Abstract
A novel porin, OmpG, is produced in response to a chromosomal mutation termed cog-192. Molecular characterization of cog-192 revealed that it is a large chromosomal deletion extending from the 3′ end of pspA through to the 5′ end of an open reading frame located immediately upstream of ompG. As a result of this 13.1-kb deletion, the expression of ompG was placed under the control of the pspA promoter. Characterization of OmpG revealed that it is quite different from other porins. Proteoliposome swelling assays showed that OmpG channels were much larger than those of the OmpF and OmpC porins, with an estimated limited diameter of about 2 nm. The channel lacked any obvious solute specificity. The folding model of OmpG suggests that it is the first 16-stranded β-barrel porin that lacks the large external loop, L3, which constricts the channels of other nonspecific and specific porins. Consistent with the folding model, circular dichroism showed that OmpG contains largely a β-sheet structure. In contrast to other Escherichia coli porins, there is no evidence that OmpG exists as stable oligomers. Although ompG DNA was present in all E. coli strains examined so far, its expression under laboratory conditions was seen only due to rare chromosomal mutations. Curiously, OmpG was constitutively expressed, albeit at low levels, in Salmonella, Shigella, and Pseudomonas species.
The outer membrane of gram-negative bacteria provides a barrier against noxious agents in the environment. Escherichia coli K-12 contains a set of outer membrane porin proteins that form channels allowing an influx of nutrients (for reviews, see references 3, 21, and 25). Two porins, OmpF and OmpC, often regarded as the classical porins, allow the flow of water-soluble solutes with molecular weights of around 500 or less (20, 25). Although these general pores lack any pronounced solute specificity, they are weakly cation selective. The PhoE porin is structurally similar to the classical porins and forms general diffusion pores with a preference for anionic solutes (26). Other porins can specifically facilitate the uptake of substrates such as maltodextrins (LamB [40]) and nucleosides (Tsx [15]). OmpA was initially thought to play only a structural role, but recent in vitro studies have shown that this protein also displays weak channel activity (37). Similarly, TolC has been shown to form channels in vitro (3).
In the last decade, publications pertaining to structure-function analysis of the porins provided a better understanding of their molecular anatomy. A major breakthrough came with the three-dimensional structural resolutions of OmpF (6) and PhoE (6, 10). The monomeric subunit of each of these homotrimeric porins consists of 16-stranded antiparallel β barrels enclosing the transmembrane pore. The pore entrance is narrowed by long external loops, and the middle of the channel is constricted by the inward folding of loop L3, while its cross section increases abruptly just after this constriction zone (facing the periplasm). Proof that the constricted area defines the filtering ability of the channels came from genetic studies (2, 16, 17).
The classical porin proteins exist as trimers that display unusually stable structural properties (28). They form trimers that resist denaturation by strong anionic detergents such as sodium dodecyl sulfate (SDS). This high stability results from hydrophobic and electrostatic interactions between the subunits (6). Porin proteins do not contain long stretches of hydrophobic residues; instead, every alternate residue facing the lipid bilayer is hydrophobic. The presence of negatively charged residues on the outer surface allows these exposed surfaces to interact with the negatively charged groups of lipopolysaccharide via divalent cation bridges (25).
Porin proteins are critical for bacterial growth. Porin-deficient strains grow slowly and accumulate mutations that permit expression of new porin proteins. This finding led to the isolation of E. coli strains that acquired the ability to synthesize a new porin protein, NmpC (4). The origin of NmpC was linked to a defective prophage, QSR. Similarly, another porin, Lc, encoded by a lambdoid bacteriophage, was isolated (8). The OmpG porin was discovered among mutants that gained increased outer membrane permeability in the absence of OmpF and LamB (18). However, unlike NmpC and Lc, OmpG is not encoded by a prophage genome. Normally LamB is required for transport of maltodextrins, sugars that are excluded from OmpF and OmpC channels because of their large size. Thus, a LamB− OmpF+ OmpC+ strain displays a maltodextrin-minus (Dex−) phenotype. Strains with this composition revert to Dex+ at a low frequency (10−9 to 10−10). The mutations were mapped to four loci: ompF (2), ompC (16, 17), imp (30), and cog (18). cog (for control of ompG) mutations resulted in the appearance of a new outer membrane protein, OmpG. Growth and uptake experiments showed that OmpG displays a porin-like activity. Genetic experiments indicated that cog itself may code for a negative regulator of OmpG expression (18).
In this report, we provide data showing that OmpG defines a novel class of porin protein whose sequence, biochemical properties, and channel properties are distinct from those of known bacterial porins.
MATERIALS AND METHODS
Media.
Luria broth and MacConkey media (both from Gibco) were prepared as described previously (33). The amount of maltodextrins (Pfsanstiehl) added to the MacConkey medium differed between preparations of the maltodextrin stock solution. The stock solution of maltodextrins was titered by determining the amount of maltodextrins needed to yield red colonies for an OmpG+ strain (RAM123) and white colonies for an OmpG− strain (DME553).
Bacterial strains and lambda phages from Kohara library.
The laboratory E. coli K-12 strains used in this study were derived from DME553 (ΔlamB106 ΔompF80). RAM123 is a Dex+ derivative of DME553 that produces OmpG due to the cog-192 mutation (18). RAM194 was derived from RAM123 by transducing the ΔompC178 allele through a linked Tn10, thus making the resulting strain OmpC−. HS3169 (malK::Tn10) was obtained from Howard Shuman. Strains K1342 [λpsp3.2; Φ(pspA′-lacZ)] and K1472 (ΔpspABC::kan) were kindly provided by Peter Model. All other E. coli strains were obtained from E. coli Genetic Stock Center. Salmonella typhimurium LT2 (ompD+) and CH338 (ompD) were kindly provided by Ken Sanderson. Shigella flexneri serotype 2a (ATCC 9473) was obtained from the American Type Culture Collection. Pseudomonas aeruginosa PAO1 was from our laboratory strain collection. Clinical isolates of Salmonella strains were made available to us by Micah Williams. Lambda clones 250 through 260 from Kohara phage library (12) and E. coli LE392 for their propagation were kindly provided by Frederick Blattner.
Protein methods.
