ABSTRACT
Myeloproliferative neoplasms (MPNs) are a class of rare hematological malignancies that result in the overproduction of myeloid lineage cells. These malignancies result in increased cytokine production and inflammation, which correlate with worsened symptom burden and prognosis. Other than bone marrow transplantation, there is no cure for myeloproliferative neoplasms. As such, treatments focus on reducing thrombotic risk, inflammation, and symptom burden. Because current pharmacological treatments carry significant side effects, there is a need to explore low-risk therapies that may modulate inflammation and alleviate symptom burden. One potential way to achieve this is adherence to a Mediterranean (MED) diet, which is rich in anti-inflammatory foods, reduces inflammatory biomarkers, and beneficially alters the gut microbiome. We performed a 15-week clinical trial of 28 individuals with MPN who were randomized to dietary counseling based on either a Mediterranean diet or standard U.S. Guidelines for Americans. Our primary objective was to determine whether MPN patients could adopt a Mediterranean eating pattern when supported with dietician counseling. As exploratory endpoints, we investigated the impact of diet and inflammation on the gut microbiome. Using shotgun metagenomic sequencing, we found that microbiome diversity and composition were stable throughout the study duration in both cohorts. Furthermore, we discovered significant differences in the microbiomes between MPN subtypes, such as increased beta-dispersion in subjects with myelofibrosis. Lastly, we found several significant correlations between the abundance of multiple bacterial taxa and cytokine levels. Together, this study provides insight into the interaction between diet, inflammation, and the gut microbiome.
IMPORTANCE
The gut microbiome serves as an interface between the host and the diet. Diet and the gut microbiome both play important roles in managing inflammation, which is a key aspect of myeloproliferative neoplasm (MPN). Studies have shown that a Mediterranean (MED) diet can reduce inflammation. Therefore, we longitudinally characterized the gut microbiomes of MPN patients in response to Mediterranean or standard 2020 US Guidelines for Americans dietary counseling to determine whether there were microbiome-associated changes in inflammation. We did not find significant changes in the gut microbiome associated with diet, but we did find several associations with inflammation. This research paves the way for future studies by identifying potential mechanistic targets implicated in inflammation within the MPN gut microbiome.
KEYWORDS: gut microbiome, diet, myeloproliferative, neoplasm, Mediterranean
INTRODUCTION
Myeloproliferative neoplasms (MPNs) are a group of hematological malignancies defined by somatic mutations that activate JAK/STAT signaling in hematopoietic stem cells (1, 2). This results in an overproduction of myeloid lineage cells. Clinically, MPNs are divided into three clinical phenotypes: polycythemia vera (PV), essential thrombocythemia (ET), and myelofibrosis (MF). PV is characterized by an elevated red blood cell mass. Elevations in platelets and white blood cells are also common. Subjects with ET have elevated platelets but rarely have increased red or white blood cells. MF is characterized by reticulin fibrosis in the bone marrow, and often cytopenia. MF can develop from a “burn out” phase following PV or ET, termed post-PV or post-ET MF, or without a preceding diagnosis of PV or ET, termed primary myelofibrosis.
One feature of MPN is increased inflammatory cytokine abundance, which correlates with worsened symptom burden and disease prognosis (3, 4). MPN symptom burden can be severe, and many individuals experience fatigue, early satiety, abdominal discomfort, night sweats, pruritus, bone pain, fever, and unintentional weight loss. Other than bone marrow transplantation, there is no cure for MPN, and management focuses on reducing thrombotic risk and alleviating symptom burden. Current pharmacological treatments for MPN include JAK inhibitors, such as ruxolitinib, but these often carry significant side effects, like immunosuppression, weight gain, and increased skin cancer (5). Consequently, there is a need to explore low-risk alternatives for MPN management.
One method to non-pharmacologically manage MPN is through the consumption of a Mediterranean (MED) diet, which emphasizes the intake of extra virgin olive oil, fruits, vegetables, whole grains, legumes, fish, nuts, and seeds. Specifically, a Mediterranean diet contains three to nine servings of vegetables, one half to two servings of fruit, 1 to 13 servings of cereals, and up to eight servings of olive oil daily (6). Adherence to a Mediterranean diet eating pattern can be quantified using a 14-point Mediterranean Diet Adherence Score (MEDAS) (7). A MED diet has been shown to reduce inflammation by lowering C-reactive protein and interleukin (IL)-6 levels and is associated with reduced obesity, cardiovascular disease, and cancer risk (8–11). Adherence to a MED diet has been found to alter the gut microbiome, which is the collection of bacteria, fungi, viruses, and other microorganisms living within the large intestine (12–17). Mechanistically, the dietary fiber and unsaturated fat in the MED diet are fermented by gut microbes, resulting in the production of anti-inflammatory metabolites (18). However, it remains to be seen whether a MED diet can be strategically used to manipulate the gut microbiota to promote health by reducing inflammation in MPN.
We performed a randomized clinical trial to investigate whether registered dietician counseling of individuals with MPN can alter their eating patterns toward a Mediterranean style. Subjects were randomly assigned to (i) MED diet counseling supplemented with complementary extra virgin olive oil (MED cohort) or (ii) diet counseling following the standard US Guidelines for Americans (USDA cohort) supplemented with grocery certificates. The study length was 15 weeks, consisting of a 2-week pre-intervention observation, 10 weeks of active dietary counseling, and 3 weeks of post-counseling follow-up. A description of the education-based MED intervention, patient satisfaction, diet adherence, symptom burden, and cytokine concentrations is available in a companion manuscript (19). As a key exploratory endpoint, we investigated whether a MED diet could produce a microbiome-mediated reduction in inflammation. Blood and stool samples were collected to measure cytokine levels and assess gut microbiome composition, respectively. Survey data were collected to assess the feasibility of a MED diet intervention among MPN patients and symptom burden was tracked using the MPN Symptom Assessment Form (MPN-SAF). In this manuscript, we detail the association of the gut microbiota with diet, MPN subtype, and cytokine concentrations.
RESULTS
Cohort description and study synopsis
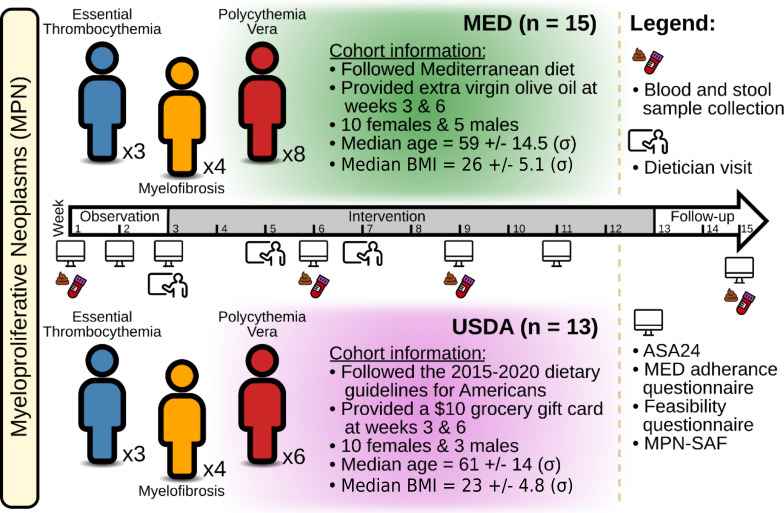
In all, 28 subjects with MPN were recruited for this study (Fig. 1). The MED cohort had 15 individuals, while the USDA cohort had 13 individuals. Within the MED cohort, three subjects had ET, four had MF, and eight had PV. Within the USDA cohort, three subjects had ET, four had MF, and six had PV. The median age for the MED cohort was 59 ± 14.5 (σ) years, while the median age for the USDA cohort was 61 ± 14 (σ) years. The median BMI for the MED cohort was 26 ± 5.1 (σ) years, and the median BMI for the USDA cohort was 23 ± 4.8 (σ) years. Both groups had 10 females each, with 5 and 3 males in the MED and USDA cohorts, respectively. The study took place over 15 weeks and had an active intervention period from weeks 3 to 12. Baseline blood and stool samples were collected at week 1, followed by additional sampling during the active intervention at weeks 6 and 9. Follow-up samples were also taken after the intervention’s end at week 15. Throughout the study, six unannounced surveys and 24-hour food recalls (ASA24) were collected to measure diet compliance and symptom burden was assessed using the MPN-SAF, which grades the 10 most clinically relevant symptoms of MPN patients (20). Table 1 provides a detailed description of each subject’s characteristics.
Fig 1.
