Abstract
The phosphoenolpyruvate-dependent phosphotransferase system (PTS) plays a major role in the ability of Escherichia coli to migrate toward PTS carbohydrates. The present study establishes that chemotaxis toward PTS substrates in Bacillus subtilis is mediated by the PTS as well as by a methyl-accepting chemotaxis protein (MCP). As for E. coli, a B. subtilis ptsH null mutant is severely deficient in chemotaxis toward most PTS carbohydrates. Tethering analysis revealed that this mutant does respond normally to the stepwise addition of a PTS substrate (positive stimulus) but fails to respond normally to the stepwise removal of such a substrate (negative stimulus). An mcpC null mutant showed no response to the stepwise addition or removal of d-glucose or d-mannitol, both of which are PTS substrates. Therefore, in contrast to E. coli PTS carbohydrate chemotaxis, B. subtilis PTS carbohydrate chemotaxis is mediated by both MCPs and the PTS; the response to positive stimulus is primarily McpC mediated, while the duration or magnitude of the response to negative PTS carbohydrate stimulus is greatly influenced by components of the PTS and McpC. In the case of the PTS substrate d-glucose, the response to negative stimulus is also partially mediated by McpA. Finally, we show that B. subtilis EnzymeI-P has the ability to inhibit B. subtilis CheA autophosphorylation in vitro. We hypothesize that chemotaxis in the spatial gradient of the capillary assay may result from a combination of a transient increase in the intracellular concentration of EnzymeI-P and a decrease in the concentration of carbohydrate-associated McpC as the cell moves down the carbohydrate concentration gradient. Both events appear to contribute to inhibition of CheA activity that increases the tendency of the bacteria to tumble. In the case of d-glucose, a decrease in d-glucose-associated McpA may also contribute to the inhibition of CheA. This bias on the otherwise random walk allows net migration, or chemotaxis, to occur.
In enteric bacteria, chemotaxis toward many carbohydrate attractants is dependent upon components of the phosphoenolpyruvate (PEP)-dependent phosphotransferase system (PTS) (1, 9, 15). This carbohydrate transport system consists of an autophosphorylating histidine kinase, EnzymeI, a common phosphocarrier protein, HPr, and a number of substrate-specific transporters, the EnzymeII complexes. At the expense of PEP, EnzymeI autophosphorylates on a histidine residue and transfers this phosphoryl group to a histidine residue on HPr. HPr-P then donates this phosphoryl group to a carbohydrate-specific EnzymeII complex. The carbohydrate substrate is the final phosphoryl group acceptor, as it is transported into the cell and is concomitantly phosphorylated by EnzymeII (13).
Chemotaxis is also controlled by a phosphoryl transfer cascade. CheA, in response to an attractant- or repellent-bound receptor (methyl-accepting chemotaxis protein [MCP]), alters its rate of autophosphorylation appropriately to transiently increase or decrease the intracellular CheY-P pool and thereby modulate swimming behavior (4, 16). In enteric bacteria, increased CheY-P leads to tumbling (19). In Bacillus subtilis, increased CheY-P leads to smooth swimming (3). In enteric bacteria, chemotaxis toward PTS substrates requires CheA, CheY, EnzymeI, and HPr but does not depend on the presence of an MCP (12, 18). These observations have led investigators to suggest that the changes in the phosphorylation state of PTS components that accompany carbohydrate transport regulate CheA activity (10).
Recent work has provided the following model for the role of the PTS in chemotaxis toward its substrates in Escherichia coli. As the bacteria encounter a PTS carbohydrate, HPr dephosphorylates EnzymeI faster than the latter protein can be rephosphorylated. The resulting increase in unphosphorylated EnzymeI and the resulting decrease in PEP both function to decrease the rate of CheA autophosphorylation. This is believed to lead to a transient decrease in the CheY-P pool that suppresses tumbling, allowing the bacteria to move up the carbohydrate gradient (10).
This article describes studies on the process of carbohydrate chemotaxis in B. subtilis. In particular, we provide evidence that McpC is absolutely required for any response to all of the PTS carbohydrates tested. This is surprising considering the fact that McpC has previously been shown to also mediate chemotaxis toward eight different amino acids (11). McpA has previously been shown to partially mediate chemotaxis toward glucose (7). This result is confirmed in the present study with the use of direct behavioral assays. Our results suggest the existence of a multidimensional signaling mechanism involving both the PTS and specific MCPs, an unprecedented finding in the study of the molecular control of bacterial carbohydrate chemotaxis.
MATERIALS AND METHODS
Construction of mutants.
Plasmid pDG491 (5), containing a chloramphenicol acetyltransferase cassette in a segment of DNA ∼6.0 kb from ptsH, was used to transform GM1341, a strain containing an in-frame deletion of ptsH. Chloramphenicol-resistant colonies (OI3306) were selected on TBAB plates (1% Bacto Tryptose, 0.3% Bacto Beef Extract, 0.5% NaCl). Chromosomal DNA was isolated from this strain and used to transform OI1085 (wild type). Again, colonies were selected for chloramphenicol resistance. About 60% of these clones were unable to grow on minimal medium plates containing d-mannitol (a PTS substrate) as the sole carbon source (diagnostic for ptsH null mutants). This is strain OI3302 (ΔptsH).
