ABSTRACT
Stomach acid provides a significant innate barrier to the entry of the food-borne pathogen Listeria monocytogenes into the human gastrointestinal tract. A key determinant of acid resistance in this bacterium is the conserved glutamate decarboxylase system, GadD2 (encoded by the gadT2D2 operon), which helps to maintain the intracellular pH during exposure to gastric acid. In this study, we identified a premature stop codon in a gene located immediately downstream of the gadT2D2 operon that was highly linked to an acid-sensitive phenotype. When this open reading frame was restored through homologous recombination, an acid-resistant phenotype was restored. Through a series of genetic, transcriptomic, and survival experiments, we established that this gene, which we designated gadR, encodes a transcriptional regulator of the gadT2D2 operon. GadR belongs to the RofA family of regulators, primarily found in streptococci, where they are involved in regulating virulence. The data further showed that gadR plays a critical role in the development of acid resistance in response to mild acid exposure, a response that is known as the adaptive acid tolerance response (ATR). A deletion analysis of the gadT2D2 promoter region identified two 18-bp palindromic sequences that are required for the GadR-mediated induction of gadT2D2, suggesting that they act as binding sites for GadR. Overall, this study uncovers a new RofA-like regulator of acid resistance in L. monocytogenes, which plays a significant role in both growth phase-dependent acid resistance and ATR and accounts for previously observed strain-to-strain differences in survival at low pH.
IMPORTANCE
The ability to survive the acidic conditions found in the stomach is crucial for the food-borne pathogen Listeria monocytogenes to gain access to the mammalian gastrointestinal tract. Little is currently known about how acid resistance is regulated in this pathogen and why this trait is highly variable between strains. Here, we used comparative genomics to identify a novel RofA-family transcriptional regulator, GadR, that controls the development of acid resistance. The RofA family of regulators was previously found only in a small group of bacterial pathogens, including streptococci, where they regulate virulence properties. We show that gadR encodes the dominant regulator of acid resistance in L. monocytogenes and that its sequence variability accounts for previously observed differences between strains in this trait. Together, these findings significantly advance our understanding of how this important pathogen copes with acid stress and suggest a potential molecular target to aid its control in the food chain.
KEYWORDS: Listeria monocytogenes, glutamate decarboxylase system, acid stress response, adaptive acid tolerance response, RALP, RofA
INTRODUCTION
The bacterium Listeria monocytogenes is a well-studied member of the Bacillota phylum (formerly the Firmicutes) due to its impact on food safety and its behavior in the host as a facultative intracellular pathogen. Ingestion of food contaminated with this bacterium is associated with a risk of infection, termed listeriosis, especially in immunocompromised individuals, where the mortality rate can be as high as 30% (1–3). The organism is particularly problematic for producers of ready-to-eat foods as it has the ability to grow at refrigeration temperatures and in some foods preserved at low pH and/or low water activity (4–6). Its robust response to acid stress also increases the risk that the pathogen can survive transit of the acidic conditions in the stomach and thereafter invade the epithelium of ileum (7, 8), which is the first stage of pathogenesis during an infection (3). While much has been learned about the mechanisms that contribute to the acid stress response in this pathogen (4, 9–11), the regulatory mechanisms that control the expression of these systems are much less well understood.
Almost three decades ago, an adaptive response to acid was described in L. monocytogenes, whereby a mild acid stimulus (pH 4.0–6.0) triggers the development of high levels of acid resistance to normally lethal acid conditions (pH 3.0) (12, 13). This response, which was named the adaptive acid tolerance response (ATR), requires de novo protein synthesis but the mechanisms underpinning its regulation have never been properly elucidated. Interestingly, entry to the stationary phase was also found to promote acid resistance in L. monocytogenes independently of pH (12). The discovery that Sigma B (SigB), an alternative sigma factor that controls the general stress response in L. monocytogenes, plays an important role in acid resistance (14–17) initially suggested that it might play a role in regulating the stationary phase acid resistance and the ATR. SigB is activated in the stationary phase, it responds to mild acid stress, and it plays a role in transcribing key components involved in protection against acid stress, including the glutamate decarboxylase and arginine deiminase pH homeostasis systems (17–19). Furthermore, mutants lacking SigB have an acid-sensitive phenotype in the stationary phase and a reduced ATR (14, 16, 19). However, sigB mutants are still capable of developing acid resistance in the stationary phase and inducing a significant ATR (16, 19), suggesting that other regulatory factors must be involved in regulating the acid stress response of this pathogen.
Two glutamate decarboxylases (GadD2 and GadD3) are present in most strains of L. monocytogenes, and a third (GadD1) is present in some lineage II strains (Fig. 1A). The corresponding genes for these decarboxylases are encoded at three distinct genetic loci (20–23). They contribute to acid stress protection by helping to maintain the intracellular pH, because the decarboxylation reaction consumes protons (24–26). Two of these systems are genetically coupled with glutamate/gamma-aminobutyrate (Glu/GABA) antiporters (GadD1T1 and GadT2D2) that allow uptake of glutamate in exchange for the product of the decarboxylation reaction, GABA. GadD3 is not co-expressed with an antiporter and appears to use intracellular glutamate independently of a Glu/GABA exchange mechanism (27, 28). GadD1T1 is reported to contribute to growth under acidic conditions (21), whereas GadT2D2 is the dominant system with respect to acid resistance (23, 29). Little is known about the transcriptional regulation of these two systems, although the transcription of gadD3 is under SigB control (30, 31). SigB also controls the expression of succinate semialdehyde dehydrogenase (Lmo0913), which is required in the GABA-shunt pathway that generates succinate and Glu from GABA and α-ketoglutarate (15, 32). There are significant strain-to-strain differences in the behaviors of the GAD systems in L. monocytogenes (23, 28). In particular, the well-studied lab strains EGD-e and 10403S, both of which belong to lineage II serotype 1/2a, produce and secrete different amounts of GABA in response to acidification. Specifically, 10403S secretes GABA in response to acidification, whereas EGD-e does not, although gadD1T1, gadT2D2, and gadD3 are conserved in these two strains (23). Furthermore, the deletion of each of the three GAD systems from these two strains produces different phenotypes with respect to acid resistance; notably, deletion of gadD2 produces an acid-sensitive phenotype in 10403S but has no effect on EGD-e (23). It is striking that the well-studied lab strain EGD-e is especially sensitive to acid, the genetic basis for which has remained unclear (23, 28, 29).
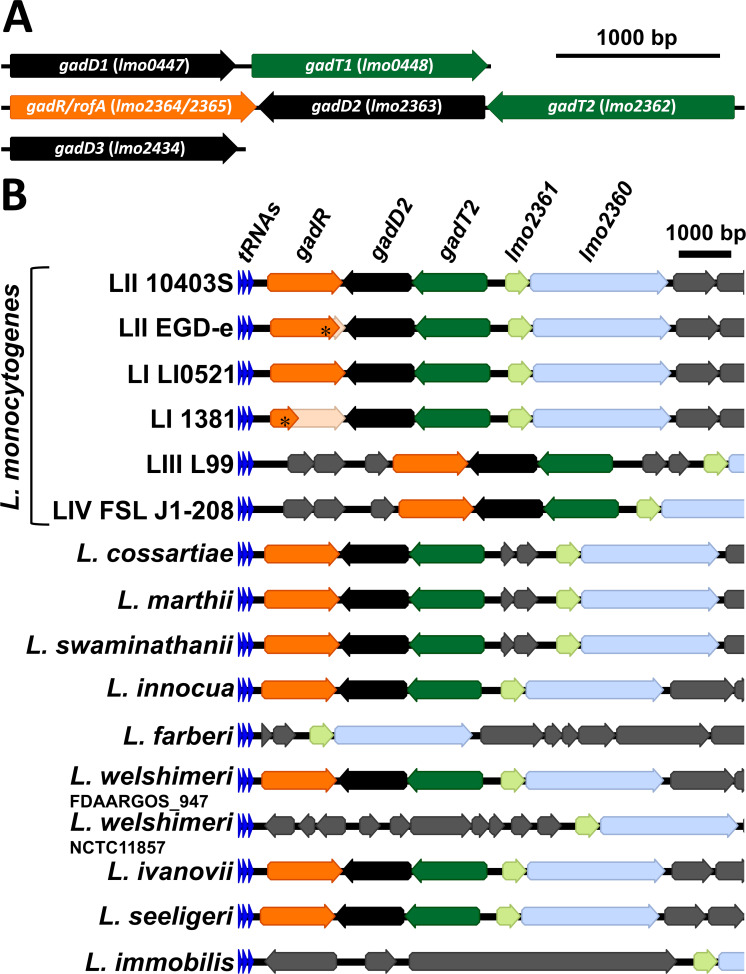
Fig 1.
The gadT2D2-gadR gene cluster is highly conserved in the Listeria senso stricto clade. (A) Operon structures of gadD1T1, gadT2D2-gadR, and gadD3 are depicted, using sequences extracted from strain 10403S. (B) Gene arrangements downstream of the conserved tRNA-Gly are depicted for all species from Listeria senso stricto clade species reported by Carlin et al. (33). L. monocytogenes lineages I–IV are abbreviated as LI–LIV. The PMSCs in gadR are highlighted with asterisks and the untranslated regions of gadR are faded.
We have recently described the phylogenetic and phenotypic characterization of a collection of 168 lab, food, environmental, and clinical isolates of L. monocytogenes with respect to their acid tolerance (growth at pH 4.9), acid resistance (survival at pH 2.3), and salt tolerance (growth in NaCl 0.8–1.5 M) (34). This collection is both phylogenetically and phenotypically diverse, with wide variabilities in acid tolerance and acid resistance across individual strains. Some phenotypic outliers were identified as harboring lesions in the sigB operon, which explains compromised general stress response and acid-sensitive phenotypes (34). One strain identified in that study, a lineage I strain designated 1381, displayed reduced acid tolerance (no growth at pH 4.9) and reduced acid resistance (poor survival at pH 2.3) although there was no evidence of mutations affecting the SigB system (34). We have recently shown that the growth defect in strain 1381 at low pH, but not the poor survival phenotype (at pH 2.3), is caused by a mutation in the mntH gene, which encodes a manganese transporter belonging to the NRAMP family (35). However, this mutation does not account for the decreased acid resistance in this strain (35). Thus, the genotype of this strain could help provide new insights into the mechanisms underpinning acid resistance in L. monocytogenes.
