Abstract
The hepatitis E virus (HEV) is a widespread human infection that causes mainly acute infection and can evolve to a chronic manifestation in immunocompromised individuals. In addition to the common strains of hepatitis E virus (HEV-A), known as Paslahepevirus balayani, pathogenic to humans, a genetically highly divergent rat origin hepevirus (RHEV) can cause hepatitis possessing a potential risk of cross-species infection and zoonotic transmission. Rocahepevirus ratti, formerly known as Orthohepevirus C, is a single-stranded RNA virus, recently reassigned to Rocahepevirus genus in the Hepeviridae family, including genotypes C1 and C2. RHEV primarily infects rats but has been identified as a rodent zoonotic virus capable of infecting humans through the consumption of contaminated food or water, causing both acute and chronic hepatitis cases in both animals and humans. This review compiles data concluding that 60% (295/489) of RHEV infections are found in Asia, being the continent with the highest zoonotic and transmission potential. Asia not only has the most animal cases but also 16 out of 21 human infections worldwide. Europe follows with 26% (128/489) of RHEV infections in animals, resulting in four human cases out of twenty-one globally. Phylogenetic analysis and genomic sequencing will be employed to gather global data, determine epidemiology, and assess geographical distribution. This information will enhance diagnostic accuracy, pathogenesis understanding, and help prevent cross-species transmission, particularly to humans.
Keywords: hepatitis E, Rocahepevirus ratti, Orthohepevirus C, RHEV, HEV-C, rodent, zoonosis
1. Introduction
The hepatitis E virus (HEV) is one of the leading causes of hepatitis worldwide. Generally, is presented as an acute infection that improves without treatment after several weeks. Clinical course includes asymptomatic infection, mild-to-moderate liver dysfunction, or fulminant hepatitis. Chronic infections have been reported among immunocompromised individuals like solid organ transplant recipients [1].
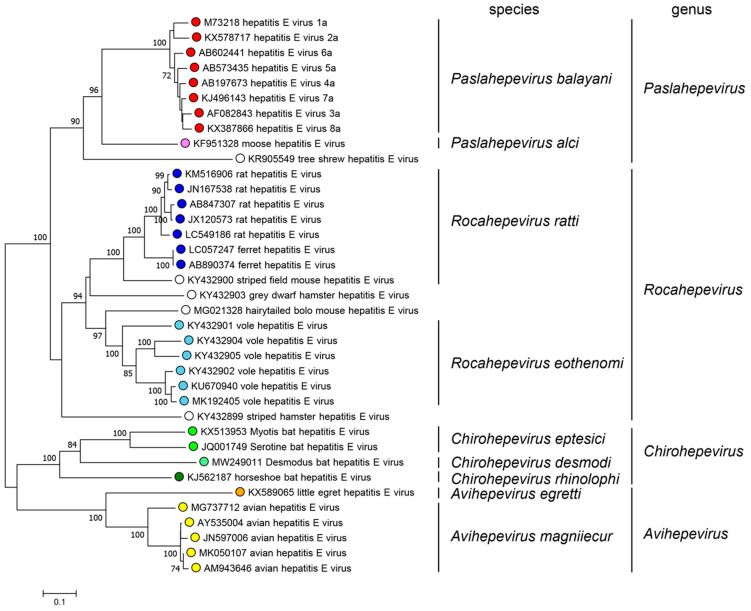
The HEV is a single-stranded RNA virus belonging to the Hepeviridae family, which included three genera until 2022: Orthohepevirus (infecting mammalian species), Orthohepevirinae (infecting unclassified species), and Piscihepevirus (infecting fish species). Orthohepevirus C (rat hepatitis E virus) also referred as HEV-C, was classified as a member of the Orthohepevirus genus within the Hepeviridae family, sharing a close genetic relationship with other hepatitis E viruses, including HEV-A, HEV-B, and HEV-D (Figure 1) [2,3].
Figure 1.
Phylogenetic tree of members of the family Hepeviridae, subfamily Orthohepevirinae (ORF1 polyprotein residues 1–450) (https://ictv.global/report/chapter/hepeviridae/hepeviridae, accessed on 31 October 2023).
The current nomenclature for the former Orthohepevirus C is Rocahepevirus ratti (RHEV), as the International Committee on Taxonomy of Viruses (ICTV) has reclassified the Orthohepevirus genus. The other virus pathogen of the Hepeviridae family that infects humans is the Paslahepevirus balayani, previously known as Orthohepevirus A (HEV-A). HEV-A virus exhibits eight genotypes (1–8) and it is primarily transmitted through the fecal-oral route, associated with contaminated water or food, and mainly prevalent in regions with inadequate hygiene practice. While Paslahepevirus balayani is recognized as the causative agent of hepatitis E in humans, Rocahepevirus ratti represents an emerging zoonotic pathogen primarily infecting rats and other rodent species, this infectious agent can potentially infect humans increasing the concern in terms of public health [4,5].
The genomic sequence analysis of RHEV has revealed new cases involving zoonotic transmission, as rodents are susceptible to genotype 1 (HEV-C1) infection [6]. This virus is primarily associated with rat populations, which can shed the agent in their feces and urine when infected, leading to environmental contamination. The actual transmission route for human infection is unknown, but different risk factors have been proposed, such as exposure to contaminated environments, contact with infected animals, or the consumption of contaminated food or water [7].
Since the RHEV discovery in rats in 2010 in Germany, this virus has been detected in multiple countries across the world in Asia, Europe, and North America [8]. Although the mechanism of RHEV infection in humans is not entirely clear, studies indicate that the virus may cause mild to moderate acute hepatitis, with symptoms including fever, fatigue, nausea, and abdominal pain. Occasionally, there are cases where the RHEV infection may progress to chronic hepatitis or liver cirrhosis, particularly among immunocompromised individuals. Molecular and serological diagnostic techniques are typically employed for identifying RHEV infection [4].
Considering the emerging nature of the Rocahepevirus ratti as a human pathogen, there is a pressing need to evaluate the existing literature on this virus, including its phylogenetic and genomic analysis, epidemiology and its existing geographic distribution, pathogenesis, and diagnosis available. The objective of this review is to help to better understand the potential risks associated with RHEV infection and to provide valuable information on devising effective measures for prevention and control of this emerging cross-species infection and zoonotic pathogen.
2. Materials and Methods
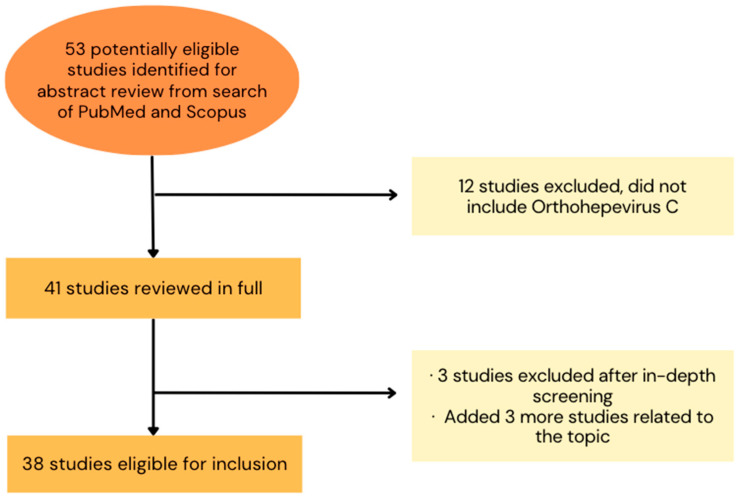
A bibliographic review was conducted to investigate the detected cases of Rocahepevirus ratti as a newly emerging virus worldwide, including its diagnostic data, in both humans and animals. For this purpose, the search was carried out using Pubmed and Scopus with the following keywords: “Orthohepevirus C”, “Rocahepevirus ratti”, “HEV-C1”, “HEV- C2”, “rodent”, and “zoonosis”. This search aimed to identify the different research developed in human and animal health related to RHEV for a better understanding of this infection. All studies considered were published worldwide in a period spanning from December 2000 to October 2023, in any available language (Figure 2).
Figure 2.
Selection of studies about Rocahepevirus ratti, including its epidemiology, 2000–2023. This selection was made for inclusion of studies in the bibliographic review. It was necessary to consider the fact that Orthohepevirus C has been renowned as Rocahepevirus ratti in recent studies (2022 and 2023).
