Abstract
In Saccharomyces cerevisiae, the transition from the G1 phase of the mitotic cycle into S phase is controlled by a set of G1 cyclins that regulate the activity of the protein kinase encoded by CDC28. Yeast cells regulate progress through the G1/S boundary in response to nutrients, moving quickly through G1 in glucose medium and more slowly in poorer medium. We have examined connections between glucose and the level of the message encoding Cln3, a G1 cyclin. We found that glucose positively regulates CLN3 mRNA levels through a set of repeated AAGAAAAA (A2GA5) elements within the CLN3 promoter. Mutations in these sequences reduce both transcriptional activation and specific interaction between CLN3 promoter elements and proteins in yeast extracts. Creation of five point mutations, replacing the G’s within these repeats with T’s, in the CLN3 promoter substantially reduces CLN3 expression in glucose medium and inhibits the ability of the cells to maintain a constant size when shifted into glucose.
Saccharomyces cerevisiae cells coordinate growth and proliferation so that cells growing under a variety of conditions are able to maintain an almost constant average size. Cell growth and division are thus in some way tethered. Were it not so, an increase in growth rate in the absence of a corresponding increase in the rate of cell division would generate larger cells. Conversely, a decrease in growth rate in the absence of a corresponding lengthening of the cell cycle would produce very small cells. Yeast cells adjust the length of the cell cycle in different media by varying the length of the G1 phase (11, 12). In rich media containing glucose, yeast cells grow rapidly and have a short G1 phase, whereas in a poor carbon source such as ethanol, yeast cells prolong the cell cycle by spending more time in G1. As nutrients are depleted, G1 lengthens until the cells eventually arrest in G1 (25). While cell size clearly plays a role in governing the length of G1, the effects of nutrients on cell cycle length cannot be explained solely by a model in which nutrients affect the time required to reach a threshold size for budding. This can be inferred from the fact that G1 length is strongly influenced by nutrients in mother cells, which have already reached critical size (10b, 15, 16).
Progress through G1 requires the association of the Cdc28 kinase with one of three G1 cyclins, Cln1, Cln2, or Cln3. Mutation in all three G1 cyclins or inactivation of Cdc28 leads to cell cycle arrest in late G1 (9). Recent observations suggest that Cln3 is an upstream regulator of other G1 cyclins (5, 23, 24). It has been shown that transcriptional manipulation of CLN3 alters the duration of G1: overexpression of CLN3 shortens G1, while deletion of CLN3 leads to a delay in G1 (2, 18). Recently, work in several laboratories has demonstrated nutrient regulation of Cln3/Cdc28 kinase activity. Nutrients regulate Cln3 protein levels through effects on transcription, translation, and protein stability (8, 10, 19, 20). CLN3 mRNA levels are high in log-phase cells rapidly growing on glucose and decline as cells enter oxidative growth in ethanol. Carbon source regulation of CLN3 transcription involves a mechanism that is independent of growth and does not require the Ras-cyclic AMP pathway (19). Changes in CLN3 mRNA seen in response to different carbon sources are reflected in Cln3 protein levels and Cln3/Cdc28 kinase activity (10).
Addition of glucose to post-log-phase cells can produce a 5- to 10-fold increase in CLN3 mRNA levels within 5 min, indicating that the cells are in some way responding to a glucose signal. Several transcriptional responses to glucose have been studied, including the induction of glycolytic enzymes, repression of genes necessary for growth in nonfermentable carbon sources, and repression of genes involved in respiration (13). Thinking that regulation of CLN3 by glucose might involve a known glucose signalling pathway, we have tested several mutations in glucose repression and induction pathways for alterations in the regulation of CLN3. These experiments have produced negative results: CLN3 induction is normal in these mutants. This indicates that the mechanism of glucose regulation of CLN3 is probably distinct from previously characterized processes. In addition, although the Mcm1 protein is thought to play a role in cell cycle-specific variations in CLN3 transcript levels (17), we have found that temperature-sensitive mcm1 mutants have normal glucose induction of CLN3 at the restrictive temperature (10a). This, taken together with the finding that induction of CLN3 by glucose is cell cycle independent (19), indicates that glucose induction of CLN3 is also distinct from the cell cycle-dependent transcriptional regulation reported by McInerny et al. (17).
We have therefore examined the CLN3 promoter in order to identify regions that regulate transcription in response to glucose. Here we report the presence of repeated sequences within the CLN3 promoter that are important for transcriptional induction by glucose. Mutations in these sequences reduce both transcriptional activation and specific interactions between CLN3 promoter elements and proteins in yeast extracts. Southwestern blots identify a 69-kDA protein that specifically binds to these repeated sequences.
MATERIALS AND METHODS
Strains and media.
The yeast strains used in this work were DS10 (MATahis3-11,15 leu2-3,112 lys1 lys2 ura3-52 trp1Δ) and TG3 (isogenic to DS10 except for a CLN3 deletion [3]). These strains were transformed with various plasmids as indicated. Cells were grown either in YEPD containing 1% yeast extract, 2% Bacto Peptone, and 2% glucose or in synthetic complete medium containing 6.7-g/liter yeast nitrogen base (Difco) supplemented with adenine, uracil, amino acids, and 2% glucose (21). Cell size measurements were performed with a Coulter Counter Channelizer using a 70-μm aperture calibrated with latex beads.
RNA preparation and Northern blotting.
Total yeast RNA was isolated as described previously (6). The RNA samples were separated by formaldehyde gel electrophoresis and transferred to a GeneScreen Plus membrane (New England Nuclear). To ensure uniform loading and transfer of RNA, ethidium bromide was added to the samples, and the ethidium-stained rRNA was visualized on the blots under UV illumination. Blots were also probed with a radiolabeled 0.6-kb SacI fragment from U2 to confirm uniform loading. Northern blots were probed with a 1.1-kb EcoRI fragment from URA3 or a 1.8-kb BamHI fragment from CLN3. All probes were radiolabeled to a specific activity of 109 cpm/μg by using [α-32p]dCTP (3,000 μCi/nmol) and random priming.
CLN3 promoter-reporter fusion constructs.