In the present study, we used comparative genomics to help further elucidate the genetic basis for the reduced acid resistance observed in this strain. Our findings reveal the presence of a hitherto unknown transcriptional regulator, GadR, that plays a crucial role in the regulation of acid resistance by modulating the expression of the GadT2D2 in this pathogen. The presence of a mutation within the gadR gene is shown to solely account for the differences in acid resistance between the two well-studied lab strains, EGD-e and 10403S. Overall, this study identifies a key regulatory component of adaptive acid resistance in L. monocytogenes and raises the possibility that acid resistance, and thus virulence, might be controlled through a strategy that specifically targets this regulator.
RESULTS
Identification of a RofA-like regulator that positively influences acid resistance
The acid-sensitive and acid-intolerant CC2 strain 1381 is closely related to the chromosomally sequenced reference strain LI0521 (35), facilitating an investigation into the genetic basis of these unusual phenotypes. There are four genes intact in strain LI0521, which are truncated by the presence of premature stop codon (PMSC) in strain 1381, including mntH (35). Among the other three is a putative RofA-like transcriptional regulator, located downstream of and oriented convergently with the acid-resistance operon gadT2D2 (Fig. 1). Notably, this gene was also truncated in the widely studied lab strain EGD-e (L374*) and thus mis-annotated as two open reading frames (ORFs), lmo2365 and lmo2364 in that strain (36). Bioinformatic analysis revealed that this gadT2D2-rofA gene cluster is conserved in most species from Listeria sensu stricto clade (Fig. 1B) (33) but absent from the Listeria sensu lato clade (data not shown). These observations suggested that this RofA-like transcriptional regulator might be the cognate regulator of the gadT2D2 operon and, therefore, we designated the full ORF as gadR. Interestingly, in addition to strains 1381 and EGD-e, CC7 strain 1147 and all five CC18 strains from the previously characterized strain collection (n = 168) are predicted to be gadR- based on the presence of PMSCs within the ORF (data not shown). All these gadR- strains survived poorly at pH 2.3 (34), suggesting a possible role of GadR in acid resistance.
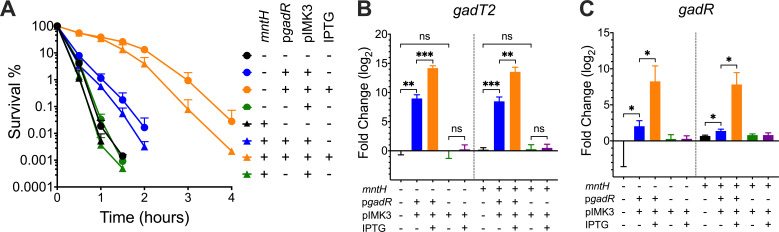
To test whether GadR influences acid resistance and gadT2 transcription, full-length gadR was cloned from CC2 strain 1380 and introduced into strain 1381 using an IPTG-inducible integrative expression vector pIMK3, generating construct pIMK3::gadR (37) (Table 1). The acid resistance of strain 1381 pIMK3::gadR during the stationary phase was enhanced in an IPTG-induced fashion compared to strain 1381 (Fig. 2A). The transcript levels of gadT2 were positively correlated with the presence of a functional copy of gadR (from pIMK3::gadR) (Fig. 2B and C). The pIMK3 vector did not affect acid resistance or gadT2 transcription (Fig. 2A and B). These results demonstrated that GadR positively influences both acid resistance and gadT2 transcription. When the pIMK3::gadR plasmid was transformed into the mntH+ strain 1381R1 (35) (Table 1), the positive effects on both acid resistance and gadT2 transcription were very similar to that observed in the mntH- parental strain (Fig. 2A and B), suggesting that the effect of GadR on acid resistance is independent of MntH.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Reference/source |
|---|---|
| Strains | |
| Escherichia coli One Shot TOP10 | Invitrogen |
| L. monocytogenes 1381 | Wu et al. (34) |
| L. monocytogenes 1381R1 | Wu et al. (34) |
| L. monocytogenes 1381 pIMK3 | This study |
| L. monocytogenes 1381 pIMK3::gadR | This study |
| L. monocytogenes 1381R1 pIMK3 | This study |
| L. monocytogenes 1381R1 pIMK3::gadR | This study |
| L. monocytogenes 10403S | Boor |
| L. monocytogenes 10403S ΔgadR | This study |
| L. monocytogenes 10403S ΔgadT2D2 | This study |
| L. monocytogenes 10403S ΔgadT2D2R | This study |
| L. monocytogenes 10403S ΔsigB | Boor |
| L. monocytogenes 10403S ΔgadR ΔsigB | This study |
| L. monocytogenes EGD-e | Piveteau |
| L. monocytogenes EGD-e gadR+ | This study |
| L. monocytogenes EGD-e ΔsigB | Marinho et al. (38) |
| L. monocytogenes EGD-e gadR+ ΔsigB | This study |
| L. monoctyogenes EGD-e gadR+ PgadT2_V1-V10 | This study |
| Plasmids | |
| pIMK3; IPTG-controlled gene expression Phelp::lacOid; Kanr | Monk et al. (37) |
| pMAD; Eryr; Ampr | Arnaud et al. (39) |
| pIMK3::gadR (pJW2); Kanr | This study |
| pMAD gadR+ (pJW3); Eryr; Ampr | This study |
| pMAD ΔgadR (pJW15); Eryr; Ampr | This study |
| pMAD ΔgadT2D2 (pJW16); Eryr; Ampr | This study |
| pMAD ΔgadT2D2R (pJW17); Eryr; Ampr | This study |
| pMAD ΔsigB (COB926); Eryr; Ampr | Marinho et al. (38) |
| pEX-K168-EGD-e_gadR_reversion; Kanr | Eurofins Genomics |
| pEX-A128-10403S_gadR_deletion; Ampr | Eurofins Genomics |
Fig 2.
GadR positively influences acid resistance and gadT2 transcription, independently of the manganese transporter MntH. (A) Stationary phase cultures of strains 1381 (mntH-) and 1381R1 (mntH+) either with or without gadR complementation (pgadR) and 1 mM IPTG induction were challenged at pH 2.3 and survival was recorded over 4 h. Transcription of gadT2 (B) and gadR (C) was measured for these strains after overnight incubation for 18 h. Transcript levels relative to the gadR- parent strain 1381 are presented. Three independent experiments were performed with technical triplicates (A) or duplicates (BC). Statistically significant differences were determined by paired t test (two-tailed) (ns, not significant; *P < 0.05; **P < 0.01; and ***P < 0.001).
GadR controls acid resistance by activating GadT2D2 expression independently of SigB
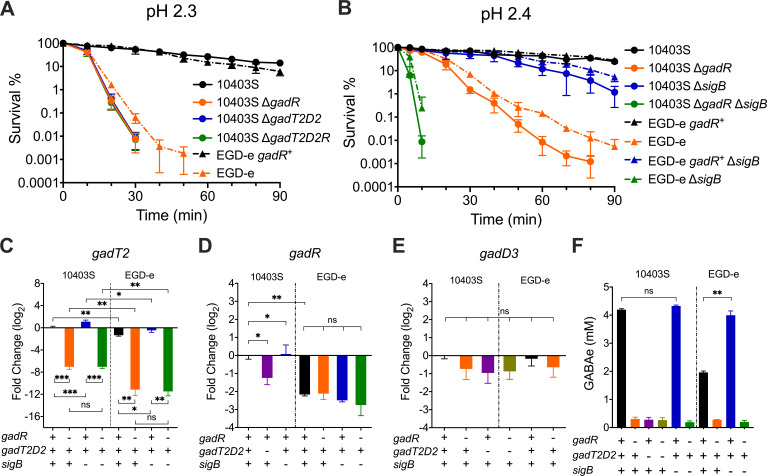
To test whether altered gadT2D2 transcription is exclusively responsible for GadR-mediated acid resistance, three mutant strains were constructed in a genetic background where the gadR was intact (lab strain 10403S); they carried deletions either in the gadR gene (ΔgadR), the gadT2D2 locus (ΔgadT2D2), or both (ΔgadT2D2R) (Table 1). The ability of these strains to survive at pH 2.3 was compared following growth to the stationary phase. All three mutants exhibited a very similar decrease in acid resistance compared to the parental strain 10403S (Fig. 3A), suggesting that GadT2D2 expression is solely responsible for the GadR-mediated acid resistance. The GadR-mediated acid resistance in strain EGD-e (gadR-) was restored by repairing the point mutation in gadR (*374L) in the chromosome by homologous recombination (Fig. 3A and B). These observations demonstrated a prominent role for GadR in acid resistance and indicated that the gadR frameshift mutation in EGD-e explains the intrinsic acid sensitivity of this reference strain. To examine the relative contributions of GadR and the general stress response regulator SigB to acid resistance, ΔsigB was introduced into strains 10430S and EGD-e with or without gadR (Table 1), and the acid resistance of strains lacking either or both the gadR and sigB was compared to the strains that have functional SigB and GadR in both genetic backgrounds (strains EGD-e and 10403S). The strains EGD-e gadR- ΔsigB and 10403S ΔgadR ΔsigB were extremely acid sensitive and, therefore, the cultures were grown to the stationary phase and exposed to pH 2.4 (rather than pH 2.3). In both genetic backgrounds, GadR made a more significant contribution than SigB to acid resistance, while the absence of both regulators resulted in extreme acid sensitivity (Fig. 3B). These results suggested that the combined effects of GadR and SigB account for most, if not all, of the acid resistance of L. monocytogenes at pH 2.4 in stationary phase. To elucidate possible crosstalk between GadR and SigB, the transcript levels of gadT2, gadR, and the SigB-dependent gadD3 during stationary phase were measured for these strains (Fig. 3C through E). The results suggested that SigB has limited, if any, influence on GadR-mediated gadT2 transcription. gadR transcription showed minor variations across these strains suggesting that it is controlled neither by GadR nor by SigB (Fig. 3D). The transcription of gadD3 was confirmed to be SigB-dependent and was unaffected by gadR deletion (Fig. 3E), suggesting that GadR does not influence SigB activity and does not control gadD3 transcription. To establish if the GadR-mediated gadT2 transcription is associated with altered Glu/GABA antiporter activity, the ability to export GABA into the medium was examined for these strains during the stationary phase following an acid challenge at pH 3 for 1 h (which is not lethal to strains carrying either gadR or sigB, Fig. S1). The results showed that significant extracellular GABA was only detectable in the presence of both gadR and gadT2D2 (Fig. 3F). Taken together, these results demonstrated that GadR promotes acid resistance by activating the expression of the GadT2D2 independently of SigB.