Cell envelopes were prepared by the French press lysis method as described previously (19). Proteins were analyzed by electrophoresis through SDS-polyacrylamide (11%) gels (SDS-PAGE) (14). The following methods were used for OmpG purification. When OmpG was needed for raising antibodies or protein sequencing, envelopes obtained from a strain lacking OmpA, OmpC, OmpF, and LamB but producing OmpG from a plasmid were treated with 0.5% SDS in 50 mM Tris-HCl (pH 8.0) for 1 h at 37°C. SDS-solubilized proteins were removed by centrifugation at 18,000 × g for 30 min. The pellet was resuspended in the SDS sample buffer (2% SDS, 10% glycerol, 5% β-mercaptoethanol, 50 mM Tris-HCl [pH 6.8]) and solubilized by boiling for 5 min. Solubilized samples were analyzed by SDS-PAGE, and the gel was stained with Coomassie blue. The OmpG band was excised and electroeluted in a Bio-Rad electroelutor. Eluted OmpG samples were concentrated by ethanol precipitation at −20°C. The pellet was resuspended in 20 mM HEPES (pH 7.4) or double-distilled H2O for protein sequence determination.
For proteoliposome swelling assays, OmpG was partially purified from envelopes by a two-step detergent solubilization method. Envelopes lacking all major outer membrane proteins except OmpG were resuspended in 0.5% n-octylglucoside (OG)–10 mM HEPES (pH 7.4) and incubated for 30 min at 37°C. The pellet recovered by centrifuging samples at 436,000 × g for 30 min was resuspended in 1.0% OG–10 mM HEPES (pH 7.4). After incubation for 30 min at 37°C, samples were centrifuged as described above, and the supernatant containing primarily OmpG was recovered. Extract containing 0.2 μg of protein was used for reconstitution of proteoliposomes as described previously (13) except that (i) a 30:2 mixture of acetone-washed phosphatidylcholine (Avanti Polar Lipids) and dicetylphosphate (Sigma) was used instead of E. coli phospholipids and (ii) dextran T-40 (Pharmacia) was used instead of dextran T-20. Proteoliposomes containing the dextran were diluted into iso-osmotic solutions of various solutes, and the permeation rates were measured by the initial rates of decrease of optical density of proteoliposome suspension (13).
OmpG for circular dichroism measurement was purified as follows. The outer membrane fraction of RAM194 was isolated by sucrose density centrifugation and extracted with 2% octyl-polyoxyethylene (Alexis Biochemicals). The extract was applied to a Bio-Rad DEAE-5PW high-pressure liquid chromatography column, which was eluted with a 0 to 0.5 M linear gradient of NaCl in 10 mM Tris-HCl (pH 8.0)–1 mM EDTA–0.2% Lubrol PX. The fraction containing essentially pure OmpG was dialyzed against the buffer just mentioned but without NaCl, and its spectrum was measured, after concentration by ultrafiltration with Amicon PM-10 membrane, with an Aviv model 62DS circular dichroism spectrometer.
Antibodies were raised as described previously (19). For protein sequence analysis, electroeluted OmpG in double-distilled H2O was acidified with trichloroacetic acid and dissolved with acetonitrile. Approximately 100 pmol of OmpG protein was spotted on a fiberglass disk (Beckman) and sequenced in a Proton 2090 protein sequencer (Beckman). For Western blot analysis, protein samples analyzed by PAGE were transferred to Immuno-Lite membranes (Bio-Rad), using a Mini Trans-Blot electrophoretic transfer cell (Bio-Rad). The Western blot analysis was carried out with solutions obtained from Bio-Rad and a 1:5,000 dilution of the OmpG antiserum.
DNA and RNA methods.
ompG+ plasmid clones were obtained by transforming an OmpG− strain with a Sau3A chromosomal gene bank, prepared from an ompG+ strain, and selecting for Dex+ (red) colonies on MacConkey-maltodextrin-ampicilliln (50 μg/ml) medium. Confirmation that the Dex+ phenotype was conferred by the expression of OmpG from a plasmid was obtained by retransforming the plasmid into an OmpG− strain and observing the Dex phenotype of transformants. cog+ plasmid clones were also obtained from a Sau3A chromosomal gene bank. The DNA sequence of cloned DNA was determined by the dideoxy-chain termination method (31). Sequence analysis was carried out with software packages from Genetics Computer Group Inc. (University of Wisconsin, Madison) and DNAMAN (Lynnon BioSoft). Lambda phages were purified through CsCl isopycnic gradients, and DNA was extracted as described previously (29). Chromosomal DNA and total RNA were isolated by a kit obtained from Amersham Life Science. Restriction enzyme digestions were carried out as instructed by the manufacturer. Southern and Northern analyses were carried out by standard protocols (29). Nucleic acids were cross-linked to Quantum Yield filters (Promega) in a Stratalinker (Stratagene). Nonradioactive hybridization probes were prepared by using a random-primer fluorescein labeling kit (Du Pont-NEN).
Nucleotide sequence accession number.
Nucleotide sequences of the ompG gene and an upstream open reading frame (orf1) were assigned GenBank accession no. ECU49400.
RESULTS
Genetic mapping and cloning of ompG.
Genetic mapping of ompG was facilitated by the isolation of linked transposon insertions. To obtain such insertions, a transposon (Tn10) insertion pool was prepared in a wild-type strain. The Tcr (from Tn10) marker from this pool was transduced into an ompG+ (Dex+ Cog−) strain so as to obtain insertion mutations that prevented OmpG expression (Dex− phenotype). These Tn10s were genetically mapped and found to be located at the 29-min region of the chromosome where cog had been previously mapped (18). Once ompG sequencing was completed (see below), ompG-specific primers were used to confirm the genetic location of ompG by using Kohara clones encompassing the 29-min region of the chromosome (12). Of the several Kohara clones used, two covering min 29.5 to 29.7 of the E. coli chromosome yielded ompG-specific fragments. Also, Southern blot analysis of Kohara clones with an ompG-specific probe confirmed its genetic location in the chromosome (data not shown).
The Tn10 insertion mutations resulting in a Dex− (OmpG−) phenotype were found to be recessive, which allowed procurement of an ompG+ (Dex+) clone by complementation using an E. coli gene library. The original ompG+ plasmid clone contained a 5.5-kb chromosomal DNA insert. Subsequent subcloning showed that a 2.6-kb DNA fragment contains sufficient information for OmpG expression. The expression of OmpG from these plasmid clones was not repressible by either chromosomal or plasmid-borne Cog (data not shown). Gene dosage experiments showed that the lack of repression was not due to a possible titration of the repressor by an excessive number of operator sites. A reorientation of the ompG+ DNA insert with respect to the plasmid-borne Tcr gene promoter resulted in no detectable OmpG expression, showing that the ompG+ clone lacked promoter sequences and possibly the site needed for Cog-mediated repression.