A series of CLN3 promoter fragments were generated by PCR and restriction digests and then inserted into the multiple cloning site of yeast 2μ plasmid pCA205 (a gift from Cathy Atchinson) upstream of URA3, which served as a reporter gene. The URA3 coding region in pCA205 is preceded by only 24 bp of the untranslated URA3 sequence containing one transcriptional start site and no known upstream activating sequences.
The −726 to +18 fragment was amplified by PCR using a primer (5′-TGCGACCGTGATGATACCGA-3′) upstream of the SacI site at −726 as an upstream primer and a primer (5′-TAATTATGGGATCCTTCAATATGGCC-3′) into which a BamHI site (underlined) was introduced as the downstream primer. The product was then cut with BamHI and SacI and inserted into the SacI/BamHI sites of pCA205. A similar strategy was used to subclone the −549 to +18 fragment into pCA205 by using the same downstream primer and an upstream primer (5′-CTCACTGTAATGATCAAGTTAC-3′) into which a BclI site was introduced. After digestion with BclI and BamHI, the PCR product was inserted into the BamHI site of pCA205. The −726 to −143 fragment was generated by using the primer described above upstream of the SacI site and a downstream primer (5′-ATTGGAGATCTGAGATTGCG-3′) with an introduced BglII site. After digestion with SacI and BglII, the PCR product was inserted into the SacI/BamHI site of pCA205. The −726 to −250 fragment was amplified by using an upstream primer (5′-GAGACACCTGCAGAGGCTACATTA-3′) and a primer (5′-GGTTTAAGTCTGCAGAGGAGACTAC-3′) containing an artificial PstI site downstream. The PCR product was digested with SalI (native site at −931) and PstI was then inserted into pBSK+. Digestion with SacI (−726) and PstI released the fragment from the plasmid for subcloning into SacI- and PstI-digested pCA205. The plasmid containing the −726 to −414 fragment was produced by deletion of the XhoI/BamHI segment from the −726 to +18 plasmid. After digestion, the ends were filled in with Klenow and ligated. The plasmid carrying the −549 to −414 segment was generated by digesting the −549 to +18 pCA205 plasmid with XhoI and BamHI. Again, the ends were ligated after Klenow filling. The −726 to −549 plasmid was made by using a primer (5′-GAGACACCTGCAGAGGCTACATTA-3′) upstream of the naturally occurring SacI (−726) site and a downstream primer (5′-GTA ACT TGA TCA TTA CAG TGA G-3′) flanking the naturally occurring BclI site. The fragment was cut with SacI and BclI and inserted into the SacI/BamHI site of pCA205. The plasmid containing the −626 to −570 promoter fragment was generated by annealing two synthetic oligonucleotides (5′-AATTCAAGAAAAAAAAAAAAGAAAAAGTGAAAAATTATCAGGCAAGAAAAAGAAATTAC-3′ and 5′- GATCGTAATTTCTTTTTCTTGCCTGATAATTTTTCACTTTTTCTTTTTT TTTTTCTT-3′) such that the resulting four-base overhangs on the ends were compatible with EcoRI and BamHI overhangs. This double-stranded oligonucleotide was then cloned into the EcoRI/BamHI site of pCA205. The corresponding plasmid with the −626 to −570 fragment carrying mutations in the A2GA5 repeats was made in the same manner, by using the following oligonucleotides (the substitutions are underlined): 5′-AATTCAAAAAAAAAAAAAAAAAAAAAGTGAAAAATTATCAGGCAAAAAAAAAAAATTAC-3′ and 5′- GATCGTAATTTTTTTTTTTTGCCTGATAATTTTTCACTTTTTTTTTTTTT TTTTTTT-3′. Both plasmids were sequenced to ensure proper positioning of the inserts. All plasmid constructs were sequenced to ensure that no mutations were generated by PCR. Plasmid copy number varied by less than 25% (data not shown).
Gel shift assays.
Yeast cells (DS10) were grown in YEPD medium (500 ml) to an A660 of 6. The cells were resuspended in fresh medium and, after incubation at 30°C for 30 min, collected by centrifugation at 2,000 rpm for 20 min. Under these conditions, transcription of CLN3 is high. The cells were resuspended in protein extraction buffer containing 0.2 M Tris (pH 8.0), 0.4 M ammonium sulfate, 10 mM MgCl2, 1 mM EDTA, 10% glycerol, 2 mM β-mercaptoethanol, 1-μg/ml pepstatin A, 0.2-mg/ml phenylmethylsulfonyl fluoride, and 20-μg/ml aprotinin. Frozen cell pellets were prepared by immediately dripping the cell suspension into liquid nitrogen. Cell disruption was performed by extensive chopping of the cell pellets in a household mini-chopper. A fine powder was obtained and allowed to thaw on ice. Cellular debris were removed by centrifugation at 20,000 rpm for 20 min at 4°C. The supernatant was precipitated with 0.534 volume of 3.8 M ammonium sulfate for 30 min at 4°C. The precipitated proteins were collected by centrifugation at 20,000 rpm for 20 min. Pellets were resuspended in 500 μl of protein buffer (20 mM HEPES [pH 8.0], 100 mM KCl, 1 mM MgCl2, 0.2 mM dithiothreitol [DTT], 5% glycerol, 0.5 mg of phenylmethylsulfonyl fluoride) and dialyzed for 3 h against two 500-ml changes of the same buffer. Insoluble materials were removed by 5 min of centrifugation at 14,000 rpm in a microcentrifuge at 4°C. The supernatant was immediately frozen on dry ice and stored at −70°C. Protein concentration was determined by using a Bio-Rad kit.
Binding reactions were carried out in a 20-μl reaction volume containing 20 mM HEPES (pH 8.0), 0.1% Nonindet P-40, 5% glycerol, 1 mM DTT, 50 mM KCl, 0.5 mM MgCl2, 2 μg of poly(dI-dC), 2 to 15 fmol of labeled DNA probe, and 4 to 8 μg of crude yeast extract. The protein extract was first incubated on ice with a molar excess of the unlabeled competitor (when added) for 5 min and then incubated for 15 min at room temperature in the presence of the labeled probe. Samples were subjected to electrophoresis on a 4% polyacrylamide gel (acrylamide/bisacrylamide ratio, 29:1) in 0.5× TBE buffer (Tris-borate [pH 8.0], 1 mM EDTA) after the gel had been prerun for 1 h at room temperature. The gel was dried on Whatman paper for 1 h at 80°C and analyzed by using a Molecular Dynamics PhosphorImager.
