Abstract
A combined physical and genetic map of the genome of strain SG24 of Vibrio cholerae O139 Bengal, a novel non-O1 strain having epidemic potential, has been constructed by using the enzymes NotI, SfiI, and CeuI. The genome of SG24 is circular, and the genome size is about 3.57 Mb. The linkages between 47 NotI and 32 SfiI fragments of V. cholerae SG24 genomic DNA were determined by combining two approaches: (i) identification of fragments produced by enzyme I in fragments produced by enzyme II by the method of fragment excision, redigestion, and end labeling and (ii) use of the linking clone libraries generated from the genome of classical O1 strain 569B. The linkages between nine CeuI fragments were determined primarily by analyses of partial fragments of the CeuI-digested genome. More than 80 cloned homologous and heterologous genes, including several operons, have been positioned on the physical map. The map of the SG24 genome represents the second map of a V. cholerae genome, and a comparison of this map with that of classical O1 strain 569B revealed considerable diversity in DNA restriction sites and allowed identification of hypervariable regions. Several genetic markers, including virulence determinant genes, are in different positions in the SG24 and 569B genomes.
Vibrio cholerae, a noninvasive, gram-negative bacterium, is the causative agent of the diarrheal disease cholera. The specificity of the somatic O antigen of V. cholerae resides in the polysaccharide moiety of the lipopolysaccharide present in the outer membrane, which forms the basis of the serological classification of this organism (42). The V. cholerae strains causing epidemic cholera have, until recently, been confined to serogroup O1, which consists of two biotypes, classical and El Tor. The classical biotype was responsible for cholera epidemics till 1961, when the El Tor biotype displaced it. V. cholerae strains other than O1, which are collectively called non-O1 vibrios, can cause only sporadic infections and are believed to lack the potential to cause epidemics (30). One of the two events, the more alarming one, has dominated the global cholera scenario in the present decade; this was the unprecedented emergence in late 1992 in India of a novel strain of V. cholerae which does not agglutinate with O1 polyvalent antiserum but has epidemic and endemic potential, a phenomenon that has never occurred in the recorded history of cholera (1, 13, 36). Strains isolated from different parts of India and Bangladesh during the epidemic were found to be of clonal origin (5, 6) and were classified as new serovar O139, synonym Bengal. The other event was the dramatic and unexpected reappearance of epidemic cholera caused by V. cholerae O1 El Tor in South America in January 1991, after a 100-year absence on that continent (21). These two events have necessitated a renewed look into all aspects of the organism that are related to pathogenesis. The epidemic caused by V. cholerae O139 persisted for about a year (31, 32) and was again displaced by El Tor. Several lines of evidence have, however, suggested that O139 originated from the El Tor biotype (4, 6, 10, 13, 43) by the acquisition of a 35-kb DNA segment which replaced most of the O1 antigen-encoding rfb gene cluster of the recipient strain (8, 14). Thus, serogroup O139 combines the virulent properties of epidemic strains with the outer appearance of nonepidemic strains.
By using restriction enzymes which have a single site in either the core region or the direct repeat sequence (RS) of the CTX genetic element (27), it was shown that the genomes of most of the O139 strains have two copies of the CTX genetic element in tandem connected by two RSs (6). The chromosomal location of the CTX genetic element in an O139 strain is the same as that reported for El Tor vibrios. The organization of the virulence gene cassettes in different O139 strains showed genetic heterogeneity in the population. While most of the epidemic O139 strains have two copies of the CTX genetic element, in some strains the number of elements has been amplified and in at least one strain a copy of the element has been deleted (6).
The genomes of El Tor strains isolated immediately before and after an O139 outbreak showed extensive restriction fragment length polymorphism (RFLP) among themselves and with the genome of O139 (33, 46). In late 1996, the appearance of a V. cholerae O139 strain having altered antibiotic sensitivity compared to that of the O139 previously seen (29) has complicated the epidemiological scenario of V. cholerae and has necessitated an examination of possible rearrangements in the genome underlying such rapid changes in phenotypic traits, which are unexpected in well-characterized clonal strains within such a short period. In view of the fact that the genetic basis of V. cholerae tropism and pathogenesis is still mostly unknown, comparative genome mapping studies to appraise the extent of genome diversity will be of interest, particularly since the emergence of new variants of this organism having epidemic potential with altered genotypes or phenotypes is turning out to be widespread rather than exceptional (20). The physical map of a classical O1 strain has been constructed (12, 25), and there was previously no second map for comparison of the genomes of V. cholerae strains in more detail. It is in this context that the present report describes the construction of a macrorestriction map of the genome of O139 by use of the enzymes NotI, SfiI, and CeuI. About 80 homologous and heterologous genes and operons have been positioned on the physical map. A comparison of the V. cholerae O139 genome with that of classical O1 revealed several gross differences.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
V. cholerae classical O1 strain 569B, El Tor O1 strain CO457, and O139 Bengal strain SG24, which were used in this study, were obtained from the National Institute of Cholera and Enteric Diseases, Calcutta, India. V. cholerae cells were grown in a gyratory shaker at 37°C in nutrient broth containing 0.1 M NaCl (pH 8.0) and maintained as described previously (38). Escherichia coli cells were grown in a gyratory shaker at 37°C in Luria-Bertani broth with appropriate antibiotics whenever required.
Preparation of high-molecular-weight genomic DNA and enzyme digestion.
Agarose plugs of cells in the logarithmic phase of growth were prepared and digested with restriction enzyme NotI, SfiI, or CeuI as described previously (25, 39). For CeuI digestion, five or six sliced agarose pieces were incubated in 2× buffer supplied by the manufacturer. After 15 min, the buffer was replaced with 100 μl of fresh 1× buffer containing 10 μg of bovine serum albumin and 4 U of the enzyme, and incubation was continued at 37°C for 3 h. For partial digestion with CeuI, six agarose pieces were digested with 1 U of the enzyme in 100 μl of the reaction mixture for 2 h at 37°C.
PFGE and hybridization experiments.