Nucleotide sequence analysis of the ompG region.
The nucleotide sequence of the entire 2.6 kb of insert DNA was determined and found to match perfectly the published sequence (5). Two ORFs (orf1 and orf2) within the ompG+ DNA insert were identified; orf2 corresponded to OmpG. orf2 starts 157 bp downstream from the end of orf1 and encodes a 301-residue-long protein. The amino-terminal end of Orf2 contains a typical signal sequence motif of 21 amino acid residues. A 14-amino-acid NH2-terminal sequence of the purified OmpG was identical to residues 22 to 35 deduced from DNA sequence of orf2, confirming that OmpG is coded by orf2.
OmpG is a novel porin protein.
At the amino acid sequence level, OmpG shows no significant homology with other proteins in the data bank. Direct alignment of the OmpG sequence with the OmpF and OmpC sequences displayed a low overall random identities of 22% (16 alignment gaps) and 16% (13 alignment gaps), respectively. That these identities are only superficial became apparent when we compared the primary sequence of OmpA with those of structurally and functionally unrelated proteins, OmpF and OmpC. Here again, the overall random identities were 22% (17 alignment gaps) between OmpA and OmpF and 17% (11 alignment gaps) between OmpA and OmpC. Finally, a low overall random identity of 17% (10 alignment gaps) was observed between OmpG and OmpA. On the other hand, between the two related proteins OmpF and OmpC, a striking 64% overall identity was observed with only seven alignment gaps.
Immunoblot analysis of filters containing purified E. coli OmpG, OmpC, OmpF, and LamB proteins showed no cross-reactivity of polyclonal OmpG antibodies with these proteins (data not shown). Furthermore, blotting of envelopes containing OmpA also gave negative signals. As expected, antibodies reacted strongly with either the purified OmpG protein or OmpG present in envelopes. Thus, OmpG is novel, as it shares neither sequence similarity nor antigenic overlap with the previously characterized outer membrane proteins of E. coli.
In certain respects, however, OmpG is related to other outer membrane proteins of E. coli. (i) Apart from its signal sequence, OmpG lacks any long hydrophobic stretches. (ii) OmpG, like some other porin proteins, lacks cysteine residues in its mature portion. (iii) The last residue of OmpG is phenylalanine, which is thought to be important for the proper insertion or assembly of outer membrane proteins (36). (iv) Like many outer membrane proteins, OmpG contains a preponderance of negatively charged residues (50 Asp and Glu, compared to 22 Arg and Lys), with an estimated pI of 4.4.
Circular dichroism and folding patterns of OmpG.
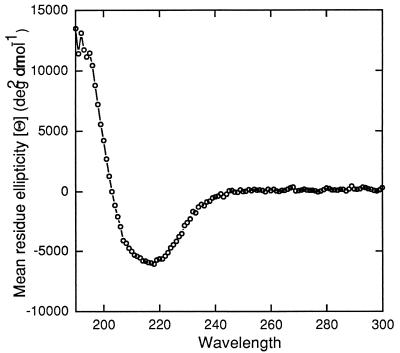
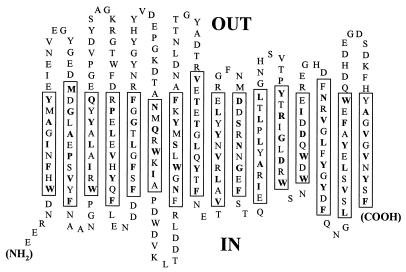
To gain a better understanding of the secondary structure of OmpG, detergent-solubilized OmpG retaining its full channel activity was analyzed by circular dichroism (Fig. 1). The ellipticity pattern showed a minimum of a fairly small magnitude, close to 210 nm. This showed that in contrast to OmpA (38), there was little contribution from α-helical structures and that OmpG had a largely β-sheet structure, similar to the classical porins. Based on the folding prediction methods of Schirmer and Cowan (predicts transmembrane stretches [32]) and Paul and Rosenbusch (predicts turns [27]), the mature portion of OmpG is predicted to have 16 transmembrane segments (Fig. 2, boxed regions). The amino acids at alternate positions within transmembrane segments are primarily hydrophobic residues that are predicted to have their side chains oriented outward toward the lipid environment. In general, the internal loops are smaller than those present on the outside, where there is a distinctive absence of an especially large loop, L3, a hallmark of the classical porins, where such a loop is shown to narrow the middle of the channel (6). The predicted OmpG folding model can now be tested by genetic and molecular means.
FIG. 1.
Circular dichroism analysis of purified OmpG. Purification and measurement were carried out as described in Materials and Methods. The magnitude of mean residue ellipticity (about −5,000 deg2 dmol−1) is significantly smaller than that found with OmpA or P. aeruginosa OprF (−9,000 to −10,000 deg2 dmol−1 [38]), two outer membrane proteins that contain substantial α-helical domains. The spectrum also does not show the two sharp negative peaks at 208 and 220 nm, which are characteristic of α helices.
FIG. 2.
Predicted folding pattern of OmpG. The OmpG amino acid sequence was analyzed by using computer programs based on the algorithms of Schirmer and Cowan (32) and Paul and Rosenbusch (27). Transmembrane β strands were predicted whenever a high value of hydrophobicity on alternate residues in a 10-residue stretch was found, and this prediction was strengthened by the presence of turn-promoting sequences on either side. Most of the transmembrane segments could be assigned unequivocally, but the presence of the 12th and 13th segments is less convincing, as the hydrophobicity of the alternate residues is not high.
OmpG is a heat-modifiable protein.
