | 168 | 2 | 0.5 | ≤0.25 | |||
| 39 | UB | 72 | 0.5 | 8≤ | 16≤ | |||
| Strains sensitive to quinolones (n = 38) | N | A | D | |||||
N, asparagine; A, alanine; Y, tyrosine; D, aspartic acid; K, lysine; P, proline; G, glycine. Orange indicates resistance to all three quinolones and green indicates resistance to one or two quinolones; blue indicates the corresponding mutations. UB, Ulaanbaatar capital city; Go, Umnugovi; Ke, Khentii; Kh, Khuvsgul; MIC, minimal inhibitory concentration; LVF, levofloxacin; MOXI, moxifloxacin; CIP, ciprofloxacin.
2.4. Rifabutin Resistance
Within the rpoB gene of the 229 sequences assessed for rifabutin resistance, we found the previously reported mutations D530N and R701C in strain UB217 (MIC 4.0 µg/mL), while strain Ke9 had multiple hypothetical mutations. In addition, we found the previously reported V538I substitution in two strains (UB75 and Kh67). However, these strains were sensitive to RFB with MICs of 0.12 and 0.06 µg/mL, respectively. There were multiple putative mutations noted, such as S273P, E470G, A1173T, L2196P, and A2710V, which are shown in Table 5 and Supplementary Table S2 (green letters).
Table 5.
rpoB gene amino acid substitutions associated with rifabutin-resistant H. pylori strains.
| No. | Strain Name | MIC µg/uL | rpoB Mutation | |||||
|---|---|---|---|---|---|---|---|---|
| E470 | D530 | V538 | R701 | L2196 | A2710 | |||
| 1 | Ke9 | 8 | G | P | V | |||
| 2 | UB217 | 4 | N | C | ||||
| 3 | UB75 | 0.12 | I | |||||
| 4 | Kh67 | 0.06 | I | |||||
| Sensitive stains (n = 217) < 0.12 | E | D | V | R | L | A | ||
E, glutamic acid; N, asparagine; D, aspartic acid; V, valine; R, arginine; L, leucine; A, alanine; G, glycine; I, isoleucine; C, cysteine. Green indicates resistance. Blue indicates the corresponding, previously reported mutation; light green indicates new hypothetical mutations. UB, Ulaanbaatar capital city; Ke, Khentii; Kh, Khuvsgul.
3. Discussion
In this study, we aimed to find new drugs to treat MDR H. pylori strains. To do this, we selected 91 representative resistant strains based on resistance status from a Mongolian population and found very high resistance rates to CIP (43%) and MOXI (42%), similar to rates for LVF (44%) previously reported. A recent retrospective study from China, as a neighboring country, revealed that the quinolone resistance rate is similar: around 48% within over 3300 clinical isolates [19]. Next-generation sequencing data from 39 quinolone-resistant and 38 quinolone-sensitive H. pylori strains showed that D91 and N87 gyrA gene mutations commonly existed in the resistant strains, in line with previous studies [4,10,14,19]. We also identified two mutations (V119I and N660D) that were significantly associated with resistance in strains without any well-known mutations. In contrast, we found that H. pylori strains were highly sensitive to both FUR and RFB, with sensitivity rates of 100% and 99.5%, respectively, even in MDR strains. Therefore, while the three quinolone antibiotics do not appear to be viable alternative antibiotic treatments in Mongolia, FUR and RFB are promising candidates.
Despite their efficacy in the present study, RFB- and/or FUR-based antibiotic treatments are only appropriate for individuals who do not respond to more than one standard treatment course, rather than as a first-line treatment. According to the literature, multiple codons of the rpoB gene contain mutations resulting in strong resistance to antibiotics; these usually lie in the codon positions 149, 524–545, 585–586, and 701 [14,18,20]. Although we found some mutations in these locations, such as D540N and R701C in strain UB217, there were no such mutations in strain Ke9; however, three hypothetical mutations (E470G, L2196P, and A2710V) were identified. Moreover, we found a previously described mutation (D538I) in two RFB-sensitive strains (UB75 and Kh67), with MICs of 0.12 µg/mL and 0.06 µg/mL, respectively. The two RFB-resistant strains, Ke9 and UB217, were resistant to five and seven antibiotics, respectively; however, both strains were sensitive to FUR and MINO (Table 3). This suggests that there are still antibiotics able to effectively treat MDR H. pylori strains; however, appropriate use of these drugs is required to prevent the development of H. pylori strains resistant to them, as FUR-resistant strains have begun to be reported [21].