Study design. A total of 28 individuals with MPNs were enrolled in the study. Participants were randomly assigned to dietary counseling following either a MED diet (n = 15) or a conventional American diet (USDA, n = 13). The study was 15 weeks long and had a 2-week observation period, a 10-week intervention period, and a 3-week follow-up period. Blood and stool samples were collected at weeks 1, 6, 9, and 15. At weeks 3, 5, and 7, participants met with a dietician and were informed about the core components of each diet and how to follow it. On weeks 1, 2, 3, 6, 9, 11, and 15, subjects were asked to fill out 24-hour dietary recalls (ASA24), MED adherence and feasibility questionnaires, and an MPN-SAF.
TABLE 1.
Subject characteristics
| Subject | Diet | MPN | Treatment | Mutation | Age | Sex | BMI |
|---|---|---|---|---|---|---|---|
| 2 | USDA | PV | Ruxolitinib (Jakafi) | JAK2 | 71 | M | 30 |
| 3 | USDA | MF | Observation only | MPL | 63 | F | 23 |
| 5 | USDA | MF | Hydroxyurea (Hydrea), interferon (Pegasys) | JAK2 | 63 | F | 22 |
| 7 | USDA | PV | Interferon (Pegasys) | JAK2 | 44 | F | 25 |
| 9 | USDA | ET | Observation only, aspirin | JAK2 | 57 | M | 26 |
| 10 | USDA | PV | Observation only, phlebotomy | JAK2 | 21 | F | 24 |
| 12 | USDA | PV | Hydroxyurea (Hydrea) | JAK2 | 77 | M | 36 |
| 14 | MED | PV | Aspirin, interferon (Pegasys) | JAK2 | 34 | F | 32 |
| 15 | MED | PV | Hydroxyurea (Hydrea) | JAK2 | 68 | F | 29 |
| 16 | MED | ET | Aspirin, hydroxyurea (Hydrea), interferon (Pegasys) | JAK2 | 70 | F | 39 |
| 17 | MED | PV | Eliquis and phlebotomy | JAK2 | 58 | F | 36 |
| 18 | MED | PV | Hydroxyurea (Hydrea), prednisone | JAK2 | 66 | M | 24 |
| 19 | USDA | ET | Aspirin | JAK2 | 61 | F | 32 |
| 20 | MED | ET | Aspirin, hydroxyurea (Hydrea) | CALR | 71 | M | 26 |
| 21 | MED | MF | Aspirin, hydroxyurea (Hydrea) | CALR | 25 | F | 22 |
| 22 | MED | MF | Observation only, Chinese herbs | JAK2 | 71 | F | 25 |
| 23 | MED | PV | Anagrelide (Agrylin), ruxolitinib (Jakafi) | JAK2 | 54 | F | 25 |
| 24 | MED | MF | Ruxolitinib (Jakafi) | JAK2 | 67 | F | 21 |
| 25 | MED | PV | Observation only, aspirin, other | JAK2 | 59 | F | 24 |
| 26 | MED | MF | Observation only, Aspirin | JAK2 | 53 | M | 28 |
| 28 | MED | PV | Aspirin, interferon (Pegasys) | JAK2 | 40 | M | 26 |
| 29 | MED | ET | Aspirin, hydroxyurea (Hydrea) | JAK2 | 70 | F | 22 |
| 30 | MED | PV | Observation only, aspirin | JAK2 | 50 | M | 27 |
| 31 | USDA | ET | Aspirin, hydroxyurea (Hydrea) | JAK2 | 66 | F | 22 |
| 32 | USDA | PV | Aspirin, hydroxyurea (Hydrea), other | JAK2 | 67 | F | 21 |
| 33 | USDA | PV | Aspirin, hydroxyurea (Hydrea) | JAK2 | 57 | F | 22 |
| 34 | USDA | MF | Other | JAK2 | 51 | F | 22 |
| 35 | USDA | MF | Observation only | JAK2 | 58 | F | 20 |
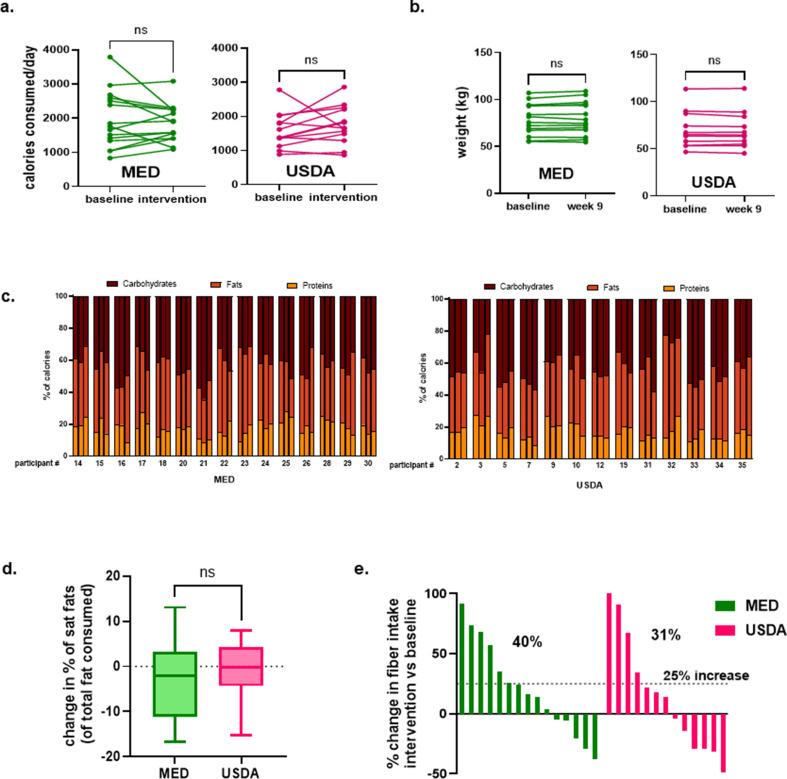
Impact of dietary intervention on consumed calories, body weight, macronutrients, and fiber intake
Participants completed 24-hour diet recalls at weeks 1, 2, 3, 6, 9, 11, and 15. Data from weeks 1 and 2 were used to calculate average baseline intake and data from weeks 6, 9, and 11 were used to calculate average intake during the active intervention. Neither group had a significant change in total calorie intake during the active intervention period (Fig. 2a). A significant change in body weight was also not observed at week 9 when compared to baseline (Fig. 2b). A visual representation of the percentage of calories from carbohydrates, protein, and fat is shown in Fig. 2c. We investigated how each diet intervention impacted the percentage of calories from saturated fat. There was a mean change in the percentage of calories from saturated fat of −1.19% and −2.25% in the USDA and MED groups, respectively, which was not statistically significant (Fig. 2d). We also measured the change in fiber intake during the active intervention period. Thirty-one percent of the USDA group increased fiber intake by at least 25% and 40% of the MED group increased fiber intake by at least 25% (Fig. 2e).
Fig 2.
Impact of dietary intervention on consumed calories, body weight, macronutrients, and fiber intake. (a) Average calories consumed in each group at baseline (average of weeks 1 and 2) and during active intervention (average of weeks 6, 9, and 11). (b) Change in weight at week 9 compared to enrollment. (c) Visualization of the percentage of calories from carbohydrates, proteins, and fat each participant consumed at baseline (left bar) during active intervention (center bar), and follow-up period (right bar). (d) Change in percentage of calories from saturated fats during intervention (average of weeks 6, 9, and 11) vs baseline (average of weeks 1 and 2). (e) Waterfall plot of change in fiber intake during the active intervention period vs baseline. Each bar represents a participant. Significance testing was performed using a t-test.
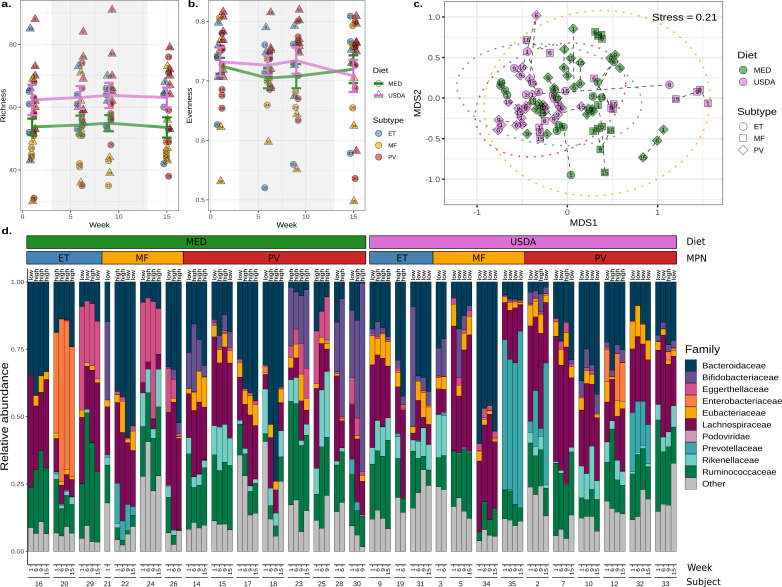
Gut microbiome diversity and composition are stable during the Mediterranean diet intervention
We began our investigation by examining how the MED diet impacts gut microbiome diversity. Analysis of species richness estimates using a linear mixed-effects model (LME) demonstrated that USDA and MED groups did not significantly differ over time after accounting for pre-intervention differences (LME P-value = 0.48, Fig. 3a). Analyses of species evenness estimates also showed no differences between diet groups (LME P-value = 0.65, Fig. 3b). Sub-setting species richness and evenness comparisons to include only samples from participants highly adherent to a Mediterranean style eating pattern and those least adherent to a Mediterranean style eating pattern during the intervention also did not reveal significant differences (LME richness P-value = 0.48, LME evenness P-value = 0.73).