To ensure the accuracy and reliability of the research, all available articles were individually evaluated for inclusion. In the study of the animals that contracted the infection, not only those with the complete sequence were considered, data from those with the partial genomic sequence were included after verifying their association with the studied virus’s genomic sequence. Authors of the studies were contacted to clarify any ambiguity or provide additional information. For the genomic sequence, data were collected from studies in which virus RNA was sequenced using Sanger sequencing or NGS. Additionally, public sequence databases like European Nucleotide Archive and GenBank were queried for each case. The working maps were constructed by using a free open source, Mapchart tool (https://www.mapchart.net/world.html, accessed on 31 October 2023) using the collected data from the generated tables.
The research methodology employed in this review ensures the selection of reliable and accurate data and aims to provide valuable insights into the detected cases and diagnostic information of RHEV globally.
3. Phylogenetic Analysis and Genomic Sequence
The Hepeviridae family is divided into two subfamilies: Orthohepevirinae, divided into four genera: Paslahepevirus (former Orthohepevirus A) the main cause of hepatitis E, which embraces eight different genotypes affecting a variety of species including humans, pigs, wild boar, deer, rabbits, and camels [7]; Rocahepevirus (previous Orthohepevirus C) which infects the Rodentia and Soricomorpha order (rats and ferrets), as well as Carnivora; Chirohepevirus (old Orthohepevirus D) which infects Chiroptera order (bats); and Avihepevirus (previous Orthohepevirus B) which infects and circulates in the avian order (birds); the second subfamily is Parahepevirinae containing one single genus, Piscihepevirus [9] (Table 1). RHEV and HEV-A are two highly divergent viruses, their genomes only share 50–60% genomic identity, differing on their transmission and pathogenicity, with RHEV infections being related to less severe disease outcomes [7].
Table 1.
| Family | Subfamily | Genus/Species | Genotype | Host Range/Reservoir |
|---|---|---|---|---|
| Hepeviridae | Orthohepevininae | Paslahepevirus balayani | HEV-1 | human |
| HEV-2 | human | |||
| HEV-3 | human, pig, wild boar, deer, rabbit, camel | |||
| HEV-4 | human, pig, wild boar, | |||
| HEV-5 | wild boar | |||
| HEV-6 | wild boar | |||
| HEV-7 | camel | |||
| HEV-8 | camel | |||
| Paslahepevirus alci | moose | |||
| Avihepevirus | aHEV (avian HEV) |
chicken | ||
| Rocahepevirus ratti | HEV-C1 | rat, shrew, kestrel, falcon, fox |
||
| HEV-C2 | ferret, mink | |||
|
Rocahepevirus
eothenomi |
vole | |||
| Chiropteranhepevirus | bat | |||
| Parahepevirinae | Piscihepevirus | trout, salmon |
The HEV virus presents eight main genotypes within the Paslahepevirus genus [4]. Genotypes 1 and 2 transmission is through the consumption of contaminated water by feces, mainly in developing countries. This situation has triggered zoonotic transmission by these genotypes, due to the contamination of the environment with the pathogen from animal feces or urine. As well as the transmission through the ingestion of raw or undercooked meat of the infected animal host, which predominates in genotypes 3 to 8 [5,6]. Within the RHEV, there are two main genotypes, HEV-C1 and HEV-C2, showing a divergence of 44%. In addition, two extra tentative genotypes (C3 and C4) have been proposed within RHEV species [3].
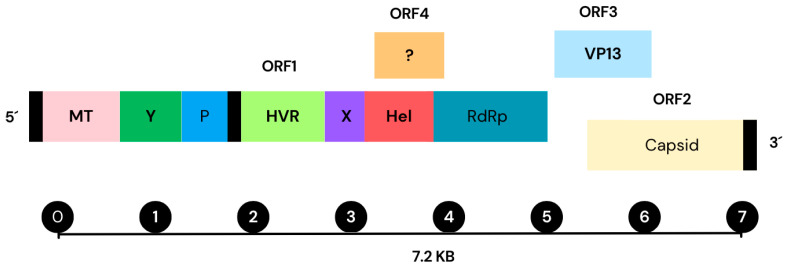
According to the genomic sequence analysis, the HEV-A virus possesses a single- stranded RNA genome positive-sense approximately 7.2 kb in length. The genome encodes three open reading frames (ORFs) which participate in the viral replication and transcription [9,10] (Figure 3). ORF1 contains a large polyprotein that is processed into several non- structural proteins, including RNA-dependent RNA polymerase (RdRp), helicase, and protease. ORF2 codifies a protein for the viral capsid, while ORF3 produces a minor protein responsible for the virion assembly and release. The genomic structure of HEV-A resembles other hepeviruses, like the RHEV [11]. Recent studies have identified a new open reading frame (ORF4), located within the ORF1 region of the HEV-A genome. This ORF4 encodes a 20 kDa protein with 139–158 amino acids span. Despite its discovery, the role of the ORF4 in the HEV-A or RHEV viral replication and pathogenesis remain unclear [12]. Investigating the ORFs of the RHEV virus is crucial to understand the genetic structure of the virus, the pathogenesis, replication, transcription and translation mechanisms, and how they adapt into the host to develop more effective strategies against this pathogen [11]. Additionally, despite a number of studies on the genomics of HEV-A and RHEV, there is still a limited knowledge on the evolutionary history and functions of the different regions. Comparing the genomic sequence of RHEV with other members of the Hepeviridae family, as well as to other related viruses, such as the Flaviviridae family, can provide valuable insights into the evolutionary history of the virus and how it has evolved to infect different host species [2]. Such comparisons can identify regions of similarity in the different strains that can be targeted to develop antiviral drugs or utilized as diagnostic markers for detection. Conducting phylogenetic analysis and genomic sequencing of RHEV is crucial for understanding how the virus evolves.
Figure 3.
Hepatitis E virus (HEV-A) genome structure, including its open reading frames (ORFs) and proteins. Contains a single-stranded positive-sense RNA molecule approximately 7.2 kb long and is presented in the genome of the virus of HEV-A. It displays a 7-methylguanosine RNA at the 5′ end and poly-A at the 3′ terminus. All HEV strains contain three open reading frames (ORFs): ORF1, ORF2, and ORF3 (an additional ORF4(?) region is found which functionality is not totally understood). ORF1 produces non-structural polyproteins for viral replication and transcription such as (Met), Y-domain, (PCP), (HVR), (Hel), and (RdRp). ORF2 encodes the capsid protein, while ORF3 encodes a multifunctional phosphoprotein, known as VP13. These proteins from ORF2 and ORF3 exhibit partial overlap and are translated from a subgenomic RNA with 2.2 kb in length. Additionally, ORF4 gives rise to an internal ribosome entry site-like protein (IRES) to respond to the stress generated in the endoplasmic reticulum (ER). ORF4 is an enhancer of viral replication. (GenBank accession number AF444002.1) [10,13,14].
4. Epidemiology and Geographical Distribution
4.1. Human Infections Associated with Rocahepevirus ratti
At this moment, up to 21 cases of HEV-C1 infection have been reported in humans through RNA detection in different samples. This data includes sixteen patients in Hong Kong, three patients in Spain, one patient in Central Africa, and one patient in France. RHEV initial detection occurred in rats from Germany and Vietnam [3], but no human cases have been found in Germany [15] or Hungary [16] after investigation. In Spain, a comprehensive study conducted by Rivero-Juarez et al. revealed the presence of Rocahepevirus ratti infection in three human patients. Two of these cases were located in the southern region, while one was identified in the northern part of Spain (Table 2).
Table 2.
Investigation of human hepatitis caused by Rocahepevirus ratti virus in different countries.
| Continent | Location [Reference] |
Screened Patients | Description of Patients Investigated |
HEV-C1 RNA Target | HEV-C1 Cases |
|---|---|---|---|---|---|
| Asia | Hong Kong 2022 [17] | 53 | Elderly/Immunocompromised | ORF-1 | 8/53 (15.1%) |
| Hong Kong 2021 [18] | 2860 | Abnormal liver function (n = 2201) Immunocompromised (n = 659) |
ORF-1 | 6/2201 (0.27%) 1/659 (0.15%) |
|
| Hong Kong 2018 [19] | 518 | Persistent Hepatitis (n = 52) | RdRp (ORF-1) | 1/52 (1.9%) | |
| Africa | D.R.Congo or Gabon * [20] |
1 | Acute Hepatitis. HEV-IgG (+), HEV-IgM (+/−) |
RdRp (ORF-1) | 1 |
| Europe | Spain [4] | 267 | Acute hepatitis unknown etiology (n = 169) Acute HEV infection (n = 98) |
RdRp (ORF-1) | 2/169 (1.2%) 1/98 (1.0%) |
| Germany [15] | 200 | HEV suspicion (2000–2020) HEV-IgM (+) or (+/−); HEV RNA (−) |
RdRp (ORF-1) | none | |
| France [21] | 224 | Individuals at risk of chronic HEV infection | RdRp (ORF-1) | none | |
| France [22] | 1 | HEV-IgM (+) | Pan-pathogen metagenomics |
1 | |
| Hungary [16] | 1439 | HEV-IgM (+) (n = 162) | Unknown | none |
* The country source of infection is not clear. Abbreviations: +: positive; −: negative; +/− borderline; none: none results were found; RdRp: RNA-dependent RNA-polymerase.