According to the Maastricht VI/Florence consensus report, developing countries tend to use quadruple therapy for H. pylori more frequently than developed countries because of higher rates of CLR resistance. Mongolia has high CLR resistance (29.9%), with H. pylori eradication rates of 68.5–97.6% [22], meaning eradication failure varies between 2.4% and 31.5%, depending on the treatment regimen [22]. In cases of H. pylori eradication failure, an RFB triple regimen (150 mg or 300 mg bis in die (BID, i.e., twice a day) or quarter in die (QID, i.e., four times a day)) is usable with AMX (1000 mg, BID) and a PPI (standard dose, BID) for 10–14 days. For FUR, triple or quadruple regimens (100–200 mg, BID) are useable with AMX (1000 mg, BID) or tetracycline (750 mg, BID) and a PPI (standard dose, BID) with or without bismuth subcitrate (220 mg, BID) for 7–14 days [7,10]. Therefore, we should think about the limitations of the above antibiotics. For instance, RFB is quite expensive, and sometime causes myelotoxicity, myalgia, rashes, etc., although these symptoms are rare [23].However, it is important to note that, in accordance with previous studies, RFB- and FUR-containing therapies should be recommended for individuals who have not responded to multiple (two or more) previous therapies [23,24,25,26,27].
4. Conclusions
Overall, our results suggest that in patients who do not respond to first- and/or second-line antibiotic regimens for H. pylori infection, an alternative treatment regimen with rifabutin and/or furazolidone may successfully treat infection and reduce the risk of further complications, such as gastric cancer. These drugs have no cross-resistance to metronidazole and are effective in populations with high metronidazole resistance. Our results also suggest that if patients do not respond to a particular antibiotic, further treatment with antibiotics from the same group (e.g., quinolones) should not be attempted because resistance is likely. Although finding drugs to eradicate MDR H. pylori is important, it is key that doctors ensure patients take rifabutin and furazolidone appropriately in order not to allow the development of strains resistant to these antibiotics. To ensure the widespread eradication of H. pylori, further studies on the adverse effects of rifabutin and furazolidone, and the potential of resistance to these drugs in susceptible countries, are required.
5. Materials and Methods
5.1. Study Population
In total, 1004 volunteers from Mongolia with dyspeptic symptoms participated in the study between November 2014 and August 2016. This sample was drawn from Ulaanbaatar (Mongolia’s capital city, in the central region), Khuvsgul province (in the north), Umnugovi province (in the south), Uvs province (in the West), and Khentii province (in the east). An upper gastrointestinal endoscopy was performed, and a biopsy sample was collected from the antrum for H. pylori culture; the specimen was placed in the transfer medium and then immediately frozen. Participants with a history of partial or total gastrectomy, or who had taken H2-receptor blockers or PPIs within four weeks of the endoscopy, were excluded from the study. Samples for culture were transferred to Ulaanbaatar and immediately stored at −80 °C. Later, all samples were transferred for H. pylori culture and further analysis at the Department of Environmental and Preventive Medicine, Oita University Faculty of Medicine, Japan.
5.2. Isolation and Culture
From biopsy samples, H. pylori samples were cultured on H. pylori-selective plates (Nissui Pharmaceutical Co., Ltd., Tokyo, Japan) and incubated at 37 °C under microaerophilic conditions (10% O2, 5% CO2, and 85% N2) for 3–7 days. Small, round, purple colonies typical of H. pylori were checked for Gram staining and tested for oxidase, catalase, and urease, as previously described [14].
5.3. Antibiotic Susceptibility Test
At first, we selected 91(25%) strains randomly from a total of 360 previous MIC determined strains. All tests were performed using the serial two-fold agar dilution method with MOXI, CIP, FUR, and RFB to determine the minimal inhibitory concentration (MIC), as previously described [14]. Briefly, bacteria were sub-cultured in Mueller Hinton II Agar medium (Becton Dickinson, New York, NY, USA) supplemented with 5% defibrinated horse blood. The bacterial suspension was adjusted to OD 0.1 using a OD590 spectrophotometer (UV-1800, Shimadzu, Kyoto, Japan), and a 48-pin inoculator was used to inoculate the culture plate with twofold serial dilutions of the test antibiotics. After 3–7 days of incubation (H. pylori microaerophilic conditions), the MIC of each antibiotic was determined as the minimal dilution at which bacterial colonies were no longer produced, according to the Clinical & Laboratory Standards Institute’s 11th edition of “Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically” (M07). H. pylori 26695 and DMSO were used as positive and negative controls and MICs were tested twice for each strain. Amoxicillin was used to compare the current results with those previously determined for these H. pylori strains; there was 97.5% agreement between results [14]. The reported clinical breakpoints for the antibiotics used in this study are 1.0 µg/mL for CIP and MOXI [27], 4.0 µg/mL for FUR [27,28,29], and 1.0 µg/mL for RFB [27]; each of these breakpoints was also used in this study.