Fig 3.
GadR modulates acid resistance by activating GadT2D2 expression independently of SigB. Cultures of L. monocytogenes WT or mutant strains were grown to the stationary phase and challenged at pH 2.3 (A) or pH 2.4 (B). EGD-e carries a frameshift in the gadR gene, whereas the ORF was restored to full-length by homologous recombination in the strain EGD-e gadR+. Transcription of gadT2 (C), gadR (D), and gadD3 (E) in these strains after overnight incubation for 18 h was calculated relative to the WT strain 10403S. The ability of these strains to secrete GABA into the extracellular environment is shown (F). Three independent experiments were performed with technical triplicates (A, B, and F) or duplicates (C, D, and E). Statistically significant differences were determined by paired t test (two-tailed) (ns, not significant; *P < 0.05; **P < 0.01; and ***P < 0.001).
Adaptive acid resistance is highly GadR-dependent
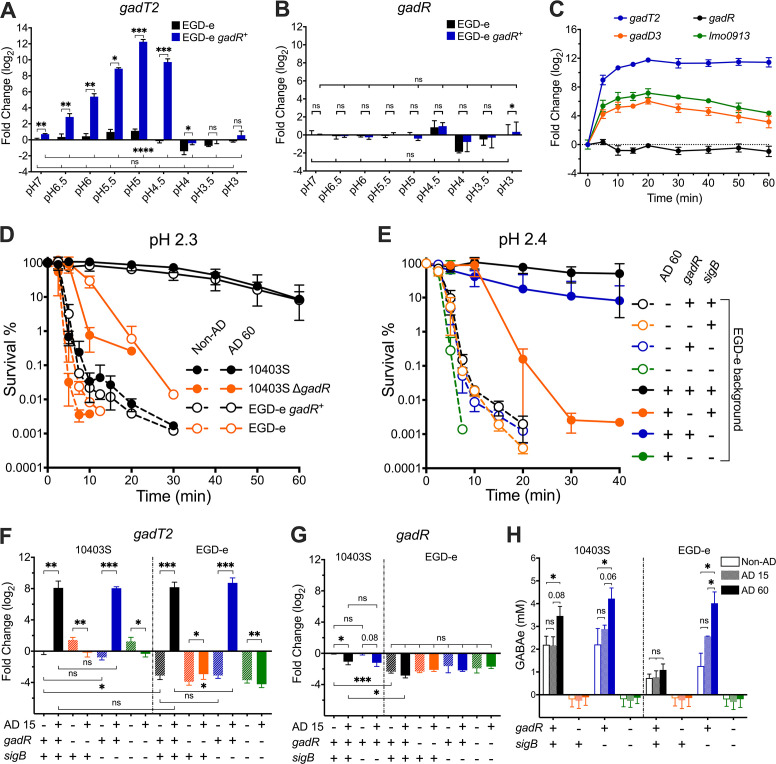
Since GadR contributes significantly to acid resistance in the stationary phase, we reasoned that it might also participate in regulating the ATR in L. monocytogenes. To test this, the transcription of gadT2, gadR, gadD1, and gadD3 was measured in exponentially grown cultures of strains EGD-e wildtype (WT) (gadR-) and the isogenic gadR+ derivative after 15 min exposure to a range of low pH conditions (pH 3–6.5, compared to pH 7). While gadT2, gadD1, and gadD3 were all acid-inducible with a peak at pH 5, only the induction of gadT2 was GadR-dependent (Fig. 4A; Fig. S2). gadR transcription was unchanged regardless of the pH, likely indicating that the gadT2 induction might involve post-transcriptional regulation of gadR (Fig. 4B). pH 5 activates gadT2 transcription to the greatest extent and this activation appeared to be rapid and continuous (Fig. 4C). The transcription of the SigB-dependent gadD3 and lmo0913 was also induced, however, to a lesser extent (Fig. 4C). These data demonstrated that GadR was responsible for the activation of gadT2 transcription in response to mildly acidic stress.
Fig 4.
Adaptive acid resistance is GadR-dependent. The transcript levels of gadT2 (A) and gadR (B) in exponential phase cultures of EGD-e WT (gadR-) or gadR+ strains with or without a 15 min exposure to pH 3.0–6.5 are shown, expressed relative to untreated EGD-e WT stain. (C) The differential transcription of gadT2, gadR, gadD3, and lmo0913 in response to pH 5.0 exposure was monitored throughout a 60-min period in strain EGD-e gadR+. Cultures of L. monocytogenes WT or mutant strains grown to exponential phase with (AD 60) or without (non-AD) a 60 min pH 5.0 adaptation were challenged in pH 2.3 (D) or pH 2.4 (E). Transcription of gadT2 (F) and gadR (G) in exponential phase cultures of L. monocytogenes WT or mutant strains with or without 15 min exposure to pH 5.0 (AD 15) are calculated and shown relative to the untreated 10403S WT strain. (F) The GABA exported into the medium was measured using exponential phase cultures either untreated (non-AD) or exposed to pH 5.0 for 15 min (AD 15) or 60 min (AD 60). Three independent experiments were performed with technical triplicates (D, E, and H) or duplicates (A, B, C, F, and G). Statistically significant differences between measurements were determined using a paired t test (two-tailed) (ns, not significant; *P < 0.05; **P < 0.01; and ***P < 0.001). Statistically significant differences across a group of samples were determined by one-way ANOVA (ns, not significant; *P < 0.05; **P < 0.01; and ***P < 0.001).
To test the involvement of GadR in the ATR, the ability of strains 10403S and EGD-e to survive at pH 2.3 with or without GadR was measured before (non-AD) and after pH 5 adaption for 60 min (AD 60). All strains from the exponential phase without pH 5 adaptation exhibited low levels of acid resistance. Strains 10403S WT and EGD-e gadR+ developed a strong ATR after adaptation (Fig. 4D and E), while strains 10403S ΔgadR and EGD-e WT (gadR-) exhibited greatly reduced levels of acid resistance after pH 5 treatment (Fig. 4D). Since EGD-e is the most widely studied strain, the relative contributions of gadR and sigB to ATR were examined using strain EGD-e WT (gadR-) and its isogenic mutants (gadR+, ΔsigB, gadR+ ΔsigB). Because of the extreme acid-sensitive phenotype of strain gadR- ΔsigB, the ability to survive at pH 2.4 (rather than pH 2.3) was measured to compare the ATR. The ATR was only slightly reduced following sigB deletion in strain EGD-e gadR+, but it was essentially absent when sigB was deleted in strain EGD-e (gadR-) (Fig. 4E). In accordance with these observations, gadT2 was strongly induced by pH 5 adaption for 15 min (AD 15) in both genetic backgrounds independently of SigB (Fig. 4F). gadR transcription appeared to be slightly downregulated following adaptation in strain 10403S; however, it remained unchanged in the EGD-e genetic background (Fig. 4G), further suggesting that GadR activation by mild acidification was likely achieved through post-transcriptional regulation. Consistent with previous phenotypic and transcriptional observations, Glu/GABA antiporter activity was only present and induced in the gadR+ strains (Fig. 4H), following an acid challenge at pH 3.25 for 1 h, conditions which are not lethal to strains carrying either gadR or sigB (Fig. S1). Interestingly, the ability to secrete GABA was notably higher in the absence of sigB in EGD-e strains (Fig. 3F and 4H), although these differences were not reflected in the transcriptional or acid-resistance measurements (Fig. 3B and 5E). Taken together, these results suggested that exposure to pH 5 induces a strong GadR-dependent ATR in L. monocytogenes.
Fig 5.