Fig 3.
Gut microbiome diversity and composition are stable during the Mediterranean diet intervention. (a) Microbial richness and (b) evenness estimates of fecal samples collected at weeks 1, 6, 9, and 15. The shaded background indicates the active dietary intervention period for both diet groups. The mean richness or evenness for each group is represented with a colored line, with the error bars reflecting the standard error. Each point is labeled centrally with the individual of origin. (c) Non-metric multidimensional scaling of Bray-Curtis dissimilarities produced from compositional microbiome data. Points are colored by diet and shaped by the MPN subtype. A 95% confidence interval was drawn around each MPN subtype (blue = ET, yellow = MF, and red = PV). Dashed lines connect samples taken from the same individual, and the week of collection is labeled centrally within each point. (d) A taxa bar plot of the top 10 most abundant microbial families across individuals, time, diet, and MPN subtypes. Each sample is labeled “high” or “low” and refers to MED adherence for the week.
Next, we examined the microbial composition, or beta-diversity, of the fecal samples. Species composition analysis using non-metric multidimensional scaling (NMDS) and permutational multivariate analysis of variance (PERMANOVA) showed that there were significant differences associated with MED and USDA groups pre-intervention (Fig. 3c; PERMANOVA R2 = 0.057, P-value = 0.046, Table S1). Therefore, we stratified our PERMANOVA analysis to investigate whether gut microbiome composition changed over time within each individual. This produced non-significant results, showing that microbiome composition was stable throughout the study (Fig. 3d; PERMANOVA R2 = 0.007, P-value = 0.76, Table S2). Next, PERMANOVA was performed on each MPN subtype to examine whether a specific subtype responded to the diet intervention more than others. No changes were detected in ET (PERMANOVA R2 = 0.046, P-value = 0.63), MF (PERMANOVA R2 = 0.026, P-value = 0.60), or PV (PERMANOVA R2 = 0.016, P-value = 0.77) subtypes over time (Table S3). Consequently, no microbial species were significantly different between MED and USDA groups after adjusting for pre-existing compositional differences.
Characterization of the functional metagenome demonstrated no significant differences between diets as measured by microbial gene richness (LME P-value = 0.65, Fig. S1A) and gene evenness (LME P-value = 0.19, Fig. S1B) after accounting for pre-intervention differences. PERMANOVA analysis indicated that there were no significant changes over time within each individual (PERMANOVA R2 = 0.009, P-value = 0.29, Table S4). No differentially abundant genes were found between MED and USDA groups.
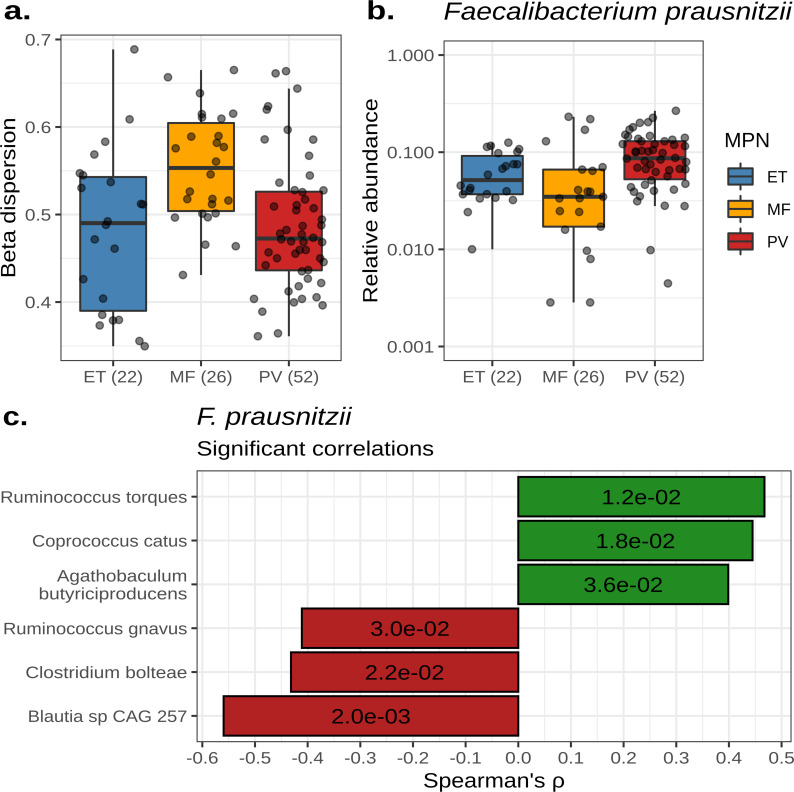
Individuals with myelofibrosis have reduced microbial diversity and altered composition
Previous research has demonstrated significant differences in the microbiomes associated with healthy individuals and those with MPN (21). Therefore, we characterized the microbiome between PV, ET, and MF subtypes further. Using species richness estimates, we observed a significant reduction in the number of unique microbes when comparing individuals with MF to PV (linear mixed-effects P-value = 0.028, Fig. S2A), and a non-significant reduction when comparing MF to ET (linear mixed-effects P-value = 0.056, Fig. 3a). Species abundance distribution, or evenness, was also reduced in MF, but was not significant compared to PV (LME P-value = 0.12) and ET subtypes (LME P-value = 0.47, Fig. 3b).
With respect to beta-diversity, samples from MF were more dissimilar from each other, resulting in a trend toward increased beta-dispersion when compared to ET (LME P-value = 0.089) and PV (LME P-value = 0.056, Fig. S2C; Fig. 4a). Conversely, PV and ET samples tended to cluster together (LME P-value = 0.895, Fig. S2C; Fig. 4a). PERMANOVA demonstrated that the individual of origin significantly explained about 51% of the variance observed in the microbiome, while the MPN subtype significantly explained approximately 6.0% of variance (PERMANOVA P-value = 0.001 for both, Table S5). For comparison, age, sex, and BMI explained 3.3%, 3.3%, and 2.4% of the variance associated with microbiome composition, respectively (PERMANOVA P-values = 0.001, Table S5). The analysis of microbial composition between MPN subtypes showed a reduction in the abundance of Faecalibacterium prausnitzii in MF subjects when compared to those with PVor ET (ANCOM2 P < 0.05, Fig. 4b). Microbes most correlated with F. prausnitzii abundance included Ruminococcus torques, Coprococcus catus, Agathobaculum butyriciproducens, Ruminococcus gnavus, Clostridium bolteae, and Blautia sp. CAG-257 (Fig. 4c).
Fig 4.
Individuals with myelofibrosis have reduced microbial diversity and altered composition. (a) A box plot showing the beta-dispersion of each MPN subtype calculated from taxonomic Bray-Curtis dissimilarities. (b) The relative abundance of Faecalibacterium prausnitzii across MPN subtypes. For (A) and (B), the number of samples per subtype is labeled parenthetically and the center line within each box defines the median. Boxes define the upper and lower quartiles and whiskers define 1.5× the interquartile range. (c) A bar plot showing the Spearman correlation coefficients of microbes significantly correlated with F. prausnitzii abundance. P-values for each correlation are labeled within each bar.
Within the functional metagenome data, we detected a significant reduction in the number of unique microbial genes within MF subjects when compared to PV (LME P-value = 0.016, Fig. S3A), but not ET (linear mixed-effects P-value = 0.244, Fig. S3A). There was no significant difference in the gene evenness among MPN subtypes (Fig. S3B). NMDS ordination demonstrated that the functional metagenome compositions of MF samples tended to be more disparate from each other when compared to PV (LME P-value = 0.34) and ET (LME P-value = 0.19, Fig. S3C and D). In addition, the MPN subtype significantly explained about 6.7% of the variance observed in functional metagenome composition (PERMANOVA P-value = 0.001, Table S6), while the individual of origin was associated with about 51% of the variance (PERMANOVA P-value = 0.001, Table S6). Differential abundance analysis produced no significantly different genes between MPN subtypes after FDR correction.
