ABSTRACT
p-Cresol sulfate (pCS) and indoxyl sulfate (IS), gut microbiome-derived metabolites, are traditionally associated with cardiovascular disease (CVD) risks in the setting of impaired kidney function. While pharmacologic provision of pCS or IS can promote pro-thrombotic phenotypes, neither the microbial enzymes involved nor direct gut microbial production have been linked to CVD. Untargeted metabolomics was performed on a discovery cohort (n = 1,149) with relatively preserved kidney function, followed by stable isotope-dilution mass spectrometry quantification of pCS and IS in an independent validation cohort (n = 3,954). Genetic engineering of human commensals to produce p-cresol and indole gain-of-function and loss-of-function mutants, followed by colonization of germ-free mice, and studies on host thrombosis were performed. Systemic pCS and IS levels were independently associated with all-cause mortality. Both in vitro and within colonized germ-free mice p-cresol productions were recapitulated by collaboration of two organisms: a Bacteroides strain that converts tyrosine to 4-hydroxyphenylacetate, and a Clostridium strain that decarboxylates 4-hydroxyphenylacetate to p-cresol. We then engineered a single organism, Bacteroides thetaiotaomicron, to produce p-cresol, indole, or both metabolites. Colonizing germ-free mice with engineered strains, we show the gut microbial genes for p-cresol (hpdBCA) and indole (tryptophanase) are sufficient to confer a pro-thrombotic phenotype in vivo. Moreover, human fecal metagenomics analyses show that abundances of hpdBCA and tryptophanase are associated with CVD. These studies show that pCS and IS, two abundant microbiome-derived metabolites, play a broader potential role in CVD than was previously known. They also suggest that therapeutic targeting of gut microbial p-cresol- and indole-producing pathways represent rational targets for CVD.
IMPORTANCE
Alterations in gut microbial composition and function have been linked to numerous diseases. Identifying microbial pathways responsible for producing molecules that adversely impact the host is an important first step in the development of therapeutic interventions. Here, we first use large-scale clinical observations to link blood levels of defined microbial products to cardiovascular disease risks. Notably, the previously identified uremic toxins p-cresol sulfate and indoxyl sulfate were shown to predict 5-year mortality risks. After identifying the microbes and microbial enzymes involved in the generation of these uremic toxins, we used bioengineering technologies coupled with colonization of germ-free mice to show that the gut microbial genes that generate p-cresol and indole are sufficient to confer p-cresol sulfate and indoxyl sulfate formation, and a pro-thrombotic phenotype in vivo. The findings and tools developed serve as a critical step in both the study and targeting of these gut microbial pathways in vivo.
KEYWORDS: gut microbes, uremic toxins, p-cresol sulfate, indoxyl sulfate, cardiovascular disease, mortality
INTRODUCTION
Cardiovascular disease (CVD) remains the leading cause of mortality worldwide; however, despite significant advances in preventive CVD treatments and prevention efforts, there remains a significant “residual CVD risk,” with numerous atherothrombotic events and cardiovascular mortality risk occurring even among optimally treated individuals (1–4). The search for additional causative factors beyond traditional CVD risk factors remains a significant area of investigation. One promising area in this regard is the gut microbiome (5, 6). Differences in gut microbial composition and function have been associated with CVD, including production of some bioactive metabolites (5, 7–10). While most of studies on gut microbes and CVD are correlative and associative, multiple mechanistic studies are identifying gut microbial metabolites as direct contributors to CVD (7, 11–16). p-Cresol sulfate (pCS) and indoxyl sulfate (IS), products derived from microbial fermentation of the aromatic amino acids tyrosine and tryptophan, respectively, are associated with CVD mortality risk in individuals with chronic kidney disease (CKD) and end-stage renal disease (17–19).
Termed “uremic solutes” because levels of pCS and IS accumulate as renal function declines, these compounds are also considered “uremic toxins” because they have been implicated in mediating adverse phenotypes in the setting of renal disease (20). The synthesis of both pCS and IS are also “metaorganismal,” since each is co-synthesized by an initial (obligate) gut microbiota-dependent process, followed by the host enzymatic transformations that facilitate metabolite excretion. Each molecule begins as a byproduct of bacterial fermentation of aromatic amino acids in a protein-rich diet. For IS production, tryptophan from the diet can be degraded into indole by microbial tryptophanase genes encoded in the genome of a variety of gut commensals, as we recently showed (21). The gut microbiota-derived indole, once absorbed via the portal circulation, can then be oxidized by CYP2E1 and sulfated by SULT1A1, both endogenous host enzymes. Synthesis of pCS follows a similar overall trajectory. Dietary protein-derived tyrosine is metabolized by gut microbes via various mechanisms to yield p-cresol, which, following absorption into the host, can then undergo sulfation by SULT1A1 to yield pCS (22, 23). While the host enzymes in these metaorganismal biochemical transformations are well-known and characterized, the gut microbes and the microbial genes utilized to produce the precursor molecules in these metaorganismal pathways remain poorly understood.
Our interest in pCS and IS in the present studies began as a result of untargeted metabolomics investigations, which identified microbial precursor of pCS as candidate molecule whose circulating (blood) levels predict incident CVD risks among subjects with normal renal function. Through subsequent targeted mass spectrometry analysis on an independent cohort, we now show that pCS and IS are associated with overall mortality in individuals with preserved kidney function. We then identify a missing link in the microbial biosynthetic pathway of p-cresol and take advantage of this finding to engineer strains of Bacteroides thetaiotaomicron that produce p-cresol, indole, or both. By colonizing germ-free mice with these strains, we confirm that microbial genes responsible for production of either p-cresol or indole lead to formation of pCS and IS, respectively. We then further demonstrate that this is sufficient to confer within the host, formation of either pCS or IS, and induction of a pro-thrombotic phenotype. Finally, in human metagenomics analyses, we show the fecal abundances of microbial hpdBCA and tryptophanase genes are independently associated with CVD. Our work, thus, demonstrates that microbiota targeting therapies aimed at reducing levels of these uremic solutes/toxins should be explored, and suggests potential microbial enzymatic transformations as rational therapeutic targets for treatment of residual CVD risk.