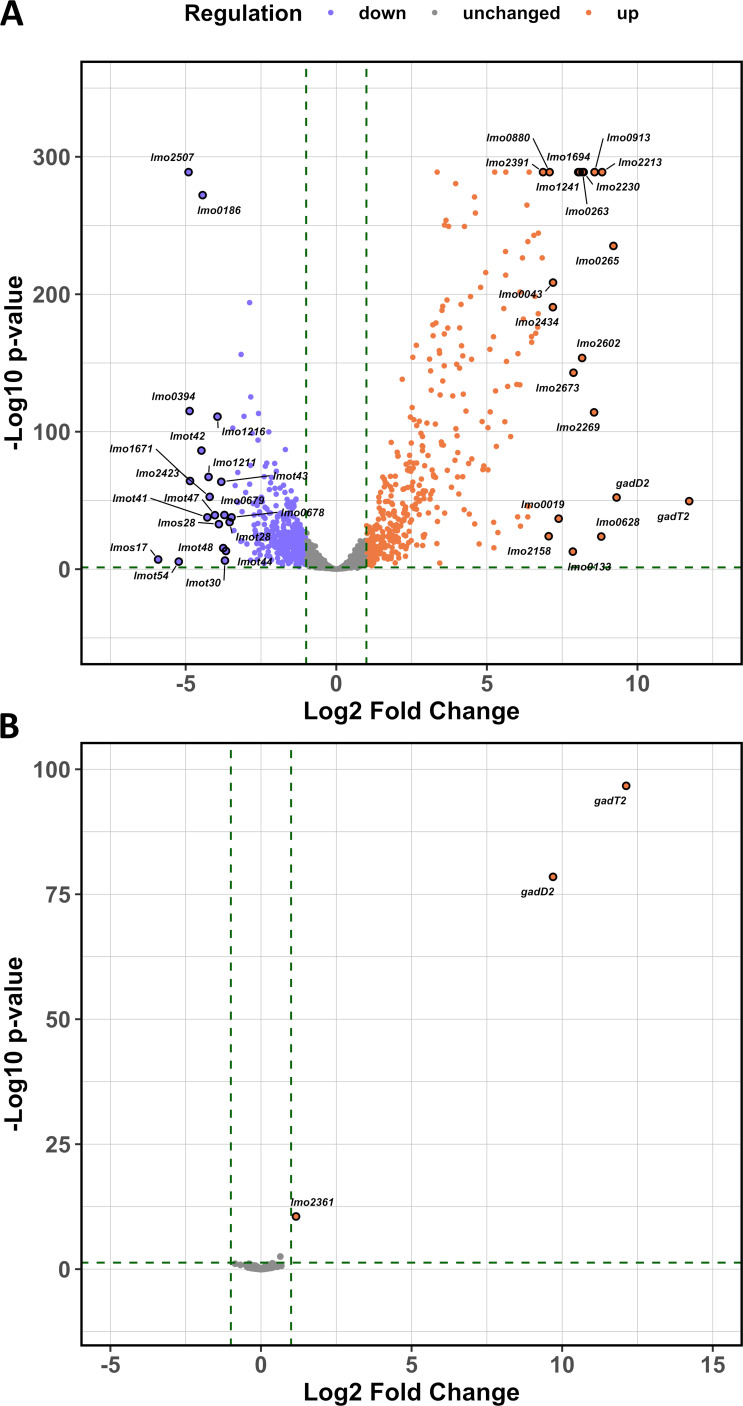
GadR is a dedicated regulator of the gadT2D2 operon during acid adaptation. (A) Global gene transcription following a 15 min pH 5.0 treatment was measured using RNA-seq, with values expressed relative to the untreated exponential phase culture of strain EGD-e gadR+. The 20 most upregulated/downregulated genes are labeled. (B) GadR-dependent genes were detected by comparing global gene transcription of strain EGD-e gadR+ to the WT EGD-e strain (gadR-) after 15 min at pH 5.0. Genes showing differential transcription are labeled. Three independent sequencing reactions were carried out. Genes with differential transcription greater than twofold and with P-value < 0.05 were considered significantly differentially regulated (marked by dotted lines on the Volcano plots).
Two 18 bp-palindromes in PgadT2 are essential for GadR-dependent ATR
Although gadT2D2 alone confers GadR-mediated acid resistance, it remained unknown whether GadR controls the expression of genes that do not directly contribute to acid resistance. To test this, the global gene transcription during exponential growth with or without a 15 min pH 5 treatment was compared between strains EGD-e WT (gadR-) and gadR+. No gene was significantly differentially regulated without acid adaption between these two strains (Fig. S3). In contrast, 382 genes were upregulated and 374 genes were downregulated in response to pH 5 treatment in strain EGD-e gadR+ (Fig. 5A; Table S2), but only the acid induction of the gadT2D2 operon and an adjacent gene lmo2361 (encodes Rrf2 family transcriptional regulator) appeared to be dependent on GadR (Fig. 5B). Moreover, gadT2D2 were co-transcribed as a transcriptional unit and they were the most upregulated genes (11.7 and 9.3 log2 fold changes, respectively) in response to pH 5 treatment across the genome (Fig. 5A; Table S2), supporting a prominent role for GadR in the ATR. These data demonstrated that in response to mild acid stress GadR exclusively influences the transcription initiated at the lmo2361-gadT2 intergenic region.
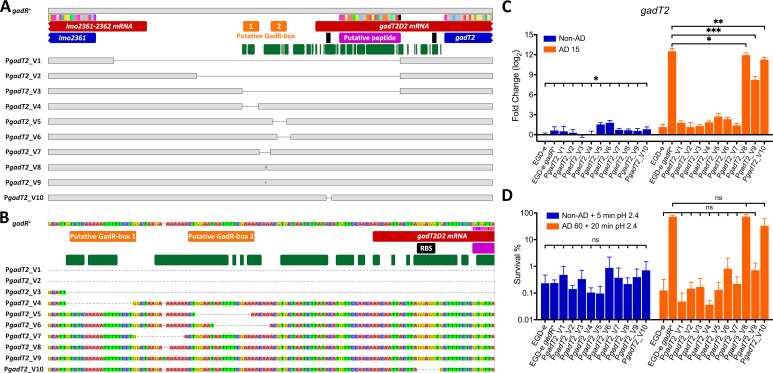
To identify the sequence motif(s) required for GadR-mediated transcriptional regulation, PgadT2 (defined as the sequence between the coding sequences of gadT2 and the upstream adjacent gene) was carefully examined. Since the genes upstream from gadT2 were not conserved across the Listeria sensu stricto clade (Fig. 2), PgadT2 sequence alignment was performed for Listeria sensu stricto spp. in order to search for the conserved region, which is likely responsible for GadR-mediated gadT2 regulation (Fig. 6A). The transcription start site (TSS) of gadT2 was mapped at ~145 bp upstream of start codon using the RNA-seq data, a prediction that was also supported by previous results (40). One notable feature that was conserved in all sequences was a pair of imperfect palindromic sequences that shared sequence similarity within the conserved region of PgadT2 and upstream of TSS (Fig. 6A and B). To test whether these palindromic sequences influence GadR-mediated gadT2 transcription, a panel of mutants (designated V1–V10) (Table 1) with various mutations in PgadT2 was constructed (Fig. 6A and B) and tested for gadT2 transcription as well as acid resistance at exponential phase with or without pH 5 treatment. Deletion of the conserved region in PgadT2 (V1, V2, and V3) abolished the GadR-mediated ATR (Fig. 6). Disruption of either one of these two putative GadR-boxes (V4, V5, and V6) or the spacer region (V7) results in the absence of GadR-mediated ATR (Fig. 6). While a Δ1 bp deletion in the spacer between the two palindromic sequences (V8) was tolerable for GadR-mediated ATR, a 1 bp insertion in spacer region (V9) results in moderately reduced gadT2 induction and absence of GadR-mediated adaptive acid resistance (Fig. 6). The results also suggested that the sequence to the 5′ of these palindromes is responsible for lmo2361 transcription (Fig. S4). Further examination of PgadT2 revealed a putative peptide of 23 aa, proceeded by a consensus ribosome binding site (RBS) (GGAGG) (Fig. 6A and B), although deletion of this RBS (V10) did not significantly affect the GadR-mediated ATR (Fig. 6). Taken together, these data suggest that the pair of 18-bp palindromes in PgadT2 are likely to serve as GadR-binding boxes that are essential for the GadR-mediated ATR in L. monocytogenes.
Fig 6.
The promoter region of gadT2 (PgadT2) has two 18-bp palindromes required for GadR-mediated activation of gadT2D2 and induction of acid resistance. The intergenic sequence between the lmo2361 and gadT2 ORFs is depicted (A), with the nucleotide sequences shown for the 120 bp region targeted for mutagenesis (B). The sequences are annotated as follows: coding sequences are shown in blue; transcripts deduced from RNA-seq analysis are shown in red; putative short translation frames are shown in pink; the putative GadR-boxes are shown in orange; predicted RBSs are shown in black; the 100% conserved bases across Listeria sensu strico spp. genomes that have gadT2D2-gadR gene cluster (n = 13, Fig. 1) are annotated with green boxes underneath. The mutants, labeled PgadT2_V1 to PgadT2_V10, were constructed in an EGD-e gadR+ background and their positions are indicated (A and B). (C) The transcription of gadT2 was measured for the panel of PgadT2 mutants during the exponential phase either with (AD 15) or without (non-AD) a 15 min pH 5.0 treatment and expressed relative to the untreated WT EGD-e strain (gadR-). (D) The ability of these strains to survive at pH 2.4 was determined using exponential phase cultures either with (AD 60) or without (non-AD) a 60 min pH 5 treatment. Three independent experiments were performed with technical triplicates (C) or duplicates (D). Statistically significant differences between conditions were determined by paired t test (two-tailed) (ns, not significant; *P < 0.05; **P < 0.01; and ***P < 0.001). Statistically significant differences across a group of samples were determined by one-way ANOVA (ns, not significant; *P < 0.05; **P < 0.01; and ***P < 0.001).
DISCUSSION
In this study, a hitherto unknown RofA-like regulator, GadR, was predicted by comparative genomics to contribute to acid resistance in L. monocytogenes. We have provided phenotypic, transcriptional, and biochemical evidence that GadR has a major impact on the adaptive acid resistance of L. monocytogenes by activating gadT2D2 expression in response to a mild acid stimulus. Through transcriptomic analyses, we showed that GadR is a dedicated regulator of gadT2D2 and identified two putative GadR-binding boxes that are essential for GadR-mediated acid resistance. The data presented here also help to explain the previously reported differences in GAD activity (23, 28) and acid resistance (34, 41) between strains of L. monocytogenes. The results suggest that adaptive acid resistance in this pathogen is largely controlled by GadR and SigB, the regulator of the general stress response. Overall, the study elucidates a key regulatory mechanism for adaptive acid resistance and helps to clarify the previously unresolved questions.