Cytokine levels are correlated with microbiome diversity and composition
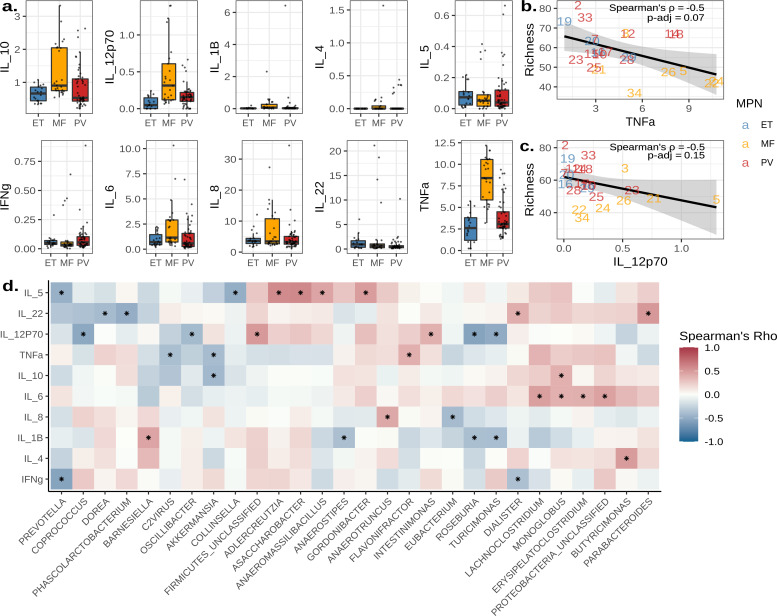
After subsequent analysis of MPN subtypes and their gut microbiomes, we next asked if the diversity or composition of the microbiome was associated with the concentrations of 10 plasma cytokines. Comparison of cytokine concentrations between MPN subtypes revealed a significant increase in TNFα and IL-12p70 in subjects with MF when compared to ET (Tukey’s test; TNFα P-adj <0.001 and IL-12p70 P-adj = 0.016) and PV (Tukey’s test; TNFα P-adj = 0.002 and IL-12p70 P-adj = 0.022, Fig. 5a). IL-6, IL-8, and IL-10 concentrations were elevated in subjects with MF but were not statistically significant (Fig. 5a). Microbial richness was most negatively correlated with TNFα (Spearman’s ρ = −0.50, P-adj = 0.07, Fig. 5b) and IL-12p70 (Spearman’s ρ = −0.45, P-adj = 0.15, Fig. 5c).
Fig 5.
Cytokine levels are correlated with microbiome diversity and composition. (a) Box plots displaying the concentration of cytokines measured in pg/mL across MPN subtypes. The center line within each box defines the median, boxes define the upper and lower quartiles, and whiskers define 1.5× the interquartile range. (b and c) Scatter plots of TNFα (b) and IL-12p70 (c) concentrations in pg/mL correlated with species richness estimates. Points are labeled by the individual of origin and colored by MPN subtypes. A line represents the mean, and the shaded area delineates the 95% confidence interval. (d) A heat map of microbial genera whose abundances significantly correlated with cytokine concentrations. Asterisks denote significant correlations (P < 0.05).
Next, we compared cytokine concentrations with microbial abundances at the genus level, resulting in 34 significant correlations (Fig. 5d). Notable correlations included associations with TNFα vs Flavonfractor (Spearman’s ρ = 0.39, P = 0.038), IL-12p70 vs Roseburia (Spearman’s ρ = −0.55, P = 0.002), and IL-8 vs Eubacterium (Spearman’s ρ = −0.41, P = 0.032, Fig. S4). Similarly, we compared cytokine concentrations with functional pathway abundances, producing 162 significant correlations (Fig. S5). Notable correlations included TNFα vs 4-deoxy-L-threo-hex-4-enopyranuronate degradation (Spearman’s ρ = −0.42, P = 0.038), TNFα vs β-(1,4)-mannan degradation (Spearman’s ρ = −0.48, P = 0.019), and IL-12p70 vs GDP-mannose biosynthesis (Spearman’s ρ = −0.59, P = 0.003, Fig. S6).
DISCUSSION
Our goal with this manuscript was to (i) assess whether a MED diet altered the gut microbiome of subjects with MPN and (ii) investigate the association between diversity and composition of the gut microbiome and levels of plasma cytokines. In a separate manuscript, we describe the feasibility of a MED diet intervention in the MPN subject population, changes in macronutrients associated with the dietary intervention, and the interaction between diet adherence, symptom burden, and cytokine concentrations (19). Here, we report that microbial diversity or composition did not significantly change during a MED diet intervention over a 10-week active dietary intervention period. Instead, we found the MPN subtype played a greater role in determining microbiome diversity and composition. Individuals with ET and PV had more similar microbial compositions, while those with MF were more disparate. Furthermore, a reduction in microbial diversity correlated with elevated TNFα and IL-12p70 concentrations in subjects with MF. These differences in cytokine concentrations were associated with the abundances of 34 microbial genera and 162 metabolic pathways, further establishing a role for the gut microbiome in inflammation and MPN.
With respect to diet-mediated changes in the microbiome, there are multiple potential explanations as to why the microbiomes of individuals remained stable throughout the dietary intervention. The first is intervention duration. Long-term adherence to a Mediterranean diet has been demonstrated to reduce the incidence of cardiovascular disease, Alzheimer’s disease, colorectal cancer, diabetes, and obesity (10, 11). Studies performing MED diet interventions have ranged from 6 weeks to 7 years (14–17, 22–30). Of studies examining gut microbiome composition, the 6-week MED diet intervention performed by Marlow et al. yielded no significant differences in gut microbiome composition or CRP levels in subjects with Crohn’s disease (30). Comparatively, Haro et al. conducted a MED diet intervention in a cohort of 20 obese men and observed differences in microbiota composition after 1 year (17). Similarly, Nagpal et al. conducted a MED diet intervention in a non-human primate model over the course of 2.5 years, and a significant difference in the microbiome was observed between macaques who consumed a Western diet vs a MED diet (16). Because this was a feasibility study, and the primary objective was to assess whether patients with MPN could change their diet with dietician counseling, a relatively short duration was chosen. Our data and others suggest that longer MED dietary intervention periods are needed to detect changes in gut microbiome composition.
Another consideration in the successful manipulation of gut microbiomes with diet is the presence of specific microbial taxa, functions, or enterotypes. Stratification of microbiomes into enterotypes has previously revealed enterotype-specific predictors of dietary intervention response (31). Klimenko et al. found that the strongest predictor of whether an individual would respond to a dietary intervention was the average number of genes per microbe (31). A negative correlation between the average number of genes per microbe and alpha diversity was found, suggesting that more diverse communities are formed by specialist microbes with fewer genes (31). The microbiomes associated with industrialized countries, like the United States, often have reduced diversity and a higher abundance of Bacteroides when compared to non-industrialized countries (32). Many Bacteroides are generalists, meaning they contain more genes and wider metabolic potentials than specialist taxa (33). The predominance of generalist taxa has been known to contribute to microbiome stability (34). Therefore, it is plausible that the microbiomes of industrialized individuals have evolved to resist perturbations, such as those caused by antibiotic usage or short-term dietary changes. Our samples contained relatively high abundances of Bacteroides, so it is possible that the microbiomes of these individuals were resistant to short-term dietary changes as reflected by the non-significant changes in diversity, composition, and function over time.
A final factor that could have affected the stability of our microbiomes is the strength of dietary intervention. Due to differences in agriculture and food processing, a MED diet in the United States is different from a MED diet in the Mediterranean region. This can affect the number of antibiotic and prebiotic compounds found in each diet. One prebiotic component of the MED diet that can influence gut microbiome composition is extra virgin olive oil (EVOO). EVOO is rich in polyphenols and oleic acid, which have been demonstrated to have anti-oxidative and anti-inflammatory properties (35, 36). Over 90% of polyphenols are digested and metabolized in the colon by the gut microbiota (37). Dietary supplementation of EVOO in humans has been shown to promote the growth of beneficial microbes like Bifidobacterium and lactic acid-producing bacteria (35, 38). In rodent models, consumption of EVOO results in an increased abundance of Bifidobacterium, Lactobacillus, and Clostridium (39, 40). Our future dietary interventions in MPN patients will include an educational module on the benefits of EVOO, which we anticipate will lead to increased EVOO consumption and a change in gut microbiome composition.