RESULTS
Uremic solutes are associated with overall mortality in individuals with preserved kidney function
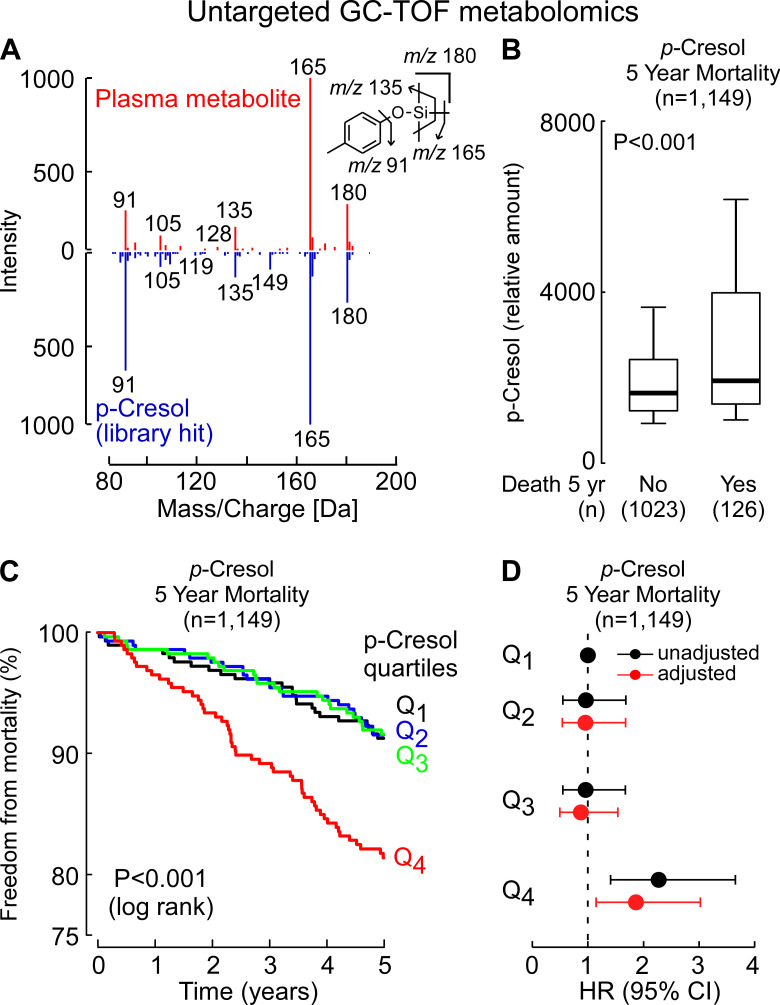
Plasma samples from individuals undergoing elective diagnostic cardiac evaluation with longitudinal follow-up (n = 1,149; Table S1) were analyzed using untargeted gas chromatography time-of-flight mass spectrometry (GC-MS-TOF) as described under Materials and Methods. A derivatized analyte consistent with p-cresol (Fig. 1A), the microbiome-derived metabolite and precursor to pCS, was noted to be significantly higher among subjects who died over the ensuing period of follow-up (5 years) compared to those that did not (Fig. 1B; P < 0.0001). Kaplan-Meier survival analyses also suggested that levels of the candidate ion identified as p-cresol were associated with incident mortality risk in this cohort (Fig. 1C). Subsequent Cox proportional hazards regression with time-to-event analysis showed subjects with elevated (fourth quartile [Q4]) levels had higher mortality risk compared to those with low (first quartile [Q1]) levels, even after adjustment for traditional risk factors (hazard ratio [HR] 95% confidence interval [CI] for incident [5 years] death risk of HR = 1.86 [1.15–3.03], P = 0.01) (Fig. 1D).
Fig 1.
Untargeted metabolomics reveals that p-cresol is associated with overall mortality. (A) Comparison of electron ionization spectra of the compound detected in plasma and an authentic standard of the trimethylsilyl derivative of p-cresol. (B) Relative plasma levels of p-cresol in sequential stable subjects undergoing elective diagnostic cardiac evaluation. Subjects (n = 1,149; discovery cohort) were divided into groups as indicated based on whether or not they died during the 5-year follow-up. In the box-whisker plot, the upper and lower boundaries of the box represent the 25th and 75th percentiles, the median is marked by a horizontal line inside the box, and the whiskers represent 10% and 90% measured values. (C) Kaplan-Meier estimates of the risk of incident death by quartile of relative amounts of p-cresol from the untargeted analysis. (D) Forest plots showing overall mortality within 5 years among test subjects according to the quartiles for the relative level of p-cresol (black), or a multivariable Cox model for hazard ratio that includes adjustments for age, sex, current smoking, high-density lipoprotein, low-density lipoprotein, triglyceride level, systolic blood pressure, diabetes mellitus, and high-sensitivity C-reactive protein (adjusted, red). Symbols represent hazard ratios and the 95% CIs are indicated by the line length.