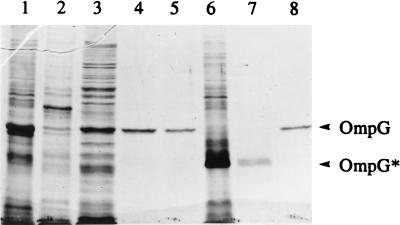
We have previously reported that unlike OmpF and OmpC, OmpG is not strongly associated within the cell envelope, as it can be completely solubilized with 1% SDS or Triton X-100 in the absence of high salt concentrations (18), a condition that does not allow solubilization of the classical porins. However, under milder solubilization conditions, such as treatment with 0.5% SDS or OG, the majority of OmpG remained associated with the membrane (Fig. 3). Interestingly, under these solubilization conditions when envelopes were analyzed by SDS-PAGE without heating, OmpG did not migrate at its denatured position of 34,000 mol wt but instead appeared as a faster-migrating species of 28,000 mol wt (Fig. 3, lane 7). Immunoblot analysis confirmed that this faster-migrating polypeptide corresponds to OmpG (data not shown). The treatment of envelopes with 1% OG led to the complete solubilization of OmpG without altering its heat-modifiable property. However, subsequent removal of OG by dialysis led to loss of this property (Fig. 3, lane 8). As shown below, both dialyzed and undialyzed preparations of OmpG were competent in forming active channels in proteoliposome swelling assays. The dialysis therefore appears to make the OmpG more susceptible to complete denaturation by SDS, presumably by removing stabilizing factors.
FIG. 3.
SDS-PAGE analysis of envelopes and detergent-solubilized OmpG. Lane 1, untreated envelopes obtained from a strain lacking OmpF, OmpC, LamB, and OmpA; lane 2, pellet after OG extraction; lane 3, supernatant of 0.5% OG extraction; lane 4, supernatant of 0.5 and 1% OG extraction; lane 5, same as lane 4 but dialyzed against 10 mM HEPES (pH 7.4) buffer; lane 6, unheated envelopes; lane 7, same as lane 4 but unheated; lane 8, same as lane 5 but unheated. Positions of denatured OmpG and heat-modifiable OmpG (OmpG*) are shown. Samples present in lanes 1 to 5 were heated by boiling for 5 min prior to SDS-PAGE analysis.
In vitro channel properties of OmpG.
In vivo growth and uptake experiments showed that OmpG has a porin-like activity that allows the influx of larger solutes than do the classical porins (18). Proteoliposome swelling assays (22, 23) were carried out to confirm the in vivo data.
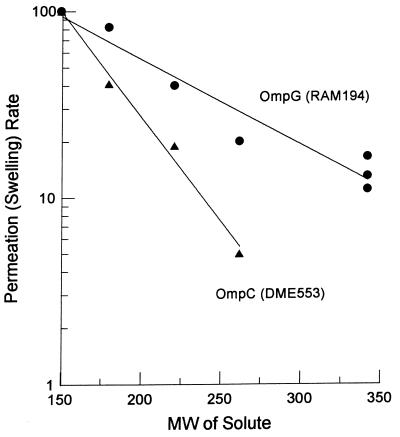
Proteoliposomes were reconstituted from phosphatidylcholine and unfractionated outer membrane fragments from a strain lacking OmpF, OmpC, and LamB but containing OmpG. The influx of sugars was measured by the osmotic swelling of these vesicles. Several conclusions can be made from these experiments (Fig. 4). (i) OmpG produces fairly efficient channels. The rate of influx for a pentose, l-arabinose, was similar to that seen for proteoliposomes reconstituted with similar amounts of OmpC. Thus, at least for sugars, OmpG functions as well as the classical trimeric porins of E. coli. (ii) OmpG channels behaved strikingly differently from OmpF and OmpC when disaccharides were used. OmpF and OmpC showed disaccharide diffusion rates around 1 to 2% of the rate of l-arabinose diffusion (also see reference 24). In contrast, OmpG channels behaved as though they were much larger, allowing the diffusion of disaccharides at rates corresponding to 15 to 30% of the rates of l-arabinose. This behavior is very similar to that of the P. aeruginosa porin OprF, where a limiting pore diameter of 2.0 nm has been calculated (42). OprF, however, is different from OmpG in its low efficiency. Isolated OmpG (purified by solubilization in OG and fractionated by ion-exchange chromatography) and OmpF preparations were also used in proteoliposome swelling assays. The results validated the above finding, showing that OmpG channels were functionally larger than those of OmpF when a disaccharide (sucrose) and a trisaccharide (raffinose) were used. Calculations by the approach of Nikaido and Rosenberg (22) show that the dependence of permeability on solute size fits best with a pore diameter of about 20 Å. Since the distance between neighboring β strands is 4.5 Å, a β barrel with 16 transmembrane strands will have a diameter of 23 Å, measured between α-carbon atoms. With the amino acid side chains protruding into the interior of this barrel, the 20-Å average diameter is consistent with our model.
FIG. 4.
Rates of penetration of various sugars through the OmpG channel. Proteoliposomes containing OmpG extract were made as described in Material and Methods, and portions of the suspension were diluted into iso-osmotic solutions of sugars to determine the rates of osmotic swelling of the vesicles. The rates were normalized to the swelling rate in l-arabinose, which was taken as 100%. The sugars used (and, in parentheses, their molecular weights) were l-arabinose (150), d-glucose (180), N-acetylglucosamine (221), 2,3-diacetamido-2,3-dideoxy-d-glucose (262), lactose (342), sucrose (342), and maltose (342). As a control, proteoliposomes containing OmpC extract were used.
Dialyzed and undialyzed preparations yielded very similar solute diffusion rates, suggesting that the loss of heat modifiability (Fig. 3) did not result from the denaturation of the protein. Using maltose, lactose, sucrose, trehalose, and (GIY)4 peptide in proteoliposome swelling assays, we failed to see any evidence of solute specificity for OmpG channels.
Outer membrane permeability of OmpG+ strains.
We have shown above that OmpG produces a larger channel than OmpF or OmpC. Furthermore, OmpG is expressed in strain RAM194 at a level similar to that of the classical porins in other K-12 strains (18). We might expect that an abundant expression of a large-channel porin would make the cells hypersensitive to inhibitors and antibiotics. We therefore tested the susceptibility of strains DME553 (OmpC+), RAM123 (OmpC+ OmpG+), and RAM194 (OmpG+) to hydrophilic (ampicillin and chloramphenicol) and hydrophobic (novobiocin and rifampin) antibiotics. Cells expressing OmpG showed hypersensitivity to hydrophilic antibiotics: paper disks (7-mm diameter) presoaked with 10 μg of ampicillin or 5 μg of chloramphenicol gave inhibition zones of 13 mm in DME553 but around 18 mm in RAM123 or RAM194. In contrast, only a slight increase in sensitivity toward hydrophobic antibiotics was noted in cells expressing OmpG. These results further corroborated the above finding that the channel formed by OmpG is larger than those formed by the classical porins.