Pulsed-field gel electrophoresis (PFGE) of enzyme-digested DNA was carried out in a Pulsaphor Plus System with a hexagonal electrode array (Pharmacia, Uppsala, Sweden) as described previously (25, 33, 39). Phage λ multimeric DNA and yeast chromosomal DNA were used as molecular mass markers. Preparation of 32P-labeled DNA probes and Southern blot hybridization conditions were as described previously (25, 39). End labeling of DNA fragments following NotI, SfiI, or CeuI digestion was done by incubating the agarose blocks in a buffer containing Klenow enzyme and [α-32P]dCTP and subjecting them to PFGE followed by autoradiography.
Isolation, redigestion, and radiolabeling of DNA fragments.
After PFGE, restriction fragments of genomic DNA digests were excised from low-melting-point agarose gels under long-wavelength UV light, and the agarose slice containing the DNA fragment was digested with a second enzyme and labeled as described previously (25).
RESULTS AND DISCUSSION
Restriction fragment analysis and genome size.
Previous studies on the V. cholerae genome (6, 25, 39) showed that the enzymes NotI and SfiI digest the genomic DNA into a small number of large fragments that can be resolved by PFGE. To construct the physical map of the genome of V. cholerae O139, intact genomic DNA of strain SG24 in agarose blocks was digested with NotI, SfiI, and CeuI and the resulting fragments were separated by PFGE. The endonuclease CeuI, which cleaves the genomic DNAs of all of the organisms examined so far at the 23S rRNA of the rrn operon and nowhere else in the genome (22, 26), has nine sites in the genome of V. cholerae O1 and O139 (33).
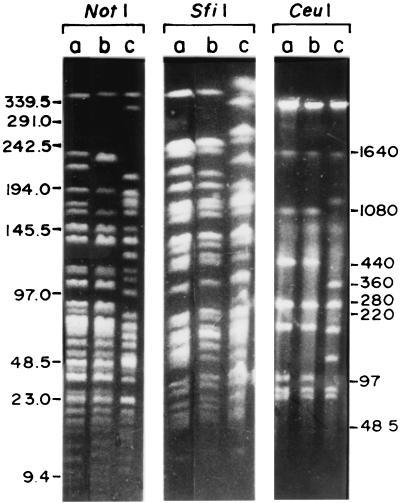
Ethidium bromide-stained gels of PFGE-separated genomic DNA fragments of strain SG24 showed that NotI, SfiI, and CeuI digestion yielded 35, 28, and 8 fragments, respectively (Fig. 1). For comparison, the NotI, SfiI, and CeuI digestion profiles of the genomes of El Tor O1 strain CO457, which displaced O139, and classical O1 strain 569B were also examined. To identify the smaller fragments that are not resolved in stained gels, the restriction fragments were end labeled with [α-32P]dCTP in agarose slices before PFGE and autoradiograms were subsequently examined. In addition to those identified in stained gels, 11 NotI fragments, 4 SfiI fragments, and 1 CeuI fragment were detected in the autoradiograms (Fig. 2). The NotI and SfiI digestion profiles of the genomes of SG24, CO457, and 569B exhibited distinct RFLP. The polymorphism is more pronounced between SG24 and 569B. The CeuI digestion profiles of SG24 and El Tor strain CO457 were identical (Fig. 1), which was expected since O139 originated from El Tor. Parenthetically, the CeuI digestion profiles of El Tor strain VC44, a strain isolated immediately before the O139 epidemic, and SG24 exhibited RFLP (33).
FIG. 1.
PFGE separation of NotI-, SfiI-, and CeuI-digested genomic DNAs of V. cholerae CO457 (lanes a), SG24 (lanes b), and 569B (lanes c). The enzyme-digested DNA was separated on 1% FastLane Agarose (FMC) with pulse time ramping between 5 and 25 s for 22 h at 10 V/cm and 3°C for NotI and SfiI. For the separation of CeuI fragments, electrophoresis was carried out by using pulse times of 5 (4 h), 10 (4 h), 25 (4 h), and 100 (12 h) s at 10 V/cm and 3°C. The gels were stained with ethidium bromide. Marker molecular sizes, in kilobases, are shown in the margins.
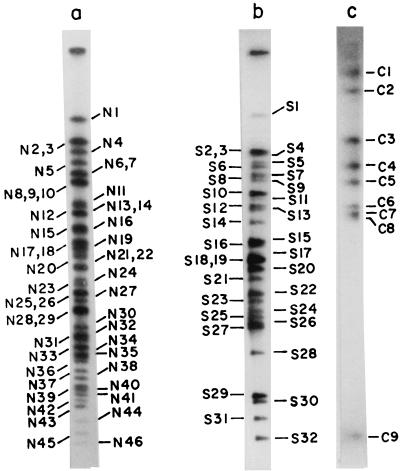
FIG. 2.
Autoradiogram of PFGE-separated NotI (lane a), SfiI (lane b), and CeuI (lane c) fragments of genomic DNA of V. cholerae O139 strain SG24. DNA in an agarose block was digested with the enzyme, end labeled with [α-32P]dCTP, and electrophoresed as described in the legend to Fig. 1. The gels were dried and exposed to X-ray film. Restriction fragments were named on the basis of the enzymes that generated the fragments (N, NotI; S, SfiI; C, CeuI) and were numbered on the basis of size in descending order.
Restriction fragments were named on the basis of the enzymes that generated the fragments (N, NotI; S, SfiI; C, CeuI) and were numbered on the basis of size in descending order (Fig. 2). NotI fragments N2 and N3, fragments N6 and N7, fragments N8, N9, and N10, fragments N13 and N14, fragments N17 and N18, fragments N21 and N22, fragments N25 and N26 and SfiI fragments S2 and S3 and fragments S18 and S19 are the same in size. The genome size of strain SG24 was estimated to be about 3.57 Mb (Table 1), as opposed to the 2.2 Mb reported previously (6). Better resolution of DNA fragments following enzyme digestion and refinement of PFGE methods by altering various parameters allowed us to resolve several previously reported NotI fragments as doublets and triplets (Table 1). Thus, the genome size of O139 is close to that of classical O1 strains (25).
TABLE 1.