A B. subtilis mcpA ptsH null mutant strain was also constructed. The HindIII/BamHI fragment from pDW4 (7) containing a portion of mcpA was cloned into pBluescript. The HindIII/BamHI fragment from this plasmid was then cloned into pHV501 (integration plasmid conferring erythromycin resistance [5a]) and used to transform OI3302. Colonies were selected for chloramphenicol and erythromycin resistance as well as for an inability to grow on d-mannitol minimal plates (OI3304 ΔmcpA ΔptsH). The mcpA null mutation in this strain was also verified with an in vivo methylation assay (7).
The full-length B. subtilis cheA gene was engineered to include an NdeI site at the start codon and a BamHI site following the stop codon. This fragment was gel purified in a low-melting-point TAE (0.4 M Tris-acetate, 1 mM EDTA [pH 8.0]) agarose gel and cloned into pT7-7 (17) to generate a vector expressing the native CheA protein (pLG104). This plasmid was introduced into E. coli RP3098 (ΔflhD ΔflhA) harboring pGp1-2, encoding the T7 RNA polymerase, to create strain OI3245.
Purification of native B. subtilis CheA and EnzymeI.
Native B. subtilis CheA was purified from strain OI3245. Cells were grown in T7 expression broth (2% Bacto Tryptone, 1% yeast extract, 0.5% NaCl, 0.2% [wt/wt] glycerol, 50 mM potassium phosphate [pH 7.2], 50 μg of kanamycin per ml) to an optical density at 595 nm of 1.5 at 30°C. The temperature was raised to 42°C for 45 min and reduced to 37°C for an additional 90 min. The cells were harvested and washed with ice cold buffer A (20 mM Tris [pH 7.5], 20% [wt/wt] sucrose, 1 mM EDTA [30 ml of culture per liter]). The cells were pelleted and washed in ice-cold distilled water (30 ml of culture per liter). Following centrifugation at 8,000 × g for 15 min, the cells were resuspended in ice-cold buffer P (0.1 mM NaCl, 3 mM KCl, 20 mM Na2HPO4 [pH 7.5], 0.5 mM phenylmethylsulfonyl fluoride, 1 μg of leupeptin per ml, 20 μg of aprotinin per ml [8 ml of culture per liter]). The cell suspension was sonicated, and RNase A and DNase I (Sigma Chemical Co.) were added to a final concentration of 40 μg/ml. After a 15-min incubation at room temperature, the solution was diluted with an equal volume of fresh buffer P and centrifuged at 13,000 × g for 30 min at 4°C.
Solid ammonium sulfate was slowly added to the supernatant at 4°C with stirring to 20% saturation. The solution was left to stir for an additional hour at 4°C and then centrifuged at 3,000 × g for 40 min (4°C). Solid ammonium sulfate was added to the resulting supernatant to 50% saturation. After 1 h of stirring at 4°C, the solution was centrifuged as before and the pellet was resuspended in 8 ml of 20 mM Tris, pH 7.5. This solution was dialyzed overnight against 4 liters of the same buffer at 4°C and applied to a Mem-Sep 1010 Cartridge Anion Exchanger (Millipore) equilibrated in the same Tris buffer and driven by a Waters 510 HPLC pump. The column was eluted with a gradient of 0 to 0.5 M NaCl in the same Tris buffer at a flow rate of 5 ml/min (total run time was 40 min).
CheA-containing fractions were pooled, concentrated, and dialyzed at 4°C against 4 liters of 20 mM Tris (pH 7.5)–5 mM MgCl2. The CheA solution was then applied to a 2-ml Hi-Trap Blue column (Pharmacia) and eluted with a step gradient of 0 to 2 M NaCl in the same Tris buffer in 100 mM increments. CheA-containing fractions were concentrated and applied to a Shodex KW-803 gel filtration column (Shoko Co., Ltd.) equilibrated in TKMD buffer (50 mM Tris [pH 8.0], 5 mM MgCl2, 50 mM KCl, 0.2 mM dithiothreitol, 10% [wt/wt] glycerol) at a flow rate of 0.25 ml/min. Purified CheA was stored in small aliquots at −70°C. All proteins were quantified with the Coomassie protein assay reagent (Pierce) and bovine serum albumin as a standard.
B. subtilis EnzymeI was purified as previously described (14) and was judged to be about 90% pure through sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie staining.
Western blotting to confirm the identity of CheA.