Two radiolabeled DNA probes were used. The first was the −726 to −549 fragment from the corresponding pCA205 construct released with EcoRI and SalI. The second probe (−622 to −603) was a double-stranded synthetic oligonucleotide made by annealing the oligonucleotides (5′-GGGCAAGAAAAAAAAAAAGAAAAAG-3′ and 5′-GGGCTTTTTCTTTTTTTTTCTTG-3′) corresponding to the region from −622 to −603 and producing non-CLN3 GGG overhangs for fill-in labeling with Klenow. Both probes were radiolabeled with [α-32P]dCTP and Klenow fragment. Competition studies were performed with a molar excess of the unlabeled probe fragment or other double-stranded oligonucleotides, as indicated.
Southwestern blotting.
The protocol described by Bassel-Duby et al. (1) was used with minor modifications. Protein extracts (50 μg) were separated by electrophoresis on a sodium dodecyl sulfate–7.5% polyacrylamide gel and transferred to nitrocellulose. To confirm uniform loading, the blot was stained with Ponceau S. The air-dried nitrocellulose paper was immersed in binding buffer containing 25 mM HEPES (pH 8.00), 60 mM KCl, 1 mM EDTA, 1 mM DTT, and 6 M guanidine hydrochloride and gently shaken for 10 min at 4°C. To renature the proteins, guanidine hydrochloride was gradually removed by repeating the same procedure eight times, each time with a concentration of guanidine hydrochloride twofold lower than that used in the previous wash. After the final wash without guanidine hydrochloride, the nitrocellulose was incubated for 1 h (room temperature) in 1× binding buffer containing 5% nonfat milk and 5-μg/ml salmon sperm DNA. The filter was then incubated in binding buffer with 0.25% nonfat milk and 106 cpm of the radiolabeled probe per ml either with or without competitor, as indicated. The probe for the Southwestern experiments was the −726 to −549 fragment described above for the gel shift experiments. Binding was carried out overnight. The filter was washed four times for 7 min each with 1× binding buffer at room temperature. The blot was analyzed by using a Molecular Dynamics PhosphorImager.
Site-directed mutagenesis.
A PCR-based mutagenesis strategy, using Pfu DNA polymerase to minimize errors, was used to generate a CLN3 promoter fragment corresponding to positions −931 to −414 in which the G’s at positions −620, −608, −583, −577, and −455 were replaced with A’s. CEN-based plasmid pKL001 (14), containing an epitope-tagged copy of the CLN3 gene driven by the normal CLN3 promoter, served as a template. First, a PCR fragment was made by using an upstream oligonucleotide flanking the XhoI site in plasmid pKL001 (5′-GGTACCGGGCCCCCCCTCGAGGTCGAC-3′; the XhoI site is underlined) and a downstream primer (−627 to −569) with four of the mutations (5′-GCAGGCTTGGTAATTTATTTATTGCCTGATAATTTTTCACTTTTTA TTTTTTTTTTATTGAAAG-3′; mutated bases are in boldface underlined). The product (positions −931 to −569) obtained with this first PCR step was then gel purified and used as the upstream primer in a second PCR with an oligonucleotide (5′-GATTAAAAGCTCGAGGAAAGTACAGATATACAAATTATAAATAGGTAGGAGGAATAAAAAAAAAAG-3′) in which the C at position −455 was replaced with a T (underlined) as a downstream primer. The final PCR product was digested with XhoI and used to replace the corresponding XhoI fragment in pKL001, replacing the wild-type region of the CLN3 promoter between positions −931 and −414, with a fragment containing the mutations. The resulting plasmid, pKL001ΔG, was sequenced to confirm the presence of the mutations and the orientation of the fragment. pKL001ΔG and the original wild-type pKL001 plasmid were transformed into TG3 cells carrying a deletion in CLN3 so that the only copy of CLN3 in the cell was that which was carried on the single-copy plasmid.
Immunoprecipitation-Western blotting.
Immunoprecipitation-Western blotting using epitope-tagged protein Cln3 was done as described by Tyers et al. (24). For each immunoprecipitation assay point, 5 mg of extract protein was used. Monoclonal antibody 12CA5 was used in the immunoprecipitations and as the primary antibody after blotting. An ECL detection kit (Amersham) was used to develop the Western blots.
RESULTS
To identify elements within the CLN3 promoter that are involved in glucose regulation of transcription, we tested the ability of CLN3 promoter fragments to drive the expression of URA3 as a reporter gene. Fragments of various lengths from the region of the CLN3 gene 5′ of the open reading frame, as indicated in Fig. 1, were inserted into the polylinker of 2μ vector pCA205, a gift from Cathy Atchinson. This places the sequences to be tested upstream of a URA3 gene that contains the URA3 coding region with only a minimal portion of the promoter, 24 bp in length. To test the constructs, yeast cells carrying the plasmids were grown to post-log phase in selective media. Glucose was added back to the cells, and the ability of the promoter fragments to enhance transcription of URA3 was assessed with Northern blots.