Sizes of restriction fragments generated by cleavage of the chromosome of V. cholerae O139 strain SG24
| Fragment no. | Size (kb)a
|
||
|---|---|---|---|
| NotI | SfiI | CeuI | |
| 1 | 370 | 380 | 1,400 |
| 2 | 240 | 264 | 1,000 |
| 3 | 240 | 264 | 460 |
| 4 | 190 | 257 | 245 |
| 5 | 166 | 220 | 170 |
| 6 | 150 | 210 | 103 |
| 7 | 150 | 180 | 87 |
| 8 | 136 | 175 | 80 |
| 9 | 136 | 170 | 6 |
| 10 | 136 | 145 | |
| 11 | 116 | 140 | |
| 12 | 114 | 135 | |
| 13 | 108 | 130 | |
| 14 | 108 | 110 | |
| 15 | 92 | 90 | |
| 16 | 89 | 85 | |
| 17 | 84 | 80 | |
| 18 | 84 | 75 | |
| 19 | 78 | 75 | |
| 20 | 72 | 60 | |
| 21 | 66 | 55 | |
| 22 | 66 | 50 | |
| 23 | 58 | 45 | |
| 24 | 54 | 40 | |
| 25 | 49 | 32 | |
| 26 | 49 | 30 | |
| 27 | 44 | 28 | |
| 28 | 38 | 15 | |
| 29 | 38 | 10 | |
| 30 | 30 | 9 | |
| 31 | 23 | 8 | |
| 32 | 22 | 7 | |
| 33 | 20 | ||
| 34 | 18 | ||
| 35 | 17 | ||
| 36 | 16 | ||
| 37 | 14 | ||
| 38 | 13 | ||
| 39 | 12 | ||
| 40 | 11 | ||
| 41 | 10 | ||
| 42 | 9.5 | ||
| 43 | 9 | ||
| 44 | 8 | ||
| 45 | 7.5 | ||
| 46 | 7 | ||
| 47 | 2.4 | ||
Totals: NotI, 3,570 kb; SfiI, 3,574 kb; CeuI, 3,551 kb.
CeuI cleavage map of the V. cholerae O139 genome.
CeuI digestion of the V. cholerae SG24 genome produced nine fragments ranging from 1,400 to 6 kb (Fig. 2; Table 1). It has been reported that, as in other organisms, CeuI has a site only in the rrn operon of V. cholerae (33). Thus, there are nine rrn operons in the V. cholerae O139 genome. The CeuI map of the SG24 genome was constructed primarily from the analyses of 10 partial fragments of the CeuI-digested genome (Table 2). The linkage between the CeuI fragments of the genome of V. cholerae SG24 is thus C2-C1-C3-C8-C5-C7-C6-C9-C4-C2, and the genome is circular (see Fig. 5). The CeuI linkage map of the genome of O139 is different from that of classical O1 strain 569B (see Fig. 6).
TABLE 2.
Partial digestion products of the SG24 genome generated by the enzyme CeuI
| Partial fragment (size [kb]) | Possible composition (size [kb]) |
|---|---|
| P1 (113) | C6, C9 (109) |
| P2 (250) | C5, C8 (250) |
| P3 (355) | C4, C6, C9 (354) |
| P4 (465) | C4, C6, C7, C9 (441) |
| P5 (550) | C3, C8 (540) |
| P6 (600) | C4, C5, C6, C7, C9 (611) |
| P7 (680) | C4, C5, C6, C7, C8, C9 (691) or C3, C8, C5 (710) |
| P8 (1,250) | C2, C4 (1,245) |
| P9 (1,900) | C1, C3 (1,860) |
| P10 (2,450) | C1, C2 (2,400) |
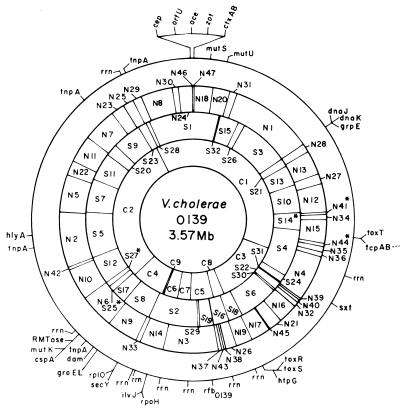
FIG. 5.
Combined physical and genetic map of the genome of V. cholerae O139 strain SG24 determined by using enzymes NotI, SfiI, and CeuI. The restriction fragments are numbered on the basis of size (Table 1). An asterisk denotes tentative assignment. The genetic markers are described in Table 4. The positioning of the genetic markers in a particular fragment is arbitrary, and the positions of the markers in a single fragment do not reflect their true order in the chromosome. CeuI sites were taken as the positions of the rrn operons.
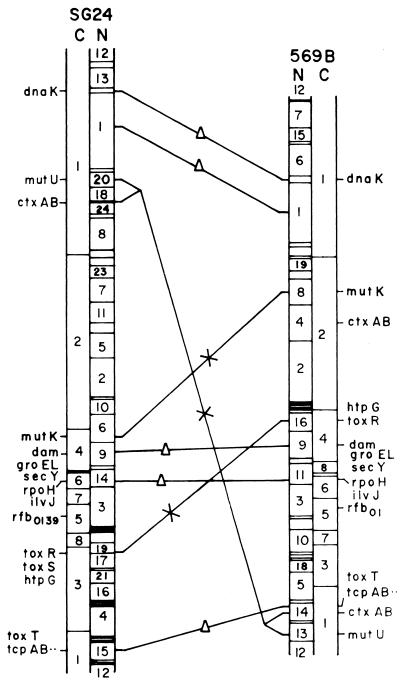
FIG. 6.
Comparison of the chromosome maps of V. cholerae O139 strain SG24 and V. cholerae classical O1 strain 569B. For a better comparison, the circular maps were linearized. A triangle on a line connecting the two genomes indicates a conserved region, and regions in the chromosomes that have undergone rearrangement are indicated by an X on a connecting line. C and N represent CeuI and NotI maps, respectively.
NotI and SfiI cleavage maps of the V. cholerae O139 genome.
The linkages between 47 NotI and 32 SfiI fragments of V. cholerae SG24 genomic DNA were determined by combining three approaches. These included (i) identification of fragments produced by enzyme I in fragments produced by enzyme II by the method of fragment excision, redigestion, and end labeling, (ii) use of the linking clone libraries generated from the genome of classical O1 strain 569B (26), and (iii) analysis of partial digestion products.