A CheA–glutathione S-transferase (GST) fusion protein (6) was purified (1.5 mg) and subjected to SDS-PAGE. The gel was Coomassie stained, and the CheA-GST fusion protein was excised from the gel. The gel slice was sent to CoCalico Biologicals, Inc. (Reamstown, Pa.), for antibody production in New Zealand rabbits. Both immune and preimmune sera were tested for the ability to react with both the native CheA and the CheA-GST fusion proteins in Western blots. Cellular extracts from OI3245 and OI3246 (6), both induced and uninduced (∼50 μg of protein), were fractionated by SDS-PAGE and transferred to polyvinylidene difluoride nitrocellulose with Tris-methanol buffer. A dilution of 1:50,000 was used for the rabbit anti-GST–CheA primary antibody in this experiment. The secondary antibody used in this experiment was a goat anti-rabbit immunoglobulin G antibody conjugated to alkaline phosphatase at a dilution of 1:7,500. The Western blots were developed through the addition of 5-bromo-4-chloro-3-indolylphosphate and nitroblue tetrazolium chloride to the polyvinylidene difluoride membrane (Millipore).
Capillary assays.
Strains were streaked on TBAB plates (with antibiotic where appropriate) and grown overnight at 30°C. Single colonies were resuspended in 2 ml of TBr (1% Bacto Tryptone, 0.5% NaCl) and diluted 1:50 in minimal medium (7) containing 20 mM d-glucitol as the only carbon source. d-Glucitol was used in place of glycerol in these experiments since the ptsH null mutant strains (OI3302, OI3304) were found to grow poorly in glycerol minimal medium (4a). Cultures were grown for 4 h and washed in chemotaxis buffer (7) containing 100 μg of chloramphenicol per ml. The cells were finally resuspended in chemotaxis buffer with chloramphenicol to an optical density at 600 nm of 0.001 and tested for chemotaxis toward a number of carbohydrates, as previously described (7). The data presented in Table 1 are averages of two experiments done in triplicate. The reported errors are standard deviations of the values obtained in the six trials. The numbers reported for each carbohydrate in Table 1 are the numbers of bacteria that migrated into the capillary tube containing the carbohydrate at the indicated concentration.
TABLE 1.
Capillary assays measuring responses of che and pts mutants to various carbohydrates
| Effector | Concn (M) | No. of bacteria ± SEMa
|
||||
|---|---|---|---|---|---|---|
| OI1085 (wild type) | OI3055 (ΔmcpA) | OI3302 (ΔptsH) | OI3304 (ΔmcpA ΔptsH) | OI3280 (ΔmcpC) | ||
| d-Glucose | 3.2 × 10−4 | 1,450 ± 340 | 1,465 ± 253 | 1,711 ± 378 | 71 ± 28 | 0 |
| 1.0 × 10−4 | 852 ± 325 | 1,190 ± 213 | 820 ± 96 | 17 ± 6 | ND | |
| 3.2 × 10−5 | 268 ± 114 | 530 ± 123 | 363 ± 155 | 20 ± 16 | ND | |
| α-Methyl-d-glucoside | 1.0 × 10−4 | 566 ± 66 | 830 ± 83 | 12 ± 10 | 12 ± 2 | 0 |
| d-Mannitol | 3.2 × 10−5 | 978 ± 285 | 940 ± 242 | 2 ± 2 | 15 ± 9 | 0 |
| d-Fructose | 3.2 × 10−5 | 926 ± 160 | 1,213 ± 260 | 3 ± 2 | 6 ± 3 | 0 |
| Asparagine | 1.0 × 10−5 | 2,953 ± 124 | 3,128 ± 360 | 1,151 ± 248 | 1,057 ± 143 | 5,026 ± 830 |
ND, not done.
Phosphorylation of EnzymeI and in vitro inhibition of CheA.
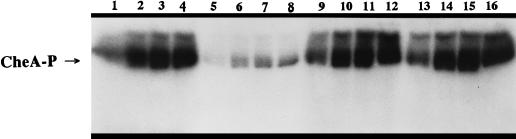
Purified B. subtilis EnzymeI (15 μg) was incubated with or without 2 mM PEP in a solution of 50 mM potassium phosphate (pH 6.5), 0.1 mM EDTA, 0.2 mM dithiothreitol, and 5 mM MgCl2 at 37°C for 20 min in a total volume of 50 μl. EnzymeI or EnzymeI-P (1.5 μg) was added to 2.4 μg of CheA in TKMD buffer (60 μl total volume). EnzymeI-P added in these experiments refers to the addition of 5 μl of the reaction mixture described above to a separate reaction vessel containing CheA. [γ-32P]ATP (0.1 mM, ∼10,000 cpm/pmol) was added, aliquots were removed at the indicated times (see the legend to Fig. 4), and reactions were stopped with the addition of 10 μl of 4× SDS-EDTA buffer. These reaction mixtures were separated by SDS-PAGE and exposed to film for 5 h. CheA-P present at each time point was quantified by excising the radiolabelled bands from the gel and subjecting them to scintillation counting.
FIG. 4.