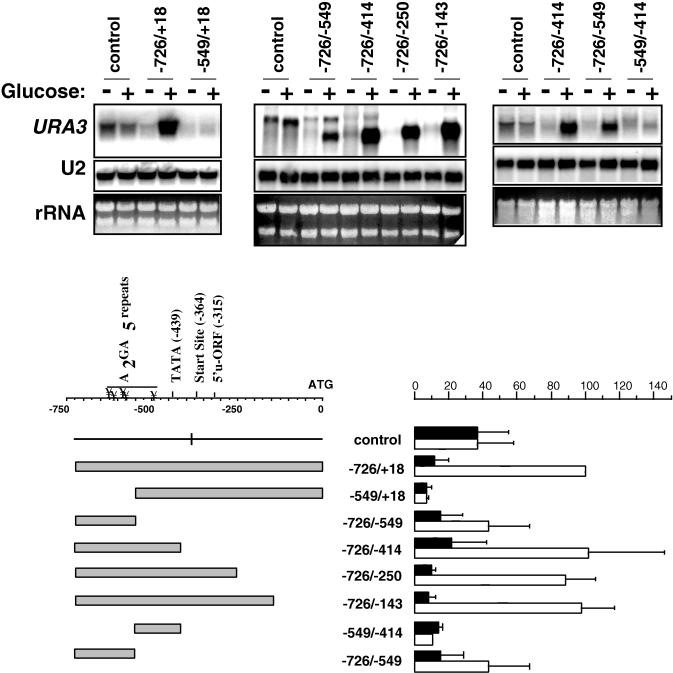
FIG. 1.
Glucose stimulation of CLN3 promoter-URA3 reporter constructs. DS10 cells were transformed with a set of plasmids carrying CLN3 promoter fragments of various lengths, as indicated, and grown in synthetic complete medium to post-log phase (optical density at 2 to 2.5). Cells were collected before and 20 min after transfer to fresh medium for RNA preparation. RNA (7.5 μg/lane in the left and center panels and 15 μg/lane in the right panel) was run on an agarose gel for Northern blotting with a URA3 probe. The bars represent the averages of PhosphorImager data for the URA3 reporter from at least three separate experiments for each construct, normalized to the −726 to +18 signal in fresh medium. Dark bars represent post-log-phase cells, and light bars indicate cells in fresh medium. Error bars represent standard deviations. The CLN3 transcriptional start site, the TATA box, the A2GA5 regulatory elements, and the upstream open reading frame (u-ORF) identified by Polymenis and Schmidt (20) are designated on the map.
As shown in Fig. 1, the vector alone with no CLN3 insert expresses some of the URA3 message, which is driven by a cryptic promoter within 2μ sequences upstream of the polylinker (data not shown). It is important to note that this basal level of transcription is glucose independent. Insertion of a fragment (−726 to +18) corresponding to the 726 bp immediately upstream of the CLN3 coding sequence caused URA3 transcription to become strongly glucose dependent. In this construct, transcription was initiated from the normal CLN3 transcriptional start site, as evidenced by primer extension mapping (data not shown) and the fact that the message decreases in size with 3′ deletion of the insert (Fig. 1). In the control plasmid, transcriptional initiation takes place within 2μ sequences approximately 350 bp upstream of the URA3 ATG. The addition of the full-length CLN3 promoter places the CLN3 initiation site (at −364) approximately the same distance from the ATG. For this reason, the control message and that produced by the −726 to +18 construct are approximately the same size. As 3′ deletions are made from the CLN3 promoter segment, the CLN3 initiation site moves closer to the ATG, leading to progressively smaller messages.
While the region extending 726 bp upstream from the CLN3 translational start produced strong glucose induction of the reporter gene, 5′ deletion of 177 bp from this fragment, leaving the region from −549 to +18 driving the promoter, led to loss of the glucose response. In contrast, truncation of the promoter from the 3′ end produced relatively little effect on the URA3 transcript level, such that the −726 to −549 element alone remained strongly glucose inducible. However, while the −726 to −549 fragment confers glucose-dependent transcription, there is a decrease in the URA3 message when driven by the −726 to −549 fragment compared to the −726 to −414 fragment. This may suggest that removal of the region between −549 to −414 caused loss of some glucose-responsive elements. On the other hand, the difference in transcriptional activity between the two fragments could also be due to the fact that the −726 to −549 element lacks the CLN3 TATA box. Further analysis of the −549 to −414 element shows that although this region may play a role in the strong glucose response manifested by the −726 to −414 fragment, it cannot enhance transcription on its own (Fig. 1).
Several of the inserts appeared to block basal expression of the URA3 message, compared to the control without an insert. This may be due to inhibitory sequences or transcriptional termination of this message somewhere within the insert. Our interest is in sequences that are able to alter transcription in response to glucose; therefore, we have not investigated this further. Additionally, some of the inserts appeared to allow continued expression of the basal transcript. For example, the −726 to −549 fragment produced a prominent glucose-inducible band with a transcriptional start site mapping to within the short URA3 untranslated sequence (not shown), as well as a less prominent band that seems to correspond to the message produced in the absence of an insert. For our purposes, we have concentrated on the more prominent lower bands that are clearly glucose inducible and have not included the less prominent upper bands in the quantitation.
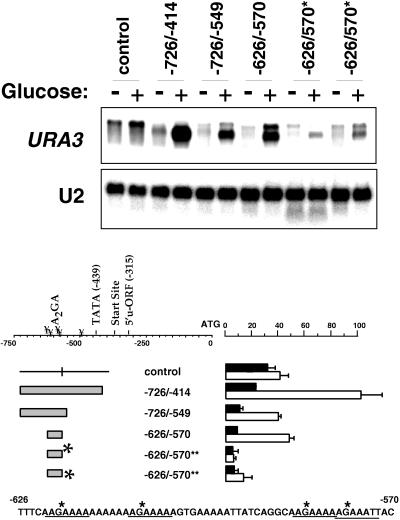
A closer examination of the −726 to −414 element shows the presence of four repeats of the eight-base sequence AAGAAAAA (A2GA5), three in the forward direction at positions −622, −610, and −585 and one inverted in the antiparallel direction at position −460. In addition, there is a similar sequence, AAGAAATT, at position −579. To investigate the significance of these repeated sequences for the transcriptional activity of the −726 to −414 fragment, smaller constructs containing these sequences were tested for the ability to enhance transcription upon addition of glucose. We found that a 57-bp oligonucleotide corresponding to the region of the CLN3 promoter from −626 to −570 is sufficient to produce a substantial glucose response (Fig. 2). This fragment contains three complete sets of the repeated sequence A2GA5 and AAGAAATT. Interestingly, when the G’s in these four repeated sequences are mutated to A’s, this fragment loses its transcriptional activity, as shown for two independent yeast transformants carrying this construct in the last four lanes of Fig. 2. This indicates that the repeated sequences play an important role in driving glucose-dependent transcription.