In China, particularly in Hong Kong, three studies have been carried out to investigate the role of Rocahepevirus ratti. The most recent of these studies was published in 2022 and focused on a cohort of 53 patients from the Microbiology Department of Queen Mary Hospital. This study was conducted between 1 August 2019 and 31 December 2020, and the results indicated a total of eight cases with positive RNA HEV-C1 [17]. This study was a continuation of a prior investigation, which analyzed 2201 sera from patients with liver function abnormalities between 1 January 2017 and 31 July 2019 (group 1), and 659 cases in a second group, composed of transplant recipients and patients with solid organ neoplasms, hematologic neoplasms, autoimmune disorders, and other immunosuppressive conditions between 1 January 2019 and 30 June 2019 (group 2). The previous study identified six cases of HEV-C1 RNA positivity in group 1 and one case in group 2, leading to a total of seven cases [18]. Notably, the first confirmed human infection caused by RHEV, originating from rats, was identified in Hong Kong in 2017. The study examined a patient who was Anti-HEV IgG positive and had borderline Anti-HEV IgM results. This patient was found to have persistent Hepatitis E after receiving a liver transplant. For this research, a total of 518 solid organ transplant recipients, including kidney, liver, lung, and heart transplants, were evaluated. These recipients exhibited persistent hepatitis, defined as an elevation of alanine aminotransferase (ALT) greater than 1.5 times the upper limit of the reference level for a continuous period of more than six weeks, from 1 January 2014, or the date of transplant (whichever was later) until 31 December 2017 [19] (Table 3).
Table 3.
Overview of human infections linked to rat HEV-C1 virus (Rocahepevirus ratti) (N = 21).
| Patient | Underlying Disease | Clinical Outcome | Potential Source of the Infection | Location (Year) |
GenBank Acc. Nº | Reference |
|---|---|---|---|---|---|---|
| 56-year-old man | Liver transplantation, HBV carrier, immunocompromised |
Persistent hepatitis | Rodent droppings |
Hong Kong (2017) | MG813927 | [19] |
| 71-year-old female | Rheumatoid arthritis, immunocompromised |
Acute hepatitis | Unknown | Hong Kong (2017) | MN450851 | [18] |
| 67-year-old man | Kidney transplantation, immunocompromised |
Persistent hepatitis | Unknown | Hong Kong (2018) | MN450852 | [18] |
| 74-year-old man | Kidney transplantation, HBV carrier, immunocompromised | Persistent hepatitis | Unknown | Hong Kong (2018) | MN450853 | [18] |
| 81-year-old man | Prostate cancer | Acute hepatitis | Unknown | Hong Kong (2019) | MN450856 | [18] |
| 73-year-old man | None, immunocompetent | Acute hepatitis | Unknown | Hong Kong (2019) | MN450857 | [18] |
| 67-year-old man | Metastatic cancer | Subclinical | Unknown | Hong Kong (2018) | MN450858 | [18] |
| 43-year-old man | HIV infection | Persistent hepatitis | Unknown | Hong Kong (2019) | MN450854 | [18] |
| 49-year-old man | None, immunocompetent | Severe acute hepatitis | Trip to Africa | D.R.Congo or Gabon (2019) | MK050105 | [20] |
| 62-year-old man | Metastatic oral cancer | Acute hepatitis | Raw food consumption |
Spain (2020) | OK082152 | [4] |
| 30-year-old man | None, immunocompetent | Acute hepatitis | Cleaning staff | Spain (2018) | OK082153 | [4] |
| 54-year-old man | None, immunocompetent | Acute hepatitis | Unknown | Spain (2019) | OK082154 | [4] |
| 83-year-old female | Hypertension, tuberculosis | Persistent hepatitis | Unknown | Hong Kong (2020) * | NA * | [17] |
| 18-year-old man | HSCT, immunocompromised |
Persistent hepatitis | Unknown | Hong Kong (2020) * | NA * | [17] |
| 61-year-old man | Hypertension | Acute hepatitis | Unknown | Hong Kong (2020) * | NA * | [17] |
| 71-year-old man | Liver transplant, immunocompromised |
Persistent hepatitis | Unknown | Hong Kong (2020) * | NA * | [17] |
| 89-year-old female | Pancreatic cancer | Acute hepatitis | Unknown | Hong Kong (2020) * | NA * | [17] |
| 79-year-old man | Hypertension, diabetes | Acute hepatitis | Unknown | Hong Kong (2020) * | NA * | [17] |
| 59-year-old man | Chronic myeloid leukemia | Acute hepatitis | Unknown | Hong Kong (2020) * | NA * | [17] |
| 61-year-old man | Kidney transplantation, immunocompromised |
Cirrhosis | Trip to India | France (2017) | OP610066 | [22] |
* Patients studied between 1 August 2019 and 31 December 2020; Abbreviations: NA: Not Available.
Another reported case of RHEV infection in humans involved a Canadian citizen who worked for the United Nations (UN) and was admitted to a hospital returning home in 2017 with severe acute hepatitis. Before his hospitalization, he had worked at various UN facilities in the Democratic Republic of Congo and Gabon from January to 12 March 2017. The exact origin of his infection remains uncertain (Gabon or DRC). Nevertheless, this case is noteworthy as the first and only case with origins from the African continent, where hepatitis E is more prevalent than in other continents. In this case, the infection is believed to have been transmitted through zoonosis, as it is RHEV [20] (Table 3).
Recent studies have been conducted, revealing also the transmission of RHEV to humans in Europe. A recent study, conducted in Spain, examined 267 cases and evaluated two cohorts: one comprising 169 patients without HEV infection, and the other consisting of 98 patients diagnosed of HEV infection (either HEV RNA or HEV-IgM positive). In the first cohort, two cases of HEV-C1 RNA-positive samples were detected, whereas one case was found in the second cohort (HEV-IgM(+)). Therefore, the first RHEV recorded cases in Europe were these three human infections identified in Spain [4]. A fourth European case was discovered in France thanks to metagenomics testing, revealing the presence of RHEV in liver/blood samples of a male who developed cirrhosis after resolving HBV infection (loss of HBsAg and non detectable DNA). This patient of Indian origin underwent renal transplantation in 2006 and despite successful treatment for hepatitis B virus (HBV) reactivation in 2008, he developed cirrhosis in 2016, leading to a double kidney-liver transplantation. Post-transplantation, the patient experienced unexplained liver abnormalities. In 2022, a pan-pathogen non-specific metagenomics method revealed the presence of RHEV in liver samples since 2017, indicating a persistent infection despite undetectable HBV DNA, so the patient’s cirrhosis was attributed to RHEV [22] (Table 3).
It is worth noting that some studies have been conducted where no positive cases were detected. A study conducted in Germany involved the evaluation of 200 patients who tested positive or borderline positive for HEV-IgM but negative for HEV RNA. This study did not yield any RHEV positive cases [15]. Similarly, in France, 224 individuals at risk of developing chronic HEV infection were assessed, and none of them tested positive for RHEV RNA [21]. Similarly, to a group of 163 individuals in Hungary with HEV-IgM(+) marker, where no RHEV was found [16].
There is scarce information regarding RHEV seroprevalence as antibody detection is not highly specific, despite amino acid homology of only 55 [8,23]. HEV and RHEV antibodies may cross-react so once specific tools could be developed, it will be possible to get a better knowledge of the circulation of RHEV as subclinical infections may be frequent. In a first attempt to study RHEV seroprevalence in Germany, sera of healthy forestry workers showed a stronger response to HEV-C1 antigen than to HEV-3 antigen [24]. A similar pattern of greater immune response of IgG and IgM against HEV-C1 antigen was observed in a number of patients in Vietnam after febrile illness [25]; and a third study using the immunodominant E2 peptide sequence of HEV-C1 established the antigenic basis for designing the first validated EIA/immunoblots in human populations [26]. Therefore, this method described in Hong Kong has allowed to estimate RHEV seroprevalence of 0,92%, indicating endemic exposure of the population in this region [27].