Inhibition of CheA autophosphorylation by EnzymeI-P in vitro. The assay was performed as described in Materials and Methods. Lanes 1 to 4 are 10-s time points in the presence of EnzymeI. Lanes 5 to 8 are 10-s time points in the presence of EnzymeI-P. Lanes 9 to 12 are 10-s time points in the presence of 0.2 mM PEP. Lanes 13 to 16 are 10-s time points in the presence of EnzymeI buffer without PEP. All time courses are shown from the earliest time point (10 s) to the latest time point (40 s).
RESULTS
The ptsH null mutant responds abnormally only to negative PTS carbohydrate stimulus.
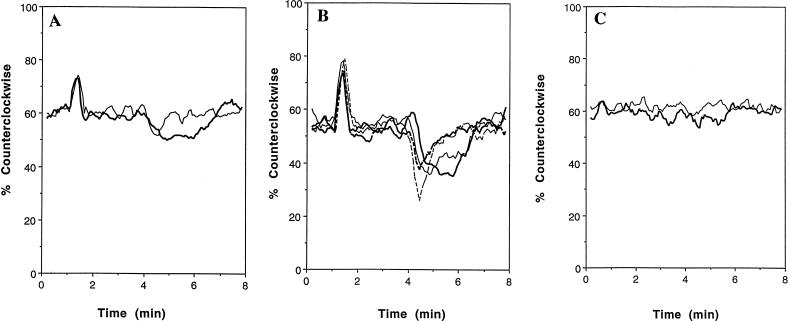
Strains OI1085 (wild type) and OI3302 (ΔptsH) were tested for the ability to carry out chemotaxis toward a number of PTS carbohydrates with a capillary assay. Chloramphenicol was added to the cells subjected to this assay to ensure that there was no induction of genes involved in PTS carbohydrate chemotaxis in the wild-type strain during the capillary assay, which could possibly lead to artifacts (since the ptsH null mutant strains cannot import PTS carbohydrates and induce expression of the same genes). As the data indicates, there is sufficient chemotaxis by uninduced cells to compare the abilities of the wild type and the various ptsH null mutants to perform chemotaxis toward these carbohydrates. The ptsH null mutant was found to be defective in chemotaxis toward all tested PTS substrates except d-glucose (Table 1). We predicted that this strain would therefore not respond to positive PTS substrate stimulus (would not increase its smooth swimming bias upon addition of the carbohydrate). Tethering analysis revealed, however, that the ptsH null mutant was exciting and adapting normally to the addition of d-mannitol (positive stimulus) (Fig. 1A). The chemotactic defect in this strain was in its prompt adaptation to the removal of d-mannitol (negative stimulus). While the wild-type strain required nearly 3 min to adapt to the removal of d-mannitol, the ptsH null mutant strain was able to adapt to this stimulus in just under 1 min (Fig. 1A).
FIG. 1.
Behaviors of tethered cells toward various carbohydrates. The average counterclockwise rotation of 30 cells for each strain was determined with a computer program. The average values for 4-s intervals were plotted against time. (A) Response of tethered OI1085 (wild type) (heavy line) and OI3302 (ΔptsH) (thin line) to the PTS carbohydrate d-mannitol. d-Mannitol (3.2 × 10−5 M) was added at 1 min and removed at 4 min. (B) Response of tethered OI1085 (wild type) (heavy solid line) OI3055 (ΔmcpA) (thin solid line), OI3302 (ΔptsH) (thin broken line), and OI3304 (ΔmcpA ΔptsH) (heavy broken line) to the PTS carbohydrate d-glucose. d-Glucose (3.2 × 10−5 M) was added at 1 min and removed at 4 min. (C) Response of tethered OI3280 (ΔmcpC) to d-glucose (heavy line) and d-mannitol (thin line). d-Glucose (3.2 × 10−5 M) or d-mannitol (3.2 × 10−5 M) was added at 1 min and removed at 4 min.
d-Glucose chemotaxis is a multidimensional process in B. subtilis.
The B. subtilis mcpA null mutant (OI3055) had previously been shown to be slightly deficient in chemotaxis toward glucose (7), an observation that was confirmed by using glycerol-grown cells in a capillary assay (data not shown). To investigate this defect further, we constructed an mcpA ptsH null mutant (OI3304) and tested it for chemotaxis toward d-glucose in a capillary assay along with the wild type and ptsH null mutant. Unlike the results obtained with other PTS carbohydrates, the ptsH null mutant showed normal chemotaxis toward d-glucose. In fact, OI1085 (wild type), OI3055 (ΔmcpA), and OI3302 (ΔptsH) all showed normal chemotaxis toward d-glucose when grown in the presence of d-glucitol (see Materials and Methods). Strain OI3304 (ΔmcpA ΔptsH), however, was severely deficient in d-glucose chemotaxis (Table 1).