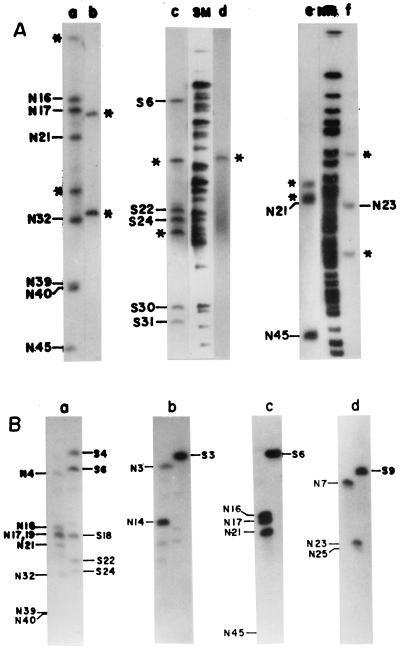
The CeuI linkage map was used as the skeleton to determine the linkages between the NotI and SfiI fragments of the genome of V. cholerae SG24. To identify linkages between different NotI or SfiI fragments, individual PFGE-separated CeuI fragments were excised from the gel, digested with NotI or SfiI, end labeled, and subjected to PFGE (Fig. 3A). Alternatively, the gel-excised CeuI fragments were used as probes for Southern blot hybridization of the NotI- or SfiI-digested genome of V. cholerae SG24 (Fig. 3B). Both of these approaches allowed clubbing of NotI and SfiI fragments that are linked. For example, when gel-excised CeuI fragment C3 was digested with the enzyme NotI, end labeled, resolved by PFGE, and autoradiographed, nine NotI fragments (N17, N45, N21, N16, N32, N40, N39, and two flanking fragments arising from N4 and N19; Fig. 3A, lane a), and seven SfiI fragments (S6, S24, S30, S22, S31, and two flanking fragments, S4 and S18; Fig. 3A, lane c) lit up, showing that these fragments are linked. The NotI or SfiI fragments overlapping the two ends of C3 were determined by hybridizing fragment C3 with the PFGE-resolved NotI- or SfiI-digested genomic DNA (Fig. 3B, part a). The order in which these fragments are present in the genome was determined by using position-specific probes, such as linking clones (Table 3), NotI or SfiI fragments that are included in the CeuI fragment, and cloned-gene probes (Table 4). Similar analyses were performed with gel-excised CeuI, NotI, and SfiI fragments.
FIG. 3.
(A) Identification of NotI or SfiI fragments of the V. cholerae SG24 genome in isolated CeuI fragments. CeuI fragments C3 (lanes a and c) and C6 (lanes b and d) were completely digested with NotI or SfiI, end labeled, and separated by PFGE. Identification of NotI fragments in isolated SfiI fragments. SfiI fragments S6 (lane e) and S9 (lane f) were completely digested with the enzyme NotI, end labeled, and separated by PFGE. NotI (NM)- or SfiI (SM)-digested, end-labeled SG24 genomic DNA was used as a marker for identification of linked fragments. Asterisks denote the flanking DNA fragments. (B) Assignment of the flanking NotI or SfiI fragments (shown in panel A) of V. cholerae SG24 genomic DNA in isolated CeuI fragments. Radiolabeled CeuI fragment C3 (a) or C6 (b) was used as the probe for Southern hybridization with the NotI- or SfiI-digested genome of V. cholerae SG24. For identification of the flanking NotI fragments in isolated SfiI fragments of the genomic DNA of strain SG24, radiolabeled SfiI fragment S6 (c) or S9 (d) was used as the probe for Southern hybridization with the NotI- or SfiI-digested SG24 genome. Hybridizations were carried out at 60°C. The filters were washed under stringent conditions as described in Materials and Methods.
TABLE 3.
Linkages between NotI and SfiI fragments of the genome of strain SG24 of V. cholerae O139 determined by using linking clones or restriction fragments
| Clone or restriction fragment |
Linkage |
|---|---|
| NLB2 | N12-N27 |
| NLB33 | N23-N25 |
| NLB39 | N13-N27 |
| NLB42 | N7-N23 |
| NLB5 | N6-N10 |
| NLB51 | N1-N28 |
| NLB92 | N2-N5 |
| NLB29 | N5-N22 |
| NLB130 | N4-N36 |
| NLS5 | N18-N20 |
| NLS16 | N24-N30 |
| N2a | S5-S12 |
| N5 | S5-S7 |
| N7a | S9-S20 |
| N11 | S11-S20 |
| C6 | N3-N14 |
| C8 | N19-N26 |
| NLH35 | N10-N42 |
| NLH39 | N19-N26 |
| NLH8 | N16-N21 |
| NLH30 | N4-N39 |
| NLH44 | N8-N29 |
| NLH108 | N13-N27 |
| NLH117 | N9-N33 |
| NLH127 | N1-N31 |
| NLH132 | N15-N34 |
| S5 | N2-N5 |
| S7 | N5-N22 |
| S10 | N12-N27 |
| S11 | N11-N22 |
| S12 | N10-N42 |
| S13 | N13-N27 |
| S20 | N7-N11 |
| S23 | N25-N29 |
Fragment excised from PFGE-separated NotI and CeuI double-digested genomic DNA.
TABLE 4.