The conservation of gadR-gadT2D2 in Listeria sensu stricto spp. (Fig. 1) points to the possibility that this acid-resistance mechanism is important for the fitness of these species in the gastrointestinal (GI) tract since they are commonly isolated from feces (42). However, the occurrence of loss-of-function mutation in gadR in strains EGD-e and 1381 raises the question of how common loss-of-function mutations in gadR are in L. monocytogenes. An inspection of the rate of PMSC occurrence in the gadR open reading frame per 100 bp in 40,080 sequenced L. monocytogenes genomes reveals that it is exceptionally high (Fig. S5). The PMSC rate in gadR is comparable to that of the inlA invasion gene, which has been reported to be highly susceptible to mutations resulting in truncation of the protein (43) and is an order of magnitude higher than the PMSC rates in gadT2 and gadD2 (Fig. S5). This finding suggests that allelic variation in gadR could be a major determinant of differences in acid resistance between L. monocytogenes strains. It is interesting to speculate about the evolutionary pressures that might lead to the selection of mutations in gadR despite its importance in acid resistance. One possibility is that the GadT2D2-mediated reaction might be detrimental/dispensable under conditions where the availability of glutamate is limited or where maintenance of a high intracellular pool of glutamate is critical. Glutamate is present at a high concentration in the cytoplasm of L. monocytogenes where it contributes to osmoregulation and acts as a counter-ion for potassium (44). It is conceivable, therefore, that under prolonged conditions of osmotic stress, it might be advantageous to dispense with GadT2D2 by inactivating the main regulator. Alternatively, environments outside the mammalian host that rarely experience acidification might allow genetic drift of the gadR sequence, in the absence of positive selection for its function. Indeed, 8.4% of LII genomes encode truncated versions of GadR, while this number is only 1.1% for LI genomes and even lower in the hypervirulent CCs: 0.3% in CC1, 1.1% in CC2, 0.2% in CC4 and CC6 (45). Notably, a considerable percentage of strains from the phylogenetic groups that are prevalent in food is predicted to carry loss-of-function mutations in gadR, e.g., CC18 (86.3%), CC7 (31.4%), and CC9 (19.5%) (data not shown).
GadR shares significant sequence similarity with RofA, which was identified in Streptococcus pyogenes nearly three decades ago as a positive regulator of protein F (46). RofA is the founding member of a group of regulators called RALPs (RofA-like Proteins) that are found exclusively in Group A streptococci and that are a subset of the Mga superfamily of regulators (47, 48). Like the RALPs and Mga, GadR has two putative helix-turn-helix (HTH) DNA binding domains in the N-terminal half of the protein (Uniprot accession numbers: HTH_Mga, PF08280, and Mga, PF05043). Both HTH domains of Mga are necessary for DNA binding and activation of the Mga regulon (49). The presence of the two putative HTH domains in GadR suggests their involvement in binding to one or both of the predicted palindromic GadR-boxes upstream of gadT2. The predicted GadR-boxes span from −80 to −32 bp relative to the TSS of gadT2, and GadR-box 2 likely overlaps with −35 box of the gadT2 promoter, which is among the reported features of Mga superfamily transcription factors (50, 51). This finding suggests that when the GadR-box 2 is occupied by GadR, the gadT2D2 operon would be repressed, whereas occupancy of the GadR-box 1, following an acid stimulus, would lead to derepression of the operon. Whether GadR dimerization occurs remains to be established, although it is interesting to note that the GadR-mediated transcriptional activation of gadT2 by acid is sensitive to the spacing between the two GadR-boxes; deletion of the spacer or insertion of 1 bp from it significantly reduces gadT2 transcription following acid shock (Fig. 6B and C; V7 and V9, respectively). Based on our results, the likely scenario is that gadR is constitutively expressed regardless of the extracellular pH (Fig. 4) and that some post-transcriptional modification is induced by mild acid stress that facilitates/alters its interaction with the regulatory region. Two phosphotransferase regulatory domains (PRDs) and a phosphotransferase system enzyme IIB-like domain are predicted at the C-terminal of GadR, similar to Mga and RofA, and these regulators were recently classified as PRD-containing virulence regulators (52). It is possible that the PRDs in GadR are involved in acid stress sensing and regulation of the activity of GadR. Further biochemical characterization of the GadR protein is underway to investigate this possibility.
While the GadR-mediated expression of GadT2D2 is shown to be a major factor in the development of an ATR, the global transcriptome analyses following pH 5.0 treatment reveal the complexity of the cellular response to acid. This acid stimulus was shown previously to trigger SigB activation (53) and indeed, ~36% of upregulated genes (n = 140) were SigB-dependent or proceeded by putative SigB promoter sequence (Table S2). Moreover, the SigB regulon accounts for ~86% (n = 60) of the 70 most upregulated genes (Table S2), including genes involved in known acid stress response (e.g., arginine/agmatine deiminase) and virulence-associated mechanisms (bile resistance, glutathione biosynthesis, and internalin expression) (20, 54–58). These data further demonstrate the importance of SigB in acid adaption. We have recently demonstrated that trace metal (Mn2+ and Zn2+) homeostasis under acid stress is critical for an adequate stress response (35). In accordance with this, the expression of Mn2+ importers (lmo1424 and lmo1847-1849) and a putative Zn2+ exporter (lmo2231) was induced, while the expression of putative Zn2+ importers was repressed (lmo1445-1447 and lmo1671) (Table S2). Several upregulated genes are involved in carbohydrate uptake and metabolism (Table S2) indicating a general shift in carbon metabolism during growth in acidic conditions. Expression of multiple transcriptional regulators (e.g., lmo2241, lmo2551, lmo0815, and lmo2494) was also affected, and these might be partially responsible for the SigB-independent differentially regulated genes (Table S2).
The identification of GadR as the principal regulator of the GadT2D2 system and the characterization of global transcriptomic response to acid stress exposure help to understand the overall physiological response to mild acid stress in L. monocytogenes. Exposure to mild acid stress serves as a signal to the rapidly growing cells to prepare for a harsher environment. The most prominent actions involve rapid GadR- and SigB-mediated transcriptional response. GadR specifically promotes high expression of GadT2D2, which serves to protect against potentially lethal acid stress by helping to neutralize intracellular pH through glutamate decarboxylation. The total GAD activity is possibly supported by SigB-mediated gadD3 expression, while other proton-consuming mechanisms (e.g., arginine/agmatine deiminase) might also contribute to the overall acid resistance (20). SigB as the general stress response regulator promotes an array of mechanisms that contribute to intracellular pH maintenance (Table S2) (19, 30). Our data suggest that there is an additive (rather than synergistic) effect in acid resistance between GadR and SigB. Besides intracellular pH maintenance, the cells also exquisitely manage metal homeostasis and accelerate carbon source utilization under acid stress, presumably to avoid metal intoxication and to acquire energy. The acid exposure also serves as a signal for host entry for the bacterium to express apparatus that are essential for GI tract survival, intestinal epithelial adhesion, and virulence factor activation (19) as supported by our transcriptomic data (Table S2).
Overall, this study sheds new light on the regulatory mechanisms that underpin the acid stress response of this important food-borne pathogen. It further highlights the central role that glutamate decarboxylation plays in adaptive acid resistance. Further studies are underway to address some of the most important outstanding questions. Chief among these is the nature of the signal detected by GadR in response to acidification. It could be either acid pH itself or some secondary effect of reduced pH on the physiology of the cell. It seems likely that some post-transcriptional modification of gadR is required to activate it, but further work will be needed to clarify this. It will be interesting to learn whether GadR plays a role in colonizing the mammalian host, in particular, if it contributes to surviving the transition through the acidic conditions in the stomach. If it proves to be critical for this early stage of the infectious cycle, it might make a good diagnostic target for identifying strains of concern in food-processing environments. Once the regulation of GadR is fully elucidated, it might also be a useful molecular target to help reduce the survival of this pathogen in acidic environments.
MATERIALS AND METHODS
Strains and culturing conditions
L. monocytogenes and Escherichia coli TOP10 strains and plasmids used in this study are listed in Table 1. L. monocytogenes strains were grown in brain heart infusion (BHI) (LAB M LAB048) at 37° C with agitation at 150 rpm unless otherwise specified. For stationary phase culture, an isolated L. monocytogenes colony was inoculated to 5 mL BHI broth in 50 mL centrifuge tubes and incubated for 18 h. To prepare exponential phase culture, overnight culture of L. monocytogenes was washed twice with fresh BHI broth and inoculated to 5 mL BHI broth in 50 mL centrifuge tubes to achieve initial OD600nm = 0.05 and incubated for ~3 h until mid-exponential phase (OD600nm = 0.4). E. coli strains were grown in Luria-Bertani (Sigma). The following antibiotics were added to the medium where specified: kanamycin 75 µg · mL−1 (kan), ampicillin 100 µg · mL−1 (amp), chloramphenicol 10 µg · mL−1 (chl), and erythromycin 2 µg · mL−1 (ery).