4.2. Animal Infections Associated with Rocahepevirus ratti
Regarding the discovery and dissemination of RHEV on a global scale, including regions such as Europe, Asia, America, and Africa, the Table 4 provides comprehensive data concerning the prevalence of cases, categorized by continents and individual countries.
Table 4.
Global data of animal infections associated with Rocahepevirus ratti (N = 489).
| Continent | Asia | n | Europe | n | America | n | Africa | n |
|---|---|---|---|---|---|---|---|---|
| China | 159 | Germany | 42 | USA | 37 | Kenya | 1 | |
| Indonesia | 117 | Czech Republic | 20 | Canada | 21 | |||
| Japan | 14 | Lithuania | 9 | Brazil | 7 | |||
| Vietnam | 5 | Romania | 9 | |||||
| Belgium | 8 | |||||||
| Great Britain | 8 | |||||||
| Hungary | 7 | |||||||
| France | 5 | |||||||
| Country RHEV infections |
Austria | 4 | ||||||
| Spain | 4 | |||||||
| Switzerland | 4 | |||||||
| Denmark | 3 | |||||||
| Greece | 2 | |||||||
| Netherlands | 2 | |||||||
| Italy | 1 | |||||||
| Total infections | 295 | 128 | 65 | 1 |
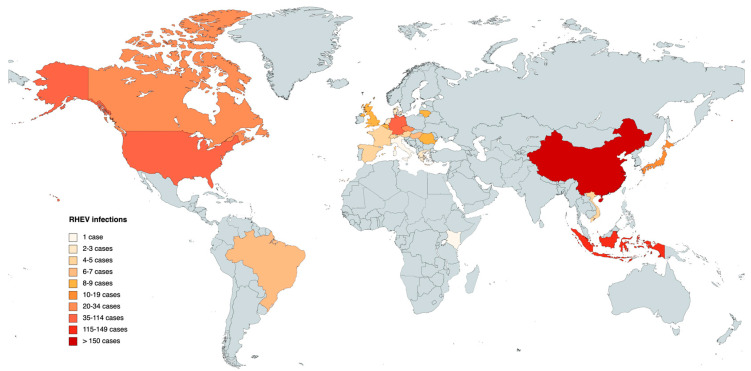
Until 2023, a cumulative total of 489 infections have been reported in different animals (mainly rodents), notably revealing Asia as the continent with the highest incidence of infected animals, accounting for a total of 295 documented instances. China emerges as the country with the highest reported cases within the Asian region. Europe, on the other hand, stands as the second-highest affected continent, with a combined total of 128 confirmed cases distributed across 15 different countries. In America, the presence of RHEV has only been detected in the United States, Canada, and Brazil, providing a combined total of 65 reported infections. In Africa, a solitary case has been identified, specifically in Kenya (Table 4).
The global map in Figure 4 corresponds to the data presented in Table 4, offering a comprehensive review of the results pertaining the different countries. The data are differentiated by RHEV animal infections, and the colored regions on the map represent the locations where detected animal infections have been reported.
Figure 4.
Map of animal infections caused by Rocahepevirus ratti in different countries. The darker the color, the greater the number of infections.
The first identification of hepevirus associated with rodents was made back to 2010 when a nested broad-spectrum reverse transcription polymerase chain reaction (RT-PCR) was employed in Germany. Subsequent genome sequencing revealed substantial genetic disparities between this virus and the human HEV strain. After thorough examination, it was classified as Rocahepevirus ratti and assigned the genotype HEV-C1. Following its initial discovery in Germany, RHEV was subsequently found in a wide range of animal species (Table 5).
Notably, the Rodentia genus exhibited the highest prevalence of detected infections, encompassing wild Norway rats (Rattus norvegicus) across 22 countries, black rats (Rattus rattus) in 16 countries, which included 12 European nations (Austria, Belgium, Czech Republic, Denmark, France, Germany, Greece, Hungary, Italy, Lithuania, Spain, and Switzerland), as well as Tanezumi rats (Rattus tanezumi) in Vietnam, where infections were observed.
According to these findings the widespread presence of HEV-C1 among rat populations across the four continents, Asia, Europe, America, and Africa. Furthermore, infections were identified in the Cricetidae family, including the Asian musk shrew (Suncus murinus). HEV-C2 genomes were also detected in ferrets and minks.
Beyond these classifications, there have been reports of unclassified rodent RHEV strains, which include Chevrier’s field mouse (Apodemus chevrieri), Pere David’s mole (Eothenomys melanogaster), hairy-tailed vole mouse (Necromys lasiurus), delicate vole mouse (Calomys tener), among others [7]. It is remarkable that RHEV infections are not limited to species within the Rodentia order; there have also been documented cases of Rocahepevirus ratti in the Soricomorpha order in two countries, China and Kenya, as well as in members of the Carnivora order, comprising six different species, found in countries such as Denmark, the Netherlands, the United States, Japan, China, Germany, and Spain. Additionally, two species from the order Falconiformes, the Common Kestrel (Falco tinnunculus) and the Red-footed Falcon (Falco vespertinus), were also identified as infected by RHEV in Hungary (Table 5).
4.3. Investigation of Rocahepevirus ratti in Wastewater Systems
Sewage surveillance is recognized as a powerful tool to gather information on the epidemiology of infectious diseases in the served population. The presence of RHEV has been explored in sewage treatment plants at different locations in Sweden, Italy, and Spain, allowing to get a better view of its epidemiology.
In Gothenburg (Sweden), a comprehensive study monitored influent and effluent wastewater for enteric viruses by qPCR, at the Rya treatment plant over a year. Seasonal variations in viral concentrations were observed in incoming wastewater, correlating with the number of diagnosed patients. Different HEV strains previously identified in drinking water, including two novel strains similar to those infecting rats and humans, were identified in the effluents [28]
Another study in Abruzzo (Italy), detected viral RNA of RHEV in 43.9% of sewage collected from 14 wastewater treatment plants, by using broadly reactive primers for hepevirus. These strains exhibited genetic variability and a clear geographic and wastewater treatment plant-related pattern. This study suggested that RHEV was a significant component of wastewater microbiota in the region [29]
Finally, a longitudinal study was carried out in Cordoba (southern Spain) from 2021 to 2023 using samples from patients with acute hepatitis, specimens from rodents, and wastewater to evaluate the correlation with human cases. The results showed that while RHEV was detected in almost all wastewater samples, there was no correlation between clinical cases and wastewater detection for both HEV and RHEV [30]
These studies highlight the importance of monitoring wastewater for RHEV. Untreated wastewater collects viruses excreted by both humans and synanthropic animals, including rodents, thereby providing a comprehensive overview of the viral strains circulating. Despite the reduction in viral concentrations during wastewater treatment, the presence of novel strains concerns the potential public health impact. Further research in this area is essential to understand the epidemiology and impact of RHEV infections on human health.
Table 5.
Summary of animal infections associated with rat origin HEV-C1 virus (Rocahepevirus ratti).
