Abstract
The 70-kDa heat shock protein (Hsp) family in all Drosophila species includes 2 environmentally inducible family members, Hsp70 and Hsp68. Two-dimensional gel electrophoresis revealed an unusual pattern of heat shock– inducible proteins in the species of the virilis group. Trypsin fingerprinting and microsequencing of tryptic peptides using ProteinChip Array technology identified the major isoelectric variants of Hsp70 family, including Hsp68 isoforms that differ in both molecular mass and isoelectric point from those in Drosophila melanogaster. The peculiar electrophoretic mobility is consistent with the deduced amino acid sequence of corresponding hsp genes from the species of the virilis group.
INTRODUCTION
The stress response induces the rapid synthesis of heat shock proteins (Hsps) in virtually all organisms to protect cellular proteins against denaturation (Lindquist and Craig 1988; Feder and Hofman 1999). The Hsps and other molecular chaperones have been widely studied in many fields of biology and numerous publications are available on their molecular and physiological functions (Feder and Krebs 1997). Molecular studies of Hsps in eukaryotic organisms indicate a high degree of conservation during evolution, especially for the best studied 70-kDa protein family (Lindquist and Craig 1988; Hightower 1991; Sorensen et al 2003). This family plays an important role in protein chaperoning and acquired thermotolerance (Garbuz et al 2002, 2003; Feder and Hofman 1999). Two distinct inducible molecular chaperones of the Hsp70 family occur in various Drosophila species. The first, Hsp70, is prone to duplication during evolution, with 2–3 additional copies in addition to the ancestral pair of inversely oriented genes depending on the species (Bettencourt and Feder 2001, 2002). The second, Hsp68, is usually encoded by a single gene distinct from the Hsp70 locus both in D melanogaster (Lindquist and Craig 1988) or in the virilis group species (Evgen'ev et al 1978; Ranz et al 1999). Most molecular and genetic investigations of the Hsp70 family focus on the Hsp70 locus; few address the role of Hsp68 in stress resistance and heat hardening (McColl et al 1996). Our previous investigations reported variation in thermotolerance and the heat shock response in the species of the virilis group of Drosophila (Garbuz et al 2002, 2003). The virilis group comprises 12 species living in greatly differing habitats (Patterson and Stone 1952; Throckmorton 1982; Spicer 1992). After heat shock, the low-latitude and probably ancestral species D virilis exceeds the high-latitude species D lummei in basal thermotolerance, the temperature threshold for heat shock factor activation, and other parameters, whereas the desert species D novamexicana differs from the mesic (pertaining to moderate habitats) species D texana in the same manner for many other traits. Hsp70 is quantitatively the major Hsp in all virilis group species and strains examined (Garbuz et al 2003). Two-dimensional electrophoresis of 35S-labeled proteins has shown that more thermotolerant species of the group (ie, D virilis and D novamexicana) synthesized higher levels of Hsps after severe heat shock than species from the moderate and cold climatic zones (D lummei and D montana). The genetic basis of stress resistance in closely related species is likely to be complex, and these differences are manifest in all major classes of Hsps that are distinguishable in 2-dimensional electrophoresis (Garbuz et al 2003).
Detailed examination of the Hsp70 family members revealed that the differences between Hsp70s of D virilis and other species (eg, D lummei) are qualitative as well as quantitative (Garbuz et al 2003). Inducible members of Hsp70 family are represented by various isoforms, not all of them recognized by an inducible Hsp70-specific antibody. This study investigates these isoelectric variants in several species of the virilis group of Drosophila. Therefore, we used microsequencing and trypsin fingerprinting to identify the individual members of Hsp70 family.