5.4. DNA Extraction and Next-Generation Sequencing
A commercially available DNA purification kit (Qiagen, Hilden, Germany) was used to extract DNA from H. pylori on day 3 of culture. DNA was quantified using a Quantus Fluorometer (Promega, Madison, WI, USA), and sequenced using the MiSeq platform (Illumina, Inc., San Diego, CA, USA). Sequences were aligned with gyrA and gyrB for samples treated with CIP and MOXI and rpoB for samples treated with RFB based on standard H. pylori 26695 sequences (GenBank accession number NC_000915.1) using the CLC Genomics Workbench (v22.0, QIAGEN). Finally, genetic mutations in antimicrobial-related genes in each H. pylori strain were identified from the DNA sequences obtained under MiSeq sequencing.
5.5. Statistical Analysis
SPSS Statistics version 25.0 (SPSS Inc., Chicago, IL, USA) was used for statistical analysis in our study. The chi-square test and Fisher’s exact test were used; p < 0.05 indicated a significant difference.
5.6. Nucleotide Sequence Accession Numbers
All nucleotide sequences analyzed in this study were deposited in the DDBJ Center Data Bank of Japan; accession numbers for the gyrA and gyrB genes are LC782993–LC789146, and for the rpoB gene LC789147–LC783375, respectively.
Acknowledgments
We appreciate the support of those who assisted in collecting samples, funding the study, and giving advice. We thank Philippa Gunn, D.Phil. for editing a draft of this manuscript. We thank Namsrai Rinchensengee and Duger Davaadorj for data curation.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms11122852/s1. Table S1: Genome parts of gyrA and gyrB genes from 91 representative, sequenced strains are available via accession number LC782993–LC789146. Table S2: Genome parts of rpoB gene from all sequenced strains are available via accession number LC789147–LC783375.
Author Contributions
Conceptualization: A.K. and Y.Y. Data acquisition: A.K. and B.G. Analysis: A.K. and J.A. Funding acquisition: J.A., T.M. and Y.Y. Resources: A.K., B.S., B.G., D.A. and K.O. Project administration: K.O. and Y.Y. Visualization: A.K. and J.A. Writing—original draft: A.K. Writing—review and editing: J.A. and Y.Y. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Ethical approval was given by the Ethics Committee of the Ministry of Health (#03, 11 September 2015), the Ethics Committee at Mongolian National University of Medical Sciences (N13-02/1A, 11 June 2015), and the Ethics Committee at Oita University Faculty of Medicine (P-12-10, 17 January 2013 and #1660, 19 July 2019).
Informed Consent Statement
Informed, written consent was obtained from all individuals and participants enrolled in the study by their own volition and without monetary incentive.