Molecular techniques
Plasmids and primers used in this study are listed in Tables 1 and 2; Table S1. To complement gadR in strains 1381 and 1381R1, the full-length gadR coding sequence was cloned (Phusion, ThermoFisher) from closely related CC2 strain 1380. The PCR product and expression vector pIMK3 (37) were digested (FastDigest, ThermoFisher) and ligated (T4 DNA ligase, Roche). Three microliters of ligation mixture was used in the thermo-shock transformation of E. coli TOP10 chemically competent cells, and the cells were plated on LBkan plates after recovery and incubated overnight at 37°C. A transformant carrying the correct insert in the plasmid was grown overnight in LBkan to propagate the plasmid for purification. Both purified plasmid (pJW2) and empty vector (pIMK3) were electroporated into electrocompetent cells of L. monocytogenes strains 1381 and 1381R1 as previously described (19, 37). To construct EGD-e gadR+ and 10403S ΔgadR, two pMAD derivatives were constructed each with a synthesized (Eurofins) 600-bp insert containing 300 bp upstream and 300 bp downstream of the mutation site with restriction sites introduced on both ends. For EGD-e gadR+, the gadR nonsense mutation in strain EGD-e was reverted in the insert (*374L) and several silent mutations were also introduced to facilitate the discrimination of mutant from WT using PCR (19). The constructed plasmids were transferred to electrocompetent L. monocytogenes EGD-e or 10403S cells and spread onto BHIery plates as previously described (19, 37). The mutation was achieved by a two-step integration (19, 59). The mutant and WT were discriminated by PCR. For introducing the sigB deletion, the previously constructed pMAD ΔsigB was transferred to strains 10430S ΔgadR and EGD-e gadR+ (38). To introduce ΔgadT2D2R and ΔgadT2D2 in strain 10403S and to create mutant panel EGD-e gadR+ PgadT2_V1-V10, plasmids used for mutagenesis were constructed by splicing by overlap extension (SOE). Deletion of the entire PgadT2 sequence was first introduced to EGD-e gadR+ to obtain EGD-e gadR+ PgadT2_V1. Various lengths of PgadT2 were then complemented to EGD-e gadR+ PgadT2_V1 by two-step integration to obtain EGD-e gadR+ PgadT2_V2-V10.
TABLE 2.
Primers used in this study
| Primer name | Primer sequence (5′–3′) |
|---|---|
| Cloning | |
| EGD-e_gadR+_F | GCTTAGGGATCCTCGACTAATTTTAG |
| EGD-e_gadR+_R | GACATGGAATTCCGGAATAATAG |
| 10403S_ΔgadR_F | ATATGGATCCGTATACTATGATACCA |
| 10403S_ΔgadR_R | ATATGCGTCGACCCACTTACCAAT |
| ΔgadTD2_up_PstI_F | ATATCTGCAGGTGATGGCGAAAAATCCGAA |
| ΔgadTD2_up_EcoRI_R | ATATGAATTCAATGCGTTTGCTGCGAATAG |
| ΔgadTD2_down_BamHI_F | ATATGGATCCATTTCAGGTGGAACAGGAGC |
| ΔgadTD2_down_PstI_R | ATATCTGCAGCGATAATCAACAATCCGGCG |
| ΔgadRTD2_down_BamHI_F | ATATGGATCCCTGGACTCAAAATCCTGTGC |
| ΔgadRTD2_down_PstI_R | ATATCTGCAGACCCTCTCCATAAAATTGCAAC |
| RT-qPCR | |
| 16S_RT_F | TGGGGAGCAAACAGGATTAG |
| 16S_RT_R | TAAGGTTCTTCGCGTTGCTT |
| lmo2362_RT_F | ATCCAACATTTGCCACTTCC |
| lmo2362_RT_R | AAGAAGATTGCGGCAAAACC |
| lmo2365_RT_F | ACTTCTCGGGTGACGGT |
| lmo2365_RT_R | CTCCTCCACATTCGTAACAAAA |
| lmo2434_RT_F | CTGAGGAAGAAAGCACGAGT |
| lmo2434_RT_R | TTTTTCTCGAGCGTTTCTGC |
| lmo0447_RT_F | TACCGGTGTTTGGCTCTTTT |
| lmo0447_RT_R | CATGATTTGCTCAGCTTCCG |
| lmo0913_RT_F | CCTGATTGGGCAAAAATGGA |
| lmo0913_RT_R | ATCTTCTTGCTTCTTCCGCA |
| lmo2361_RT_F | GACGATGTTGTACACCCAGA |
| lmo2361_RT_R | TCTTCAGGGTCTTTAGCAAGC |
| lmo2230_RT_F | TGGGCGAAAAGACTTTCACT |
| lmo2230_RT_R | TGGAAATTTTGGTGCAGTTTCA |
Transcriptional analysis
Transcriptional analysis was performed as previously described with minor adjustments (19). Briefly, for stationary phase transcription analysis, RNA samples were taken directly from overnight culture. In acid adaption analysis, 5 M HCl was added to exponential phase culture (OD600nm = 0.4) to acidify the media to pH 6.5, 6.0, 5.5, 5.0, 4.5, 4.0, 3.5, and 3.0 (the volumes of 5 M HCl required were pre-determined). RNA samples were taken for transcription analysis after 15 min of exposure to acid stress. Alternatively, the exponentially growing culture was acidified to pH 5 and incubated at 37°C. RNA samples were taken at 0, 5, 10, 15, 20, 30, 45, and 60 min for transcription analysis. To extract RNA, 1 mL of culture was mixed with 5 mL RNALater (Sigma) and incubated at ambient temperature (~18°C) for RNA preservation. All subsequent RNA extraction and cDNA synthesis steps were carried out as previously described (34). Cells were then recovered by centrifugation and resuspended in the RNA Lysis buffer RLT (Qiagen RNeasy minikit). The mixture was transferred to lysis matrix B tubes for mechanical lysis (40 s at 6 m · s−1, twice). The rest of the RNA extraction procedures followed the manufacturer’s instructions. The resulting RNA samples were treated with DNase (Turbo, Invitrogen). The quantity and quality of RNA samples were examined using Nanodrop before reverse transcription (SuperScript III, Invitrogen) was carried out (13 µL reaction, ~0.5 µg RNA applied). qPCR (LightCycler 480 SYBR Green I Master, Roche) was performed with 100× diluted cDNA samples in a Roche LightCycler 480 system (34). All primers for gene expression analysis were designed to anneal to regions that were conserved across strains to be analyzed (Table 2). Three independent experiments were carried out each with two technical repeats. Relative gene transcriptions were calculated using Q-gene (60) with 16S as the reference gene.
Acid survival experiments
The ability of L. monocytogenes strains to survive in lethal acidic conditions was tested as previously described with minor adjustments (61). To examine the acid resistance at the stationary phase, 100 µL stationary phase culture was mixed with 900 µL BHI pH 2.3 or pH 2.4 in 1.5 mL microcentrifuge tubes and incubated at 37°C statically. Samples were taken at indicated time points, serial diluted (10×, in phosphate-buffered saline), and 10 µL of cell suspensions from each dilution was then spotted onto BHI agar to quantify viable cells. Acid resistance was determined as the percentage of survival at each time point relative to the starting cell count. Three independent experiments were carried out, each with triplicates. To examine the GadR-mediated adaptive acid stress response, 100 µL exponential phase culture with/without acid adaption for 60 min was mixed with 900 µL BHI pH 2.3 or pH 2.4 in 1.5 mL microcentrifuge tubes. The samples were incubated and analyzed following the same procedures as described for stationary phase acid resistance determination.
GABA assay
The abilities of strains 10403S and EGD-e and their derivatives to produce extracellular GABA at the stationary phase and exponential phase with/without acid adaption were examined using a previously established method with minor adjustments (62). For stationary phase culture, 4 mL overnight culture was resuspended in 2 mL BHIchl pH 3 and incubated for 1 h at 37°C. For adaptive acid stress response, 2 mL culture was taken from exponential phase cultures that were exposed under pH 5 stress for 0, 15, and 60 min, then resuspended in 200 µL BHIchl pH 3.25, and incubated for 1 h at 37°C. Following this incubation, the samples were spun down (10,000 g⋅ 5 min) and the supernatant was collected for analysis. To quantify the GABA in the supernatant collected, 5 µL sample was added to 95 µL freshly prepared reaction master mix (80 mM Tris buffer, 750 mM sodium sulfate, 10 mM dithiothreitol, 1.4 mM NADP+, 2 mM α-ketoglutarate, and 0.1 g ⋅ L−1 GABase) and incubated at 37°C for 1 h in a temperature-controlled plate reader with OD340nm measured every 60 s for 3 h. Standard curves were made by analyzing 1–10 mM GABA solution (made with BHI pH 3 or pH 3.25). OD340nm measured at 1 h was inspected for GABAe calculation. Three independent experiments were carried out, each with duplicates.
Transcriptomic analysis
RNA samples for transcriptomic analysis were extracted as mentioned above and sent to Novogene for RNA sequencing and subsequent bioinformatic analysis following a standard procedure. Briefly, for library construction, ribosomal RNA was removed from total RNA with the Illumina Ribo-Zero Plus rRNA Depletion Kit. After fragmentation, the first strand of cDNA was synthesized using random hexamer primers. During the second strand cDNA synthesis, dUTPs were replaced with dTTPs in the reaction buffer. The directional library was ready after end repair, A-tailing, adapter ligation, size selection, USER enzyme digestion, PCR amplification, and purification with AMPure XP beads. The library was checked with Qubit and real-time PCR for quantification and bioanalyzer for size distribution detection. Quantified libraries were pooled and sequenced on Illumina platforms, according to effective library concentration and data amount required. The sequencing was performed in the Illumina NovaSeq6000 using a sequencing strategy based on paired-end reads with a sequencing length of 150 bp per read (PE150). The raw reads were mapped to reference genome EGD-e, and log2 fold changes were calculated by comparing the normalized read counts between two given groups. Raw sequencing data are available from NCBI SRA under project number: PRJNA947476.
Statistics
All statistical analyses were performed in Prism 8.
ACKNOWLEDGMENTS
The authors are grateful to colleagues in the Bacterial Stress Response Group at the University of Galway, Professor Cormac Gahan (APC Microbiome, University College Cork, Ireland), Professor Jörgen Johansson (Department of Molecular Biology, Umeå University, Sweden), and Dr. Lilliana Radoshevich (Department of Microbiology and Immunology, University of Iowa, USA) for helpful discussions.
This project was supported by the Irish Department of Agriculture, Food and the Marine (17/F/244) and by the Science Foundation Ireland Frontiers for the Future Programme (21/FFP-P/10078). J.W. was also supported by the Irish Higher Education Authority funded Cost Extensions for Research Disrupted by COVID-19.
Contributor Information
Conor P. O'Byrne, Email: conor.obyrne@universityofgalway.ie.