RESULTS AND DISCUSSION
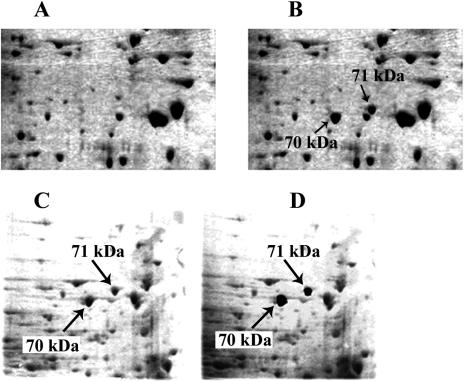
Immunoblotting with antibody 7FB, which in D melanogaster reacts only with inducible Hsp70s, detects various proteins in D virilis and other species investigated (Fig 1A; Table 1). Interestingly, the number of detected isoforms varies with species and strain. Thus D virilis strain 9 has 3 such isoforms, which presumably correspond to Hsp70, whereas strain 1433 has only 1. Furthermore, D virilis strain 9 also exhibits 2 inducible isoforms (vs 1 in D virilis strain 1433) having slightly higher molecular mass (71 kDa), not recognizable by 7FB, but recognizable by antibody 7.10.3, which reacts with all Hsp70 family members in most Drosophila species examined, D lummei and other species of the group examined in this study exhibit only 1 such higher molecular weight isoform, which characteristically has a more acidic isoelectric point than presumptive Hsp70 (Fig 1B). The inducible nature of this isoform was also revealed by silver staining (Fig 2A,B) and 35S-labeling experiments (data not shown). We hypothesized that this particular group of proteins corresponds to D melanogaster Hsp68. To test this hypothesis, we excised the 70-kDa protein spot (recognized by 7FB) and 71-kDa protein spot (inducible but not recognized by 7FB) and performed fingerprinting and microsequencing analysis. These experiments used D virilis strain 1433, in which both inducible proteins of interest (70 kDa and 71 kDa) are represented by single isoforms, which are well separated on the gel and easy to isolate (Fig 2A,B).
Fig 1.
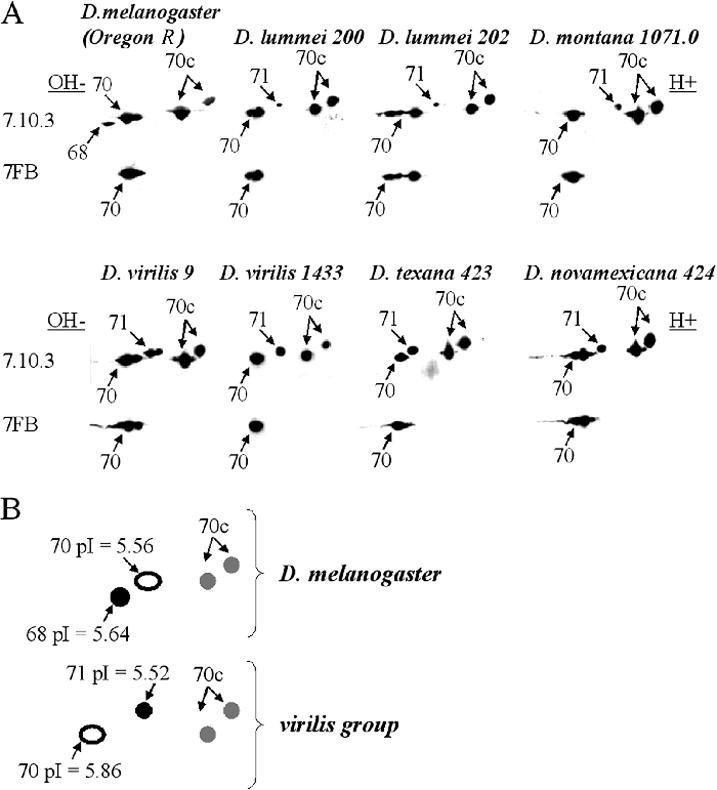
Major isoelectric variants of the heat shock protein 70 (Hsp70) family in various species of Drosophila. (A) Immunoblots of Hsp70 family members in strains and species. Primary antibody 7FB recognizes only the 70-kDa–inducible Hsp70 family member in Drosophila melanogaster, and primary antibody 7.10.3 recognizes all Hsp70 members (see Materials and Methods). Heat shock was 38.5°C for 30 minutes, with 3 hours recovery at 25°C before lysis. Arrows show the position of unidentified members of Hsp70 family with molecular masses indicated. Two-dimensional gel electrophoresis and other procedures applied were as described (O'Farrell et al 1977; Ulmasov et al 1992; Zatsepina et al 2001; Garbuz et al 2003). The position of major Hsps was determined by both immunoblotting and staining of gels with silver or Coomassie G-250 (Creighton 1990). After sodium dodecyl sulfate–polyacrylamide gel electrophoresis of fly lysates prepared as above, the proteins were transferred to nitrocellulose membrane (Hybond ECL; Amersham, Chalfont