Data Availability Statement
Data are contained within the article and supplementary materials.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analysis, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Funding Statement
This research was funded by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan (18KK0266, 19H03473, 21H00346 and 22H02871 to Y.Y.), (17K09353, 21K07898 to J.K.) and (18K16182, 21K08010 to T.M.) and partially supported by the Research Center for GLOBAL and LOCAL Infectious Diseases, Oita University (2021B13 to K.O.). A.K. and B.S. were doctoral students supported by the Japanese Government (Monbukagakusho: MEXT) Scholarship Programs for 2020 and 2019. Kh. Ayush, and B. Saruuljavkhlan were doctoral students supported by the Japanese Government (Monbukagakusho: MEXT) Scholarship Programs for 2020, and 2019.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Savoldi A., Carrara E., Graham D.Y., Conti M., Tacconelli E. Prevalence of Antibiotic Resistance in Helicobacter pylori: A Systematic Review and Meta-analysis in World Health Organization Regions. Gastroenterology. 2018;155:1372–1382.e17. doi: 10.1053/j.gastro.2018.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dang B.N., Graham D.Y. Helicobacter pylori infection and antibiotic resistance: A WHO high priority? Nat. Rev. Gastroenterol. Hepatol. 2017;14:383–384. doi: 10.1038/nrgastro.2017.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tacconelli E., Carrara E., Savoldi A., Harbarth S., Mendelson M., Monnet D.L., Pulcini C., Kahlmeter G., Kluytmans J., Carmeli Y., et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018;18:318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 4.Tuan V.P., Narith D., Tshibangu-Kabamba E., Dung H.D.Q., Viet P.T., Sokomoth S., Binh T.T., Sokhem S., Tri T.D., Ngov S., et al. A Next-Generation Sequencing-Based Approach to Identify Genetic Determinants of Antibiotic Resistance in Cambodian Helicobacter pylori Clinical Isolates. J. Clin. Med. 2019;8:858. doi: 10.3390/jcm8060858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.WHO . International Classification of Disease 11th Revision: Helicobacter pylori. World Health Organization; Geneva, Switzerland: 2021. [Google Scholar]
- 6.Kwon D.H., Dore M.P., Kim J.J., Kato M., Lee M., Wu J.Y., Graham D.Y. High-level beta-lactam resistance associated with acquired multidrug resistance in Helicobacter pylori. Antimicrob. Agents Chemother. 2003;47:2169–2178. doi: 10.1128/AAC.47.7.2169-2178.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malfertheiner P., Megraud F., Rokkas T., Gisbert J.P., Liou J.M., Schulz C., Gasbarrini A., Hunt R.H., Leja M., O’Morain C., et al. Management of Helicobacter pylori infection: The Maastricht VI/Florence consensus report. Gut. 2022;71:1724–1762. doi: 10.1136/gutjnl-2022-327745. [DOI] [PubMed] [Google Scholar]
- 8.Aldred K.J., Kerns R.J., Osheroff N. Mechanism of quinolone action and resistance. Biochemistry. 2014;53:1565–1574. doi: 10.1021/bi5000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Correia S., Poeta P., Hébraud M., Capelo J.L., Igrejas G. Mechanisms of quinolone action and resistance: Where do we stand? J. Med. Microbiol. 2017;66:551–559. doi: 10.1099/jmm.0.000475. [DOI] [PubMed] [Google Scholar]
- 10.Tshibangu-Kabamba E., Yamaoka Y. Helicobacter pylori infection and antibiotic resistance—From biology to clinical implications. Nat. Rev. Gastroenterol. Hepatol. 2021;18:613–629. doi: 10.1038/s41575-021-00449-x. [DOI] [PubMed] [Google Scholar]
- 11.Moore R.A., Beckthold B., Wong S., Kureishi A., Bryan L.E. Nucleotide sequence of the gyrA gene and characterization of ciprofloxacin-resistant mutants of Helicobacter pylori. Antimicrob. Agents Chemother. 1995;39:107–111. doi: 10.1128/AAC.39.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mori H., Suzuki H., Matsuzaki J., Masaoka T., Kanai T. Acquisition of double mutation in gyrA caused high resistance to sitafloxacin in Helicobacter pylori after unsuccessful eradication with sitafloxacin-containing regimens. United Eur. Gastroenterol. J. 2018;6:391–397. doi: 10.1177/2050640617737215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bolor-Erdene M., Namdag B., Yamaoka Y., Jav S. Antibiotic resistance of Helicobacter pylori in Mongolia. J. Infect. Dev. Ctries. 2017;11:887–894. doi: 10.3855/jidc.8619. [DOI] [PubMed] [Google Scholar]