A MED diet is also typically higher in dietary fiber when compared to a typical USDA diet. Dietary fiber is fermented by the gut microbiota to produce short-chain fatty acids, such as acetate, propionate, and butyrate. Butyrate is critical for gut health, as it is the primary source of energy for colonocytes and reduces inflammation by stimulating the production of T-regulatory cells and IL-10-producing cells (41). In this study, a minority of participants increased their fiber intake during the intervention period. As such, we did not find that the abundances of butyrate-producing bacteria differed between diets. Instead, we saw a reduction in the butyrate-producing microbe, F. prausnitzii, in subjects with MF. We also noted significant positive correlations between F. prausnitzii, Agathobaculum butyriciproducens, and Coprococcus catus abundances. A. butyriciproducens is a butyrate-producing microbe, while C. catus produces both butyrate and propionate (42). We also observed broader, community-wide differences between subjects with ET, PV, and MF. Notably, the microbiome composition of MF subjects was more dissimilar to each other when compared to ET and PV. Our previous work comparing the gut microbiome composition of healthy and MPN subjects similarly showed that individuals with MF had increased beta-dispersion when compared to ET and PV (21). These results describe a phenomenon known as the “Anna Karenina principle” for animal microbiomes, which states that stressors affect microbiomes in unpredictable ways, leading to increased community beta-dispersion (43, 44).
One likely stressor resulting in higher MF beta-dispersion is the increased concentration of pro-inflammatory cytokines. Inflammation has been known to negatively affect the gut microbiome. Supporting this notion, we found that TNFα and IL-12p70 were significantly increased in MF subjects, which negatively correlated with species richness overall. We found IL-12p70 negatively correlated with the genus Roseburia, which are butyrate-producing microorganisms known to alleviate inflammation by promoting T-regulatory cell differentiation (45, 46). We also observed a significant negative correlation with Eubacterium and the pro-inflammatory cytokine, IL-8. Eubacterium also produces butyrate and has been shown to lessen inflammation by promoting IL-10 production (45, 47). TNFα plasma concentrations were found to be negatively correlated with the abundance of the 4-deoxy-L-threo-hex-4-enopyranuronate degradation pathway, which aids in the degradation of uronic acids, which includes pectin fiber. Another fiber whose degradation pathway was negatively correlated with TNFα was β-(1,4)-mannan.
Taken together, it is possible that the increased inflammation observed in individuals with MPN, particularly MF, is exacerbated by the lack of dietary fiber and short-chain fatty acid production. The MED diet has been previously shown to promote the growth of F. prausnitzii specifically; therefore, future experiments could attempt to restore microbial short-chain fatty acid production to reduce inflammation. When designing dietary interventions, however, special attention should be given to the intervention duration and the ability of existing gut microbes to use and respond to prebiotic compounds. This may ensure that the desired outcomes are achieved, allowing us to manipulate the gut microbiome to promote health and ameliorate disease.
MATERIALS AND METHODS
Recruitment of subjects
Patients were recruited between October 2018 and September 2019. Participants were included if they were over the age of 18 with a previous diagnosis of a Philadelphia chromosome-negative MPN (including PV, ET, and MF), had an ECOG score of 2 or less, a life expectancy of greater than 20 weeks, had internet access, an email address, and could read and understand English. Any type of previous or current therapy was also allowed. Participants were excluded if they were pregnant or planning on becoming pregnant, lost more than 10 pounds or 10% of their body weight in the last 6 months, or were allergic to nuts and olive oil. In all, 47 participants were screened. Five did not meet the inclusion criteria, and an additional 11 subjects were excluded due to incomplete survey data during the observation period. In all, 31 subjects were randomly assigned to a diet, but two withdrew participation and one failed to provide sufficient survey data. The final number of study participants was 28, with 15 belonging to the MED cohort and 13 belonging to the USDA cohort.
Collection of dietary intervention feasibility, adherence, and symptom burden data
During the first week of the intervention period, each participant met individually with a dietician to learn about the central components of their assigned diet. There were follow-up dietician visits during weeks 5 and 7. Participants were emailed educational materials on their respective diets weekly during the 10-week active intervention period. Furthermore, participants in the MED cohort were given 750 mL of extra virgin olive oil, and those in the USDA cohort were given a $10 grocery gift card at weeks 3 and 6. Throughout the study, participants were required to fill out 4 unannounced surveys given during weeks 1, 2, 3, 6, 9, 11, and 15. The first survey measured dietary intervention feasibility and asked, “how easy is it for you to follow this diet, with 1 being very easy to follow and 10 being very difficult to follow?” The second survey measured MED diet adherence. For this, the established 14-item Mediterranean diet adherence score (MEDAS) was used (7). Adherence to a MED diet was defined as a “high” for the week if a score of >8/14 was obtained. Next, we asked subjects to complete 24-hour food recalls using the Automated Self-Administered 24-hour Dietary Assessment Tool (ASA24), providing detailed information about calories consumed, macronutrients, and dietary fiber. Lastly, symptom burden was assessed via the MPN-SAF, which grades the 10 most clinically relevant MPN symptoms (20). Surveys were administered through email.
Blood collection and cytokine measurements
Peripheral blood was drawn on weeks 1, 3, 6, and 15 in tubes containing ethylenediaminetetraacetic acid. Plasma was obtained by centrifuging 3–4 mL of blood for 10 min at 2,500 rpm, aliquoted, and stored at −80°C. Frozen plasma was sent to Quanterix in Billerica, MA for analysis. A Human CorPlex 10 Cytokine Array kit #85–0329 (IL-12p70, IL-1B, IL-4, IL-5, IFNɣ, IL-6, IL-8, IL-22, TNFα, and IL-10) was used according to the manufacturer’s protocol and analyzed using a Quanterix SPX imager system on-site at Quanterix Headquarters in Billerica, MA.
Fecal sample collection
To perform gut microbiome analysis, four stool samples were requested from each participant over the course of the 15-week trial. The samples were collected by the participants themselves using Zymo RNA/DNA shield fecal collection tubes (Cat. #R1101) during weeks 1, 6, 9, and 15. Samples were returned in person or by mail. In total, 103 samples were collected. Samples were stored at −80°C once returned.
DNA extraction
Fecal samples stored in a DNA/RNA shield were thawed on ice, vortexed to homogenize, then DNA from 1,000 uL of fecal slurry was extracted using the ZymoBiomics DNA Miniprep Kit (Cat. #D4300) according to the manufacturer’s protocol. Bead lysis during the extraction was performed at 6.5 m/s for 5 min total (MPBio FastPrep-24).
Shotgun library preparation and sequencing
Libraries for shotgun sequencing were prepared using the Illumina DNA prep kit (Cat. # 20018705), using an adapted low-volume protocol (48). In summary, we reduced the amount of DNA used per sample to a maximum of 5 uL or 50 ng (whichever was reached first). Tagmentation was performed according to the manufacturer’s protocol, but volumes were reduced to 1 uL of bead-linked transposome and tagmentation buffer each. Next, 1.25 uL of 1 uM i5 and i7 indices were added to each sample and annealed via polymerase chain reaction using 10 uL of KAPA HiFi HotStart ReadyMix (Cat. # 7958935001). Afterward, libraries were combined, size selected, and cleaned using 56 and 14.4 uL of sample purification beads according to the low-volume protocol. Positive and negative sequencing controls were included during the library preparation using the ZymoBIOMICS Microbial Community DNA Standard (Cat. #D6305) and purified water, respectively. The quality of libraries was assessed with the Quanti-iT PicoGreen dsDNA (Cat. #P7589) for quantity and Agilent Bioanalyzer High Sensitivity DNA Analysis (Cat. #5067–4626) for fragment size. Libraries were shipped overnight on dry ice to Novogene Corporation Inc. (Sacramento, CA) to be sequenced using Illumina’s Hiseq 4000. An average of 2,819,107 ± 670,543 (σ) paired-end reads per sample, 150 base pairs long, were obtained.
OTU table generation
Raw data were first cleaned to remove sequencing adapters and artifacts using the BBMap v38.79 script “bbduk.sh” with the flag “ref = adapters,artifacts” (49). BBMap’s “demuxbyname.sh” was used to demultiplex sequences using the default parameters. Quality filtering of sequences was performed using PRINSEQ ++ v1.2 with the following parameters: -trim_left 5 -trim_right 5 - min_len 100 -trim_qual_right 28 - min_qual_mean 25 (50). Quality checking was done with FastQCv0.11.8 on default parameters. This resulted in a mean and standard deviation of 2,731,886 ± 648,042 paired-end reads, respectively. Human-derived reads were removed using BowTie2 v2.4.5 using the default parameters and hg38 as the reference genome, which produced an average of 2,498,159 ± 960,477 (σ) reads per sample (51). Taxonomic assignment of sequence data was performed using MetaPhlAn v3.0.14 with the default parameters and the CHOCOPhlAn v2019.01 database (52).