Nancy E. Freitag, University of Illinois Chicago, Chicago, Illinois, USA
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/mbio.01716-23.
Figures S1-S5; Tables S1 and S2.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. European food safety authority, European centre for disease prevention and control . 2017. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food‐borne outbreaks in 2016. EFS2 15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Swaminathan B, Gerner-Smidt P. 2007. The epidemiology of human Listeriosis. Microbes Infect 9:1236–1243. doi: 10.1016/j.micinf.2007.05.011 [DOI] [PubMed] [Google Scholar]
- 3. Lecuit M. 2020. Listeria monocytogenes, a model in infection biology. Cell Microbiol 22:e13186. doi: 10.1111/cmi.13186 [DOI] [PubMed] [Google Scholar]
- 4. NicAogáin K, O’Byrne CP. 2016. The role of stress and stress adaptations in determining the fate of the bacterial pathogen Listeria monocytogenes in the food chain. Front Microbiol 7:1865. doi: 10.3389/fmicb.2016.01865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kallipolitis B, Gahan CG, Piveteau P. 2020. Factors contributing to Listeria monocytogenes transmission and impact on food safety. Curr Opin Food Sci 36:9–17. doi: 10.1016/j.cofs.2020.09.009 [DOI] [Google Scholar]
- 6. van der Veen S, Moezelaar R, Abee T, Wells-Bennik MHJ. 2008. The growth limits of a large number of Listeria monocytogenes strains at combinations of stresses show serotype- and niche-specific traits. J Appl Microbiol 105:1246–1258. doi: 10.1111/j.1365-2672.2008.03873.x [DOI] [PubMed] [Google Scholar]
- 7. Giannella RA, Broitman SA, Zamcheck N. 1972. Gastric acid barrier to ingested microorganisms in man: studies in vivo and in vitro . Gut 13:251–256. doi: 10.1136/gut.13.4.251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Merrell DS, Camilli A. 2002. Acid tolerance of gastrointestinal pathogens. Curr Opin Microbiol 5:51–55. doi: 10.1016/s1369-5274(02)00285-0 [DOI] [PubMed] [Google Scholar]
- 9. Dorey A, Marinho C, Piveteau P, O’Byrne C. 2019. Role and regulation of the stress activated sigma factor sigma B (σB) in the saprophytic and host-associated life stages of Listeria monocytogenes. Adv Appl Microbiol 106:1–48. doi: 10.1016/bs.aambs.2018.11.001 [DOI] [PubMed] [Google Scholar]
- 10. O’Byrne CP, Karatzas KAG. 2008. The role of sigma B (σB) in the stress adaptations of Listeria monocytogenes: overlaps between stress adaptation and virulence. Adv Appl Microbiol 65:115–140. doi: 10.1016/S0065-2164(08)00605-9 [DOI] [PubMed] [Google Scholar]
- 11. Guerreiro DN, Arcari T, O’Byrne CP. 2020. The σB-mediated general stress response of Listeria monocytogenes: life and death decision making in a pathogen. Front Microbiol 11:1505. doi: 10.3389/fmicb.2020.01505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Davis MJ, Coote PJ, O’Byrne CP. 1996. Acid tolerance in Listeria monocytogenes: the adaptive acid tolerance response (ATR) and growth-phase-dependent acid resistance. Microbiology (Reading) 142:2975–2982. doi: 10.1099/13500872-142-10-2975 [DOI] [PubMed] [Google Scholar]
- 13. O’Driscoll B, Gahan CG, Hill C. 1996. Adaptive acid tolerance response in Listeria monocytogenes: isolation of an acid-tolerant mutant which demonstrates increased virulence. Appl Environ Microbiol 62:1693–1698. doi: 10.1128/aem.62.5.1693-1698.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wiedmann M, Arvik TJ, Hurley RJ, Boor KJ. 1998. General stress transcription factor σB and its role in acid tolerance and virulence of Listeria monocytogenes. J Bacteriol 180:3650–3656. doi: 10.1128/JB.180.14.3650-3656.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Abram F, Starr E, Karatzas KAG, Matlawska-Wasowska K, Boyd A, Wiedmann M, Boor KJ, Connally D, O’Byrne CP. 2008. Identification of components of the sigma b regulon in Listeria monocytogenes that contribute to acid and salt tolerance. Appl Environ Microbiol 74:6848–6858. doi: 10.1128/AEM.00442-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ferreira A, Sue D, O’Byrne CP, Boor KJ. 2003. Role of Listeria monocytogenes σB in survival of lethal acidic conditions and in the acquired acid tolerance response. Appl Environ Microbiol 69:2692–2698. doi: 10.1128/AEM.69.5.2692-2698.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Becker LA, Cetin MS, Hutkins RW, Benson AK. 1998. Identification of the gene encoding the alternative sigma factor σB from Listeria monocytogenes and its role in osmotolerance. J Bacteriol 180:4547–4554. doi: 10.1128/JB.180.17.4547-4554.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guerreiro DN, Wu J, Dessaux C, Oliveira AH, Tiensuu T, Gudynaite D, Marinho CM, Boyd A, García-Del Portillo F, Johansson J, O’Byrne CP. 2020. Mild stress conditions during laboratory culture promote the proliferation of mutations that negatively affect sigma B activity in Listeria monocytogenes. J Bacteriol 202:e00751-19. doi: 10.1128/JB.00751-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guerreiro DN, Pucciarelli MG, Tiensuu T, Gudynaite D, Boyd A, Johansson J, García-Del Portillo F, O’Byrne CP. 2022. Acid stress signals are integrated into the σB-dependent general stress response pathway via the stressosome in the food-borne pathogen Listeria monocytogenes. PLoS Pathog 18:e1010213. doi: 10.1371/journal.ppat.1010213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Arcari T, Feger M-L, Guerreiro DN, Wu J, O’Byrne CP. 2020. Comparative review of the responses of Listeria monocytogenes and Escherichia coli to low pH stress. Genes 11:1330. doi: 10.3390/genes11111330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cotter PD, Ryan S, Gahan CGM, Hill C. 2005. Presence of GadD1 glutamate decarboxylase in selected Listeria monocytogenes strains is associated with an ability to grow at low pH. Appl Environ Microbiol 71:2832–2839. doi: 10.1128/AEM.71.6.2832-2839.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cotter PD, O’Reilly K, Hill C. 2001. Role of the glutamate decarboxylase acid resistance system in the survival of Listeria monocytogenes LO28 in low pH foods. J Food Prot 64:1362–1368. doi: 10.4315/0362-028x-64.9.1362 [DOI] [PubMed] [Google Scholar]
- 23. Feehily C, Finnerty A, Casey PG, Hill C, Gahan CGM, O’Byrne CP, Karatzas K-AG. 2014. Divergent evolution of the activity and regulation of the glutamate decarboxylase systems in Listeria monocytogenes EGD-e and 10403S: roles in virulence and acid tolerance. PLoS One 9:e112649. doi: 10.1371/journal.pone.0112649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lund PA, De Biase D, Liran O, Scheler O, Mira NP, Cetecioglu Z, Fernández EN, Bover-Cid S, Hall R, Sauer M, O’Byrne C. 2020. Understanding how microorganisms respond to acid pH is central to their control and successful exploitation. Front Microbiol 11:556140. doi: 10.3389/fmicb.2020.556140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Foster JW. 2004. Escherichia coli acid resistance: tales of an amateur acidophile. Nat Rev Microbiol 2:898–907. doi: 10.1038/nrmicro1021 [DOI] [PubMed] [Google Scholar]
- 26. Schwarz J, Schumacher K, Brameyer S, Jung K. 2022. Bacterial battle against acidity. FEMS Microbiol Rev 46:fuac037. doi: 10.1093/femsre/fuac037 [DOI] [PubMed] [Google Scholar]
- 27. Karatzas K-AG, Brennan O, Heavin S, Morrissey J, O’Byrne CP. 2010. Intracellular accumulation of high levels of γ-aminobutyrate by Listeria monocytogenes 10403S in response to low pH: uncoupling of γ-aminobutyrate synthesis from efflux in a chemically defined medium. Appl Environ Microbiol 76:3529–3537. doi: 10.1128/AEM.03063-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Karatzas K-AG, Suur L, O’Byrne CP. 2012. Characterization of the intracellular glutamate decarboxylase system: analysis of its function, transcription, and role in the acid resistance of various strains of Listeria monocytogenes. Appl Environ Microbiol 78:3571–3579. doi: 10.1128/AEM.00227-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cotter PD, Gahan CGM, Hill C. 2001. A glutamate decarboxylase system protects Listeria monocytogenes in gastric fluid. Mol Microbiol 40:465–475. doi: 10.1046/j.1365-2958.2001.02398.x [DOI] [PubMed] [Google Scholar]
- 30. Guerreiro DN, Boyd A, O’Byrne CP. 2022. The Stressosome is required to transduce low pH signals leading to increased transcription of the amino acid-based acid tolerance mechanisms in Listeria monocytogenes Access Microbiol 4:acmi000455. doi: 10.1099/acmi.0.000455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Raengpradub S, Wiedmann M, Boor KJ. 2008. Comparative analysis of the σB-dependent stress responses in Listeria monocytogenes and Listeria innocua strains exposed to selected stress conditions. Appl Environ Microbiol 74:158–171. doi: 10.1128/AEM.00951-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Feehily C, O’Byrne CP, Karatzas KAG. 2013. Functional γ-aminobutyrate shunt in Listeria monocytogenes: role in acid tolerance and succinate biosynthesis. Appl Environ Microbiol 79:74–80. doi: 10.1128/AEM.02184-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Carlin CR, Liao J, Hudson LK, Peters TL, Denes TG, Orsi RH, Guo X, Wiedmann M. 2022. Soil collected in the great smoky mountains National Park yielded a novel Listeria sensu stricto species L. swaminathanii. Microbiol Spectr 10:e0044222. doi: 10.1128/spectrum.00442-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wu J, NicAogáin K, McAuliffe O, Jordan K, O’Byrne C, Dudley EG. 2022. Phylogenetic and phenotypic analyses of a collection of food and clinical Listeria monocytogenes isolates reveal loss of function of sigma B from several clonal complexes. Appl Environ Microbiol 88:e0051-22. doi: 10.1128/aem.00051-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wu J, McAuliffe O, O’Byrne CP. 2023. Manganese uptake mediated by the NRAMP-type transporter MntH is required for acid tolerance in Listeria monocytogenes. Int J Food Microbiol 399:110238. doi: 10.1016/j.ijfoodmicro.2023.110238 [DOI] [PubMed] [Google Scholar]