- 14.Azzaya D., Gantuya B., Oyuntsetseg K., Davaadorj D., Matsumoto T., Akada J., Yamaoka Y. High Antibiotic Resistance of Helicobacter pylori and Its Associated Novel Gene Mutations among the Mongolian Population. Microorganisms. 2020;8:1062. doi: 10.3390/microorganisms8071062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feitosa I., Mori B., Santos A., Konishi J., Teles C., Costa A. What are the immunopharmacological effects of furazolidone? A systematic review. Immunopharmacol. Immunotoxicol. 2021;43:674–679. doi: 10.1080/08923973.2021.1979034. [DOI] [PubMed] [Google Scholar]
- 16.McOsker C.C., Fitzpatrick P.M. Nitrofurantoin: Mechanism of action and implications for resistance development in common uropathogens. J. Antimicrob. Chemother. 1994;33((Suppl. A)):23–30. doi: 10.1093/jac/33.suppl_A.23. [DOI] [PubMed] [Google Scholar]
- 17.Akada J.K., Shirai M., Fujii K., Okita K., Nakazawa T. In vitro anti-Helicobacter pylori activities of new rifamycin derivatives, KRM-1648 and KRM-1657. Antimicrob. Agents Chemother. 1999;43:1072–1076. doi: 10.1128/AAC.43.5.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heep M., Beck D., Bayerdörffer E., Lehn N. Rifampin and Rifabutin Resistance Mechanism in Helicobacter pylori. Antimicrob. Agents Chemother. 1999;43:1497–1499. doi: 10.1128/AAC.43.6.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiong M., Aljaberi H.S.M., Ansari N.K., Sun Y., Yin S., Nasifu L., Sun H., Xu T., Pan Y., Nie Z., et al. Phenotype and genotype analysis for Helicobacter pylori antibiotic resistance in outpatients: A retrospective study. Microbiol. Spectr. 2023;11:e00550-23. doi: 10.1128/spectrum.00550-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishizawa T., Suzuki H., Matsuzaki J., Muraoka H., Tsugawa H., Hirata K., Hibi T. Helicobacter pylori resistance to rifabutin in the last 7 years. Antimicrob. Agents Chemother. 2011;55:5374–5375. doi: 10.1128/AAC.05437-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zamani M., Rahbar A., Shokri-Shirvani J. Resistance of Helicobacter pylori to furazolidone and levofloxacin: A viewpoint. World J. Gastroenterol. 2017;23:6920–6922. doi: 10.3748/wjg.v23.i37.6920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Byambajav T.O., Bira N., Choijamts G., Davaadorj D., Gantuya B., Sarantuya T., Sarantuya G., Enkhtsetseg A., Erdenetsogt D., Battulga A., et al. Initial Trials With Susceptibility-Based and Empiric Anti-H. pylori Therapies in Mongolia. Front. Pharmacol. 2019;10:394. doi: 10.3389/fphar.2019.00394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gisbert J.P. Rifabutin for the Treatment of Helicobacter pylori Infection: A Review. Pathogens. 2020;10:15. doi: 10.3390/pathogens10010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gisbert J.P., Calvet X. Review article: Rifabutin in the treatment of refractory Helicobacter pylori infection. Aliment. Pharmacol. Ther. 2012;35:209–221. doi: 10.1111/j.1365-2036.2011.04937.x. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y.W., Hu W.L., Cai Y., Zheng W.F., Du Q., Kim J.J., Kao J.Y., Dai N., Si J.M. Outcomes of furazolidone- and amoxicillin-based quadruple therapy for Helicobacter pylori infection and predictors of failed eradication. World J. Gastroenterol. 2018;24:4596–4605. doi: 10.3748/wjg.v24.i40.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mohammadi M., Attaran B., Malekzadeh R., Graham D.Y. Furazolidone, an Underutilized Drug for H. pylori Eradication: Lessons from Iran. Dig. Dis. Sci. 2017;62:1890–1896. doi: 10.1007/s10620-017-4628-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miftahussurur M., Waskito L.A., Syam A.F., Nusi I.A., Siregar G., Richardo M., Bakry A.F., Rezkitha Y.A.A., Wibawa I.D.N., Yamaoka Y., et al. Alternative eradication regimens for Helicobacter pylori infection in Indonesian regions with high metronidazole and levofloxacin resistance. Infect. Drug Resist. 2019;12:345–358. doi: 10.2147/IDR.S187063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Su Z., Xu H., Zhang C., Shao S., Li L., Wang H., Wang H., Qiu G. Mutations in Helicobacter pylori porD and oorD genes may contribute to furazolidone resistance. Croat. Med. J. 2006;47:410–415. [PMC free article] [PubMed] [Google Scholar]
- 29.Kwon D.H., Lee M., Kim J.J., Kim J.G., El-Zaatari F.A., Osato M.S., Graham D.Y. Furazolidone- and nitrofurantoin-resistant Helicobacter pylori: Prevalence and role of genes involved in metronidazole resistance. Antimicrob. Agents Chemother. 2001;45:306–308. doi: 10.1128/AAC.45.1.306-308.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are contained within the article and supplementary materials.