Tethering analysis revealed that all of these null mutant strains displayed different behavioral profiles only in response to the removal of d-glucose (negative stimulus), compared to the wild type (Fig. 1B). The mcpA ptsH double null mutant showed a removal response that was similar in magnitude to that of the wild type. The duration of the removal response in this strain, however, was three times shorter than those for the wild-type and the mcpA null mutant. The ptsH null mutant showed a removal response (decrease in percent counterclockwise flagellar rotation) that was 50% greater in magnitude, but three times shorter in duration, than those of the wild type and mcpA null mutant. This 50% increase in the magnitude of the removal response is based on the fact that the wild-type and mcpA null mutants showed a 20% decrease in counterclockwise rotation (from about 55 to 35%) upon the removal of d-glucose, while the ptsH null mutant displayed a 30% decrease in counterclockwise rotation (from about 55 to 25%) to the removal of the same carbohydrate. The ptsH null mutation, therefore, has the same effect on chemotaxis toward d-mannitol and d-glucose: it allows the cells to adapt to negative stimuli roughly three times faster than the wild-type strain.
Chemotaxis toward PTS carbohydrates is mediated by MCPs in B. subtilis.
All four of the strains studied responded normally to the addition of both d-glucose and d-mannitol (Fig. 1A and B), suggesting that the response to positive PTS carbohydrate stimuli is mediated by another component, most likely an MCP. Indeed, an mcpC null mutant (OI3280) showed no response to the addition or removal of d-glucose or d-mannitol, confirming this hypothesis (Fig. 1C). It has recently been shown that McpC is required for chemotaxis toward a number of amino acids (11). The failure of strain OI3280 to respond to PTS substrates was confirmed by backcrossing its DNA against wild-type strain OI1085, with selection for erythromycin resistance. Of 64 colonies tested, all 64 showed a defect in both amino acid taxis and PTS taxis. Thus, this study confirms that McpC is also required for any response to positive or negative PTS carbohydrate stimulus.
Purification of B. subtilis CheA.
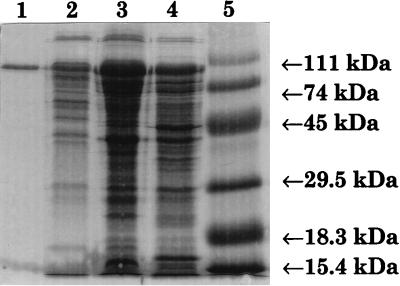
The B. subtilis cheA gene was placed under the T7 promoter in E. coli RP3098 harboring pGp1-2. Following induction at 42°C for 45 min, an overexpressed protein at a molecular mass of about 100 kDa appeared on an SDS-polyacrylamide gel (Fig. 2). This protein had a somewhat larger molecular mass than the predicted 74 kDa but reacted strongly with a rabbit anti-CheA–GST polyclonal antibody (Fig. 3) in a Western blot. Sequence analysis of the B. subtilis cheA gene revealed no other potential ribosome binding sites (E. coli or B. subtilis). For this reason, we believe that there is only one form of CheA in B. subtilis, in contrast to the two forms of CheA, long and short, that have been identified in E. coli (8).
FIG. 2.
Purification of B. subtilis CheA. CheA was purified as described in Materials and Methods. Lanes: 1, purified CheA following Hi-Trap Blue and gel filtration chromatography; 2, CheA fraction following anion-exchange chromatography; 3, CheA fraction following 50% ammonium sulfate precipitation; 4, crude cell lysate from strain OI3245; 5, prestained high-molecular-weight protein markers (Bethesda Research Laboratories).
FIG. 3.
Western blotting performed on cellular extracts from OI3245 (induced at 42°C and uninduced). The experiment was performed as described in Materials and Methods. Lane 1, cellular extract from OI3246 (CheA-GST expression strain); lane 2, cellular extract from strain OI3245 (native CheA expression strain) that had been induced at 42°C; lane 3, cellular extract from strain OI3245 that had not been induced at 42°C; lane 4, cellular extract from strain OI3245 that had been induced at 42°C but which was incubated with a 1:2,000 dilution of preimmune serum from the same rabbit. The immune serum containing anti-CheA–GST polyclonal antibody was diluted 1:50,000 for use in this experiment (lanes 1 to 3). The alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G antibody (Promega) used in this experiment was diluted 1:7,500. The blot was developed with a mixture of 5-bromo-4-chloro-3-indolylphosphate and nitroblue tetrazolium chloride.
EnzymeI-P inhibits B. subtilis CheA autophosphorylation in vitro.
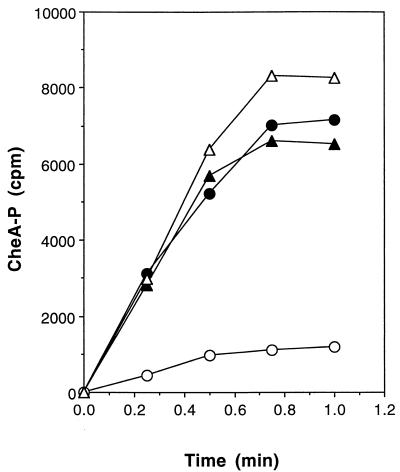
Recent studies on the E. coli chemotaxis system have shown that unphosphorylated EnzymeI specifically inhibits E. coli CheA autophosphorylation (10). To investigate whether the phosphorylation state of B. subtilis EnzymeI had any effect on B. subtilis CheA activity, we assayed for CheA autophosphorylation activity in the presence of EnzymeI, EnzymeI-P, and PEP. The purification of B. subtilis CheA and EnzymeI is described in Materials and Methods. EnzymeI-P was found to dramatically inhibit CheA autophosphorylation in vitro (Fig. 4 and 5). The effect was not due to the PEP present in the EnzymeI phosphorylation reaction, since PEP alone had no effect on CheA autophosphorylation. The unphosphorylated form of EnzymeI also had no effect on CheA autophosphorylation (Fig. 4 and 5).