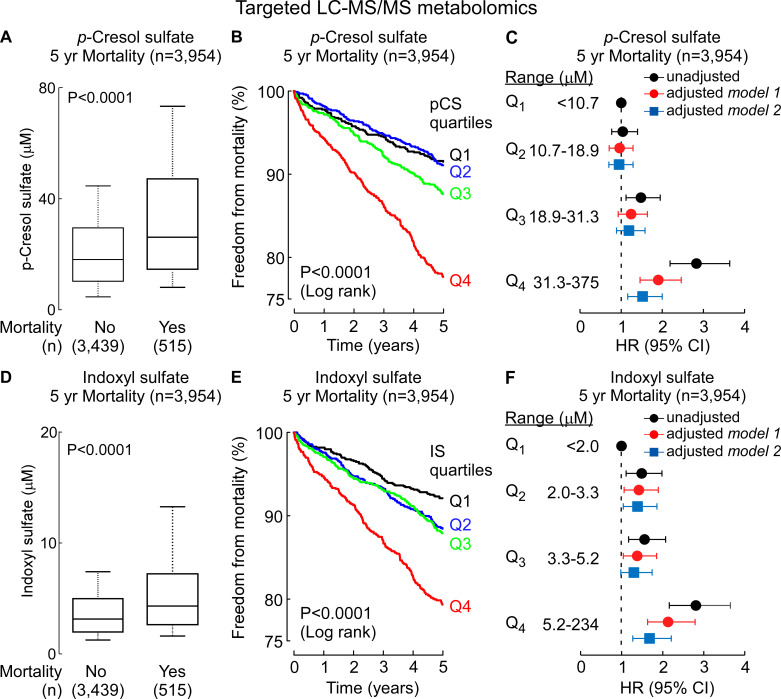
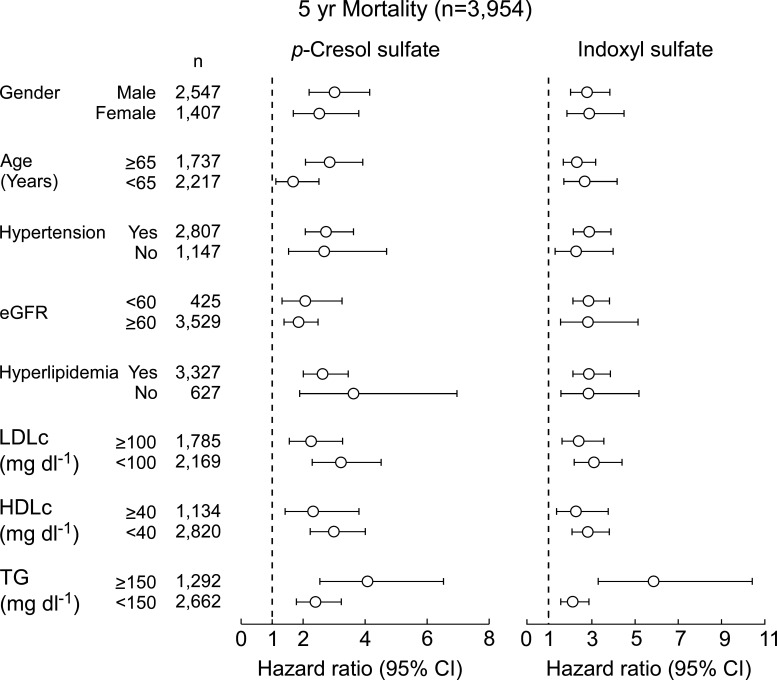
We were somewhat surprised by the p-cresol finding because our GC-TOF methods employed were not optimized for detection of a volatile compound like p-cresol. Untargeted analyses as performed are not quantitative, and given use of a sample drying step where the analyte could be lost, we reasoned that the p-cresol detected using our analytical method most probably was a product of pCS hydrolysis during sample preparation (see Materials and Methods). Given the novelty of these findings (pCS previously has been linked to CVD and mortality risks, but only among those with significantly impaired renal function like end-stage renal disease and hemodialysis), we decided to validate our observations using quantitative methods in an independent (non-overlapping) cohort (n = 3,954 subjects; Table S2) using a complementary technique: targeted stable isotope-dilution LC-MS/MS (see Materials and Methods). We also expanded our targeted LC-MS/MS analyses to also include IS, since both pCS and IS are commonly referred to as “uremic toxins” and studied concurrently in clinical observational studies (17, 24–27). As shown in Fig. 2, both pCS and IS each were significantly higher in subjects who died over the course of 5 years (P < 0.001 each, Wilcoxon rank-sum test, Fig. 2A and D). Kaplan-Meier survival analyses revealed that subjects with either high pCS or IS levels had overall poorer survival over 5 years of follow-up (Fig. 2B and E). Specifically, individuals with pCS and IS levels in the highest vs lowest quartile (Q4 vs Q1) for each compound demonstrated a significantly increased risk of incident (5 years) death (HR = 2.83 [2.19–3.64]; P < 0.0001 and HR = 2.81 [2.16–3.65]; P < 0.0001, respectively). The associations between pCS and IS and death each remained significant after adjusting for traditional risk factors, as well as following additional adjustments for renal function (HR = 1.52 [1.16–2.00]; P = 0.002 and HR = 1.68 [1.27–2.21]; P = 0.0002 for pCS and IS, respectively) (Fig. 2C and F). Furthermore, the association between pCS or IS and risk of incident (5 years) death holds true when subjects were divided into subgroups of subjects with relatively preserved kidney function (estimated glomerular filtration rate [eGFR] ≥ 60 mL/min/1.73 m2) and subjects with impairment in kidney function (eGFR < 60 mL/min/1.73 m2) (Table S3). In further sensitivity analyses, we observed the associations between pCS and IS and mortality risk remained significant in both males and females alike, younger versus older, as well as within multiple different subgroups including subjects with or without hyperlipidemia or hypertension (Fig. 3). Collectively, these data demonstrate that pCS and IS, which are generally considered to be relevant only in the setting of renal disease, are associated with mortality in the broader population of individuals with predominantly preserved renal function, as well as in the absence of traditional cardiovascular risk factors.
Fig 2.
Stable isotope-dilution LC-MS/MS analyses verify systemic levels of uremic toxins p-cresol sulfate and indoxyl sulfate are associated with incident overall mortality risks. Plasma levels of p-cresol sulfate (A) and indoxyl sulfate (D) in sequential stable subjects undergoing elective diagnostic cardiac evaluation. Subjects (n = 3,954) were divided into groups as indicated based on whether or not they experienced an incident death event within 5 years. In the box-whisker plot, the upper and lower boundaries of the box represent the 25th and 75th percentiles, the median is marked by a horizontal line inside the box, and the whiskers represent 10% and 90% measured values. Kaplan-Meier estimates and the risk of incident overall mortality ranked by quartile of (B) p-cresol sulfate and (E) indoxyl sulfate levels. Forest plots indicate the hazard ratio (95% CI) for incident (5 years) risks of overall mortality for (C) p-cresol sulfate and (F) indoxyl sulfate quartiles. Hazard ratio (unadjusted, black) and multivariable Cox model adjusted [gray; adjusted for age, sex, current smoking, high-density lipoprotein, low-density lipoprotein, triglyceride level, systolic blood pressure, diabetes mellitus, and high-sensitivity C-reactive protein (model 1) and model 1 + kidney function (model 2)]. Symbols represent hazard ratios and the 95% confidence intervals are indicated by line length.