| Order/Family | Species | Type of Detection | Year and Location | Type of HEV | GenBank Acc. Nº | Type of Genomic Sequence | Ref. |
|---|---|---|---|---|---|---|---|
| Rodentia/ Muridae |
Norway rat (Rattus norvegicus) | 2 HEV IgM+/134 sera; 1 RT-PCR+ |
2003 (Los Angeles; USA) | HEV-C1 | JF516246 | Complete/Partial | [31] |
| Norway rat (Rattus norvegicus), Black rat (Rattus rattus) |
63 RT-PCR+/508 liver samples | 2005–2016 (12 European countries) | HEV-C1 (49) | KX774641–KX774673 | Complete/Partial | [32] | |
| Norway rat (Rattus norvegicus) | 14 RT-PCR+/101 liver samples | 2008–2010 (Hamburg, Berlin, Stuttgart, Esslingen; Germany) | HEV-C1 | JN167530–JN167538 | Complete/Partial | [33] | |
| Norway rat (Rattus norvegicus) | 2 RT-PCR+/6 liver samples | July 2009 (Hamburg; Germany) | HEV-C1 | GU345042, GU345043 | Complete | [34] | |
| Norway rat (Rattus norvegicus), Tanezumi rat (Rattus tanezumi) | 5 HEV IgM+/139 sera; 1 RT-PCR+ |
2011 (Haiphong and Hanoi; Vietnam) | HEV-C1 | JN040433 (=JX120573) | Complete/Partial | [35] | |
| Norway rat (Rattus norvegicus) Greater bandicoot rat (Bandicota indica), Rattus flavipectus, Rattus rattoides losea | 59 HEV IgM+/713 sera; 12 RT-PCR+ |
December 2011–September 2012 (Zhanjiang, China) |
HEV-C1 | KC465998–KC465999 | Complete/Partial | [36] | |
| Norway rat (Rattus norvegicus), Black rat (Rattus rattus) |
35 RT-PCR+/446 liver samples | 2012 (15 states; USA) | HEV-3 (34) HEV-C1 (1) |
JQ898480–JQ898514 | Complete | [37] | |
| Norway rat (Rattus norvegicus) | 8 RT-PCR+/61 liver samples | 2014–2016 (Great Britain) | HEV-C1 | MK770165–MK770171 | Complete | [38] | |
| Norway rat (Rattus norvegicus) | 7 RT-PCR+/159 rectal swabs | 2018–2019 (Hong-Kong, China) | HEV-C1 | MN450855, MN450859– MN450864 | Complete | [18] | |
| Norway rat (Rattus norvegicus) | 1 RT-PCR+/10 fecal samples | 2017–2018 (Budapest; Hungary) | HEV-C1 | MT847624 | Complete | [3] | |
| Norway rat (Rattus norvegicus) | 21 RT-PCR+/372 liver samples | 2018–2021 (Ontario Canada) | HEV-C1 | OQ617169–OQ617185 | Partial | [39] | |
| Norway rat (Rattus norvegicus) | 9 RT-PCR+/69 liver samples | 2016–2023 (Eastern Romania) | HEV-C1 | OQ601523–OQ601531 | Partial | [40] | |
| Rodentia/ Muridae (continue) |
Black rat (Rattus rattus) | 17 RT-PCR+/116 sera | August 2011–February 2012 (Lambok Island; Indonesia) | HEV-C1 | AB725884–AB725900 | Complete | [41] |
| Black rat (Rattus rattus) | 99 RT-PCR+/369 sera | September–October 2012 (Solo, Indonesia) | HEV-C1 | AB847305–AB847406 | Complete/Partial | [42] | |
| Black rat (Rattus rattus) | 2 RT-PCR+/242 sera | 2014–2016 (Bali and Sumbawa, Indonesia) |
HEV-C1 | LC225388–LC225389 | Complete | [43] | |
| Black (Rattus rattus), Norway rats (Rattus norvegicus) |
9 RT-qPCR+/109 liver and chest fluid samples | 2014–2017 (Lithuania) | HEV-C1 | MH400712–MH400717 | Complete | [44] | |
| Black (Rattus rattus), Norway rats (Rattus norvegicus) |
5 HEV IgG+/20 fecal samples/7 tissue samples/428 sera | 2014–2017 (Japan) | HEV-C1 | LC573546 | Complete | [45] | |
| Chevrier’s field mouse (Apodemus chevrieri) | 59 RT-PCR+/202 sera | 2013–2015 (Lijiang, China) | HEV-C | MG020022–MG020025 | Complete | [6] | |
| Striped field mouse (Apodemus agrarius) | 2 RT-PCR+/2 sera | 2014 (Jilin; China) | HEV-C | KY432900 | Complete | [46] | |
| Rodentia/ Cricetidae |
Hairy-tailed mouse (Necromys lasiurus), Delicate vesper mouse (Calomys tener) | 4 RT-PCR+/109 sera 3 RT-PCR+/252 sera |
2008–2013 (Brazil) | HEV-C | MG021328 | Partial | [47] |
| Bank vole (Myodes glareolus) | 1 RT-PCR+/1206 sera | 2009–2013 (Germany) | HEV-C | MK192412 | Partial | [48] | |
| Common vole (Microtus arvalis) | 13 RT-PCR+/646 sera | 2009–2013 (Germany, Czech Republic) and 2016 (Hungary) | HEV-C | MK192405-MK192409 MK192413-MK192420 | Complete/Partial | [48] | |
| Pere David’s vole (Eothenomys melanogaster) |
4 RT-PCR+/55 sera | 2013–2015 (Lijiang, China) | HEV-C | MG020022–MG020025 | Complete | [6] | |
| Inez’s red-backed vole (Eothenomys inez) | 2 RT-PCR+/23 sera | 2014 (Shanxi; China) | HEV-C | KY432904 | Complete | [46] | |
| Chinese striped hamster (Cricetulus barabensis) |
5 RT-PCR+/60 sera | 2014 (Hebei; China) | HEV-C | KY432899 | Complete | [46] | |
| Narrow-headed vole (Microtus gregalis) | 4 RT-PCR+/44 sera | 2015 (Xinjiang; China) | HEV-C | KY432902 | Complete | [46] | |
| Grey red-backed vole (Myodes rufocanus) | 3 RT-PCR+/23 sera | 2015 (Heilongjiang; China) | HEV-C | KY432901 | Complete | [46] | |
| Gray Dwarf Hamster (Cricetulus migratorius) |
1 RT-PCR+/15 sera | 2015 (Xinjiang; China) | HEV-C | KY432903 | Complete | [46] | |
| Soricomorpha/ Soricidae |
Asian musk shrew (Suncus murinus) | 12 HEV IgM+/260 sera; 5 RT-PCR+ |
December 2011–September 2012 (Zhanjiang; China) |
HEV-C1 | KC465990–KC466001 | Complete/Partial | [49] |
| Oliver’s shrew (Crocidura olivieri) | 1 RT-PCR+/5 sera | 2014 (Kenya) | HEV-C | NA | Partial | [50] | |
| Carnivora/ Mustelidae |
Western Ferrets (Mustela putorius) American mink (Neovison vison) |
2 RT-PCR+/63 fecal samples | 2008–2010 (Denmark) 2010 (Netherlands), 2013 (USA), 2009–2013 (Japan), 2016 (China) |
HEV-C2 | AB890375-AB890379 LC177789-LC177791 JN998606-JN998607 | Complete/Partial | [51] |
| Syrian brown bear (Ursus arctos syriacus) | 2 NBS-RT-PCR+/22 sera | 20011–2016 (Germany) | HEV-C1 | MF480313 | Partial | [52] | |
| Red Foxes (Vulpes vulpes) | 1 RT-PCR+/26 fecal samples | 2020 (Hungary) | HEV-C2 | MN906015 | Complete | [53] | |
| Dog (Canis familiaris) Cat (Felis catus) |
2 HEV Ig+/296 sera; 1 cat/1 dog |
2020 (Spain) | HEV-C1 | NA | Partial | [54] | |
| Farconiformes/ Falconidae |
Common kestrel (Falco tinnunculus) and Red-footed falcon (Falco vespertinus) | 2 RT-PCR+/18 fecal samples 1 RT-PCR+/7 fecal samples | 2014 (Hungary) | HEV-C | KU670940 | Complete/Partial | [55] |
Abbreviations: +: positive; NA: Not Available.
5. Pathogenesis and Diagnosis
Rocahepevirus ratti, like Paslahepevirus balayani, can cause acute hepatitis, chronic hepatitis, and subclinical infection. HEV-C1 infections are self-limiting and less severe than HEV-A infections, characterized by lower mean peak ALT and bilirubin levels in those with intact immunity. In general, the criteria for inclusion in these investigations to identify RHEV infection include clinical and biological manifestations consistent with acute or chronic hepatitis, as well as an ALT level significantly greater than the upper limit of normal. Infections in immunocompromised individuals, on the other hand, are challenging, with 50% of HEV-C1 infections progressing to persistence, according to research by S. Sridhar et al. [18]. This may be due to the fact that immunocompromised people might be more vulnerable to HEV-C1 infections. Not only was the viral pathogenicity examined, but meningoencephalitis was detected as a sequela in an immunocompromised patient after the hepatitis had entirely cured. Therefore, the correlation between RHEV infection and neurological symptoms must be studied. Subclinical HEV-C1 infection, without changes in hepatic function tests or clinical symptoms, is concerning over the safety of blood transfusions because the virus can be transmitted through contaminated blood samples due to the high viral load of HEV-C1 in plasma. RHEV’s viral tropism is oriented towards the liver, with a special affinity for liver cells. Virus replication occurs there, leading to liver damage and the release of liver enzymes into the blood, resulting in acute and chronic hepatitis in humans and animals. Recognizing the pathogenicity is crucial for the development of therapies for the prevention and treatment of hepatitis caused by this virus [18].