FIG. 5.
The radioactive CheA-P bands shown in Fig. 4 were excised from the gel and subjected to scintillation counting. Shown are EnzymeI (•), EnzymeI-P (○), buffer control (▵), and PEP only (▴).
DISCUSSION
Does the PTS play a role in B. subtilis chemotaxis toward PTS carbohydrates? The results presented in this article clearly demonstrate that a functional PTS is critical for chemotaxis toward a number of PTS carbohydrates (Table 1). As measured in the spatial gradient of the capillary assay, a ptsH null mutant showed no chemotaxis toward α-methyl-d-glucoside, d-mannitol, or d-fructose, all of which are PTS substrates for B. subtilis. Chemotaxis toward d-glucose, however, was unaffected by the null mutation in ptsH despite the fact that it, too, is a PTS substrate. The ability to perform chemotaxis toward glucose in the absence of a functional PTS has been attributed to the additional regulation of the removal response to d-glucose by McpA (see below).
What is the nature of the defect in the chemotaxis of the ptsH null mutant strain to these carbohydrates? To address this question, we performed tethering (behavioral) assays on OI1085 (wild type) and OI3302 (ΔptsH) to measure the responses to positive (addition of the carbohydrate) and negative (removal of the carbohydrate) stimuli. Since the ptsH null mutant strain displayed no chemotaxis toward d-mannitol in the capillary assay (Table 1), we fully expected that this strain would show no response to positive stimulus with this carbohydrate. We found, however, that the ptsH null mutant strain was exciting and adapting normally to the addition of d-mannitol (Fig. 1A [first 3 min]). The defect in this strain was found to be in the duration of its response to the removal of d-mannitol (negative stimulus) (Fig. 1A [last 4 min]). Whereas the response to the removal of d-mannitol lasted nearly 3 min in the wild-type strain, the ptsH null mutant strain was able to adapt in just under 1 min. The magnitude of the removal response was the same in the two strains, however, showing about a 10% decrease in smooth swimming (percent counterclockwise flagellum rotation) upon removal of the carbohydrate. It appears, then, that the PTS plays a role only in the duration of the response to negative PTS carbohydrate stimuli in B. subtilis.
The fact that the ptsH null mutant did not migrate up the d-mannitol concentration gradient in the capillary assay while it differed from the wild type only in the duration of the response to the step removal of the carbohydrate (negative stimulus) implies that the behavior of the cell when moving down an attractant concentration gradient is the critical event that allows chemotaxis in B. subtilis. This result is reminiscent of early behavioral studies on E. coli, in which it was determined that chemotaxis occurs due to the suppression of tumbling events associated with travelling toward increasing concentrations of attractants (2). In other words, E. coli is able to perform chemotaxis by suppressing CheA activity and prolonging smooth swimming events. In the same way, it appears that suppression of CheA activity is also critical to chemotaxis in B. subtilis. The consequence of this, however, is that B. subtilis shows an increased tendency to tumble, a behavior that would become important as the cell travels toward lower concentrations of the carbohydrate. If the activation of CheA (increased periods of smooth swimming) were important to B. subtilis as it moved up the carbohydrate concentration gradient, then one would expect the ptsH null mutant, which displayed completely normal excitation and adaptation to the addition of all of the PTS carbohydrates tested, to show at least some accumulation in the capillary assay toward d-mannitol. This, however, was not observed (Table 1; Fig. 1A).
How is it that chemotaxis of the ptsH null mutant strain toward d-glucose is normal despite the fact that it is also a PTS carbohydrate? Figure 1B and C clearly shows that d-glucose chemotaxis is a multidimensional process in B. subtilis, involving McpC, McpA, and a functional PTS. The ptsH null mutant strain showed normal excitation and adaptation to the addition of d-glucose. Similar to the case with d-mannitol, this strain adapted to the removal of d-glucose nearly three times faster than the wild type. Unlike the case with d-mannitol, however, the magnitude of the response elicited by the removal of d-glucose was roughly 50% greater than that seen in the wild type (Fig. 1B). This effect on the magnitude of the removal response was offset in the ptsH null mutant strain by an additional null mutation in mcpA, a gene previously shown to be involved in chemotaxis toward d-glucose. The mcpA ptsH null mutant strain showed a response to the removal of d-glucose that was equal in magnitude but, again, threefold shorter in duration than that of the wild type (Fig. 1B). Since the ptsH null mutant strain had normal chemotaxis to d-glucose as measured in the capillary assay, we hypothesize that the additional 10% decrease in smooth swimming (50% difference in the magnitude of the response) that accompanied d-glucose removal in this strain was sufficient to offset the effect of the shorter duration of the response to the removal of d-glucose in these assays. The mcpA null mutant showed a response to the removal of d-glucose that was equal, both in magnitude and duration, to that of the wild type (Fig. 1B). We conclude from these results that McpC, McpA, and the PTS all participate in helping to bring about the response to negative d-glucose stimulus in B. subtilis. As is the case for d-mannitol, the PTS regulates the duration of the response to negative d-glucose stimulus.