FIG. 2.
A set of AAGAAAAA repeats plays a role in glucose-dependent transcription. DS10 cells carrying a set of promoter-reporter fusions were treated as described in the legend to Fig. 1. Cells were collected before and 20 min after transfer to fresh medium for RNA preparation and Northern blotting (7.5 μg/lane) with a URA3 probe. The bars represent the averages of PhosphorImager data for the URA3 reporter from at least three separate experiments for each construct, normalized to the −726 to +18 signal in fresh medium. Dark bars represent post-log-phase cells, and light bars indicate cells in fresh medium. Error bars represent standard deviations. Asterisks indicate the replacement of the indicated G’s in the AAGAAAAA repeats with A’s. The results for two independent transformants are shown for the construct carrying the substitution mutations. The sequence from −626 to −570 is shown with the positions of the substitutions marked with asterisks. u-ORF, upstream open reading frame.
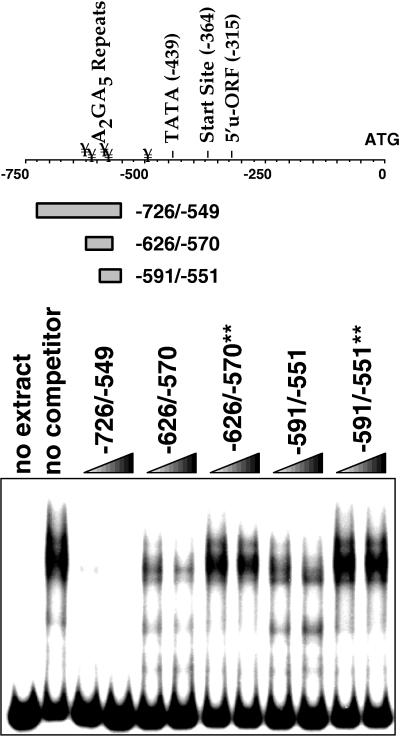
Looking for factors that mediate the glucose response, we used gel shift assays to determine whether yeast nuclear extracts contain a protein that will bind specifically to the DNA elements that drive glucose-dependent transcription. For this experiment, we used a DNA fragment corresponding to the sequence from −726 to −549 as a labeled probe. We found that this labeled fragment can form complexes with proteins in the yeast extract that can be specifically competed with a molar excess of the same fragment (Fig. 3). Shorter double-stranded oligonucleotides corresponding to positions −626 to −570 and −591 to −551, which contain four and two sets of the repeated sequence A2GA5, respectively, also competed, but less effectively. We cannot tell from these experiments whether the less complete competition with the shorter oligonucleotides represents decreased affinity due to the absence of interactions with sites that are present on the longer probe or the presence of multiple proteins, some of which bind to sites on the probe that are not represented in the shorter fragments and therefore not competed. However, it is clear that competition with the shorter fragments was dependent on the A2GA5 repeats. Again, mutation of G’s in the repeated sequences to A’s diminished the ability of these fragments to compete with the labeled fragment (competing oligonucleotides containing the mutations are indicated by asterisks).
FIG. 3.
A set of AAGAAAAA repeats plays a role in protein binding to sequences that drive glucose-dependent transcription. A double-stranded DNA fragment corresponding to the sequence between −726 and −549 was labeled, incubated with protein extract, and run on a native polyacrylamide gel as described in Materials and Methods. Lanes: 1, probe (2 fmol) with no extract; 2 to 10, probe and extract; 2, no competitor; 3, 50× unlabeled −726 to −549 fragment; 4, 100× unlabeled −726 to −549 fragment; 5, 50× unlabeled −626 to −570 fragment; 6, 100× unlabeled −626 to −570 fragment; 7, 50× unlabeled −626 to −570 fragment with A’s substituted for the G’s in the A2GA5 repeats; 8, 100× unlabeled −626 to −570 fragment with A’s substituted for the G’s in the A2GA5 repeats; 9, 50× unlabeled −591 to −551 fragment; 10, 100× unlabeled −591 to −551 fragment; 11, 50× unlabeled −591 to −551 fragment with A’s substituted for the G’s in the A2GA5 repeats; 12, 100× unlabeled −591 to −551 fragment with A’s substituted for the G’s in the A2GA5 repeats. u-ORF, upstream open reading frame.
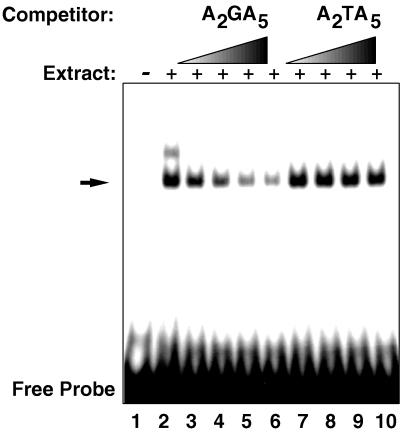
We found that a double-stranded oligonucleotide corresponding to the region between positions −622 and −603 with the sequence CCCAAGAAAAAAAAAAAGAAAAAGGG, containing two of the A2GA5 repeats (underlined), was also able to produce a DNA-protein complex (Fig. 4). In this experiment, two bands are evident, the lower labeled complex can be competed away with a molar excess of the unlabeled fragment, but this competition is abolished if T’s are substituted for the G’s in the two A2GA5 repeats. The upper band is less prominent and appears to be less specific in that it is competed by both oligonucleotides. These results indicate that the repeated sequences that are necessary for glucose induction are also important for the formation of specific DNA-protein complexes.
FIG. 4.
Protein binding to a minimal AAGAAAAA sequence. The gel shift experiment was done as described in the legend to Fig. 3, except that a double-stranded oligonucleotide with the sequence CCCAAGAAAAAAAAAAAGAAAAAGGG was used as the labeled probe. Lanes 1, probe with no extract; 2 to 10, probe and extract; 2, no competitor, 3 to 6 respectively, competition with 5-, 10-, 15-, and 20-fold excesses of unlabeled fragment; 7 to 10 respectively, competition with 5-, 10-, 15-, and 20-fold excesses of unlabeled fragment with T’s substituted for the G’s in the A2GA5 repeats. The arrow indicates the lower, specific band.