St. Giles, Bucks, UK) according to the manufacturer's protocol and reacted with monoclonal antibodies specific to the entire Drosophila Hsp70 family (7.10.3) and only inducible form of Hsp70 (7FB) as previously described (Zatsepina et al 2001). It is noteworthy that 7FB antibodies do not recognize Hsp68 either in D melanogaster or in other Drosophila species investigated. Immune complexes were detected via chemiluminescence (ECL kit; Amersham) with appropriate peroxidase-conjugated anti-rat secondary antibodies. (B) Schematic illustration of various groups of proteins belonging to Hsp70 family. Black filled circles represent inducible proteins recognized by both 7.10.3 and 7FB, open circles represent inducible proteins recognized by 7.10.3 only, and gray circles represent constitutively present proteins recognized by 7.10.3 only. Isoelectric points of the proteins of interest as well as molecular masses were calculated using DNAStar and GeneRunner programs basing on the amino acid content of D virilis Hsp70 and D lummei Hsp68 proteins deduced from the sequence of the correspondent genes (accession # AY445083 and AY395705) cloned and sequenced in our laboratory.
Table 1.
Drosophila strains examined in the present study
Fig 2.
Two-dimensional (2D) electrophoretic separation of total proteins isolated from adult flies (A) Control 25°C; (B) heat shock 38.5°C 30 minute plus 3 hours of recovery at 25°C. Silver stain. Inducible heat shock protein 70 members are indicated by arrows. Gel resulted from 2D separation of proteins (Drosophila virilis strain 1433) stained with Coomassie G-250 used for the isolation of 2 electromorphs of interest. (C) Gel before isolation. (D) The same gel with the excised spots indicated by arrows
Positive identification of these proteins used surface-enhanced laser desorption ionization–time-of-flight (SELDI-TOF) technology provided by Ciphergen Biosystems Inc, Fremont, CA. This technology combines chromatography on the chemically active Protein Chip™ array surface with laser desorption ionization TOF mass spectrometry (MS). After in-gel trypsin digestion and mass fingerprinting analysis using linear SELDI-MS, the National Center for Biotechnology Information database was searched with the Profound search engine.
This search established that, unsurprisingly, the 70-kDa spot is the Hsp70 Drosophila protein. Of 32 peptides compared with the database, 14 were a correct match, covering 31% of the Hsp70 of Drosophila. The 71-kDa protein in D virilis samples best matched Drosophila Hsp68, with a probability score of 1.00e + 00 and 8 of 22 introduced peptides matching 15% of Drosophila's Hsp68 protein sequence.