- 36. Glaser P, Frangeul L, Buchrieser C, Rusniok C, Amend A, Baquero F, Berche P, Bloecker H, Brandt P, Chakraborty T, Charbit A, Chetouani F, Couvé E, de Daruvar A, Dehoux P, Domann E, Domínguez-Bernal G, Duchaud E, Durant L, Dussurget O, Entian KD, Fsihi H, García-del Portillo F, Garrido P, Gautier L, Goebel W, Gómez-López N, Hain T, Hauf J, Jackson D, Jones LM, Kaerst U, Kreft J, Kuhn M, Kunst F, Kurapkat G, Madueno E, Maitournam A, Vicente JM, Ng E, Nedjari H, Nordsiek G, Novella S, de Pablos B, Pérez-Diaz JC, Purcell R, Remmel B, Rose M, Schlueter T, Simoes N, Tierrez A, Vázquez-Boland JA, Voss H, Wehland J, Cossart P. 2001. Comparative genomics of Listeria species. Science 294:849–852. doi: 10.1126/science.1063447 [DOI] [PubMed] [Google Scholar]
- 37. Monk IR, Gahan CGM, Hill C. 2008. Tools for functional postgenomic analysis of Listeria monocytogenes. Appl Environ Microbiol 74:3921–3934. doi: 10.1128/AEM.00314-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Marinho CM, Dos Santos PT, Kallipolitis BH, Johansson J, Ignatov D, Guerreiro DN, Piveteau P, O’Byrne CP. 2019. The σB-dependent regulatory sRNA Rli47 represses isoleucine biosynthesis in Listeria monocytogenes through a direct interaction with the ilvA transcript. RNA Biol 16:1424–1437. doi: 10.1080/15476286.2019.1632776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Arnaud M, Chastanet A, Débarbouillé M. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl Environ Microbiol 70:6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bécavin C, Koutero M, Tchitchek N, Cerutti F, Lechat P, Maillet N, Hoede C, Chiapello H, Gaspin C, Cossart P. 2017. Listeriomics: an interactive web platform for systems biology of Listeria mSystems 2:e00186-16. doi: 10.1128/mSystems.00186-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lundén J, Tolvanen R, Korkeala H. 2008. Acid and heat tolerance of persistent and nonpersistent Listeria monocytogenes food plant strains: acid and heat tolerance of L. Monocytogenes. Lett Appl Microbiol 46:276–280. doi: 10.1111/j.1472-765X.2007.02305.x [DOI] [PubMed] [Google Scholar]
- 42. Schardt J, Jones G, Müller-Herbst S, Schauer K, D’Orazio SEF, Fuchs TM. 2017. Comparison between Listeria sensu stricto and Listeria sensu lato strains identifies novel determinants involved in infection. Sci Rep 7:17821. doi: 10.1038/s41598-017-17570-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen Y, Ross WH, Whiting RC, Van Stelten A, Nightingale KK, Wiedmann M, Scott VN. 2011. Variation in Listeria monocytogenes dose responses in relation to subtypes encoding a full-length or truncated internalin A. Appl Environ Microbiol 77:1171–1180. doi: 10.1128/AEM.01564-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Patchett RA, Kelly AF, Kroll RG. 1992. Effect of sodium chloride on the intracellular solute pools of Listeria monocytogenes. Appl Environ Microbiol 58:3959–3963. doi: 10.1128/aem.58.12.3959-3963.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Moura A, Criscuolo A, Pouseele H, Maury MM, Leclercq A, Tarr C, Björkman JT, Dallman T, Reimer A, Enouf V, Larsonneur E, Carleton H, Bracq-Dieye H, Katz LS, Jones L, Touchon M, Tourdjman M, Walker M, Stroika S, Cantinelli T, Chenal-Francisque V, Kucerova Z, Rocha EPC, Nadon C, Grant K, Nielsen EM, Pot B, Gerner-Smidt P, Lecuit M, Brisse S. 2017. Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat Microbiol 2:16185. doi: 10.1038/nmicrobiol.2016.185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fogg GC, Gibson CM, Caparon MG. 1994. The identification of rofA, a positive-acting regulatory component of prtF expression: use of an mγδ-based shuttle mutagenesis strategy in Streptococcus pyogenes. Mol Microbiol 11:671–684. doi: 10.1111/j.1365-2958.1994.tb00345.x [DOI] [PubMed] [Google Scholar]
- 47. Dramsi S, Dubrac S, Konto-Ghiorghi Y, Da Cunha V, Couvé E, Glaser P, Caliot E, Débarbouillé M, Bellais S, Trieu-Cuot P, Mistou M-Y. 2012. Rga, a Rofa-like regulator, is the major transcriptional activator of the PI-2A pilus in Streptococcus agalactiae. Microb Drug Resist 18:286–297. doi: 10.1089/mdr.2012.0005 [DOI] [PubMed] [Google Scholar]
- 48. McIver KS. 2009. Stand-alone response regulators controlling global virulence networks in Streptococcus pyogenes. Contrib Microbiol 16:103–119. doi: 10.1159/000219375 [DOI] [PubMed] [Google Scholar]
- 49. McIver KS, Myles RL. 2002. Two DNA-binding domains of Mga are required for virulence gene activation in the group A streptococcus. Mol Microbiol 43:1591–1601. doi: 10.1046/j.1365-2958.2002.02849.x [DOI] [PubMed] [Google Scholar]
- 50. Hause LL, McIver KS. 2012. Nucleotides critical for the interaction of the Streptococcus pyogenes Mga virulence regulator with Mga-regulated promoter sequences. J Bacteriol 194:4904–4919. doi: 10.1128/JB.00809-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. McIver KS, Heath AS, Green BD, Scott JR. 1995. Specific binding of the activator Mga to promoter sequences of the emm and scpA genes in the group A streptococcus. J Bacteriol 177:6619–6624. doi: 10.1128/jb.177.22.6619-6624.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rom JS, Hart MT, McIver KS. 2021. PRD-containing virulence regulators (PCVRs) in pathogenic bacteria. Front Cell Infect Microbiol 11:772874. doi: 10.3389/fcimb.2021.772874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Liu Y, Orsi RH, Gaballa A, Wiedmann M, Boor KJ, Guariglia-Oropeza V. 2019. Systematic review of the Listeria monocytogenes σB regulon supports a role in stress response, virulence and metabolism. Future Microbiol 14:801–828. [DOI] [PubMed] [Google Scholar]
- 54. Cheng C, Dong Z, Han X, Sun J, Wang H, Jiang L, Yang Y, Ma T, Chen Z, Yu J, Fang W, Song H. 2017. Listeria monocytogenes 10403S arginine repressor ArgR finely tunes arginine metabolism regulation under acidic conditions. Front Microbiol 8:145. doi: 10.3389/fmicb.2017.00145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ryan S, Begley M, Gahan CGM, Hill C. 2009. Molecular characterization of the arginine deiminase system in Listeria monocytogenes: regulation and role in acid tolerance. Environ Microbiol 11:432–445. doi: 10.1111/j.1462-2920.2008.01782.x [DOI] [PubMed] [Google Scholar]
- 56. Reniere ML, Whiteley AT, Hamilton KL, John SM, Lauer P, Brennan RG, Portnoy DA. 2015. Glutathione activates virulence gene expression of an intracellular pathogen. Nature 517:170–173. doi: 10.1038/nature14029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sue D, Boor KJ, Wiedmann M. 2003. σB-dependent expression patterns of compatible solute transporter genes opuCA and lmo1421 and the conjugated bile salt hydrolase gene bile salt hydrolase gene in Listeria monocytogenes. Microbiology (Reading) 149:3247–3256. doi: 10.1099/mic.0.26526-0 [DOI] [PubMed] [Google Scholar]
- 58. Garner MR, Njaa BL, Wiedmann M, Boor KJ. 2006. Sigma B contributes to Listeria monocytogenes gastrointestinal infection but not to systemic spread in the guinea pig infection model. Infect Immun 74:876–886. doi: 10.1128/IAI.74.2.876-886.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. O’Donoghue B, NicAogáin K, Bennett C, Conneely A, Tiensuu T, Johansson J, O’Byrne C. 2016. Blue-light inhibition of Listeria monocytogenes growth is mediated by reactive oxygen species and is influenced by σB and the blue-light sensor Lmo0799. Appl Environ Microbiol 82:4017–4027. doi: 10.1128/AEM.00685-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Simon P. 2003. Q-gene: processing quantitative real-time RT-PCR data. Bioinformatics 19:1439–1440. doi: 10.1093/bioinformatics/btg157 [DOI] [PubMed] [Google Scholar]
- 61. Wu J, McAuliffe O, O’Byrne CP. 2023. Trehalose transport occurs via treb in Listeria monocytogenes and it influences biofilm development and acid resistance. Int J Food Microbiol 394:110165. doi: 10.1016/j.ijfoodmicro.2023.110165 [DOI] [PubMed] [Google Scholar]
- 62. O’Byrne CP, Feehily C, Ham R, Karatzas KAG. 2011. A modified rapid enzymatic microtiter plate assay for the quantification of intracellular γ-aminobutyric acid and succinate semialdehyde in bacterial cells. J Microbiol Methods 84:137–139. doi: 10.1016/j.mimet.2010.10.017 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1-S5; Tables S1 and S2.