Because addition of glucose to the starved cells produced a large increase in the CLN3 message, we searched for differences between DNA-protein complexes formed with extracts from post-log-phase cells and complexes formed with extracts from log-phase cells. We have been unable to identify any consistent difference in gel shift patterns between log-phase and post-log-phase extracts by using a variety of labeled probes (data not shown).
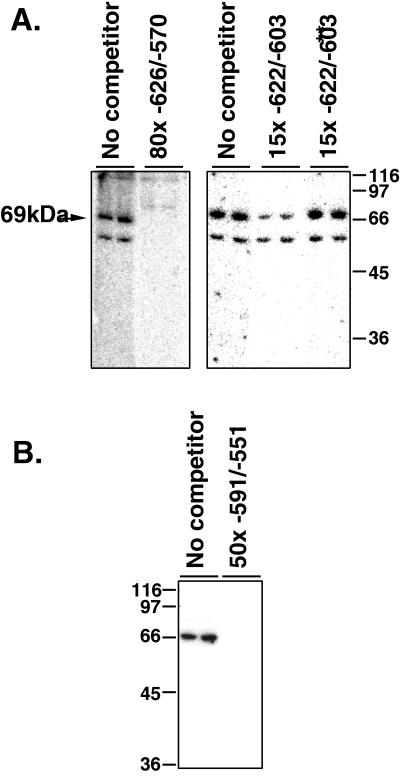
We used Southwestern blotting with a labeled double-stranded oligonucleotide probe corresponding to the region between −626 and −570 to estimate the sizes of proteins that bind to the A2GA5 repeats. This probe identified two bands that were competed away by excess unlabeled probe. One of these bands, with an apparent molecular mass of 69 kDA was competed with a 15-fold excess of the short oligonucleotide, corresponding to the region between positions −622 and −603 (CCCAAGAAAAAAAAAAAGAAAAAGGG), used in Fig. 4, containing just the two A2GA5 repeats (underlined). Competition was diminished when the unlabeled oligonucleotide was mutated to replace the G’s in the A2GA5 sequences with T’s (Fig. 5). In other experiments using a shorter double-stranded oligonucleotide as a probe, corresponding to the region between positions −591 and −551, the upper 69-kDa band was labeled, but not the lower one, suggesting that the lower band requires sequences that are in the larger probe but are not found in the smaller for binding (Fig. 5B).
FIG. 5.
Southwestern blots identify a 69-kDA protein that binds to a DNA sequence containing the A2GA5 repeats. Extracts were run on sodium dodecyl sulfate-containing gels and blotted as described in Materials and Methods. Blots were renatured, and strips containing duplicate lanes were incubated with the indicated labeled double-stranded DNA fragment as described in Materials and Methods. (A) Blots probed with double-stranded DNA corresponding to the sequence between positions −726 and −549. The left blot shows competition with an 80-fold molar excess of the unlabeled fragment. The right blot shows competition with a 15-fold molar excess of either an unlabeled double-stranded oligonucleotide (AAGAAAAAAAAAAAGAAAAA [−622/−603]) or the corresponding fragment with T’s substituted for the G’s in the A2GA5 repeats (−622/−603**). (B) Blots were probed with double-stranded DNA corresponding to the sequence between −591 and −551 and unlabeled competitor as indicated. The values beside the lanes are molecular sizes in kilodaltons.
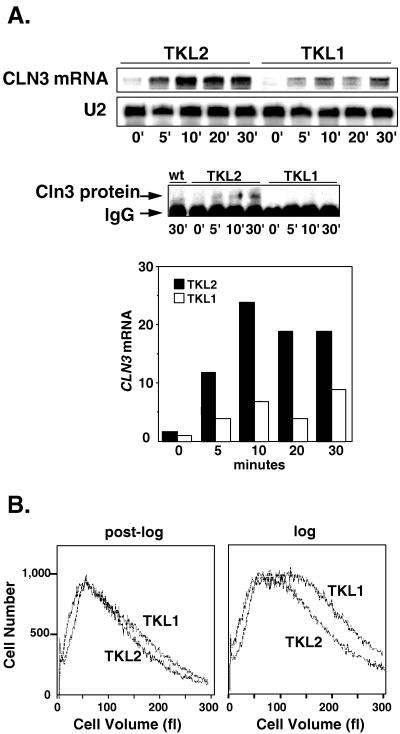
To confirm the importance of the A2GA5 repeats in the normal transcriptional regulation of CLN3 by glucose, we mutated five repeated A2GA5 sequences in the CLN3 promoter, replacing the central G’s in the A2GA5 repeats with T’s at positions −620, −608, −583, −577, and −455. A restriction fragment containing the mutated promoter sequences was then used to replace the corresponding fragment from the normal CLN3 promoter in plasmid pKL001. pKL001 is a CEN-based plasmid that contains an epitope-tagged CLN3 coding sequence driven by the CLN3 promoter (14). This plasmid was transformed into a cln3Δ strain to provide the only copy of CLN3 in the cell. We then compared the expression of CLN3 in cells carrying the mutant promoter (TKL1) with that in cells carrying the parent plasmid containing the wild-type promoter (TKL2). We found that mutations of the repeated sequences produced a substantial reduction in the ability of glucose to induce CLN3 mRNA levels (Fig. 6A). This decreased CLN3 mRNA was also reflected in lower Cln3 protein levels, as shown by the immunoprecipitation-Western blot in Fig. 6A. Cln3 protein levels are difficult to measure in cells that are not overexpressing the protein. While it is clear that the mutant promoter produces less Cln3 protein in vivo, it is difficult to estimate the magnitude of this difference because Cln3 protein levels were close to the limit of detection.
FIG. 6.