Microbiome functional potential data generation
Individual gene annotations were produced by first cross-assembling reads into contiguous sequences using MEGAHIT v1.1.1 with a minimum length of 2,500 base pairs and the flag “--k-list 31,41,51,61,71,81,91,101,111” (53). Afterward, open reading frames were assigned with Prodigal v2.6.3 and then annotated with eggNOG mapper v2.0 using the eggNOG v5.0 database (54, 55). Next, BowTie2 v2.4.5 was used to align samples to the annotated genes to obtain a table of per sample counts for each gene. Lastly, per-sample gene counts were normalized to reads per kilobase per genome equivalent using MicrobeCensus v1.1.1 on default parameters (56). For the functional annotation of metabolic pathways, we ran our quality-filtered, unassembled reads through HUMAnN v3.0.1 using the default parameters and the UniRef90 v201901b database (52). The “humann_renorm_table” and “humann_join_tables” scripts were used to create a pathway abundance table of normalized counts in copies per million.
Data analysis
Data analysis of OTUs, genes, and pathways was performed in R v4.2.1. The first step was removing microbes or genes that contaminated our sequencing controls from all samples. The Vegan v2.6–2 package was used to calculate the following metrics: richness with the “specnumber” function, evenness with the formula “diversity(x, index = “Shannon”)/log10(specnumber(x)),” Bray-Curtis beta-diversity with the “vegdist” function, PERMANOVA with the “adonis2” function, NMDS with the “metaMDS” function, and beta-dispersion with the “betadisper” function. Please see Tables S1 to 6 for PERMANOVA formulas and parameters. Significance testing of richness, evenness, and beta-dispersion was performed using linear mixed-effect models with the nlme v3.1–159 package. Significance testing of cytokine concentrations was done using an ANOVA and Tukey’s post hoc test with the “aov” and “TukeyHSD” functions. Spearman correlations were obtained using the “cor.test” and “rcorr” functions. Differential abundance of OTUs was determined with ANCOM v2.1 with the parameters: rand_formula = “~ 1 | Subject,” p_adj_method = “none,” alpha = 0.05. For gene and pathway abundances, we averaged abundances within each subject to eliminate repeated measurements and performed a Kruskal-Wallis test. When appropriate, multiple comparisons were corrected using the “p.adjust(x, method = “fdr”)” function. All code, scripts, and parameters for data processing and analysis can be found at https://github.com/Javelarb/MPN_diet_intervention.
ACKNOWLEDGMENTS
This study was partially funded by the UCI Anti-Cancer Challenge Grant P30CA062203, the MPN Research Foundation, and the UCI Microbiome Center. We would also like to acknowledge funding from the American Cancer Society, #IRG-16-187-13, the Samueli Scholar program supported by the Susan Samueli Integrative Health Institute, and the NIH-IMSD training grant GM055246, which supported J.A.-B.
AGF developed the study, recruited and consented patients, and analyzed the data; LFML recruited and consented patients, collected and analyzed the data; JN coordinated patient visits and collected clinical data; HN coordinated patient visits and collected clinical data; AO developed the study and analyzed the data; and JAB conducted a portion of the experiments and performed the data analysis. JAB wrote the manuscript with guidance from AF and KW.
Contributor Information
Katrine L. Whiteson, Email: katrine@uci.edu.
Edward G. Ruby, University of Hawaii at Manoa, Honolulu, Hawaii, USA
Ruben A. T. Mars, University Medical Center Groningen and University of Groningen, Groningen, the Netherlands
DATA AVAILABILITY
Sequencing data are available on the Sequence Read Archive under the BioProject ID PRJNA918651. The taxonomic table with corresponding metadata used to generate the source data can be found in Supplementary_file_2.csv. Larger files used in our functional metagenomic analysis are available on the Dryad Digital Repository (doi:10.7280/D1DT3F). Additional data and materials are available upon reasonable request. All code for data processing and analysis are available on GitHub.
ETHICS APPROVAL
This study was approved by the Institutional Review Board (IRB) of the University of California, Irvine (HS#2018-4521) and registered on clinicaltrials.gov (NCT03907436).
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/mbio.02308-23.
Figures S1-S6 and Tables S1-S6.
Taxonomic table and metadata used for the analysis.
Legend for File S2.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Kralovics R, Passamonti F, Buser AS, Teo S-S, Tiedt R, Passweg JR, Tichelli A, Cazzola M, Skoda RC. 2005. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med 352:1779–1790. doi: 10.1056/NEJMoa051113 [DOI] [PubMed] [Google Scholar]
- 2. Nangalia J, Massie CE, Baxter EJ, Nice FL, Gundem G, Wedge DC, Avezov E, Li J, Kollmann K, Kent DG, Aziz A, Godfrey AL, Hinton J, Martincorena I, Van Loo P, Jones AV, Guglielmelli P, Tarpey P, Harding HP, Fitzpatrick JD, Goudie CT, Ortmann CA, Loughran SJ, Raine K, Jones DR, Butler AP, Teague JW, O’Meara S, McLaren S, Bianchi M, Silber Y, Dimitropoulou D, Bloxham D, Mudie L, Maddison M, Robinson B, Keohane C, Maclean C, Hill K, Orchard K, Tauro S, Du M-Q, Greaves M, Bowen D, Huntly BJP, Harrison CN, Cross NCP, Ron D, Vannucchi AM, Papaemmanuil E, Campbell PJ, Green AR. 2013. Somatic CALR mutations in myeloproliferative neoplasms with nonmutated JAK2. N Engl J Med 369:2391–2405. doi: 10.1056/NEJMoa1312542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fleischman AG. 2015. Inflammation as a driver of clonal evolution in myeloproliferative neoplasm. Mediators Inflamm 2015:606819. doi: 10.1155/2015/606819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Craver BM, El Alaoui K, Scherber RM, Fleischman AG. 2018. The critical role of inflammation in the pathogenesis and progression of myeloid malignancies. Cancers (Basel) 10:104. doi: 10.3390/cancers10040104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tefferi A. 2012. JAK inhibitors for myeloproliferative neoplasms: clarifying facts from myths. Blood 119:2721–2730. doi: 10.1182/blood-2011-11-395228 [DOI] [PubMed] [Google Scholar]
- 6. Davis C, Bryan J, Hodgson J, Murphy K. 2015. Definition of the Mediterranean diet; a literature review. Nutrients 7:9139–9153. doi: 10.3390/nu7115459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Martínez-González MA, García-Arellano A, Toledo E, Salas-Salvadó J, Buil-Cosiales P, Corella D, Covas MI, Schröder H, Arós F, Gómez-Gracia E, Fiol M, Ruiz-Gutiérrez V, Lapetra J, Lamuela-Raventos RM, Serra-Majem L, Pintó X, Muñoz MA, Wärnberg J, Ros E, Estruch R, PREDIMED Study Investigators . 2012. A 14-item Mediterranean diet assessment tool and obesity indexes among high-risk subjects: the PREDIMED trial. PLoS One 7:e43134. doi: 10.1371/journal.pone.0043134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Smidowicz A, Regula J. 2015. Effect of nutritional status and dietary patterns on human serum C-reactive protein and interleukin-6 concentrations. Adv Nutr 6:738–747. doi: 10.3945/an.115.009415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Estruch R. 2010. Anti-inflammatory effects of the Mediterranean diet: the experience of the PREDIMED study. Proc Nutr Soc 69:333–340. doi: 10.1017/S0029665110001539 [DOI] [PubMed] [Google Scholar]
- 10. Sofi F, Abbate R, Gensini GF, Casini A. 2010. Accruing evidence on benefits of adherence to the Mediterranean diet on health: an updated systematic review and meta-analysis. Am J Clin Nutr 92:1189–1196. doi: 10.3945/ajcn.2010.29673 [DOI] [PubMed] [Google Scholar]