OmpG lacks stable oligomeric forms.
The majority of porin proteins exist as homotrimers. However, the following experiments suggest that OmpG is monomeric porin. First, we carried out two-dimensional PAGE analysis (first dimension under native and second dimension under SDS-denaturing conditions) to look for evidence that OmpG is an oligomeric porin. We could detect OmpF trimers but no oligomeric forms of OmpG. Second, we carried out cross-linking experiments using dimethylsuberimidate and formaldehyde. In these experiments, either purified envelopes, OG-solubilized extracts, or whole cells were used. SDS-PAGE followed by immunoblot analysis did not reveal any evidence of cross-linked OmpG, although oligomeric forms were visible with OmpF (data not shown).
Expression of OmpG in other gram-negative bacteria.
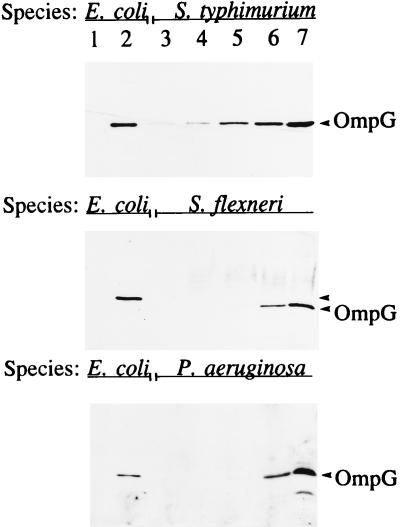
Immunoblot analysis using OmpG-specific antibodies showed the presence of a cross-reacting protein band with similar sizes in membranes of S. flexneri and S. typhimurium and even in P. aeruginosa, which does not belong to the family Enterobacteriaceae (Fig. 5). In addition to being detected in S. typhimurium LT2, OmpG expression was detected in several clinical Salmonella isolates grown under our standard laboratory conditions (data not shown). It is curious that unlike in our E. coli strains, where OmpG is not expressed, it is produced, albeit apparently at a low level, in other bacterial species. It is conceivable that the lack of OmpG expression in our contemporary K-12 strains was due to chromosomal aberrations caused by the heavy mutagenesis used to construct these strains in the first place or that optimum growth conditions for its expression have not been found. Attempts to define growth conditions under which OmpG is expressed without mutations have so far been unsuccessful. To test the first hypothesis, we examined wild-type E. coli K-12 strains 679, Y10, W1, and AB1157, E. coli B, E. coli C, and E. coli K1 for OmpG expression by Western blot analysis. We failed to see OmpG from these strains. However, all strains yielded ompG-specific DNA fragments by PCR analysis.
FIG. 5.
Immunoblot analysis of OmpG from envelopes of various gram-negative bacteria. Lanes 1 and 2 contain envelopes (1.5 μg of protein) from OmpG− and OmpG+ E. coli strains, respectively; lanes 3 to 7 contain 0.75, 1.5, 3, 6, and 12 μg of total protein, respectively. Gels were blotted with polyclonal antibodies raised against E. coli OmpG.
Unlike E. coli, S. typhimurium contains three constitutively expressed porins, OmpD in addition to OmpF and OmpC (25). Immunoblot analysis using OmpG antibodies on envelopes prepared from OmpD+ and OmpD− (ompD::Tn10) strains showed the presence of a cross-reacting protein band in both strains (data not shown). That OmpG is unrelated to OmpD was not surprising because we already knew that OmpG is unrelated to OmpF and OmpC, which show high antigenic and sequence conservation with OmpD (34, 35).
Regulation of OmpG: cloning of cog+.
By using conventional genetic mapping methods, cog-192 was mapped at 29 min on the chromosome (18). Diploid analysis revealed that cog-192 was recessive, suggesting that it may disable the activity of a repressor protein that negatively controls ompG expression. However, the low frequency by which cog-192 was isolated suggested that it is not a simple null mutation. The recessive nature of the cog-192 mutation permitted the isolation of a plasmid clone containing the wild type cog+ gene from an E. coli gene bank by genetic complementation. The DNA insert carrying cog+ was roughly 10 kb long and thus likely to contain other genes that may or may not be relevant to cog function. The original cog+ clone was reduced to half its size by removing a restriction fragment. When introduced into an OmpG+ (Dex+) strain, this shorter clone still conferred an OmpG− (Dex−) phenotype. Partial DNA sequencing from either end of the clone revealed the presence of truncated aldH on the one end and truncated pspC on the other. Four ORFs organized in the order aldH-goaG-pspF-pspA-pspB-pspC have been identified between these two genes (5, 11). Two smaller plasmid clones retaining both pspA and pspB (a 1.8-kb BglII-SalI fragment) or just pspA (an 800-bp BglII-EcoRI fragment in which the EcoRI site was created immediately downstream from pspA by site-directed mutagenesis) also complemented the cog-192 mutation. This showed a possible involvement of pspA in the regulation of OmpG expression.
Three additional observations strengthened the notion that cog-192 allows OmpG expression by somehow affecting the psp operon: (i) both cog-192 and psp map at 29 min on the E. coli chromosome, (ii) an introduction of ΔpspA::Kmr abolishes OmpG expression; and (iii) consistent with a proposal that pspA is autoregulated (41), we found that the presence of the cog-192 mutation elevates the activity of a pspA::lacZ construct 10-fold.
Molecular characterization of cog-192.