Is chemotaxis toward PTS carbohydrates mediated by MCPs in B. subtilis? The excitation and adaptation to positive stimulus seen in the ptsH null mutant strain in reaction to d-mannitol and d-glucose suggest that something other than the PTS is responsible for this, most likely an MCP(s). We therefore tested an mcpC null mutant strain for chemotaxis toward d-mannitol and d-glucose by the capillary assay. We found that this strain showed no chemotaxis toward either carbohydrate (Table 1). Tethering analysis revealed that the mcpC null mutant strain showed no response to positive or negative stimulus with d-mannitol or d-glucose, confirming the hypothesis that PTS carbohydrate chemotaxis is MCP mediated in B. subtilis. McpC is homologous to both B. subtilis and E. coli MCPs and has been determined to be involved in chemotaxis toward a number of amino acids (11). The fact that the mcpC null mutant showed no response to positive or negative PTS carbohydrate stimulus (Fig. 1C) shows that this receptor plays a major role in both responses. McpC appears to be required for any response to the stepwise addition of PTS carbohydrates. The fact that the mcpC null mutant shows no response to the stepwise removal of PTS carbohydrates, despite the fact that the PTS system and McpA are still present, indicates that the negative stimulus brought about by these two components is not strong enough to bring about a behavioral response in the absence of McpC.
By what mechanism does the PTS regulate chemotaxis in B. subtilis? Studies on the E. coli PTS and chemotaxis system have shown that carbohydrate chemotaxis is entirely MCP independent. It does, however, require a functional PTS. Specifically, it has been determined that the unphosphorylated form of E. coli EnzymeI directly inhibits E. coli CheA autophosphorylation (10). This inhibition of E. coli CheA was found to be relieved through the phosphorylation of EnzymeI. In addition, PEP was found to enhance the rate of E. coli CheA autophosphorylation. These results have led to the following working model for the role of the PTS in the enteric chemotaxis system. As the cell imports and phosphorylates a PTS substrate, there are transient decreases in the intracellular concentrations of both PEP and EnzymeI-P. Together, these two conditions are believed to lead to inhibition of CheA activity. Consequently, there is a transient decrease in CheY-P levels that suppresses tumbling and allows the cell to move up the sugar concentration gradient (10). To determine whether B. subtilis CheA autophosphorylation is regulated directly by B. subtilis EnzymeI, both proteins were purified and assayed in vitro. EnzymeI-P was found to inhibit CheA autophosphorylation dramatically (Fig. 4 and 5). EnzymeI (unphosphorylated) or its substrate, PEP, alone had no effect on the autophosphorylation activity of B. subtilis CheA in vitro.
One might conclude from these results and the results of the tethering assays (Fig. 1) that an increase in the intracellular EnzymeI-P/EnzymeI ratio may lead to an inhibition of CheA activity that could prolong the response to the removal of PTS attractants. In the ptsH null mutant, this ratio would presumably remain constant (since no phosphoryl group transfer is occurring) as the cell moves down a PTS carbohydrate gradient, removing a potential source of CheA regulation under these circumstances. In the wild type, we hypothesize that there may be a transient increase in the EnzymeI-P/EnzymeI ratio as the cell moves toward lower PTS carbohydrate concentrations and the rate of transport of the molecule decreases. This could perhaps explain the relatively long (3-min) duration of the response to the removal of PTS carbohydrates seen in the wild type but not in the ptsH null mutant.
While the response to negative PTS carbohydrate stimulus appears to be mediated by the PTS and McpC, the strength of the signal elicited by either component individually in the spatial gradient does not appear to be strong enough to allow migration into the capillary tube. Only in the case of d-glucose, where the response to negative stimulus is partially mediated by two different MCPs (McpC and McpA), is the signal sufficiently strong to override the effect of not having a functional PTS (Fig. 1C; Table 1).
This work presents evidence for the existence of a multidimensional and perhaps novel signaling system controlling carbohydrate chemotaxis in B. subtilis, involving both MCPs and components of the PTS. Further experiments are under way to help define the interactions taking place between these two complex signal transduction pathways.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grants AI203336 and GM55434 from the National Institutes of Health.