- 11. Gotsis E, Anagnostis P, Mariolis A, Vlachou A, Katsiki N, Karagiannis A. 2015. Health benefits of the Mediterranean diet: an update of research over the last 5 years. Angiology 66:304–318. doi: 10.1177/0003319714532169 [DOI] [PubMed] [Google Scholar]
- 12. De Filippis F, Pellegrini N, Vannini L, Jeffery IB, La Storia A, Laghi L, Serrazanetti DI, Di Cagno R, Ferrocino I, Lazzi C, Turroni S, Cocolin L, Brigidi P, Neviani E, Gobbetti M, O’Toole PW, Ercolini D. 2016. High-level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut 65:1812–1821. doi: 10.1136/gutjnl-2015-309957 [DOI] [PubMed] [Google Scholar]
- 13. Mitsou EK, Kakali A, Antonopoulou S, Mountzouris KC, Yannakoulia M, Panagiotakos DB, Kyriacou A. 2017. Adherence to the Mediterranean diet is associated with the gut microbiota pattern and gastrointestinal characteristics in an adult population. Br J Nutr 117:1645–1655. doi: 10.1017/S0007114517001593 [DOI] [PubMed] [Google Scholar]
- 14. Ghosh TS, Rampelli S, Jeffery IB, Santoro A, Neto M, Capri M, Giampieri E, Jennings A, Candela M, Turroni S, Zoetendal EG, Hermes GDA, Elodie C, Meunier N, Brugere CM, Pujos-Guillot E, Berendsen AM, De Groot L, Feskins EJM, Kaluza J, Pietruszka B, Bielak MJ, Comte B, Maijo-Ferre M, Nicoletti C, De Vos WM, Fairweather-Tait S, Cassidy A, Brigidi P, Franceschi C, O’Toole PW. 2020. Mediterranean diet intervention alters the gut microbiome in older people reducing frailty and improving health status: the NU-AGE 1-year dietary intervention across five European countries. Gut 69:1218–1228. doi: 10.1136/gutjnl-2019-319654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Meslier V, Laiola M, Roager HM, De Filippis F, Roume H, Quinquis B, Giacco R, Mennella I, Ferracane R, Pons N, Pasolli E, Rivellese A, Dragsted LO, Vitaglione P, Ehrlich SD, Ercolini D. 2020. Mediterranean diet intervention in overweight and obese subjects lowers plasma cholesterol and causes changes in the gut microbiome and metabolome independently of energy intake. Gut 69:1258–1268. doi: 10.1136/gutjnl-2019-320438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nagpal R, Shively CA, Appt SA, Register TC, Michalson KT, Vitolins MZ, Yadav H. 2018. Gut microbiome composition in non-human primates consuming a Western or Mediterranean diet. Front Nutr 5:28. doi: 10.3389/fnut.2018.00028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Haro C, Montes-Borrego M, Rangel-Zúñiga OA, Alcalá-Díaz JF, Gómez-Delgado F, Pérez-Martínez P, Delgado-Lista J, Quintana-Navarro GM, Tinahones FJ, Landa BB, López-Miranda J, Camargo A, Pérez-Jiménez F. 2016. Two healthy diets modulate gut microbial community improving insulin sensitivity in a human obese population. J Clin Endocrinol Metab 101:233–242. doi: 10.1210/jc.2015-3351 [DOI] [PubMed] [Google Scholar]
- 18. Bailey MA, Holscher HD. 2018. Microbiome-mediated effects of the Mediterranean diet on inflammation. Adv Nutr 9:193–206. doi: 10.1093/advances/nmy013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mendez Luque LF, Avelar-Barragan J, Nguyen H, Nguyen J, Soyfer EM, Liu J, Chen JH, Mehrotra N, Kosiorek HE, Dueck A, Himstead A, Heide E, Lem M, El Alaoui K, Mas Marin E, Scherber RM, Mesa RA, Whiteson KL, Odegaard A, Fleischman AG. 2023. The NUTRIENT trial (NUTRitional intervention among myEloproliferative Neoplasms): feasibility phase. medRxiv:2023.05.09.23289740. doi: 10.1101/2023.05.09.23289740 [DOI] [PMC free article] [PubMed]
- 20. Scherber R, Dueck AC, Johansson P, Barbui T, Barosi G, Vannucchi AM, Passamonti F, Andreasson B, Ferarri ML, Rambaldi A, Samuelsson J, Birgegard G, Tefferi A, Harrison CN, Radia D, Mesa RA. 2011. The myeloproliferative neoplasm symptom assessment form (MPN-SAF): international prospective validation and reliability trial in 402 patients. Blood 118:401–408. doi: 10.1182/blood-2011-01-328955 [DOI] [PubMed] [Google Scholar]
- 21. Oliver A, El Alaoui K, Haunschild C, Avelar-Barragan J, Mendez Luque LF, Whiteson K, Fleischman AG. 2022. Fecal microbial community composition in myeloproliferative neoplasm patients is associated with an inflammatory state. Microbiol Spectr 10:e0003222. doi: 10.1128/spectrum.00032-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shively CA, Appt SE, Vitolins MZ, Uberseder B, Michalson KT, Silverstein-Metzler MG, Register TC. 2019. Erratum: Mediterranean versus Western diet effects on caloric intake, obesity, metabolism, and hepatosteatosis in nonhuman primates. Obesity (Silver Spring) 27:1211. doi: 10.1002/oby.22529 [DOI] [PubMed] [Google Scholar]
- 23. Garcia-Rios A, Gomez-Delgado FJ, Garaulet M, Alcala-Diaz JF, Delgado-Lista FJ, Marin C, Rangel-Zuñiga OA, Rodriguez-Cantalejo F, Gomez-Luna P, Ordovas JM, Perez-Jimenez F, Lopez-Miranda J, Perez-Martinez P. 2014. Beneficial effect of CLOCK gene polymorphism rs1801260 in combination with low-fat diet on insulin metabolism in the patients with metabolic syndrome. Chronobiology International 31:401–408. doi: 10.3109/07420528.2013.864300 [DOI] [PubMed] [Google Scholar]
- 24. Kaaks R, Bellati C, Venturelli E, Rinaldi S, Secreto G, Biessy C, Pala V, Sieri S, Berrino F. 2003. Effects of dietary intervention on IGF-I and IGF-binding proteins, and related alterations in sex steroid metabolism: the diet and androgens (DIANA) randomised trial. Eur J Clin Nutr 57:1079–1088. doi: 10.1038/sj.ejcn.1601647 [DOI] [PubMed] [Google Scholar]
- 25. Delgado-Lista J, Perez-Martinez P, Garcia-Rios A, Alcala-Diaz JF, Perez-Caballero AI, Gomez-Delgado F, Fuentes F, Quintana-Navarro G, Lopez-Segura F, Ortiz-Morales AM, Delgado-Casado N, Yubero-Serrano EM, Camargo A, Marin C, Rodriguez-Cantalejo F, Gomez-Luna P, Ordovas JM, Lopez-Miranda J, Perez-Jimenez F. 2016. CORonary diet intervention with olive oil and cardiovascular PREVention study (the CORDIOPREV study): rationale, methods, and baseline characteristics. American Heart Journal 177:42–50. doi: 10.1016/j.ahj.2016.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shai I, Schwarzfuchs D, Henkin Y, Shahar DR, Witkow S, Greenberg I, Golan R, Fraser D, Bolotin A, Vardi H, Tangi-Rozental O, Zuk-Ramot R, Sarusi B, Brickner D, Schwartz Z, Sheiner E, Marko R, Katorza E, Thiery J, Fiedler GM, Blüher M, Stumvoll M, Stampfer MJ, Dietary Intervention Randomized Controlled Trial (DIRECT) Group . 2008. Weight loss with a low-carbohydrate, Mediterranean, or low-fat diet. N Engl J Med 359:229–241. doi: 10.1056/NEJMoa0708681 [DOI] [PubMed] [Google Scholar]
- 27. Mekki K, Bouzidi-bekada N, Kaddous A, Bouchenak M. 2010. Mediterranean diet improves dyslipidemia and biomarkers in chronic renal failure patients. Food Funct 1:110–115. doi: 10.1039/c0fo00032a [DOI] [PubMed] [Google Scholar]
- 28. Paniagua JA, de la Sacristana AG, Sánchez E, Romero I, Vidal-Puig A, Berral FJ, Escribano A, Moyano MJ, Peréz-Martinez P, López-Miranda J, Pérez-Jiménez F. 2007. A MUFA-rich diet improves posprandial glucose, lipid and GLP-1 responses in insulin-resistant subjects. J Am Coll Nutr 26:434–444. doi: 10.1080/07315724.2007.10719633 [DOI] [PubMed] [Google Scholar]
- 29. Salas-Salvadó J, Bulló M, Estruch R, Ros E, Covas MI, Ibarrola-Jurado N, Corella D, Arós F, Gómez-Gracia E, Ruiz-Gutiérrez V, Romaguera D, Lapetra J, Lamuela-Raventós RM, Serra-Majem L, Pintó X, Basora J, Muñoz MA, Sorlí JV, Martínez-González MA. 2014. Prevention of diabetes with Mediterranean diets: a subgroup analysis of a randomized trial. Ann Intern Med 160:1–10. doi: 10.7326/M13-1725 [DOI] [PubMed] [Google Scholar]