Homologous and heterologus genes positioned on the physical map of the genome of strain SG24 of V. cholerae O139
| Probe | Gene(s) (product or function) | Source | Reference |
|---|---|---|---|
| pCVD15 | ctxAB (cholera toxin) | V. cholerae | 18 |
| pKB370 | mutK (DNA mismatch repair), cspA (cold shock protein), rmt (RNA methyltransferase) | V. cholerae | Unpublished |
| pJT470 | mutS (DNA mismatch repair) | V. cholerae | 3 |
| pJA360 | mutU (DNA mismatch repair) | V. cholerae | Unpublished |
| pRB101 | dam (adenine methylase) | V. cholerae | 2 |
| pGS350 | rpoH (ς32) | V. cholerae | 40 |
| pOF12 | groEL (Hsp60) | E. coli | 16 |
| pSC830 | dnaK (Hsp70) | V. cholerae | Unpublished |
| pDB19 | secY (protein translocation) | V. cholerae | 7 |
| pVM7 | toxR (virulence gene activator) | V. cholerae | 28 |
| pSC18.1 | tcp (toxin-coregulated pilus) | V. cholerae | 19 |
| pEB1111 | tnpA (transposase, IS1004) | V. cholerae | 9 |
| pSXT1 | sxt (sulfamethoxazole and trimethoprim resistance) | V. cholerae | 44 |
| pKY017 | hlyA (hemolysin) | V. cholerae | 45 |
| pEB1150 | rfbO139 (O antigen) | V. cholerae | 8 |
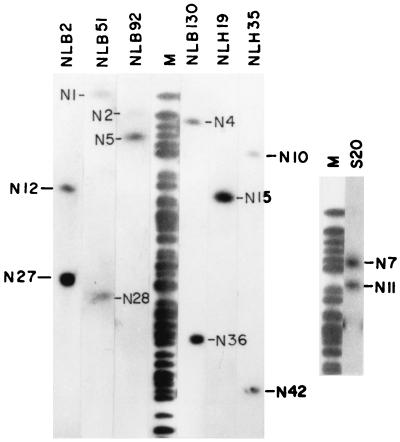
The NotI linking clone library generated from the genome of classical O1 strain 569B (25) has been extensively used to determine the linkages between NotI fragments. Some representative examples of hybridization experiments using linking clones as probes are shown in Fig. 4. The linkages between the NotI fragments of the 569B genome determined by using linking clones NLH44 and NLB29 (25) are different from those of the NotI-digested genome of SG24 (data not shown). Linking clone NLH19 linked NotI fragments N15 and N26 of the 569B genome (25), but it hybridized with only NotI fragment N15 of the SG24 genome, indicating deletion of the linked fragment (Fig. 4). Besides NotI linking clones, several SfiI and CeuI fragments of the O139 genome having a single NotI site served as NotI linking clones, and NotI and CeuI fragments having a single SfiI site were used as SfiI linking clones (Fig. 4; Table 3).
FIG. 4.
Determination of linkages between NotI fragments of SG24 genomic DNA by using a NotI linking clone library of the 569B genome and isolated SfiI fragments of the SG24 genome. Nick-translated linking clones NLB2, NLB51, NLB92, NLB130, NLH19, NLH35, and NLH44 and SfiI fragment S20 were hybridized with PFGE-separated NotI fragments of the SG24 genome. NotI-digested and end-labeled genomic DNA was used as molecular size markers (lane M) for identification of linked fragments. The NotI fragments that are linked to each other are marked.
Mapping of similar-size fragments.
To identify and position similar-size fragments on the map, a combination of the following approaches was adopted: (i) analysis of PFGE-separated genomic DNA digested with any two of the three enzymes NotI, SfiI, and CeuI, (ii) hybridization of comigrating fragments generated by one enzyme with the genomic DNA digested with a second enzyme, and (iii) use of suitable linking clones or cloned genes. For example, to position NotI fragments N2 and N3, N6 and N7, and N13 and N14, which appeared as doublets on the map, genomic DNA was digested with NotI and CeuI and double digestion products were separated by PFGE. The digestion profile revealed that one of the two fragments N2 and N3, N6 and N7, or N13 and N14 has a CeuI site(s) and the other does not (data not shown). One of the fragments in the N2-N3 doublet is located in fragment C2 (which will be referred to as N2), and the other is located in fragment C6 (which will be referred to as N3). This was confirmed by Southern blot hybridization of the NotI-digested O139 genome by using fragment C6 (Fig. 3B, part b) or C2 as the probe and by end-labeling experiments. Linking clone NLH44 hybridized with fragment N8 and the doublet N28-N29 (data not shown). When the same probe was used for the Southern hybridization of NotI-SfiI-double-digested genomic DNA, it hybridized with fragment N8 and another smaller fragment designated X (generated from either N28 or N29), indicating the presence of an SfiI site(s). The fragment in the N28-N29 doublet hybridizing with linking clone NLH44 will be referred to as N29. Linking clone NLB51 hybridized with N1 and N28 and with S3. Besides, dnaK also hybridized with N28 and S3. Linking clone NLH8 hybridized with C3 and S6 and links fragment N16 and either N21 or N22 (which will be referred to as N21). Linking clone NLB29 links fragments N5 and N22 (Table 3) and also hybridized with C2 and S7. While positioning comigrating NotI fragments on the map, it was revealed that N8-N9-N10 is a triplet (Table 1). This was confirmed by digesting the gel-excised triplet with the enzyme AscI or by hybridization using fragment C1, C2, or C4 as the probe (data not shown). Doublets N2-N3 and N6-N7 were also digested with AscI, which confirmed that they are doublets (data not shown). The other comigrating fragments were similarly positioned on the map. By combining all of the approaches, a multienzyme macrorestriction map of the genome of V. cholerae O139 strain SG24 has been constructed (Fig. 5).
Positioning of genetic markers on the physical map.
More than 80 cloned genes (Table 4) and nine rrn operons have been positioned on the physical map of the V. cholerae O139 genome by hybridization using homologous and heterologous probes (Fig. 5). The gene probes used comprised O139 antigen-specific genes, the hemolysin gene, DNA repair genes, heat shock protein genes, cholera toxin genes, trimethoprim-sulfamethoxazole resistance genes, and some other virulence determinant genes (Table 4). In the V. cholerae genome, the CeuI sites are located only in the rrn operons (33) similar to those reported for several other bacteria (22). The genes have been positioned on the physical map on fragments they hybridized with and do not represent the exact locations and orientations on the fragment. Genes hybridizing with the same fragment have been positioned on the fragment arbitrarily, and the positioning does not reflect the true order of the genes in the chromosome.
Comparison of the physical maps of O139 and classical O1 strain 569B.