Mutations in five A2GA5 elements in the CLN3 promoter decrease CLN3 expression and increase cell size in glucose. (A) Cells carrying a deletion of the chromosomal copy of CLN3 and carrying CEN plasmids expressing CLN3 from either the wild-type (wt) promoter (TKL2) or a promoter carrying five point mutations in which T’s were substituted for the G’s in the A2GA5 repeats (TKL1) were grown in synthetic medium to post-log phase (optical density of 2). Fresh glucose was added to 2% at time zero, and samples were collected at the indicated times for Northern blotting (top) and immunoprecipitation-Western blotting (bottom) as described in Materials and methods. IgG (immunoglobulin G) refers to the band produced by the 12CA5 antibodies used in the immunoprecipitations. The bar graph represents PhosphorImager data for the CLN3 mRNA for each construct, normalized to the U2 signal. The scale is arbitrary. (B) Post-log-phase cells (optical density at 660 nm of 2) and cells growing in log phase (optical density at 660 nm of 0.5) in synthetic medium were collected for size measurements with a Coulter Counter Channelizer. DS10 was used as an untagged control in the immunoprecipitation-Western blotting experiment.
The induction of CLN3 mRNA by glucose suggests that cells increase CLN3 expression in order to accelerate progress through the cell cycle. This would allow the cells to keep up with the increase in cellular growth rate that glucose produces and maintain a relatively constant cell size. We found that while the mutations in the CLN3 promoter produced little noticeable effect on post-log-phase cells, in glucose, the mutant cells were approximately twice as large as those expressing CLN3 at normal levels from the wild-type promoter (Fig. 6B). This is consistent with the lower Cln3 levels and a decreased ability to accelerate movement through the cell cycle in response to glucose.
DISCUSSION
We have previously demonstrated that CLN3 mRNA levels are regulated by carbon source and that glucose induction of CLN3 transcription does not depend on either cellular growth or progression through the cell cycle (19). By using CLN3 promoter-reporter fusion constructs, we have identified a region between positions −762 and −414 in the CLN3 promoter that can confer glucose regulation on a heterologous reporter gene. This region within the CLN3 promoter contains five copies, four exact and one partial, of the sequence AAGAAAAA. Small double-stranded oligonucleotides containing these repeated sequences are able to drive glucose-dependent expression of the reporter gene. Mutations in these elements substantially reduce transcriptional induction of both a URA3 reporter and the CLN3 gene by glucose. Mobility shift assays demonstrate specific binding of proteins from yeast extracts to DNA fragments containing the repeats. Protein binding to the labeled oligonucleotide can be competed with a molar excess of unlabeled oligonucleotides containing the above repeated sequences, but mutation of the central G’s within the repeats decreases the ability of the unlabeled DNA to compete. These results indicate the importance of the repeated elements for both induction of transcription and DNA-protein interactions. Southwestern blotting revealed a protein with an apparent size of 69 kDa that binds specifically to these sequences in a manner that depends on the intact repeats, as well as a smaller protein that binds in a manner that is not affected by changes in the repeats.
We have found no evidence to indicate that glucose regulates the binding of proteins to elements of the CLN3 promoter that confer glucose responsiveness. This suggests that glucose regulation of CLN3 involves a process that is more complex than the simple regulated binding of a transactivator, or repressor protein, to the CLN3 promoter. One important step in learning how glucose affects CLN3 expression would be to identify the protein(s) that binds to the glucose-responsive elements in the CLN3 promoter. It would be interesting to know how these proteins are affected by glucose. Another important question is whether these factors are specific to CLN3 expression or are part of a common pathway that coordinates the expression of a large group of genes in response to glucose. In this regard, a database search yielded only a few yeast promoters carrying these repeated elements. These genes did not fall into any obvious group in terms of function, nor do they all appear to be transcriptionally regulated by glucose. Therefore, we cannot say whether the features that regulate transcription of CLN3 are important in regulating other genes.
Mutation of only five bases, confined to A2GA5 repeats within the CLN3 promoter, produces cells that do not properly increase Cln3 levels in response to glucose. This decrease response clearly has functional significance, because in glucose medium, these cells become approximately twice as large as the controls that express CLN3 from the wild-type promoter. We were unable to detect an appreciable amount of Cln3 protein in the TKL1 cells; however, we believe that Cln3 protein is being expressed in these cells, because they appear smaller than cells with CLN3 completely deleted. The single-copy plasmid produced Cln3 protein levels that were, in our study, close to the limit of detection. We propose that TKL1 cells have some level of functional Cln3 protein that is too low to give a signal in our experiments. While it is clear that these elements contribute to the response to glucose, it is equally clear that these elements alone do not, by themselves, account for the cell cycle shortening that glucose produces. First of all, while mutation of the five A2GA5 repeats decreased the ability of glucose to increase CLN3 mRNA levels, this did not completely abolish the response. This may be because other elements or mechanisms that do not involve these repeated sequences remain able to increase CLN3 transcript levels in response to glucose. It is also possible that the remaining response stems from the presence of additional imperfect copies of the repeated sequence. While we have focused on the 8-bp A2GA5 sequences, there are a total of 13 copies of the shorter 6-bp sequence AAGAAA within the 1,000 bases upstream of the CLN3 coding sequence. It is possible that these also contribute to the glucose response. Secondly, Cln3 protein levels are regulated on a variety of levels in addition to transcription in response to nutrient signals. These mechanisms include posttranscriptional regulation by rich medium (20), the Ras-cyclic AMP pathway (10), protein synthesis rates (10), and nitrogen limitation (8). Finally, the CLN3 gene is believed to act in a redundant pathway, in that deletion of CLN3 is only lethal in combination with loss of other genes, most notably, BCK2 (4, 7). It seems likely that nutrients will regulate other cell cycle components in addition to CLN3.
The closest mammalian homologs to Cln3 appear to be the D-type cyclins. Both D-type cyclins and CLN3 have similarities in expression, being less affected by cell cycle position than other cyclins, and both types of cyclin appear to function in the process of ending G1. The D cyclins serve to link mitogenic signals from the cellular environment to the process of exiting G1 (22). As with the D cyclins, Cln3 levels appear to be regulated by environmental signals, in this case originating from nutrient resources. The transcriptional regulation of CLN3 that we describe appears to be but one of the multiple layers of control that allow yeast cells to modulate G1 length.
ACKNOWLEDGMENTS
We thank Fred Cross for providing plasmids.