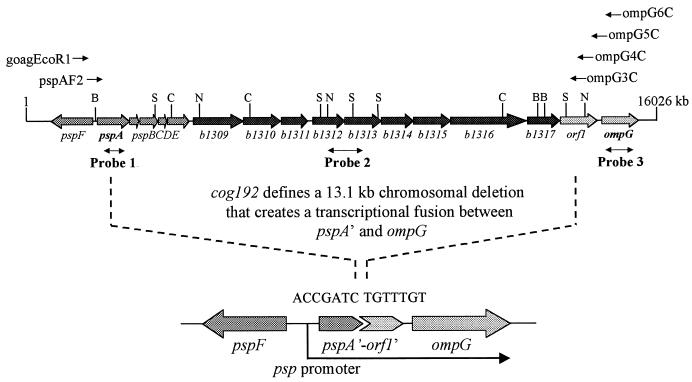
Complementation of cog-192 by plasmid clones bearing just the pspA gene provided evidence that cog-192 may define a chromosomal aberration affecting pspA gene function. When we used pspA-specific primers to amplify DNA from cog+ and cog-192 strains by PCR, only the cog+ strain yielded the pspA fragment. The psp region was also examined by Southern blot analysis using three different probes, specific to pspA, b1312 to 1313, and ompG (Fig. 6). The data revealed that in the cog-192 strain, regions corresponding to the pspA and b1312 to 1313 probes were missing (Fig. 6). The ompG-specific probe revealed a substantial alteration in the genome immediately upstream to the ompG gene (Fig. 6).
FIG. 6.
Molecular organization of the chromosomal region encompassing ompG and the psp operon. Ten uncharacterized genes (b1309 to 1317 and orf1) are present between ompG and the psp operon. These 10 ORFs as well as a significant portion of the psp operon are deleted (marked by dotted lines) in the cog-192 strain. The deletion aligns ompG with the pspA promoter; precise ends of the deletion are shown in a close-up diagram below. Double-headed arrows show DNA probes used in Southern and/or Northern blot analysis. Locations of various primers and of restriction enzyme sites are shown at the top. Abbreviations: B, BglII; C, ClaI; N, NcoI; and S, SalI.
cog-192 defines a large chromosomal deletion.
In the cog+ strain, the pspF and ompG genes are separated by about 14.0 kb. Thus, under standard PCR conditions, it may not be possible to amplify a 14.0-kb intervening DNA fragment with primers specific to pspF and ompG. However, if this distance is significantly reduced, it should become possible to PCR amplify DNA by pspF forward and ompG reverse primers. To test this, we used two forward primers that were complementary to the goaG/pspF region and four reverse primers that were complementary to the orf1/ompG region (Fig. 6). As anticipated, no DNA fragments could be amplified from the cog+ strain. On the other hand, all but one set of primers yielded DNA fragments from the cog-192 strain. Amplified DNA fragments, however, were much shorter than expected from a genome where the region between pspF and ompG was intact, showing that these two regions are now immediately adjacent to each other in the cog-192 strain.
PCR-amplified DNA from the cog-192 strain was used for nucleotide sequence analysis to pinpoint the deletion junction. The results showed that the region between pspA and orf1, including the last 369 (of 666) bp of the pspA gene and the first 685 (of 969) bp of orf1 were deleted in the cog-192 strain (Fig. 6). This showed that cog-192 defined a large chromosomal deletion of a total of 13,127 bp between the pspF and ompG genes.
cog-192 creates a transcriptional fusion between pspA′ and ompG.
As mentioned above, the expression of OmpG in the cog-192 strain was turned off by the presence of a pspA+ plasmid, indicating that PspA somehow controls OmpG expression in this strain. One explanation for this observation could be that the cog-192 deletion resulted in ompG being placed under the control of a pspA promoter. Since the pspA promoter is regulated by the pspA gene product (41), the presence of a multicopy pspA+ plasmid would be expected to have a negative effect on its activity and hence ompG expression. This possibility was tested by Northern blot analysis of ompG mRNA (Fig. 7).
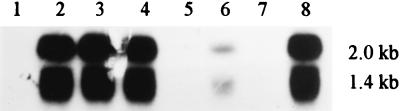
FIG. 7.
Northern blot analysis to detect ompG-specific transcripts. Total RNA (3 μg) obtained from DME553 (cog+; lanes 1 and 7), RAM123 (cog-192; lanes 2 and 8), RAM123 recA::Kmr (lane 3), RAM123 recA::Kmr/pBR322 (lane 4), RAM123 recA::Kmr/pBR322-pspFAB (lane 5), and RAM123 recA::Kmr/pBR322-pspA (lane 6) was probed with an ompG-specific probe. Molecular sizes of the two ompG-specific mRNA species are 1.4 and 2.0 kb.
No ompG-specific transcripts were detected from a cog+ strain (Fig. 7; lanes 1 and 7). In contrast, two ompG-specific transcripts were detected from the cog-192 strain (lanes 2 and 8). These transcripts were either absent (lane 5) or present in a greatly reduced amount (lane 6) when the cog-192 strain was transformed with a plasmid bearing pspFAB or pspA, respectively. These results showed that a promoter that is negatively regulated by the pspA gene product controls ompG transcription in the cog-192 strain. Northern analysis also revealed the presence of two ompG-related transcripts that were regulated in the same fashion (Fig. 7). One interpretation of this observation is that the two transcripts initiated from the same promoter but were terminated at different points.
Chromosomal deletions are necessary for OmpG expression.
We sought additional Dex+ revertants from DME553 (cog+ Dex−) to examine whether mutations other than deletions could permit OmpG expression. Dex+ revertants were obtained from 10 independent cultures. Dex+ isolates expressing OmpG were estimated to be around 1% of the total Dex+ revertants obtained, as judged by Western blotting. PCR and Southern analyses of the chromosomal DNA from five independently obtained Dex+ isolates revealed deletions upstream of ompG. In two isolates, the deletion extended beyond the entire psp operon; in three, the deletions retained pspA. As was the case for the cog-192 strain, OmpG expression in the latter three isolates was inhibited by a plasmid carrying pspA+. These results support the notion that deletions upstream of the ompG structural gene are necessary for OmpG expression under laboratory growth conditions. These deletions must create novel junctions between ompG and an upstream promoter element that may not be genetically related to ompG in the parental strain.
DISCUSSION
The molecular and biochemical analyses presented in this study showed that OmpG is a unique class of porin. Consistent with our previous in vivo findings (18), the in vitro data revealed that OmpG channels are functionally larger than those of the classical porins. The major reason for large OmpG channels may be the absence of a large external loop, L3, which in classical porins folds back into the channel and serves to restrict pore size (6). Since the distance between neighboring strands in a β sheet is 0.45 nm, the 16-membered β barrel predicted for OmpG (Fig. 2) will produce a channel with the diameter 0.45 × 16/π = 2.3 nm. Considering that side chains will narrow the channel somewhat, this value is in perfect agreement with the diameter determined by the solute size dependence of sugar flux (Fig. 6). If our model is correct, OmpG is the first porin with such a simple barrel structure.