A comparison of the physical map of the V. cholerae O139 genome constructed in the present study with that of classical O1 strain 569B (25) revealed conservation in certain regions but gross rearrangements in the genome of O139 with respect to the classical O1 genome (Fig. 6). Restriction site variability between the two genomes is another feature emerging from the comparison. A fraction of sites is conserved between the two strains. The genome of V. cholerae O139 has gained a number of NotI sites. The variable regions in the genomes of both strains might be due to exchange of genetic elements. Extensive RFLP was recorded between the genomes of classical O1, El Tor, and O139 following digestion with NotI, SfiI, or CeuI (Fig. 1) and is more predominant between the O139 and classical O1 genomes. The genome of SG24 has more NotI (total, 47) and SfiI (total, 32) sites (Table 1) than does the genome of classical O1 strain 569B (25). Although the chromosomal locations of several genes, like rpoH, the tcp gene cluster, etc., are conserved (Fig. 6), they show RFLP because of loss or creation of restriction sites.
NotI linking clones of the genome of the classical O1 strain allowed identification of regions of the O139 genome that might have undergone rearrangements. Several linking clones of the genome of classical O1 strain 569B hybridized with only one fragment of the NotI-digested O139 genome (Fig. 4), indicating that either the NotI site was lost or one of the linked NotI fragments underwent deletion in the O139 genome. While NotI linking clone NLH44 linked fragments N6 (175 kb) and N22 (42 kb) in the NotI-digested 569B genome, it showed linkages between fragments N8 (136 kb) and N29 (38 kb) of the NotI-digested SG24 genome. This clone hybridized with the SfiI-digested genomes of strains 569B and O139 in fragments S3 (280 kb) and S1 (380 kb), respectively (data not shown). Linking clone NLB29 hybridized with one similar-size NotI fragment of both the 569B and SG24 genomes, and the other fragment showed RFLP.
The copy number of direct RS IS1004 is reduced to four in the genome of O139 from eight in that of classical O1 (9). Most of the genetic markers examined exhibited RFLP in the NotI- or SfiI-digested genomic DNAs of O139 and classical O1. In classical O1 strains, the CTX genetic element is present in two copies that are separated by about 1.5 Mb in the genome (12). The genome of V. cholerae O139, which originated from El Tor, has two copies of the element in tandem, as expected (5, 6). One copy of the CTX genetic element in classical O1 strain 569B is closely linked to several other virulence determinant genes like the tcp operon and toxT and is located in NotI fragment N14 (12). The two copies of the CTX genetic element present in tandem in the O139 genome are located about 1 Mb apart from the tcp operon and the toxT gene (Fig. 5). In the genome of V. cholerae O139, the position of toxR, the positive transcriptional activator gene regulating the expression of several major virulence genes, including cholera toxin gene ctxAB, has changed from that in strain 569B and is no longer linked to the groEL, dam, and secY genes (Fig. 5 and 6). Another variable region stretches the CTX genetic element along with mutator genes mutU and mutS. The locations of the mutK, cspA, and RNA methyltransferase genes in the genome of O139 are different from those in the classical O1 genome (12). The presence of such hypervariable regions in the genome has been reported in several bacterial pathogens (11, 15, 17, 34, 35, 37, 41). However, the rfb and hlyA loci in the genomes of SG24 and 569B are conserved.
The number and location of rrn operons in the genomes of enteric bacteria appear to be highly conserved. In contrast to seven rrn operons in the genomes of the different gram-negative bacteria examined so far (22), the genomes of V. cholerae O1 and O139 have nine (33). While the number of rrn operons in strains 569B and SG24 remained the same, their positions in the chromosome have altered, suggesting major genomic rearrangements between O1 and O139 (Fig. 6). A similar observation has been reported in Salmonella typhi, an enteric pathogen causing typhoid fever. It was suggested that the genetic events responsible for rearrangement in the genome of S. typhi are presumably due to recombination between the rrn genes and not at other sites (23, 24). The role of rrn operons in imparting genome instability in V. cholerae has yet to be investigated. However, in contrast to S. typhi, where the lengths of the CeuI fragments remain unaltered in almost all of the strains studied, the sizes of the CeuI fragments of the genomes of O1 and O139 strains are different (Fig. 1). It has been reported that V. cholerae strains belonging to non-O1–non-O139 serogroups have 10 rrn operons in their genomes (33). Intraspecies variation in rrn operons has not been reported in other organisms.
Genomic rearrangements at high frequency might play a positive role in the developmental processes of bacteria. To survive in the environment while maintaining epidemic potential and to elude all of the associated onslaughts following infection, pathogenic organisms might prefer to store their virulence factors in relatively plastic regions of the chromosome, which would give them the kind of flexibility necessary to sustain their pathogenic potential.
ACKNOWLEDGMENTS
We thank M. K. Waldor, New England Medical Center, Boston, Mass., for providing plasmid pSXT1 and K. Yamamoto, Osaka University, Osaka, Japan, for plasmid pKY017.
This work was supported by research grant BT/R&D/PRO-109/15/8/96 from the Department of Biotechnology, Government of India. S.K. is the recipient of a Research Associateship from the Council of Scientific and Industrial Research, Government of India.