This work was supported by Public Health Service grant GM42406 from the National Institutes of Health.
REFERENCES
- 1.Bassel-Duby R, Hernandez M D, Gonzalez M A, Krueger J K, Williams R S. A 40-kilodalton protein binds specifically to an upstream sequence element essential for muscle-specific transcription of the human myoglobin promoter. Mol Cell Biol. 1992;12:5024–5032. doi: 10.1128/mcb.12.11.5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cross F R. DAF1, a mutant gene affecting size control, pheremone arrest, and cell cycle kinetics of Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:4675–4684. doi: 10.1128/mcb.8.11.4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cross F R. Cell cycle arrest caused by CLN gene deficiency in Saccharomyces cerevisiae resembles START-I arrest and is independent of the mating-pheremone signalling pathway. Mol Cell Biol. 1990;10:6482–6490. doi: 10.1128/mcb.10.12.6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Di Como C J, Chang H, Arndt K T. Activation of CLN1 and CLN2 G1 cyclin gene expression by BCK2. Mol Cell Biol. 1995;15:1835–1846. doi: 10.1128/mcb.15.4.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dirick L, Bohm T, Nasmyth K. Roles and regulation of Cln-Cdc28 kinases at the start of the cell cycle of Saccharomyces cerevisiae. EMBO J. 1995;14:4803–4813. doi: 10.1002/j.1460-2075.1995.tb00162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ellwood M S, Craig E A. Differential regulation of the 70K heat shock gene and related genes in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:1454–1459. doi: 10.1128/mcb.4.8.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Epstein C B, Cross F R. Genes that can bypass the CLN requirement for Saccharomyces cerevisiae cell cycle START. Mol Cell Biol. 1994;14:2041–2047. doi: 10.1128/mcb.14.3.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gallego C, Gari E, Colomina N, Herrero E, Aldea M. The Cln3 cyclin is down-regulated by translational repression and degradation during the G1 arrest caused by nitrogen deprivation in budding yeast. EMBO J. 1997;16:7196–7206. doi: 10.1093/emboj/16.23.7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hadwiger J A, Wittenberg C, Richardson H E, de Barros Lopes M, Reed S I. A family of cyclin homologs that control G1 in yeast. Proc Natl Acad Sci USA. 1989;86:6255–6259. doi: 10.1073/pnas.86.16.6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hall, D. D., D. D. Markwardt, F. Parviz, and W. Heideman. Regulation of the Cln3-Cdc28 kinase by cAMP in Saccharomyces cerevisiae. EMBO J., in press. [DOI] [PMC free article] [PubMed]
- 10a.Heideman, W. Unpublished data.
- 10b.Heideman, W., and D. D. Hall. Unpublished data.
- 11.Jagadish M N, Carter B L A. Genetic control of cell division in yeast cultured at different growth rates. Nature. 1977;269:145–147. doi: 10.1038/269145a0. [DOI] [PubMed] [Google Scholar]
- 12.Johnston G C, Pringle J R, Hartwell L H. Coordination of growth with cell division in the yeast Saccharomyces cerevisiae. Exp Cell Res. 1977;105:79–98. doi: 10.1016/0014-4827(77)90154-9. [DOI] [PubMed] [Google Scholar]
- 13.Johnston M, Carlson M. Broach, Pringle, and Jones (ed.), The molecular and cellular biology of the yeast Saccharomyces: gene expression. II. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. Regulation of carbon and phosphate utilization; pp. 193–281. [Google Scholar]
- 14.Levine K, Huang K, Cross F R. Saccharomyces cerevisiae G1 cyclins differ in their intrinsic functional specificities. Mol Cell Biol. 1996;16:6794–6803. doi: 10.1128/mcb.16.12.6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lord P, Wheals A. Variability in individual cell cycles of Saccharomyces cerevisiae. J Cell Sci. 1981;50:361–376. doi: 10.1242/jcs.50.1.361. [DOI] [PubMed] [Google Scholar]
- 16.Lord P, Wheals A. Rate of cell cycle initiation of yeast cells when cell size is not a rate-determining factor. J Cell Sci. 1983;59:183–201. doi: 10.1242/jcs.59.1.183. [DOI] [PubMed] [Google Scholar]
- 17.McInerny C J, Partridge J F, Mikesell G E, Creemer D P, Breeden L L. A novel MCM1-dependent element in the SWI4, CLN3, CDC6, and CDC47 promoters activates M/G(1)-specific transcription. Genes Dev. 1997;11:1277–1288. doi: 10.1101/gad.11.10.1277. [DOI] [PubMed] [Google Scholar]
- 18.Nash R, Tokiwa G, Anand S, Erickson K, Futcher A B. The WHI+ gene of Saccharomyces cerevisiae tethers cell division to cell size and is a cyclin homolog. EMBO J. 1988;7:4335–4346. doi: 10.1002/j.1460-2075.1988.tb03332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parviz F, Heideman W. Regulation of CLN3 mRNA levels by nutrients in Saccharomyces cerevisiae. J Bacteriol. 1998;180:225–230. doi: 10.1128/jb.180.2.225-230.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Polymenis M, Schmidt E V. Coupling cell division to cell growth by translational control of the G1 cyclin CLN3 in yeast. Genes Dev. 1997;11:2522–2531. doi: 10.1101/gad.11.19.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 22.Sherr C. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 23.Stuart D, Wittenberg C. CLN3, not positive feedback, determines the timing of CLN2 transcription in cycling cells. Genes Dev. 1995;9:2780–2794. doi: 10.1101/gad.9.22.2780. [DOI] [PubMed] [Google Scholar]
- 24.Tyers M, Tokiwa G, Futcher B. Comparison of the Saccharomyces cerevisiae G1 cyclins: Cln3 may be an upstream activator of Cln1, Cln2 and other cyclins. EMBO J. 1993;12:1955–1968. doi: 10.1002/j.1460-2075.1993.tb05845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Werner-Washburne M, Braun E, Johnston G C, Singer R A. Stationary phase in the yeast Saccharomyces cerevisiae. Microbiol Rev. 1993;57:383–401. doi: 10.1128/mr.57.2.383-401.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]