Large OmpG channels mean that a greater variety of nutrients can be taken up by the bacteria, which would provide a growth advantage to bacterial strains producing OmpG. Yet, OmpG expression has not been observed in wild-type E. coli strains obtained from laboratory collections. In contrast, OmpG expression has been observed in Salmonella and Shigella strains analyzed so far. The level of expression, however, appears to be in the range of 10 to 20% of the levels of classical porins, which may explain why the presence of this porin was not noticed earlier. Future work should address the significance of OmpG in these organisms. It is conceivable that OmpG was a functional porin in the ancestor of present day Enterobacteriaceae and that its expression was down regulated or abolished in E. coli, which confronts the constant challenge of toxic bile salts (39).
The discovery of OmpG showed the potential of bacterial genomes to code for a number of porin proteins. Of course, not all porins are expressed at the same time. Some, such as general porins, may be required under all growth conditions, although their relative levels fluctuate in response to physiological parameters. These proteins are present in large quantities (around 105 molecules/cell). However, solute diffusion through OmpF and OmpC channels is not efficient, which may have contributed to the evolution of bacteria with specialized porins, such as LamB and Tsx, which are needed only under conditions of specific nutrient availability. Solute specificity for OmpG channels and physiological parameters regulating its expression are not known.
OmpG expression was achieved under laboratory conditions where the entry of large sugars was not possible through general channels. A mutation, cog-192, permitting OmpG expression turned out to be a chromosomal deletion that removed 13.1 kb of DNA immediately upstream of ompG. This deletion placed ompG expression under the control of the pspA promoter. The regulation data contrasted with our previous hypothesis that operator or repressor mutations derepress OmpG expression (18). Indeed, the recessive nature of cog-192 and plasmid complementation data supported a repressor theory. Molecular analysis of cog mutations, however, demonstrated that regulated expression of OmpG was the result of a novel gene fusion. Jacob and colleagues (1, 9) reported deletions fusing two unrelated operons where the downstream genes were also placed under the transcriptional control of an upstream promoter for unrelated operon. Computer analysis of the nucleotide sequence upstream of ompG did not reveal the existence of a ς70-dependent promoter element. It is conceivable that ompG and 10 other ORFs sandwiched between it and the psp operon are not expressed in E. coli due to the absence of an active promoter.
Sequence homology data showed that several of these 10 uncharacterized ORFs may code for proteins involved in the transport or metabolism of sugars (5). Interestingly, orf1, located immediately upstream of ompG, shows a strikingly high identity with the ATPase component of various ABC transporters (7). These components are usually associated with other proteins dedicated to the transport of a specific substrate, and genes coding for these other components are present in the vicinity of orf1. At this point it is unclear whether orf1 and ompG are part of the same operon. Our data showed that the OmpG-mediated Dex+ phenotype is dependent on the malK gene product of the mal operon but not on orf1. Curiously, we found that orf1 sequences from E. coli B and E. coli C strains had the capacity to encode a 360-residue protein (compared to the 322-residue Orf1 from K-12). A truncation of Orf1 in E. coli K-12 was due to the presence of a natural frameshift mutation (deletion of G) close to the end of the gene. The truncation of Orf1 may render the protein nonfunctional and/or highly susceptible to proteolysis.
ACKNOWLEDGMENTS
We thank Howard Shuman and Peter Model for bacterial strains. We are grateful to Peter Model for numerous valuable discussions.
This work was supported in part by grants from the Arizona Disease Control Research Commission and from the Public Health Service (GM-RO1 48167-06) to R.M. and by Public Health Service grant AI-09644 to H.N. D.A.F. was a recipient of a predoctoral fellowship from NIGMS. J.C. was supported from a graduate fellowship from the Molecular and Cellular Biology program at ASU.
REFERENCES
- 1.Ames B N, Hartman P E, Jacob F. Chromosomal alterations affecting the regulation of histidine biosynthetic enzymes of Salmonella. J Mol Biol. 1963;7:23–42. doi: 10.1016/s0022-2836(63)80016-9. [DOI] [PubMed] [Google Scholar]
- 2.Benson S A, Occi J L, Sampson B A. Mutations that alter the pore function of the OmpF porin of Escherichia coli K-12. J Mol Biol. 1988;203:961–970. doi: 10.1016/0022-2836(88)90121-0. [DOI] [PubMed] [Google Scholar]
- 3.Benz R. Uptake of solutes through bacterial outer membranes. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier Science; 1994. pp. 397–423. [Google Scholar]
- 4.Blasband A J, Marcotte W R, Jr, Schnaitman C A. Structure of the lc and nmpC outer membrane porin protein genes of lambdoid bacteriophage. J Mol Biol. 1986;261:12723–12732. [PubMed] [Google Scholar]
- 5.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 6.Cowan S W, Schirmer T, Rummel G, Steiert M, Ghosh R, Paupit R A, Jansonius J N, Rosenbusch J P. Crystal structures explain functional properties of two E. coli porins. Nature. 1992;358:727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- 7.Higgins C F. ABC transporters: from microorganism to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 8.Highton P J, Chang Y, Marcotte W R, Jr, Schnaitman C A. Evidence that the outer membrane protein gene nmpC of Escherichia coli lies with the defective qsr′ prophage. J Bacteriol. 1985;162:256–262. doi: 10.1128/jb.162.1.256-262.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacob F, Ullman A, Monod J. Deletions fusing the lactose and purine operons of Escherichia coli. J Mol Biol. 1965;31:704–719. [Google Scholar]
- 10.Jap B K, Walian P J, Gehring K. Structural architecture of an outer membrane channel as determined by electron crystallography. Nature. 1991;350:167–170. doi: 10.1038/350167a0. [DOI] [PubMed] [Google Scholar]