Ferrets and rats have been proposed as candidate animal models to study RHEV pathogenesis. Ferrets exhibit three patterns of infection: subclinical infection, acute hepatitis, and persistent infection [56]. Induced infections of RHEV on immunosuppressed rats have shown the effect of high-dose immunosuppression to induce chronic hepatitis and viral load suppression was observed with ribavirin treatment [57].
Molecular biology techniques have facilitated the identification of RHEV RNA in liver, feces, and blood samples obtained from various animal and human species. Such techniques include RT-PCR amplification, Sanger DNA sequencing, and metagenomic tests involving next generation sequencing. The combination of these methods, along with detailed phylogenetic and sequence evaluations, have allowed sequencing of RHEV genomes [4]. The first human RHEV infection was detected in a patient with chronic hepatitis, and this case was validated by RT-PCR and sequencing of plasma, stool, saliva, and liver tissue samples.
Currently, there are no commercially available serological methods to identify antibodies against RHEV or differentiate them from antibodies against HEV-A. Due to the risk of false negatives, the diagnosis cannot be based on serological tests alone. In addition, HEV-1 to HEV-4 have not been shown to provide protection against RHEV infection. However, a new specific immunoassay for RHEV antibody detection has been developed based on HEV-C1 p241 peptide and validated in solid organ recipients and immunocompetent individuals. This tool allowed to estimate a RHEV seroprevalence of 0.92%, indicating endemic exposure in Hong Kong [27]. This peptide (HEV-C1 p241 antigen) has been shown to be an immunogenic vaccine candidate against HEV-C1 [26].
The most accurate method for detecting Rocahepevirus ratti infection is the RT-PCR method by which viral genomic RNA can be identified. The sensitivity of the method depends on the design and selection of specific primers. The molecular diagnosis of HEV-A and RHEV may be challenging as several assays have shown low sensitivity for endemic genotypes in Europe and the number of genotypes/subtypes described is increasing, therefore, without an evaluation, their sensitivity in this context is unknown. Primers used to detect human HEV may not be able to recognize the RHEV genome in patient samples, limiting the sensitivity of PCR, and only the use of specific primers for RHEV has allowed its detection in different samples. It is important to note that the genetic diversity of HEV-C1 is still unclear, which could also limit the sensitivity of the techniques [19]. A recent study conducted by Lopez-Lopez et al. showed that the use of two parallel PCR assays (using ORF1 and ORF3) targeting different regions of the viral genome leads to significant improvement in the molecular diagnosis of RHEV, which can be a useful tool to map the transmission routes of HEV-C1 and hence to prevent human infections [58].
6. Conclusions
An increasing number of cases of animals and humans infected with Rocahepevirus ratti are being detected. Some species of rats have been confirmed as natural reservoirs of RHEV, and they are considered a source of zoonotic infection. It is likely that more cases will be discovered as detection techniques improve. Understanding the virus evolution and diffusion is essential for the development of successful prevention and control measures, treatments, and vaccines. The information obtained from the phylogenetic analysis and genomic sequencing will help researchers track the development of new strains, contributing to epidemiological distribution maps. In addition, this information can also be used to develop diagnostic tests to accurately detect the virus in collected samples and evolve in the discovery of a more precise pathogenesis. These tests are needed to assist healthcare professionals in diagnosing RHEV infections and preventing the spread of the virus. This is a public health concern because it is a new pathogen that can be transmitted to other species and even to humans. Further research is needed to determine the exact route of transmission of the virus to humans, as it remains unknown.
Author Contributions
Conception and design: G.R. and S.C.; collection and assembly of data: G.R. and S.B; designed Tables and Figures and wrote the manuscript: G.R. and S.B; writing—original draft preparation: G.R and S.B.; writing—review and editing: G.R. and S.B; supervision, G.R.; revised the paper and contributed to results discussion: G.R., S.C. and S.C. All authors have read and agreed to the published version of the manuscript.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research received no external funding.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.National Institute of Diabetes and Digestive and Kidney Diseases Hepatitis Viral. [(accessed on 31 October 2023)]; Available online: https://www.niddk.nih.gov/health-information/informacion-de-la-salud/enfermedades-higado/hepatitis-viral.
- 2.Purdy M.A., Drexler J.F., Meng X.-J., Norder H., Okamoto H., Van der Poel W.H.M., Reuter G., de Souza W.M., Ulrich R.G., Smith D.B. ICTV Virus Taxonomy Profile: Hepeviridae 2022. J. Gen. Virol. 2022;103:001778. doi: 10.1099/jgv.0.001778. [DOI] [PubMed] [Google Scholar]
- 3.Reuter G., Boros Á., Pankovics P. Review of Hepatitis E Virus in Rats: Evident Risk of Species Orthohepevirus C to Human Zoonotic Infection and Disease. Viruses. 2020;12:1148. doi: 10.3390/v12101148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rivero-Juarez A., Frias M., Perez A.B., Pineda J.A., Reina G., Fuentes-Lopez A., Freyre-Carrillo C., Ramirez-Arellano E., Alados J.C., Rivero A. Orthohepevirus C Infection as an Emerging Cause of Acute Hepatitis in Spain: First Report in Europe. J. Hepatol. 2022;77:326–331. doi: 10.1016/j.jhep.2022.01.028. [DOI] [PubMed] [Google Scholar]
- 5.Schoch C.L., Ciufo S., Domrachev M., Hotton C.L., Kannan S., Khovanskaya R., Leipe D., McVeigh R., O’Neill K., Robbertse B., et al. NCBI Taxonomy: A Comprehensive Update on Curation, Resources and Tools. Database. 2020;2020:baaa062. doi: 10.1093/database/baaa062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang B., Li W., Zhou J.H., Li B., Zhang W., Yang W.H., Pan H., Wang L.X., Bock C.T., Shi Z.L., et al. Chevrier’s Field Mouse (Apodemus chevrieri) and Père David’s Vole (Eothenomys melanogaster) in China Carry Orthohepeviruses That Form Two Putative Novel Genotypes within the Species Orthohepevirus C. Virol. Sin. 2018;33:44–58. doi: 10.1007/s12250-018-0011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang B., Harms D., Yang X.L., Bock C.T. Orthohepevirus C: An Expanding Species of Emerging Hepatitis e Virus Variants. Pathogens. 2020;9:154. doi: 10.3390/pathogens9030154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johne R., Heckel G., Plenge-Bönig A., Kindler E., Maresch C., Reetz J., Schielke A., Ulrich R.G. Novel Hepatitis E Virus Genotype in Norway Rats, Germany. Emerg. Infect. Dis. 2010;16:1452. doi: 10.3201/eid1609.100444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seitz R. Hepatitis e Virus: German Advisory Committee Blood (Arbeitskreis blut), Subgroup “Assessment of Pathogens Transmissible by Blood”. Transfus. Med. Hemother. 