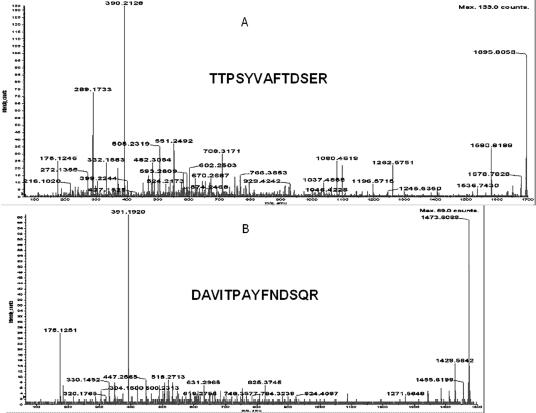
Both identifications were confirmed by microsequencing of tryptic peptides using the same type of ProteinChip™ array and a PCI-1000 interface (Ciphergen Biosystems Inc.) to a Q-star (ABI, Sciex) tandem MS instrument. In Figure 3A, the results of such analysis of the parental ion of 1473.6203 is shown together with the peptide sequence obtained. Search of the NCBI database with the Mascot search engine gave a Mowse score of 80 for this peptide and unequivocally identified it as Hsp70 of Drosophila. Six more peptides were microsequenced, and all matched Hsp70 of Drosophila.
Fig 3.
Microsequencing of the proteins which were identified as heat shock protein (Hsp)70 (A) and Hsp68 (B) using trypsin fingerprinting and database search. The determined sequences are presented in bold. Protein identification was done by trypsin fingerprinting using surface-enhanced laser desorption ionization–time-of-flight–mass spectrometry (SELDI-TOF-MS) (Ciphergen Biosystems Inc.) followed by NCBI database search using the Profound search engine and confirmed by SELDI-TOF-MS MS (Ciphergen Biosystems Inc.) analysis. All proteins used in these experiments were obtained as gel slices after two-dimensional sodium dodecyl sulfate–polyacrylamide gel electrophoresis stained with Coomassie Blue (G-250). “No protein” control gel slices were included into each experiment. Gel slices were processed exactly as recommended in Ciphergen product insert to Protein ID kit up to gel drying step (Stage B, point 7). The following changes were introduced to the subsequent steps of trypsin in-gel digestion and peptide recovery: (1) trypsin (Roche Applied Science, Basel, Switzerland) was dissolved in water to the final concentration of 1 μg/μL; (2) dried and crushed gel slices were resuspended in 50 μL of 50 mM ammonium bicarbonate buffer, pH 8.0 (freshly prepared) containing 12.5 ng/μL of trypsin and digested for 16 hours at 37°C; (3) supernatants were collected and freeze-dried in SpeedVac for 20 minutes; (4) peptides were dissolved in 10 μL of H2O, 3 μL was applied per spot of H4 ProteinChip® array (Ciphergen Biosystems Inc.), and air dried; (5) each spot of the array was washed with 5 μL of 5% acetonitrile in water 2 times and air dried; and (6) 1 μL of 20% CHCA in 50% acetonitrile–0.5% trifluoroacetic acid (TFA) was applied per spot for Protein Chip Array Reader (PBSII) SELDI-TOF (Ciphergen Biosystems Inc.) analysis. One microliter of saturated cyano-4-hydroxy-cinnamic acid (CHCA) in the same solution was added per spot of array followed by microsequencing using PCI-1000 interface (Ciphergen Biosystems Inc.) to QStar I (ABI Sciex) tandem MS instrument. PBSII was calibrated externally with All-in-1 peptide standard (Ciphergen Biosystems Inc.) spot by spot, and calibration equations for each spot were saved separately. After calibration, the same chip was reread automatically twice and calibration equations for each spot were applied, correspondingly, each time. Recalibration and change of the equation were introduced, if needed. The experimental chip was read in automatic mode using laser intensity 190 and sensitivity 9. Saved calibration equations were applied to each spot, correspondingly. All peaks with signal to noise ratio above 2.5 were labeled automatically and peptides masses were used for NCBI database search using Profound (old or new version) for trypsin fingerprinting identification. All labeled peak mass values were included into search using automatic introduction by Ciphergen ProteinChip™ software 3.0. Search was done using the following parameters: Cys modified by acrylamide, mass error 1.0 Da, and number of miscleavages −1. The same database was searched using Mascot search engine and masses of fragmented parent ions produced by PCI-1000-QStar analysis to confirm the identification done by trypsin fingerprinting analysis.