Fig 3.
Long-term mortality risk among patient subgroups. Hazard ratio for 5-year overall mortality based on the Cox proportional hazards regression analysis comparing fourth vs first (referent) quartiles (Q). Data points (open circles) in the center indicate HR and 95% CIs are represented by line length. Numbers for each subgroup are indicated by n. eGFR, estimated glomerular filtration rate; LDL-c, low-density lipoprotein cholesterol; HDL-c, high-density lipoprotein cholesterol; TG, triglyceride.
Identification of enzymes involved in p-cresol biosynthesis
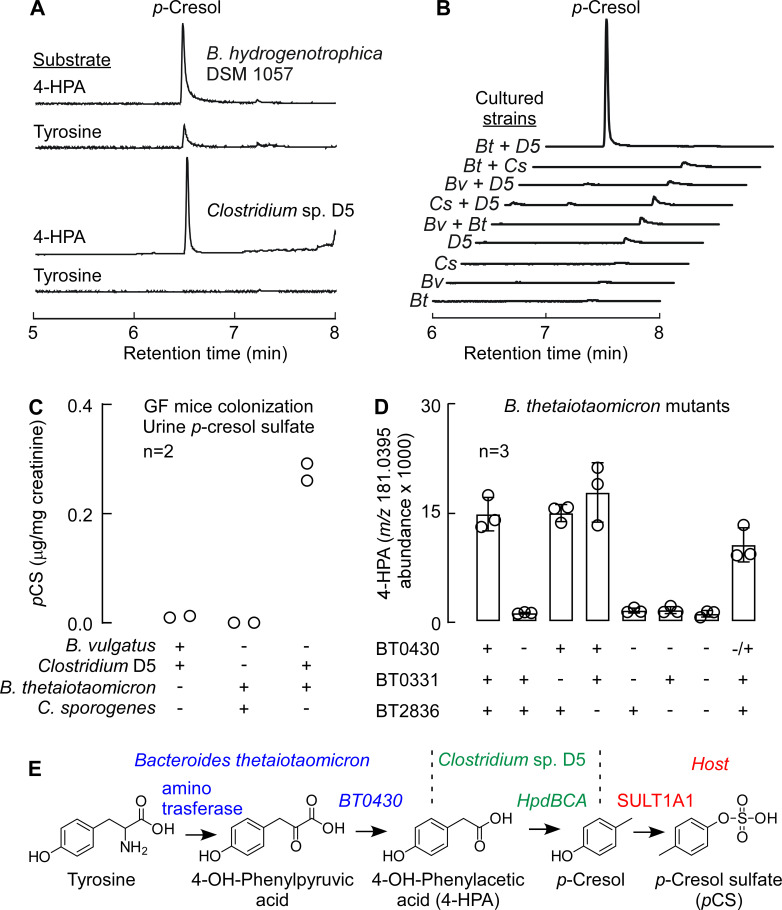
Previous work from our laboratories and others had identified the bacterial genes required for indole production (21, 28), but the biosynthetic route for p-cresol is less understood. Several human gut commensals have initially been reported to produce p-cresol from tyrosine (29), but the quantity of p-cresol produced in vitro (typically 0.1–1.0 µg/mL) cannot account for the high levels of pCS commonly detected in vivo (20 mg/mL in CKD patients). Later, Saito et al. identified several p-cresol-producing commensals by screening multiple bacterial strains in culture media supplemented with tyrosine and its metabolites (30). Previous work showed that hpdBCA in Clostridium difficile, which encodes a multi-component 4-hydroxyphenylacetate (4-HPA) decarboxylase, can convert 4-HPA (31), and to a lesser extent tyrosine (32), to p-cresol. By searching for homologs of hpdBCA in reference genomes from the National Institutes of Health (NIH) Human Microbiome Project database, we found a similar operon in Blautia hydrogenotrophica DSM 10507 and Clostridium sp. D5. We tested the capacity of each strain to produce p-cresol from tyrosine and 4-HPA under anaerobic conditions. While both strains were able to produce p-cresol robustly from 4-HPA, production from tyrosine was negligible by B. hydrogenotrophica DSM 10507, consistent with previous findings (30), and undetectable by Clostridium sp. D5 (Fig. 4A). Given that metabolic byproducts excreted by one bacteria are often utilized by other bacteria in the gut microbial community (16, 33), an alternative hypothesis was suggested: the synthesis of p-cresol could be the result of a collaborative pathway involving two microbes, where the first microbe converts tyrosine to 4-HPA and a second decarboxylates 4-HPA to p-cresol.
Fig 4.
A collaborative pathway for the production of p-cresol. (A) GC-MS chromatogram of p-cresol production from metabolites of Clostridium sp. D5 and B. hydrogenotrophica DSM 10507 in the presence of 4-HPA and tyrosine (Tyr). (B) GC-MS chromatograms from mono-cultures and co-cultures. Bt, B. thetaiotaomicron; Bv, Bacteroides vulgatus; Cs, Clostridium sporogenes; D5, Clostridium sp. D5. (C) Urine p-cresol sulfate (pCS) from germ-free mice were co-colonized with either B. vulgatus and Clostridium D5 or B. thetaiotaomicron and C. sporogenes or B. thetaiotaomicron and Clostridium sp. D5. (D) The production of 4-HPA in B. thetaiotaomicron mutants. Data bars indicate the average LC-MS ion counts (negative mode) of three biological replicates ± SD. (E) Schematic of a collaborative pathway for the production of p-cresol and p-cresol sulfate.