2015;42:247–265. doi: 10.1159/000431191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kenney S.P., Meng X.J. Hepatitis E Virus Genome Structure and Replication Strategy. Cold Spring Harb. Perspect. Med. 2019;9:a031724. doi: 10.1101/cshperspect.a031724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang B., Meng X.J. Structural and Molecular Biology of Hepatitis E Virus. Comput. Struct. Biotechnol. J. 2021;19:1907–1916. doi: 10.1016/j.csbj.2021.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bai H., Ami Y., Suzaki Y., Doan Y.H., Muramatsu M., Li T.C. Open Reading Frame 4 Is Not Essential in the Replication and Infection of Genotype 1 Hepatitis E Virus. Viruses. 2023;15:784. doi: 10.3390/v15030784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Capai L., Charrel R., Falchi A. Hepatitis e in High-Income Countries: What Do We Know? And What Are the Knowledge Gaps? Viruses. 2018;10:285. doi: 10.3390/v10060285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cancela F., Noceti O., Arbiza J., Mirazo S. Structural Aspects of Hepatitis E Virus. Arch. Virol. 2022;167:2457–2481. doi: 10.1007/s00705-022-05575-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faber M., Wenzel J.J., Erl M., Stark K., Schemmerer M. No Evidence for Orthohepevirus C in Archived Human Samples in Germany, 2000–2020. Viruses. 2022;14:742. doi: 10.3390/v14040742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pankovics P., Némethy O., Boros Á., Pár G., Szakály P., Reuter G. Four-Year Long (2014–2017) Clinical and Laboratory Surveillance of Hepatitis E Virus Infections Using Combined Antibody, Molecular, Antigen and Avidity Detection Methods: Increasing Incidence and Chronic HEV Case in Hungary. J. Clin. Virol. 2020;124:104284. doi: 10.1016/j.jcv.2020.104284. [DOI] [PubMed] [Google Scholar]
- 17.Sridhar S., Yip C.C.Y., Lo K.H.Y., Wu S., Situ J., Chew N.F.S., Leung K.H., Chan H.S.Y., Wong S.C.Y., Leung A.W.S., et al. Hepatitis e Virus Species C Infection in Humans, Hong Kong. Clin. Infect. Dis. 2022;75:288–296. doi: 10.1093/cid/ciab919. [DOI] [PubMed] [Google Scholar]
- 18.Sridhar S., Yip C.C.Y., Wu S., Chew N.F.S., Leung K.H., Chan J.F.W., Zhao P.S., Chan W.M., Poon R.W.S., Tsoi H.W., et al. Transmission of Rat Hepatitis E Virus Infection to Humans in Hong Kong: A Clinical and Epidemiological Analysis. Hepatology. 2021;73:10–22. doi: 10.1002/hep.31138. [DOI] [PubMed] [Google Scholar]
- 19.Sridhar S., Yip C.C.Y., Wu S., Cai J., Zhang A.J.X., Leung K.H., Chung T.W.H., Chan J.F.W., Chan W.M., Teng J.L.L., et al. Rat Hepatitis E Virus as Cause of Persistent Hepatitis after Liver Transplant. Emerg. Infect. Dis. 2018;24:2241–2250. doi: 10.3201/eid2412.180937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andonov A., Robbins M., Borlang J., Cao J., Hatchette T., Stueck A., Deschambault Y., Murnaghan K., Varga J., Johnston L. Rat Hepatitis e Virus Linked to Severe Acute Hepatitis in an Immunocompetent Patient. J. Infect. Dis. 2019;220:951–955. doi: 10.1093/infdis/jiz025. [DOI] [PubMed] [Google Scholar]
- 21.Parraud D., Lhomme S., Péron J.M., Da Silva I., Tavitian S., Kamar N., Izopet J., Abravanel F. Rat Hepatitis E Virus: Presence in Humans in South-Western France? Front. Med. 2021;8:726363. doi: 10.3389/fmed.2021.726363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodriguez C., Marchand S., Sessa A., Cappy P., Pawlotsky J.M. Orthohepevirus C Hepatitis, an Underdiagnosed Disease? J. Hepatol. 2023;79:2457–2481. doi: 10.1016/j.jhep.2023.02.008. [DOI] [PubMed] [Google Scholar]
- 23.Li T.-C., Ami Y., Suzaki Y., Yasuda S.P., Yoshimatsu K., Arikawa J., Takeda N., Takaji W. Characterization of Full Genome of Rat Hepatitis E Virus Strain from Vietnam. Emerg. Infect. Dis. 2013;19:115–118. doi: 10.3201/eid1901.121007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dremsek P., Wenzel J.J., Johne R., Ziller M., Hofmann J., Groschup M.H., Werdermann S., Mohn U., Dorn S., Motz M., et al. Seroprevalence Study in Forestry Workers from Eastern Germany Using Novel Genotype 3- and Rat Hepatitis E Virus-Specific Immunoglobulin G ELISAs. Med. Microbiol. Immunol. 2012;201:189–200. doi: 10.1007/s00430-011-0221-2. [DOI] [PubMed] [Google Scholar]
- 25.Shimizu K., Hamaguchi S., Ngo C.C., Li T.C., Ando S., Yoshimatsu K., Yasuda S.P., Koma T., Isozumi R., Tsuda Y., et al. Serological Evidence of Infection with Rodent-Borne Hepatitis E Virus HEV-C1 or Antigenically Related Virus in Humans. J. Vet. Med. Sci. 2016;78:1677–1681. doi: 10.1292/jvms.16-0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sridhar S., Situ J., Cai J.-P., Yip C.C.-Y., Wu S., Zhang A.J.-X., Wen L., Chew N.F.-S., Chan W.-M., Poon R.W.-S., et al. Multimodal Investigation of Rat Hepatitis E Virus Antigenicity: Implications for Infection, Diagnostics, and Vaccine Efficacy. J. Hepatol. 2021;74:1315–1324. doi: 10.1016/j.jhep.2020.12.028. [DOI] [PubMed] [Google Scholar]
- 27.Situ J., Hon-Yin Lo K., Cai J.-P., Li Z., Wu S., Hon-Kiu Shun E., Foo-Siong Chew N., Yiu-Hung Tsoi J., Sze-Man Chan G., Hei-Man Chan W., et al. An Immunoassay System to Investigate Epidemiology of Rocahepevirus ratti (Rat Hepatitis E Virus) Infection in Humans. JHEP Rep. 2023;5:100793. doi: 10.1016/j.jhepr.2023.100793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang H., Neyvaldt J., Enache L., Sikora P., Mattsson A., Johansson A., Lindh M., Bergstedt O., Norder H. Variations among Viruses in Influent Water and Effluent Water at a Wastewater Plant over One Year as Assessed by Quantitative PCR and Metagenomics. Appl. Environ. Microbiol. 2020;86:e02073-20. doi: 10.1128/AEM.02073-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palombieri A., Di Profio F., Sarchese V., Fruci P., Suffredini E., Martella V., Veneri C., Bonanno Ferraro G., Mancini P., La Rosa G., et al. Surveillance for Rat Hepatitis E in Wastewater Networks, Italy. Microbiol. Spectr. 2023;11:e02675-23. doi: 10.1128/spectrum.02675-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Casares-Jimenez M., Garcia-Garcia T., Suárez-Cárdenas J.M., Perez-Jimenez A.B., Martín M.A., Caballero-Gómez J., Michán C., Corona-Mata D., Risalde M.A., Perez-Valero I., et al. Correlation of Hepatitis E and Rat Hepatitis E Viruses Urban Wastewater Monitoring and Clinical Cases. Sci. Total Environ. 2024;908:168203. doi: 10.1016/j.scitotenv.2023.168203. [DOI] [PubMed] [Google Scholar]
- 31.Purcell R.H., Engle R.E., Rood M.P., Kabrane-Lazizi Y., Nguyen H.T., Govindarajan S., Claire M.S., Emerson S.U. Hepatitis E Virus in Rats, Los Angeles, California, USA. Emerg. Infect. Dis. 2011;17:2216–2222. doi: 10.3201/eid1712.110482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ryll R., Bernstein S., Heuser E., Schlegel M., Dremsek P., Zumpe M., Wolf S., Pépin M., Bajomi D., Müller G., et al. Detection of Rat Hepatitis E Virus in Wild Norway Rats (Rattus norvegicus) and Black Rats (Rattus rattus) from 11 European Countries. Vet. Microbiol. 2017;208:58–68. doi: 10.1016/j.vetmic.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 33.Johne R., Plenge-Bönig A., Hess M., Ulrich R.G., Reetz J., Schielke A. Detection of a Novel Hepatitis E-like Virus in Faeces of Wild Rats Using a Nested Broad-Spectrum RT-PCR. J. Gen. Virol. 2010;91:750–758. doi: 10.1099/vir.0.016584-0. [DOI] [PubMed] [Google Scholar]
- 34.Johne R., Dremsek P., Kindler E., Schielke A., Plenge-Bönig A., Gregersen H., Wessels U., Schmidt K., Rietschel W., Groschup M.H., et al. Rat Hepatitis E Virus: Geographical Clustering within Germany and Serological Detection in Wild Norway Rats (Rattus norvegicus) Infect. Genet. Evol. 2012;12:947–956. doi: 10.1016/j.meegid.2012.02.021. [DOI] [PubMed] [Google Scholar]