REFERENCES
- 1.Albert M J, Siddique A K, Islam M S, Faruque A S J, Ansaruzzaman M, Faruque S M, Sack R B. Large outbreak of clinical cholera due to Vibrio cholerae non-O1 in Bangladesh. Lancet. 1993;341:704. doi: 10.1016/0140-6736(93)90481-u. [DOI] [PubMed] [Google Scholar]
- 2.Bandyopadhyay R, Das J. DNA adenine methyl transferase encoding gene (dam) of Vibrio cholerae. Gene. 1994;140:67–71. doi: 10.1016/0378-1119(94)90732-3. [DOI] [PubMed] [Google Scholar]
- 3.Bera T K, Ghosh S K, Das J. Cloning and characterization of the mutL and mutS genes of Vibrio cholerae: nucleotide sequence of the mutL gene. Nucleic Acids Res. 1989;17:6241–6251. doi: 10.1093/nar/17.15.6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berche P, Poyart C, Abachin E, Lelivre H, Vandepitte J, Dodin A, Fournier J-M. The novel epidemic strain O139 is closely related to the pandemic strain O1 of Vibrio cholerae. J Infect Dis. 1994;170:701–704. doi: 10.1093/infdis/170.3.701. [DOI] [PubMed] [Google Scholar]
- 5.Bhadra R K, Roychowdhury S, Das J. Vibrio cholerae O139 El Tor biotype. Lancet. 1994;343:728. doi: 10.1016/s0140-6736(94)91604-7. [DOI] [PubMed] [Google Scholar]
- 6.Bhadra R K, Roychoudhury S, Banerjee R K, Kar S, Majumdar R, Sengupta S, Chatterjee S, Khetawat G, Das J. Cholera toxin (CTX) genetic element in Vibrio cholerae O139. Microbiology. 1995;141:1977–1983. doi: 10.1099/13500872-141-8-1977. [DOI] [PubMed] [Google Scholar]
- 7.Bhattacharyya D, Das J. V. cholerae analogue of E. coli secY gene: identification, cloning and characterization. Gene. 1997;196:261–266. doi: 10.1016/s0378-1119(97)00238-2. [DOI] [PubMed] [Google Scholar]
- 8.Bik E M, Bunschoten A E, Gouw R D, Mooi F R. Genesis of the novel epidemic Vibrio cholerae O139 strain: evidence for horizontal transfer of genes involved in polysaccharide synthesis. EMBO J. 1995;14:209–216. doi: 10.1002/j.1460-2075.1995.tb06993.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bik E M, Gouw R D, Mooi F R. DNA fingerprinting of Vibrio cholerae strains with a novel insertion sequence element: tool to identify epidemic strains. J Clin Microbiol. 1996;34:1453–1461. doi: 10.1128/jcm.34.6.1453-1461.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calia K E, Muratagh M, Ferraro M J, Calderwood S B. Comparison of Vibrio cholerae O139 with V. cholerae O1 classical and El Tor biotypes. Infect Immun. 1994;62:1504–1506. doi: 10.1128/iai.62.4.1504-1506.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Canard R, Saint-Joanis B, Cole S T. Genomic diversity and organization of virulence genes in the pathogenic anaerobe Clostridium perfringens. Mol Microbiol. 1992;6:1421–1429. doi: 10.1111/j.1365-2958.1992.tb00862.x. [DOI] [PubMed] [Google Scholar]
- 12.Chatterjee S, Mondal A K, Begum N A, Roychoudhury S, Das J. Ordered cloned DNA map of the genome of Vibrio cholerae 569B and localization of genetic markers. J Bacteriol. 1998;180:901–908. doi: 10.1128/jb.180.4.901-908.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cholera Working Group. Large epidemic of cholera-like disease in Bangladesh caused by Vibrio cholerae O139 synonym Bengal. Lancet. 1993;342:387–390. [PubMed] [Google Scholar]
- 14.Comstock L E, Johnson J A, Michalski J M, Morris J G, Jr, Kaper J B. Cloning and sequence of a region encoding a surface polysaccharide of Vibrio cholerae O139 and characterization of the insertion site in the chromosome of Vibrio cholerae O1. Mol Microbiol. 1996;19:815–826. doi: 10.1046/j.1365-2958.1996.407928.x. [DOI] [PubMed] [Google Scholar]
- 15.Dempsey J A F, Wallace A B, Cannon J G. The physical map of the chromosome of a serogroup A strain of Neisseria meningitidis shows complex rearrangements relative to the chromosomes of the two mapped strains of the closely related species N. gonorrhoeae. J Bacteriol. 1995;177:6390–6400. doi: 10.1128/jb.177.22.6390-6400.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fayet O, Ziegelhoffer T, Georgopoulos C. The groES and groEL heat shock gene products of Escherichia coli are essential for bacterial growth at all temperatures. J Bacteriol. 1989;171:1379–1385. doi: 10.1128/jb.171.3.1379-1385.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang Q, Hiratsuka K, Taylor D E. Variability of gene order in different Helicobacter pylori strains contributes to genomic diversity. Mol Microbiol. 1996;20:833–842. doi: 10.1111/j.1365-2958.1996.tb02521.x. [DOI] [PubMed] [Google Scholar]
- 18.Kaper J B, Lockman H, Baldini M M, Levine M M. A recombinant live oral cholera vaccine. Bio/Technology. 1984;2:345–349. [Google Scholar]
- 19.Keasler S P, Hall R H. Detecting and biotyping Vibrio cholerae O1 with multiplex polymerase chain reaction. Lancet. 1993;341:1661. doi: 10.1016/0140-6736(93)90792-f. [DOI] [PubMed] [Google Scholar]
- 20.Khetawat, G., R. K. Bhadra, S. Nandi, and J. Das. Resurgent Vibrio cholerae O139: rearrangement of CTX genetic elements and amplification of rrn operons. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 21.Levine M M. South America: the nature of cholera. Lancet. 1991;338:45–46. [Google Scholar]
- 22.Liu S-L, Hessel A, Sanderson K E. Genomic mapping with I-CeuI, an intron-encoded endonuclease specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli and other bacteria. Proc Natl Acad Sci USA. 1993;90:6874–6878. doi: 10.1073/pnas.90.14.6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu S-L, Sanderson K E. Rearrangements in the genome of the bacterium Salmonella typhi. J Bacteriol. 1995;177:3355–3357. [Google Scholar]