Figure 3B shows the results of microsequencing of 1695.8058 parental ion of the protein identified as Hsp68 by trypsin fingerprinting, single MS analysis, and SwissProt database search by Profound. The search of the NCBI database by Mascot identified this peptide as a product of the hsp68 gene of Drosophila with a Mowse score of 71. Five more peptides were microsequenced, and all belonged to Hsp68 of Drosophila. Thus, both trypsin fingerprinting and tandem MS-MS analysis unequivocally identified 2 proteins under investigation as Drosophila Hsp70 and Hsp68.
To explain the observed mobility of the 2 Hsps, we have deduced an amino acid sequence of D virilis Hsp70 and Hsp68 using the corresponding clones recently sequenced in our laboratory (Velikodvorskaya et al, personal communication). The analysis showed that the peculiar electrophoretic behavior of these proteins is easily explained by the amino acid number and content and does not represent the result of posttranslational modification. A small insertion in the sequence of the hsp68 gene of D lummei, accession #AY395705 (and presumably in all other species of the virilis group) apparently accounts for this peculiar electrophoretic behavior (Figure 1).
The induction pattern of Hsp70 family proteins in various species (Fig 1A; Table 1) demonstrates that species from moderate and cold climates (D lummei and D montana) usually produce significantly lower levels of Hsp68 after heat shock than species from high temperature zones (D virilis, D texana, and D novamexicana).
The identification of the proteins is a major challenge in proteomics. Several procedures and novel technologies have been developed, and in this study, we report novel experimental methods to identify peptides with the ProteinChip Array technique. The approach exemplified here (trypsin fingerprinting and microsequencing) is of particular use in resolving diverse constitutive and inducible forms belonging to 1 protein family (eg, Hsp70 family) when other techniques cannot identify the isoforms appearing under different conditions (Ulmasov et al 1992; Norris et al 1995). The approach is also of great value for understanding evolutionary variation among proteins, particularly when they have similar functions.
Acknowledgments
We are grateful to Dr Martin Feder of the University of Chicago for many helpful suggestions and editing of the whole manuscript.
This work was supported by grants of Russian Academy of Sciences for Basic Science 02-04-49121 and 03-04-48918 and grant provided by program of Physico-Chemical Biology of Russian Academy of Sciences to M.B.E.
REFERENCES
- Bettencourt BR, Feder ME. Hsp70 duplication in the Drosophila melanogaster species group: how and when two become five? Mol Biol Evol. 2001;18:1272–1282. doi: 10.1093/oxfordjournals.molbev.a003912.0737-4038(2001)018<1272:HDITDM>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Bettencourt BR, Feder ME. Rapid concerted evolution via gene conversion at the Drosophila hsp70 genes. J Mol Evol. 2002;2:93–102. doi: 10.1007/s00239-001-0044-7.0022-2844(2002)002<0093:RCEVGC>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Creighton TE. 1990 Protein Structure: A Practical Approach. IRL Press, Oxford, England. [Google Scholar]
- Evgen'ev M, Kolchinski A, Levin AV, Preobrazhenskaya OL, Sarkisova E. Heat-shock DNA homology in distantly related species of. Drosophila. Chromosoma. 1978;68:357–365. doi: 10.1007/BF00327170. [DOI] [PubMed] [Google Scholar]