We therefore performed studies to support the hypothesis that two microbes sequentially contributed to the overall tyrosine → p-cresol metabolic transformation within the gut. Certain common strains of Bacteroides have been reported to convert tyrosine to 4-HPA, including B. thetaiotaomicron, but not Bacteroides vulgatus (34). We therefore used these two strains in co-culture experiments in which we fed tyrosine to a pair of bacterial strains: the first strain was a 4-HPA-producing or non-producing Bacteroides species; and the second strain was Clostridium sp. D5, which converts 4-HPA to p-cresol, or Clostridium sporogenes, an hpdBCA-negative commensal. As shown in Fig. 4B, robust p-cresol production was only observed in the combination that included a 4-HPA producer and an hpdBCA-positive Clostridium (B. thetaiotaomicron and Clostridium sp. D5).
In pilot studies, we then colonized germ-free mice with the strain pairs tested in vitro for a week. As expected, robust production of p-cresol (leading to pCS in the urine) was only observed in mice colonized by B. thetaiotaomicron and Clostridium sp. D5. Mice colonized with Clostridium sp. D5 and B. vulgatus had low levels of pCS in their urine, indicating that B. vulgatus may produce small amounts of 4-HPA in vivo, or Clostridium sp. D5 may be able to produce p-cresol from tyrosine at a low level (Fig. 4C). Collectively, these data indicate that a collaborative pathway can account for p-cresol production in vivo, although we cannot exclude the possibility of an undiscovered strain that can convert tyrosine to p-cresol.
Genes involved in 4-HPA production
Next, we sought to gain insight into the collaborative pathway by identifying the genes in B. thetaiotaomicron responsible for converting tyrosine to 4-HPA. By analogy to the conversion of phenylalanine to phenylacetate, we reasoned that the production of 4-HPA likely involved two steps: the transamination of tyrosine to 4-hydroxyphenylpyruvate (4-HPP) and the oxidative decarboxylation of 4-HPP to 4-HPA. A computational search of the B. thetaiotaomicron genome revealed >10 putative aminotransferases. Since aminotransferases often have overlapping substrate specificities (35), we reasoned that a single aminotransferase may not be responsible for the production of 4-HPA, so we focused instead on identifying the oxidative decarboxylase responsible for 4-HPA production.
In our previous study on B. thetaiotaomicron, we identified three homologs for 2-oxoacid ferredoxin oxidoreductase, a family of enzymes which decarboxylate pyruvic acid into the corresponding acetic acid derivative (36). The three homologs are coded by BT0332-329, BT0430-0429, and BT2836-2837 gene clusters. To determine which of these oxidoreductases contribute to 4-HPA production, we generated clean deletions of a single gene in each cluster and measured the capacity of each mutant to produce 4-HPA. Of the three single mutants, only ΔBT0430 exhibited decreased 4-HPA production (Fig. 4D), and even this mutant produced residual levels of 4-HPA. Complementing the ΔBT0430 mutant with BT0430 gene restored the production of 4-HPA to near-wild-type levels (Fig. 4D).
Strains harboring double and triple deletions of BT0430, BT0331, and BT2836 maintained low-level 4-HPA production without affecting bacterial growth, suggesting the existence of an additional pathway for 4-HPA production, potentially involving a member of the pyruvate:ferredoxin oxidoreductase (PFOR) superfamily (37). Nevertheless, our data suggest that BT0430 is predominantly responsible for 4-HPA production in B. thetaiotaomicron. Taken together, our findings support the collaborative pathway for p-cresol proposed in Fig. 4E.
Engineering B. thetaiotaomicron to produce indole and p-cresol simultaneously
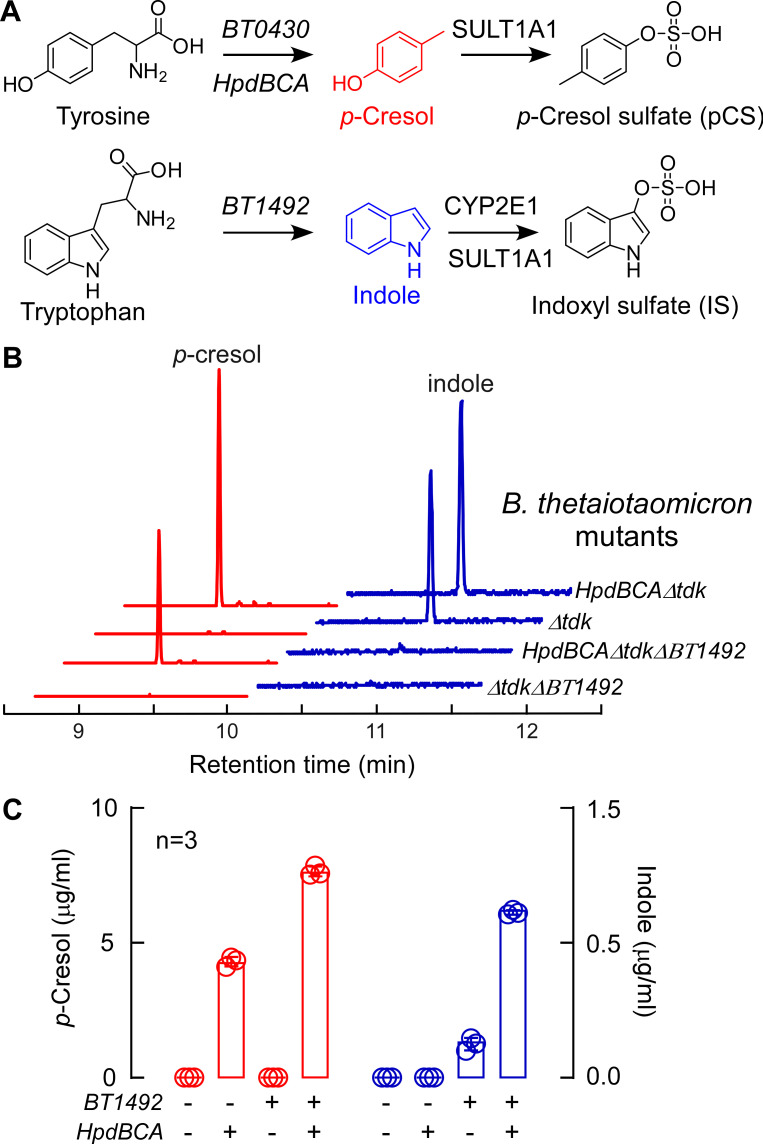
Indole and p-cresol are both connected to the potential cardiovascular sequelae of CKD and end-stage renal disease, and each metabolite is correlated with mortality risk in humans (even with preserved renal function). Since these metabolites are produced simultaneously by the microbiome, we sought to create a system that would enable us to study their effects on the host. Such a system could be complex and difficult to control if it involves multiple bacterial species. As an alternative approach, we set out to engineer a single strain of B. thetaiotaomicron to produce either p-cresol or indole alone, or both metabolites simultaneously (Fig. 5A).