- 24.Liu S-L, Sanderson K E. Highly plastic chromosomal organization in Salmonella typhi. Proc Natl Acad Sci USA. 1996;93:10303–10308. doi: 10.1073/pnas.93.19.10303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Majumder R, Sengupta S, Khetawat G, Bhadra R K, Roychoudhury S, Das J. Physical map of the genome of Vibrio cholerae 569B and localization of genetic markers. J Bacteriol. 1996;178:1105–1112. doi: 10.1128/jb.178.4.1105-1112.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marshall P, Lemieux C. Cleavage pattern of the homing endonuclease encoded by the fifth intron in the chloroplast large subunit rRNA-encoding gene of Chlamydomonas eugametos. Gene. 1991;104:241–245. doi: 10.1016/0378-1119(91)90256-b. [DOI] [PubMed] [Google Scholar]
- 27.Mekalanos J J. Duplication and amplification of toxin genes in Vibrio cholerae. Cell. 1983;35:253–263. doi: 10.1016/0092-8674(83)90228-3. [DOI] [PubMed] [Google Scholar]
- 28.Miller V L, Mekalanos J J. Synthesis of cholera toxin is positively regulated at the transcriptional level by toxR. Proc Natl Acad Sci USA. 1984;81:3471–3475. doi: 10.1073/pnas.81.11.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitra R, Basu A, Dutta D, Nair G B, Takeda Y. Resurgence of Vibrio cholerae O139 Bengal with altered antibiogram in Calcutta, India. Lancet. 1996;348:1181. doi: 10.1016/s0140-6736(05)65326-3. [DOI] [PubMed] [Google Scholar]
- 30.Morris J G. Non-O group 1 Vibrio cholerae: a look at the epidemiology of an occasional pathogen. Epidemiol Rev. 1990;12:179–191. doi: 10.1093/oxfordjournals.epirev.a036052. [DOI] [PubMed] [Google Scholar]
- 31.Mukhopadhyay A K, Garg S, Nair G B, Kar S, Ghosh R K, Pajni S, Ghosh A, Shimada T, Takeda T, Takeda Y. Biotype traits and antibiotic susceptibility of Vibrio cholerae serogroup O1 before, during and after the emergence of the O139 serogroup. Epidemiol Infect. 1995;115:427–434. doi: 10.1017/s0950268800058581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mukhopadhyay A K, Garg S, Mitra R, Basu A, Rajendran K, Dutta D, Bhattacharya S K, Shimada T, Takeda T, Takeda Y, Nair G B. Temporal shifts in traits of Vibrio cholerae strains isolated from hospitalized patients in Calcutta: a 3-year (1993 to 1995) analysis. J Clin Microbiol. 1996;34:2537–2543. doi: 10.1128/jcm.34.10.2537-2543.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nandi S, Khetawat G, Sengupta S, Majumder R, Kar S, Bhadra R K, Roychoudhury S, Das J. Rearrangements in the genomes of Vibrio cholerae strains belonging to different serovars and biovars. Int J Syst Bacteriol. 1997;47:858–862. doi: 10.1099/00207713-47-3-858. [DOI] [PubMed] [Google Scholar]
- 34.Philipp W J, Nair S, Guglielmi G, Lagranderie M, Gicquel B, Cole S T. Physical mapping of Mycobacterium bovis BCG Pasteur reveals differences from the genome map of Mycobacterium tuberculosis H37Rv and from M. bovis. Microbiology. 1996;142:3135–3145. doi: 10.1099/13500872-142-11-3135. [DOI] [PubMed] [Google Scholar]
- 35.Pyle L E, Taylor T, Flinch L R. Genomic maps of some strains within the Mycoplasma mycoides cluster. J Bacteriol. 1990;172:7265–7268. doi: 10.1128/jb.172.12.7265-7268.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramamurthy T, Garg S, Sharma R, Bhattacharya S K, Nair G B, Shimada T, Takeda T, Karasawa T, Kurazono H, Pal A, Takeda Y. Emergence of novel strain of Vibrio cholerae with epidemic potential in southern and eastern India. Lancet. 1993;341:703–704. doi: 10.1016/0140-6736(93)90480-5. [DOI] [PubMed] [Google Scholar]
- 37.Romling U, Greipel J, Tummler B. Gradient of genomic diversity in the Pseudomonas aeruginosa chromosome. Mol Microbiol. 1995;17:323–332. doi: 10.1111/j.1365-2958.1995.mmi_17020323.x. [DOI] [PubMed] [Google Scholar]
- 38.Roy N K, Das G, Balganesh T S, Dey S N, Ghosh R K, Das J. Enterotoxin production, DNA repair and alkaline phosphatase of Vibrio cholerae before and after animal passage. J Gen Microbiol. 1982;128:1927–1932. doi: 10.1099/00221287-128-9-1927. [DOI] [PubMed] [Google Scholar]
- 39.Roychoudhury S, Bhadra R K, Das J. Genome size and restriction fragment length polymorphism analysis of Vibrio cholerae strains belonging to different serovars and biotypes. FEMS Microbiol Lett. 1994;115:329–334. doi: 10.1111/j.1574-6968.1994.tb06659.x. [DOI] [PubMed] [Google Scholar]
- 40.Sahu G K, Chowdhury R, Das J. The rpoH gene encoding ς32 homology of Vibrio cholerae. Gene. 1997;189:203–207. doi: 10.1016/s0378-1119(96)00849-9. [DOI] [PubMed] [Google Scholar]
- 41.Schmidt K D, Tummler B, Romling U. Comparative genome mapping of Pseudomonas aeruginosa PAO with P. aeruginosa C, which belongs to a major clone in cystic fibrosis patients and aquatic habitats. J Bacteriol. 1996;178:85–93. doi: 10.1128/jb.178.1.85-93.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shimada T, Arakawa E, Itoh K, Okitsu T, Matsushima A, Asai Y, Yamai S, Nakazato T, Nair G B, Albert M J, Takeda Y. Extended serotyping scheme for Vibrio cholerae. Curr Microbiol. 1994;28:175–178. [Google Scholar]
- 43.Waldor M K, Mekalanos J J. ToxR regulates virulence gene expression in non-O1 strains of Vibrio cholerae that cause epidemic cholera. Infect Immun. 1994;62:72–78. doi: 10.1128/iai.62.1.72-78.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waldor M K, Tschape H, Mekalanos J J. A new type of conjugative transposon encodes resistance to sulfamethoxazole, trimethoprim, and streptomycin in Vibrio cholerae O139. J Bacteriol. 1996;178:4157–4165. doi: 10.1128/jb.178.14.4157-4165.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamamoto K, Ichinose Y, Shinagawa H, Makino K, Nakata A, Iwanaga M, Honda T, Miwatani T. Two-step processing for activation of the cytolysin/hemolysin of Vibrio cholerae O1 biotype El Tor: nucleotide sequence of the structural gene (hlyA) and characterization of the processed products. Infect Immun. 1990;58:4106–4116. doi: 10.1128/iai.58.12.4106-4116.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamasaki S, Nair G B, Bhattacharya S K, Yamamoto S, Kurazono H, Takeda Y. Cryptic appearance of a new clone of Vibrio cholerae serogroup O1 biotype El Tor in Calcutta, India. Microbiol Immunol. 1997;41:1–6. doi: 10.1111/j.1348-0421.1997.tb01165.x. [DOI] [PubMed] [Google Scholar]