- 30. Marlow G, Ellett S, Ferguson IR, Zhu S, Karunasinghe N, Jesuthasan AC, Han DY, Fraser AG, Ferguson LR. 2013. Transcriptomics to study the effect of a Mediterranean-inspired diet on inflammation in crohn's disease patients. Hum Genomics 7:24. doi: 10.1186/1479-7364-7-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Klimenko NS, Odintsova VE, Revel-Muroz A, Tyakht AV. 2022. The hallmarks of dietary intervention-resilient gut microbiome. NPJ Biofilms Microbiomes 8:77. doi: 10.1038/s41522-022-00342-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Smits SA, Leach J, Sonnenburg ED, Gonzalez CG, Lichtman JS, Reid G, Knight R, Manjurano A, Changalucha J, Elias JE, Dominguez-Bello MG, Sonnenburg JL. 2017. Seasonal cycling in the gut microbiome of the hadza hunter-gatherers of tanzania. Science 357:802–806. doi: 10.1126/science.aan4834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sriswasdi S, Yang CC, Iwasaki W. 2017. Generalist species drive microbial dispersion and evolution. Nat Commun 8:1162. doi: 10.1038/s41467-017-01265-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Matias MG, Combe M, Barbera C, Mouquet N. 2012. Ecological strategies shape the insurance potential of biodiversity. Front Microbiol 3:432. doi: 10.3389/fmicb.2012.00432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Luisi MLE, Lucarini L, Biffi B, Rafanelli E, Pietramellara G, Durante M, Vidali S, Provensi G, Madiai S, Gheri CF, Masini E, Ceccherini MT. 2019. Effect of Mediterranean diet enriched in high quality extra virgin olive oil on oxidative stress.Inflammation and gut microbiota in obese and normal weight adult subjects. Front Pharmacol 10:1366. doi: 10.3389/fphar.2019.01366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Millman J, Okamoto S, Kimura A, Uema T, Higa M, Yonamine M, Namba T, Ogata E, Yamazaki S, Shimabukuro M, Tsutsui M, Matsushita M, Ikematsu S, Masuzaki H. 2020. Metabolically and immunologically beneficial impact of extra virgin olive and flaxseed oils on composition of gut microbiota in mice. Eur J Nutr 59:2411–2425. doi: 10.1007/s00394-019-02088-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ozdal T, Sela DA, Xiao J, Boyacioglu D, Chen F, Capanoglu E. 2016. The reciprocal interactions between polyphenols and gut microbiota and effects on bioaccessibility. Nutrients 8:78. doi: 10.3390/nu8020078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Martín-Peláez S, Mosele JI, Pizarro N, Farràs M, de la Torre R, Subirana I, Pérez-Cano FJ, Castañer O, Solà R, Fernandez-Castillejo S, Heredia S, Farré M, Motilva MJ, Fitó M. 2017. Effect of virgin olive oil and thyme phenolic compounds on blood lipid profile: implications of human gut microbiota. Eur J Nutr 56:119–131. doi: 10.1007/s00394-015-1063-2 [DOI] [PubMed] [Google Scholar]
- 39. Zhao Z, Shi A, Wang Q, Zhou J. 2019. High oleic acid peanut oil and extra virgin olive oil supplementation attenuate metabolic syndrome in rats by modulating the gut microbiota. Nutrients 11:3005. doi: 10.3390/nu11123005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hidalgo M, Prieto I, Abriouel H, Villarejo AB, Ramírez-Sánchez M, Cobo A, Benomar N, Gálvez A, Martínez-Cañamero M. 2018. Changes in gut microbiota linked to a reduction in systolic blood pressure in spontaneously hypertensive rats fed an extra virgin olive oil-enriched diet. Plant Foods Hum Nutr 73:1–6. doi: 10.1007/s11130-017-0650-1 [DOI] [PubMed] [Google Scholar]
- 41. Chen J, Vitetta L. 2018. Inflammation-modulating effect of butyrate in the prevention of colon cancer by dietary fiber. Clin Colorectal Cancer 17:e541–e544. doi: 10.1016/j.clcc.2018.05.001 [DOI] [PubMed] [Google Scholar]
- 42. Reichardt N, Duncan SH, Young P, Belenguer A, McWilliam Leitch C, Scott KP, Flint HJ, Louis P. 2014. Phylogenetic distribution of three pathways for propionate production within the human gut microbiota. ISME J 8:1323–1335. doi: 10.1038/ismej.2014.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zaneveld JR, McMinds R, Vega Thurber R. 2017. Stress and stability: applying the anna karenina principle to animal microbiomes. Nat Microbiol 2:17121. doi: 10.1038/nmicrobiol.2017.121 [DOI] [PubMed] [Google Scholar]
- 44. Kaszubinski SF, Pechal JL, Smiles K, Schmidt CJ, Jordan HR, Meek MH, Benbow ME. 2020. Dysbiosis in the dead: human postmortem microbiome beta-dispersion as an indicator of manner and cause of death. Front Microbiol 11:555347. doi: 10.3389/fmicb.2020.555347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kumari M, Singh P, Nataraj BH, Kokkiligadda A, Naithani H, Azmal Ali S, Behare PV, Nagpal R. 2021. Fostering next-generation probiotics in human gut by targeted dietary modulation: an emerging perspective. Food Res Int 150:110716. doi: 10.1016/j.foodres.2021.110716 [DOI] [PubMed] [Google Scholar]
- 46. Zhu C, Song K, Shen Z, Quan Y, Tan B, Luo W, Wu S, Tang K, Yang Z, Wang X. 2018. Roseburia Intestinalis inhibits interleukin-17 excretion and promotes regulatory T cells differentiation in colitis. Mol Med Rep 17:7567–7574. doi: 10.3892/mmr.2018.8833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chung WSF, Meijerink M, Zeuner B, Holck J, Louis P, Meyer AS, Wells JM, Flint HJ, Duncan SH. 2017. Prebiotic potential of pectin and pectic oligosaccharides to promote anti-inflammatory commensal bacteria in the human colon. FEMS Microbiol Ecol 93. doi: 10.1093/femsec/fix127 [DOI] [PubMed] [Google Scholar]
- 48. Weihe C, Avelar-Barragan J. n.d. Next generation shotgun library preparation for Illumina sequencing - low volume v1. doi: 10.17504/protocols.io.bvv8n69w [DOI]
- 49. Bushnell B. 2014. BBMap: a fast, accurate, splice-aware aligner
- 50. Cantu VA, Sadural J, Edwards R. 2019. PRINSEQ++, a multi-threaded tool for fast and efficient quality control and preprocessing of sequencing datasets. Peerj Preprints. doi: 10.7287/peerj.preprints.27553v1 [DOI]
- 51. Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Beghini F, McIver LJ, Blanco-Míguez A, Dubois L, Asnicar F, Maharjan S, Mailyan A, Manghi P, Scholz M, Thomas AM, Valles-Colomer M, Weingart G, Zhang Y, Zolfo M, Huttenhower C, Franzosa EA, Segata N. 2021. Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with bioBakery 3. Elife 10:e65088. doi: 10.7554/eLife.65088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li D, Luo R, Liu CM, Leung CM, Ting HF, Sadakane K, Yamashita H, Lam TW. 2016. MEGAHIT v1.0: a fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods 102:3–11. doi: 10.1016/j.ymeth.2016.02.020 [DOI] [PubMed] [Google Scholar]
- 54. Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Huerta-Cepas J, Szklarczyk D, Heller D, Hernández-Plaza A, Forslund SK, Cook H, Mende DR, Letunic I, Rattei T, Jensen LJ, von Mering C, Bork P. 2019. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res 47:D309–D314. doi: 10.1093/nar/gky1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nayfach S, Pollard KS. 2015. Average genome size estimation improves comparative metagenomics and sheds light on the functional ecology of the human microbiome. Genome Biol 16:51. doi: 10.1186/s13059-015-0611-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1-S6 and Tables S1-S6.
Taxonomic table and metadata used for the analysis.
Legend for File S2.
Data Availability Statement
Sequencing data are available on the Sequence Read Archive under the BioProject ID PRJNA918651. The taxonomic table with corresponding metadata used to generate the source data can be found in Supplementary_file_2.csv. Larger files used in our functional metagenomic analysis are available on the Dryad Digital Repository (doi:10.7280/D1DT3F). Additional data and materials are available upon reasonable request. All code for data processing and analysis are available on GitHub.