Fig 5.
Engineered Bacteroides strains synthesize p-cresol and indole in vitro. (A) Schematic presentation of genes involved in p-cresol and indole production. (B) Representative GC-MS chromatograms of selected ions for p-cresol (red; m/z 107.1 selected ion) and indole (blue; m/z 117.1 selected ion) production in culture from engineered B. thetaiotaomicron strains. Strains with the hpdBCA operon produce p-cresol, whereas strains containing the BT1492 gene produce indole. (C) Amounts of p-cresol (red) and indole (blue) produced by cultured B. thetaiotaomicron mutants (n = 3) measured by GC-MS.
We chose Bacteroides thetaiotaomicron VPI-5482 as our chassis for engineering; this strain harbors an indole-producing tryptophanase (BT1492+) but no operon for p-cresol biogenesis (hpdBCA). We started with B. thetaiotaomicron ∆BT1492, a tryptophanase mutant previously generated in our lab (21); we refer to this indole−/p-cresol− strain as ΔtdkΔBT1492. We transferred the hpdBCA operon from C. difficile JIR 8094 into this strain under the control of a constitutive promoter derived from Bacteroides phage B124-14 (38), yielding the indole−/p-cresol+ strain HpdBCAΔtdkΔBT1492. Since the level of indole production from wild-type B. thetaiotaomicron is low, we inserted two additional copies of BT1492 (three total) to generate the indole+/p-cresol− strain. Finally, we constructed the indole+/p-cresol+ strain HpdBCAΔtdk, which harbors two additions: the hpdBCA operon from C. difficile and one additional copy of BT1492 (two total). These strains were tested for their capacity to produce p-cresol and indole in vitro. As predicted, the presence of hpdBCA and BT1492 determined the production of p-cresol and indole, respectively (Fig. 5B and C). Notably, the fact that an hpdBCA operon from Clostridium functions in B. thetaiotaomicron suggests that Bacteroides—a Gram-negative that is the most common genus of gut bacteria—could be a model host for studying a broader range of metabolites produced exclusively by the Gram-positive Firmicutes.
hpdBCA and BT1492 modulate thrombosis potential in vivo
Previous studies have shown that pharmacologic provision (i.e., added exogenously) of pCS or IS can promote pro-thrombotic phenotypes (28, 39, 40), but not in the context of direct demonstration of heightened host thrombosis by a defined gut microbial enzyme system. We therefore set out to address this gap in knowledge using the strains of Bacteroides engineered to produce individually either pCS, IS, or both metabolites. First, we validated an in vivo assay for thrombosis induced by arterial injury that was modulated by direct provision of the uremic toxins (Fig. S1A). After raising the levels of pCS and IS by injecting them as purified chemicals intraperitoneally, an FeCl3-induced carotid artery injury model was used to model thrombosis. We measured two parameters: the rate of platelet clotting following carotid artery injury, and the time to cessation of flow within the carotid artery. pCS and IS each individually induced heightened platelet thrombus formation within the injured carotid artery (Fig. S1A) and reduced the time to cessation of blood flow following injury (i.e., the occlusion time) when compared to mice treated with saline, the vehicle (Fig. S1B; P < 0.0001, Kruskal-Wallis).
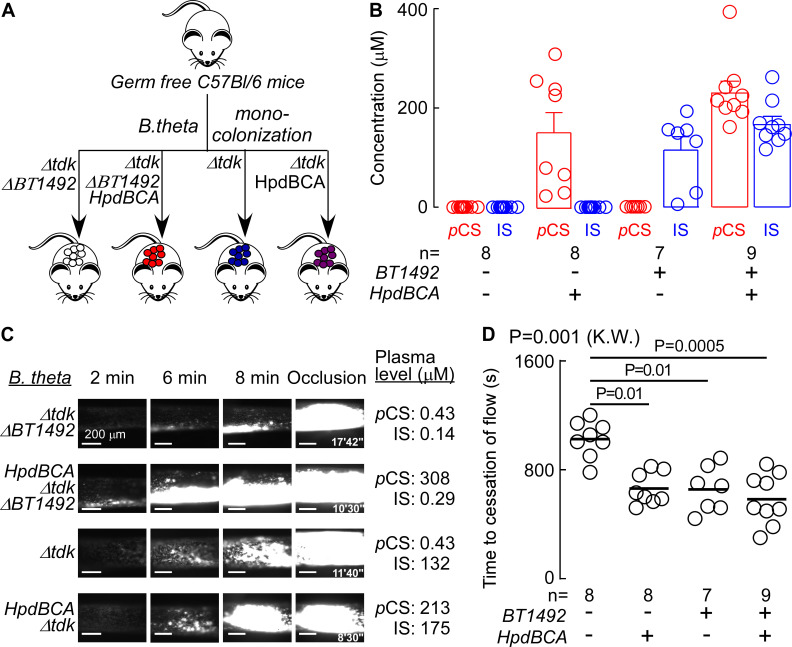
In a subsequent series of experiments, we tested whether the engineered strains of Bacteroides both contribute to pCS and IS production and foster a pro-thrombotic phenotype when transplanted into a mammalian host. We thus colonized germ-free mice with the four strains of B. thetaiotaomicron (BT1492/hpdBCA +/+, +/−, −/+, −/−) for up to 7 days and then measured both colonization efficiency and plasma levels of pCS and IS. Notably, the B. thetaiotaomicron mutants with different capacities for p-cresol and indole production did not significantly impact colonization level (Fig. S2). However, the status of BT1492 and hpdBCA determined the level of IS and pCS, respectively, demonstrating that our genetically engineered strains enable us to control the presence/absence of each metabolite independently, recapitulating the entire metaorganismal pathways (Fig. 6A and B). Finally, we tested whether the presence of a functional hpdBCA or BT1492 gene product within colonized gnotobiotic mice can modulate platelet function and thrombosis potential in vivo. Germ-free mice colonized by each strain were subjected to arterial injury. We observed that the rate of thrombus generation increased, and the time to cessation of blood flow within the injured vessel decreased, in mice harboring commensals with either a functional hpdBCA or BT1492 in their gut colonist (Fig. 6C and D). These results demonstrate that bacterially produced indole or p-cresol, converted to IS and pCS, respectively, by the host, each are capable of increasing thrombosis potential in a well-controlled setting. Notably, the presence of both (combined) loci (BT1492+/hpdBCA+) did not exacerbate the pro-thrombotic phenotype.