REFERENCES
- 1.Adler J A, Epstein W. Phosphotransferase-system enzymes as chemoreceptors for certain sugars in Escherichia coli chemotaxis. Proc Natl Acad Sci USA. 1974;71:2895–2899. doi: 10.1073/pnas.71.7.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berg H C, Brown D A. Chemotaxis in Escherichia coli analyzed by three-dimensional tracking. Nature. 1972;239:500–504. doi: 10.1038/239500a0. [DOI] [PubMed] [Google Scholar]
- 3.Bischoff D S, Ordal G W. Sequence and characterization of Bacillus subtilis CheB, a homolog of Escherichia coli CheY, and its role in a different mechanism of chemotaxis. J Biol Chem. 1991;266:12301–12305. [PubMed] [Google Scholar]
- 4.Bourret R B, Borkovich K A, Simon M I. Signal transduction pathways involving protein phosphorylation in prokaryotes. Annu Rev Biochem. 1991;60:401–441. doi: 10.1146/annurev.bi.60.070191.002153. [DOI] [PubMed] [Google Scholar]
- 4a.Deutscher, J. Personal communication.
- 5.Deutscher J, Reizer J, Fischer C, Galinier A, Saier M H, Jr, Steinmetz M. Loss of protein kinase-catalyzed phosphorylation of HPr, a phosphocarrier protein of the phosphotransferase system, by mutation of the ptsH gene confers catabolite repression resistance to several catabolic genes of Bacillus subtilis. J Bacteriol. 1994;176:3336–3344. doi: 10.1128/jb.176.11.3336-3344.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a.Ehrlich, S. D. Personal communication.
- 6.Garrity L F, Ordal G W. Activation of the CheA kinase by asparagine in Bacillus subtilis chemotaxis. Microbiology. 1995;143:2945–2951. doi: 10.1099/00221287-143-9-2945. [DOI] [PubMed] [Google Scholar]
- 7.Hanlon D W, Ordal G W. Cloning and characterization of genes encoding methyl-accepting chemotaxis proteins in Bacillus subtilis. J Biol Chem. 1994;269:14038–14046. [PubMed] [Google Scholar]
- 8.Hess F J, Oosawa K, Matsumura P, Simon M I. Protein phosphorylation is involved in bacterial chemotaxis. Proc Natl Acad Sci USA. 1987;84:7609–7613. doi: 10.1073/pnas.84.21.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lengeler J W, Auberger A-M, Mayer R, Pecher A. The phosphoenolpyruvate-dependent carbohydrate: phosphotransferase system enzymes II as chemoreceptors in chemotaxis of Escherichia coli K-12. Mol Gen Genet. 1981;183:163–170. doi: 10.1007/BF00270156. [DOI] [PubMed] [Google Scholar]
- 10.Lux R, Jahreis K, Bettenbrock K, Parkinson J S, Lengeler J W. Coupling the phosphotransferase and the methyl-accepting chemotaxis protein-dependent chemotaxis signaling pathways of Escherichia coli. Proc Natl Acad Sci USA. 1995;92:11583–11587. doi: 10.1073/pnas.92.25.11583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mueller J, Schiel S, Ordal G W, Saxild H H. Functional and genetic characterization of McpC, which encodes a third methyl-accepting chemotaxis protein in Bacillus subtilis. Microbiology. 1997;143:3231–3240. doi: 10.1099/00221287-143-10-3231. [DOI] [PubMed] [Google Scholar]
- 12.Niwano M, Taylor B L. Novel sensory adaptation mechanism in bacterial chemotaxis to oxygen and phosphotransferase substrates. Proc Natl Acad Sci USA. 1982;79:11–15. doi: 10.1073/pnas.79.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Postma P W, Lengeler J W, Jacobson G R. Phosphoenolpyruvate:carbohydrate phosphotransferase system of bacteria. Microbiol Rev. 1993;57:543–594. doi: 10.1128/mr.57.3.543-594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reizer J, Sutrina S L, Wu L-F, Deutscher J, Reddy P, Saier M H. Functional interactions between proteins of the phosphoenolpyruvate:sugar phosphotransferase systems of Bacillus subtilis and Escherichia coli. J Biol Chem. 1992;267:9158–9169. [PubMed] [Google Scholar]
- 15.Roseman S, Meadow N D. Signal transduction by the bacterial phosphotransferase system. Diauxie and the crr gene (J. Monod revisited) J Biol Chem. 1990;265:2993–2996. [PubMed] [Google Scholar]
- 16.Stock J B, Ninfa A J, Stock A M. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989;53:450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tabor S, Richardson C C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor B L, Johnson M S, Smith J M. Signaling pathways in bacterial chemotaxis. Bot Acta. 1988;101:101–104. [Google Scholar]
- 19.Welch M K, Oosawa K, Aizawa S-I, Eisenbach M. Phosphorylation-dependent binding of a signal transduction molecule to the flagellar switch of bacteria. Proc Natl Acad Sci USA. 1993;90:8787–8791. doi: 10.1073/pnas.90.19.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]