- 35.Li T.C., Yoshimatsu K., Yasuda S.P., Arikawa J., Koma T., Kataoka M., Ami Y., Suzaki Y., Mai L.T.Q., Hoa N.T., et al. Characterization of Self-Assembled Virus-like Particles of Rat Hepatitis E Virus Generated by Recombinant Baculoviruses. J. Gen. Virol. 2011;92:2830–2837. doi: 10.1099/vir.0.034835-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li W., Guan D., Su J., Takeda N., Wakita T., Li T.C., Ke C.W. High Prevalence of Rat Hepatitis E Virus in Wild Rats in China. Vet. Microbiol. 2013;165:275–280. doi: 10.1016/j.vetmic.2013.03.017. [DOI] [PubMed] [Google Scholar]
- 37.Lack J.B., Volk K., Van Den Bussche R.A. Hepatitis E Virus Genotype 3 in Wild Rats, United States. Emerg. Infect. Dis. 2012;18:1268–1273. doi: 10.3201/eid1808.120070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murphy E.G., Williams N.J., Jennings D., Chantrey J., Verin R., Grierson S., McElhinney L.M., Bennett M. First Detection of Hepatitis E Virus (Orthohepevirus C) in Wild Brown Rats (Rattus norvegicus) from Great Britain. Zoonoses Public Health. 2019;66:686–694. doi: 10.1111/zph.12581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robinson S.J., Borlang J., Himsworth C.G., Pearl D.L., Weese J.S., Dibernardo A., Osiowy C., Nasheri N., Jardine C.M. Rat Hepatitis E Virus in Norway Rats, Ontario, Canada, 2018–2021. Emerg. Infect. Dis. 2023;29:1890–1894. doi: 10.3201/eid2909.230517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Porea D., Raileanu C., Crivei L.A., Gotu V., Savuta G., Pavio N. First Detection of Hepatitis E Virus (Rocahepevirus ratti Genotype C1) in Synanthropic Norway Rats (Rattus norvegicus) in Romania. Viruses. 2023;15:1337. doi: 10.3390/v15061337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mulyanto, Depamede S.N., Sriasih M., Takahashi M., Nagashima S., Jirintai S., Nishizawa T., Okamoto H. Frequent Detection and Characterization of Hepatitis E Virus Variants in Wild Rats (Rattus rattus) in Indonesia. Arch. Virol. 2013;158:87–96. doi: 10.1007/s00705-012-1462-0. [DOI] [PubMed] [Google Scholar]
- 42.Mulyanto, Suparyatmo J.B., Andayani I.G.A.S., Khalid, Takahashi M., Ohnishi H., Jirintai S., Nagashima S., Nishizawa T., Okamoto H. Marked Genomic Heterogeneity of Rat Hepatitis E Virus Strains in Indonesia Demonstrated on a Full-Length Genome Analysis. Virus Res. 2014;179:102–112. doi: 10.1016/j.virusres.2013.10.029. [DOI] [PubMed] [Google Scholar]
- 43.Primadharsini P.P., Mulyanto, Wibawa I.D.N., Anggoro J., Nishizawa T., Takahashi M., Jirintai S., Okamoto H. The Identification and Characterization of Novel Rat Hepatitis E Virus Strains in Bali and Sumbawa, Indonesia. Arch. Virol. 2018;163:1345–1349. doi: 10.1007/s00705-018-3736-7. [DOI] [PubMed] [Google Scholar]
- 44.Simanavicius M., Juskaite K., Verbickaite A., Jasiulionis M., Tamosiunas P.L., Petraityte-Burneikiene R., Zvirbliene A., Ulrich R.G., Kucinskaite-Kodze I. Detection of Rat Hepatitis E Virus, but Not Human Pathogenic Hepatitis E Virus Genotype 1–4 Infections in Wild Rats from Lithuania. Vet. Microbiol. 2018;221:129–133. doi: 10.1016/j.vetmic.2018.06.014. [DOI] [PubMed] [Google Scholar]
- 45.Takahashi M., Kunita S., Kawakami M., Kadosaka T., Fujita H., Takada N., Miyake M., Kobayashi T., Ohnishi H., Nagashima S., et al. First Detection and Characterization of Rat Hepatitis E Virus (HEV-C1) in Japan. Virus Res. 2022;314:198766. doi: 10.1016/j.virusres.2022.198766. [DOI] [PubMed] [Google Scholar]
- 46.Wu Z., Lu L., Du J., Yang L., Ren X., Liu B., Jiang J., Yang J., Dong J., Sun L., et al. Comparative Analysis of Rodent and Small Mammal Viromes to Better Understand the Wildlife Origin of Emerging Infectious Diseases. Microbiome. 2018;6:178. doi: 10.1186/s40168-018-0554-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Souza W.M., Romeiro M.F., Sabino-Santos G., Maia F.G.M., Fumagalli M.J., Modha S., Nunes M.R.T., Murcia P.R., Figueiredo L.T.M. Novel Orthohepeviruses in Wild Rodents from São Paulo State, Brazil. Virology. 2018;519:12–16. doi: 10.1016/j.virol.2018.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ryll R., Heckel G., Corman V.M., Drexler J.F., Ulrich R.G. Genomic and Spatial Variability of a European Common Vole Hepevirus. Arch. Virol. 2019;164:2671–2682. doi: 10.1007/s00705-019-04347-1. [DOI] [PubMed] [Google Scholar]
- 49.Guan D., Li W., Su J., Fang L., Takeda N., Wakita T., Li T.C., Ke C. Asian Musk Shrew as a Reservoir of Rat Hepatitis E Virus, China. Emerg. Infect. Dis. 2013;19:1341–1343. doi: 10.3201/eid1908.130069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Onyuok S.O., Hu B., Li B., Fan Y., Kering K., Ochola G.O., Zheng X.S., Obanda V., Ommeh S., Yang X.L., et al. Molecular Detection and Genetic Characterization of Novel RNA Viruses in Wild and Synanthropic Rodents and Shrews in Kenya. Front. Microbiol. 2019;10:2696. doi: 10.3389/fmicb.2019.02696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li T.C., Yang T., Ami Y., Suzaki Y., Shirakura M., Kishida N., Asanuma H., Takeda N., Takaji W. Complete Genome of Hepatitis E Virus from Laboratory Ferrets. Emerg. Infect. Dis. 2014;20:709–712. doi: 10.3201/eid2004.131815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spahr C., Ryll R., Knauf-Witzens T., Vahlenkamp T.W., Ulrich R.G., Johne R. Serological Evidence of Hepatitis E Virus Infection in Zoo Animals and Identification of a Rodent-Borne Strain in a Syrian Brown Bear. Vet. Microbiol. 2017;212:87–92. doi: 10.1016/j.vetmic.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 53.Lanszki Z., Kurucz K., Zeghbib S., Kemenesi G., Lanszki J., Jakab F. Identification of Hepatitis E Virus in the Feces of Red Foxes (Vulpes vulpes) Animals. 2020;10:1841. doi: 10.3390/ani10101841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Caballero-Gómez J., Rivero-Juarez A., Jurado-Tarifa E., Jiménez-Martín D., Jiménez-Ruiz E., Castro-Scholten S., Ulrich R.G., López-López P., Rivero A., García-Bocanegra I. Serological and Molecular Survey of Hepatitis E Virus in Cats and Dogs in Spain. Transbound. Emerg. Dis. 2022;69:240–248. doi: 10.1111/tbed.14437. [DOI] [PubMed] [Google Scholar]
- 55.Reuter G., Boros Á., Mátics R., Kapusinszky B., Delwart E., Pankovics P. Divergent Hepatitis E Virus in Birds of Prey, Common Kestrel (Falco tinnunculus) and Red-Footed Falcon (F. vespertinus), Hungary. Infect. Genet. Evol. 2016;43:343–346. doi: 10.1016/j.meegid.2016.06.013. [DOI] [PubMed] [Google Scholar]
- 56.Li T.-C., Yang T., Yoshizaki S., Ami Y., Suzaki Y., Ishii K., Kishida N., Shirakura M., Asanuma H., Takeda N., et al. Ferret Hepatitis E Virus Infection Induces Acute Hepatitis and Persistent Infection in Ferrets. Vet. Microbiol. 2016;183:30–36. doi: 10.1016/j.vetmic.2015.11.014. [DOI] [PubMed] [Google Scholar]
- 57.Sridhar S., Wu S., Situ J., Shun E.H.-K., Li Z., Zhang A.J.-X., Hui K., Fong C.H.-Y., Poon V.K.-M., Chew N.F.-S., et al. A Small Animal Model of Chronic Hepatitis E Infection Using Immunocompromised Rats. JHEP Rep. 2022;4:100546. doi: 10.1016/j.jhepr.2022.100546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lopez-Lopez P., Frias M., Perez-Jimenez A.B., Freyre-Carrillo C., Pineda J.A., Aguilera A., Fuentes A., Alados J.C., Reina G., Ramirez-Arellano E., et al. Optimization of the Molecular Diagnosis of the Acute Hepatitis E Virus Infection. Microb. Biotechnol. 2023;16:1325–1332. doi: 10.1111/1751-7915.14247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.