- 11.Jovanovic G, Model P. The RIB element in the goaG-pspF intergenic region of Escherichia coli. J Bacteriol. 1997;179:3095–3102. doi: 10.1128/jb.179.10.3095-3102.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kohara Y, Akiyama K, Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a larger genomic library. Cell. 1987;50:495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- 13.Luckey M, Nikaido H. Specificity of diffusion channels produced by λ phage receptor protein of Escherichia coli. Proc Natl Acad Sci USA. 1980;77:167–171. doi: 10.1073/pnas.77.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lugtenberg B, Meijers J, Perters R, van der Hoek P, van Alphen L. Electrophoretic resolution of the major outer membrane proteins of Escherichia coli K-12 into four bands. FEBS Lett. 1975;58:254–258. doi: 10.1016/0014-5793(75)80272-9. [DOI] [PubMed] [Google Scholar]
- 15.Maier C, Bremer E, Schmid A, Benz R. Pore-forming activity of the Tsx protein from the outer membrane of Escherichia coli. Demonstration of a nucleoside-specific binding site. J Biol Chem. 1988;263:2493–2499. [PubMed] [Google Scholar]
- 16.Misra R, Benson S A. Isolation and characterization of OmpC porin mutants with altered pore properties. J Bacteriol. 1988;170:528–533. doi: 10.1128/jb.170.2.528-533.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Misra R, Benson S A. Genetic identification of pore domain of the OmpC porin of Escherichia coli K-12. J Bacteriol. 1988;170:3611–3617. doi: 10.1128/jb.170.8.3611-3617.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Misra R, Benson S A. A novel mutation, cog, which results in the production of a new porin protein (OmpG) of Escherichia coli K-12. J Bacteriol. 1989;171:4105–4111. doi: 10.1128/jb.171.8.4105-4111.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Misra R, Peterson A, Ferenci T, Silhavy T J. A genetic approach for analyzing the pathway of LamB assembly into the outer membrane of Escherichia coli. J Biol Chem. 1991;266:13592–13597. [PubMed] [Google Scholar]
- 20.Nakae T. Identification of the outer membrane protein of E. coli that produces transmembrane channels in reconstituted vesicle membranes. Biochem Biophys Res Commun. 1976;71:877–884. doi: 10.1016/0006-291x(76)90913-x. [DOI] [PubMed] [Google Scholar]
- 21.Nikaido H. Porins and specific diffusion channels in bacterial outer membrane. J Biol Chem. 1994;269:3905–3908. [PubMed] [Google Scholar]
- 22.Nikaido H, Rosenberg E Y. Effect on solute size on diffusion rates through the transmembrane pores of the outer membrane of Escherichia coli. J Gen Physiol. 1981;77:121–135. doi: 10.1085/jgp.77.2.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nikaido H, Rosenberg E Y. Porin channels in Escherichia coli: studies with liposomes reconstituted from purified proteins. J Bacteriol. 1983;153:241–252. doi: 10.1128/jb.153.1.241-252.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nikaido H, Rosenberg E Y, Foulds J. Porin channels in Escherichia coli: studies with beta-lactams in intact cells. J Bacteriol. 1983;153:232–240. doi: 10.1128/jb.153.1.232-240.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nikaido H, Vaara M. Molecular basis for bacterial outer membrane permeability. Microbiol Rev. 1985;49:1–32. doi: 10.1128/mr.49.1.1-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Overbeeke N, Lugtenberg B. Expression of outer membrane protein e of Escherichia coli K-12 by phosphate limitation. FEBS Lett. 1980;112:229–232. doi: 10.1016/0014-5793(80)80186-4. [DOI] [PubMed] [Google Scholar]
- 27.Paul C, Rosenbusch J P. Folding patterns of porin and bacteriorhodopsin. EMBO J. 1985;4:1593–1597. doi: 10.1002/j.1460-2075.1985.tb03822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosenbusch J P. Characterization of the major envelope protein from Escherichia coli: regular arrangement on the peptidoglycan and unusual dodecylsulfate binding. J Biol Chem. 1974;249:8019–8029. [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Sampson B A, Misra R, Benson S A. Identification and characterization of a new gene of Escherichia coli K-12 involved in outer membrane permeability. Genetics. 1989;122:491–501. doi: 10.1093/genetics/122.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schirmer T, Cowan S W. Prediction of membrane-spanning β-strands and its application to maltoporin. Protein Sci. 1993;2:1361–1363. doi: 10.1002/pro.5560020820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1984. [Google Scholar]
- 34.Singh S P, Miller S, Williams Y U, Rudd K E, Nikaido H. Immunochemical structure of the OmpD porin from Salmonella typhimurium. Microbiology. 1996;142:3201–3210. doi: 10.1099/13500872-142-11-3201. [DOI] [PubMed] [Google Scholar]
- 35.Singh S P, Upshaw Y, Abdullah T, Singh S R, Klebba P E. Structural relatedness of enteric bacterial porins assessed with monoclonal antibodies to Salmonella typhimurium OmpD and OmpC. J Bacteriol. 1992;174:1965–1973. doi: 10.1128/jb.174.6.1965-1973.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Struyve M, Moons M, Tommassen J. Carboxy-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. J Mol Biol. 1991;218:141–148. doi: 10.1016/0022-2836(91)90880-f. [DOI] [PubMed] [Google Scholar]
- 37.Sugawara E, Nikaido H. OmpA protein of Escherichia coli outer membrane occurs in open and closed channel forms. J Biol Chem. 1994;269:17981–17987. [PubMed] [Google Scholar]
- 38.Sugawara E, Steiert M, Rouhani S, Nikaido H. Secondary structure of the outer membrane proteins OmpA of Escherichia coli and OprF of Pseudomonas aeruginosa. J Bacteriol. 1996;178:6067–6069. doi: 10.1128/jb.178.20.6067-6069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thanassi D G, Cheng L W, Nikaido H. Active efflux of bile salts by Escherichia coli. J Bacteriol. 1997;179:2512–2518. doi: 10.1128/jb.179.8.2512-2518.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wandersman C, Schwartz M, Ferenci T. Escherichia coli mutants impaired in maltodextrin transport. J Bacteriol. 1979;140:1–13. doi: 10.1128/jb.140.1.1-13.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weiner L, Brissette J, Model P. Stress-induced expression of the Escherichia coli phage shock protein operon is dependent on Sigma-54 and modulated by positive and negative feedback mechanisms. Genes Dev. 1991;5:1912–1923. doi: 10.1101/gad.5.10.1912. [DOI] [PubMed] [Google Scholar]
- 42.Yoshimura H, Zelman L S, Nikaido H. Purification and properties of Pseudomonas aeruginosa porin. J Biol Chem. 1983;258:2308–2314. [PubMed] [Google Scholar]