Fig 6.
Difference in microbial genes responsible for p-cresol or indole production is associated with increased in vivo thrombosis potential. (A) Scheme illustrating microbial transplant study design. Germ-free (C57B1/6) mice were subjected to gavage with four different engineered B. thetaiotaomicron strains with different capacities for p-cresol and indole production: (ΔBT1942 (white), hpdBCA ΔBT1942 (red), Δtdk (blue), and hpdBCA Δtdk (purple). (B) Levels of pCS and IS in mouse plasma 2 days post gavage and 24 h post folic acid intraperitoneal (IP) injection at time of thrombosis model. (C, D) In vivo thrombosis potential was measured by the FeCl3-induced carotid artery injury model. Representative vital microscopy images of carotid artery thrombus formation are shown at the indicated time points following arterial injury (C), and time to cessation of blood flow in mice (D) measured in the indicated number of animals (n = 7–9). The bar represents mean time to cessation of blood. Significance was measured with a Kruskal-Wallis (K.W.) test followed with Dunn’s multiple comparisons test.
Elevated levels of tryptophanase and hpdBCA gene homologs are independently associated with atherosclerotic cardiovascular disease (ASCVD)
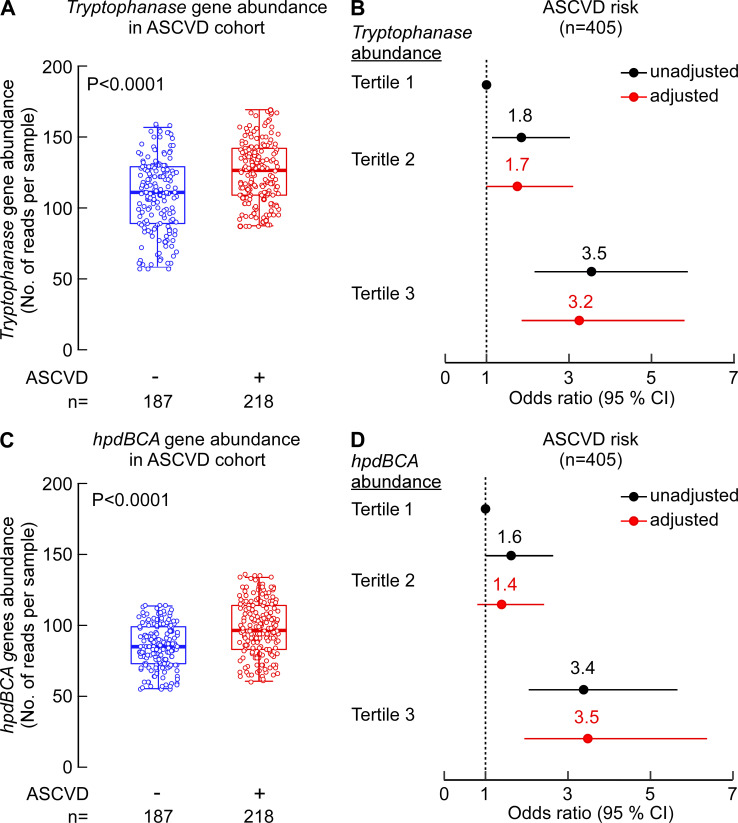
Next, we looked at the abundance of tryptophanase and hpdBCA genes in a previously published metagenomics analysis on stool samples from a cohort composed of individuals with ASCVD (n = 218) versus controls (n = 187) (41). As shown in Fig. 7, individuals with ASCVD had higher abundances of tryptophanase and hpdBCA genes when compared to controls (P < 0.0001 for both genes, Fig. 7A and C). The same trend (higher in ASCVD vs controls) was observed when tryptophanase and hpdBCA gene expressions were each normalized to median expression level of six different single copy microbial ribosomal reference genes, as outlined in Materials and Methods (P = 0.012 and P = 0.016, respectively, for tryptophanase and hpdBCA). Subsequent analysis showed subjects with elevated levels of either tryptophanase or hpdBCA genes (third tertile [(T3]) had higher prevalence of ASCVD compared to those with low (first tertile [T1]) levels, even after adjustment for traditional CVD risk factors (odd ratio [OR] 95% CI for ASCVD, OR = 3.2 [1.9–5.8], P < 0.0001 and OR = 3.5 [1.9–6.4], P < 0.0001, respectively) (Fig. 7B and C).
Fig 7.
Elevated levels of tryptophanase and hpdBCA gene homologs are associated with ASCVD. The metagenomics data from Jie at al.’s study (41) were used to determine fecal abundance of tryptophanase and hpdBCA genes in individuals with (n = 218) and without ASCVD (n = 187). (A, C) Box-whisker (5%–95%) plots of tryptophanase (A) and hpdBCA (C) gene abundance in the fecal metagenome of control individuals vs individuals with ASCVD. P-values were calculated using Wilcoxon rank-sum test. (B, D) Forest plots indicating the ASCVD prevalence according to the tertiles of tryptophanase (B) and hpdBCA (D) gene abundance. The multivariable logistic regression model for odds ratio in panels B and D included adjustments for age, sex, and hyperlipidemia. The 95% CI is indicated by line length.