- Feder M, Hofman G. Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu Rev Physiol. 1999;61:243–282. doi: 10.1146/annurev.physiol.61.1.243.0066-4278(1999)061<0243:HPMCAT>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Feder ME, Krebs RA. Ecological and evolutionary physiology of heat shock proteins and the stress response in Drosophila: complementary insights from genetic engineering and natural variation. EXS. 1997;83:155–173. doi: 10.1007/978-3-0348-8882-0_9.0071-335X(1997)083<0155:EAEPOH>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Garbuz D, Evgen'ev M, Feder M, Zatsepina O. Evolution of thermotolerance and heat shock-response: evidence from inter/ intraspecific comparison and interspecific hybridization in the virilis species group of Drosophila. I. Thermal phenotype. J Exp Biol. 2003;206:2399–2408. doi: 10.1242/jeb.00429.0022-0949(2003)206<2399:EOTAHS>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Garbuz DG, Molodsov VB, Velikodvorskaia VV, Evgen'ev MB, Zatsepina OG. Evolution of response to heat shock in genus. Drosophila. Genetica (Russ.) 2002;38:925–936. [PubMed] [Google Scholar]
- Hightower L. Heat shock, stress proteins, chaperones and proteotoxicity. Cell. 1991;66:191–197. doi: 10.1016/0092-8674(91)90611-2.0092-8674(1991)066<0191:HSSPCA>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Lindquist S, Craig EA. The heat shock proteins. Annu Rev Genet. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215.0066-4197(1988)022<0631:THSP>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- McColl G, Hoffmann AA, McKechie SW. Response of two heat shock genes to selection for knockdown heat resistance in. Drosophila melanogaster. Genetics. 1996;143:1615–1627. doi: 10.1093/genetics/143.4.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris CE, DiIorio PJ, Schultz RJ, Hightower LE. Variation in heat shock proteins within tropical and desert species of poeciliid fishes. Mol Biol Evol. 1995;12:1048–1062. doi: 10.1093/oxfordjournals.molbev.a040280.0737-4038(1995)012<1048:VIHSPW>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- O'Farrell PZ, Goodman HM, O'Farrell PH. High-resolution two-dimensional electrophoresis of basic as well as acidic proteins. Cell. 1977;12:1133–1142. doi: 10.1016/0092-8674(77)90176-3.0092-8674(1977)012<1133:HTEOBA>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Patterson JT, Stone WS. 1952 Evolution in the Genus Drosophila. Macmillan, New York. [Google Scholar]
- Ranz JM, Caceres M, Ruiz A. Comparative mapping of cosmids and gene clones from a 1.6 Mb chromosomal region of Drosophila melanogaster in three species of the distantly related subgenus. Drosophila. Chromosoma. 1999;108:32–43. doi: 10.1007/s004120050349. [DOI] [PubMed] [Google Scholar]
- Sorensen JG, Kristensen TN, Loeschcke V. The evolutionary and ecological role of heat shock proteins. Ecol Lett. 2003;6:1025–1037.1461-023X(2003)006<1025:TEAERO>2.0.CO;2 [Google Scholar]
- Spicer GS. Reevaluation of the phylogeny of the Drosophila– virilis species group (Diptera, Drosophilidae) Ann Entomol Soc Am. 1992;85:11–25.0013-8746(1992)085<0011:ROTPOT>2.0.CO;2 [Google Scholar]
- Throckmorton LH. 1982 The virilis species group. In: The Genetics and Biology of Drosophila 3d, vol 3b, ed Ashburner M, Carson HL, Thompson JN. Academic Press Inc, London, 227–296. [Google Scholar]
- Ulmasov KA, Shammakov S, Karaev K, Evgen'ev MB. Heat shock proteins and thermoresistance in lizards. Proc Natl Acad Sci U S A. 1992;89:1666–1670. doi: 10.1073/pnas.89.5.1666.0027-8424(1992)089<1666:HSPATI>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zatsepina OG, Velikodvorskaia VV, Molodtsov VB, Garbuz D, Lerman DN, Bettencourt BR, Feder ME, Evgen'ev MB. A Drosophila melanogaster strain from sub-equatorial Africa has exceptional thermotolerance but decreased Hsp70 expression. J Exp Biol. 2001;204:1869–1881. doi: 10.1242/jeb.204.11.1869.0022-0949(2001)204<1869:ADMSFS>2.0.CO;2 [DOI] [PubMed] [Google Scholar]