DISCUSSION
Patients with chronic kidney disease or end-stage renal disease are known to have elevated levels of pCS and IS, both of which are also poorly dialyzable. Moreover, the association of these “uremic toxins” with CVD and mortality risk in patients with impaired renal function has been reported in multiple studies (17–19, 42). As a consequence, pCS and IS have been studied almost exclusively in the context of kidney disease. By using untargeted metabolomics as a discovery platform on individuals with primarily preserved renal function, we unexpectedly observed that p-cresol (presumed proxy for pCS) and mortality risk might be extended to the much broader population of individuals with preserved kidney function. This led to studies using a separate validation cohort (n = 3,954) and quantitative stable isotope labeled LC-MS/MS analyses that confirmed that the uremic solutes (both pCS and IS) are associated with heightened mortality risks outside the setting of renal disease. It is also notable that in a recent multi-center, randomized, controlled, isocaloric protein intervention trial conducted in subjects (n = 151) with diabetes and preserved kidney function, a 12-week high-protein diet was shown to increase circulating levels of IS, but not pCS (43).
pCS is among the most prevalent and concentrated of circulating microbiome-derived metabolites (with average high circulating values of 15 µM in normal and 230 µM in uremic subjects [20]), yet its biosynthetic pathway was only partially characterized. By showing that p-cresol can be produced collaboratively by certain Bacteroides species (tyrosine → 4-HPA transformation) in co-culture with hpdBCA-positive Firmicutes (4-HPA → p-cresol transformation), we complete a plausible, genetically defined pathway to p-cresol. Our recent study showed that another gut microbial prevalent enzyme, phenylpyruvate decarboxylase (PPDC), can also catalyze the 4-hydroxypyruvic acid conversion into 4-HPA (37). Consequently, future studies will be needed to determine the actual contribution of each of these two enzymes (phenylpyruvate:ferredoxin oxidoreductase [PPFOR] and PPDC) in the formation of intestinal 4-HPA. In subsequent experiments, we engineered the second half of the p-cresol pathway into B. thetaiotaomicron, creating a simple system in which the production of two metabolites can be easily controlled and studied. We propose that a similar approach in the future could be used to de-orphan the pathways for hundreds of microbiome-derived metabolites and study their effects on the host.
It has previously been described that IS increases thrombotic risk through enhancement of platelet activity (44), by altering tissue factor stability (45) and by enhancing thrombus formation in animal models of vascular injury (28, 44). Elevated levels of pCS, on the other hand, promoted vascular dysfunction and remodeling (39), while both solutes exerted proinflammatory effects that have been proposed to promote vascular damage (40). Here, we showed that gut microbial activity of hpdBCA and tryptophanase genes, individually or in tandem, was sufficient to impact host thrombosis potential. Moreover, the present studies show for the first time that fecal abundances of both microbial genes, hpdBCA and tryptophanase, were independently associated with ASCVD in humans.
The present studies, and previous work in which other pro-thrombotic gut microbiota-generated metabolites are identified (e.g., trimethylamine N-oxide [TMAO], phenylacetylglutamine [PAGln] [11, 12, 46]) and pathways responsible for their production (36, 37, 47, 48), provide a platform for developing new therapeutic approaches to potentially attenuate thrombosis (and potentially broader CVD-related phenotypes) in the host. Importantly, while the physiologic effect of these metabolites can be the same (enhanced thrombosis and platelet hyper-responsiveness), this approach will allow personalized treatments tailored to the particular set of pathways present and active in each individual. We anticipate a future where individually tailored therapeutic interventions to reduce CVD risks and thrombosis potential are driven by the pattern of elevated pro-thrombotic gut microbial metabolites a subject possesses. As a proof of concept of the idea of small-molecule inhibition of a gut microbial pathway to elicit beneficial effect within the host, we recently demonstrated that a non-lethal inhibitor of the gut microbial enzyme catalyzing conversion of choline to trimethylamine (TMA)/TMAO (CutC) inhibited both atherosclerosis (48) and thrombosis potential in vivo (49).
Numerous studies have reported that changes in the gut microbial community composition are associated with a variety of diseases including CVD, renal disease, and numerous others (5, 10, 50, 51). Identifying microbial pathways responsible for the production of individual bioactive metabolites that impact the host, both harmful and beneficial, could provide a basis for designing communities that produce a suite of compounds beneficial to the host. While linking microbial products to human diseases, elucidating their biological effects, and identifying microbes and enzymes involved in their metabolism, this study and other recent work (12, 16, 21, 36, 37, 46, 49, 52–57) demonstrate that integrating clinical and mechanistic studies with bioengineering technologies, and targeted inhibition of gut microbial enzymes, is a logical path forward toward reaching a future where personalized therapeutic interventions to reduce cardiovascular or metabolic disease risks are achievable.
MATERIALS AND METHODS
Human subjects
Both the discovery cohort (n = 1,149) employed for untargeted metabolomics investigations and the independent non-overlapping validation cohort (n = 3,954) were obtained from sequential consenting subjects enrolled in the study GeneBank at the Cleveland Clinic: Molecular Determinants of Coronary Artery Disease (GATC). GeneBank is registered under ClinicalTrials.gov Identifier: NCT00590200. Fasting plasma samples were collected in EDTA tubes and immediately processed and stored at −80° C until analyzed. Adjudicated all‐cause mortality was monitored for 5 years. High-sensitivity C-reactive protein, serum creatinine, lipid profiles, and hemoglobin A1C levels were measured on the Roche Cobas platform (Roche Diagnostics).
Mice
Animal care and experimentation were consistent with NIH guidelines. C57BL/6J mice, 8–10 weeks old, were purchased from The Jackson Laboratory and maintained in facilities at the Cleveland Clinic Lerner Research Institute. Some of the studies employed germ-free C57BL/6 male mice bred at the University of Wisconsin-Madison gnotobiotic animal facilities and shipped germ-free to the Cleveland Clinic Lerner Research Institute Gnotobiotic Facility. Studies in Fig. 3 were performed using germ-free C57BL/6 mice from the Mazmanian Laboratory GF colony at California Institute of Technology.
Bacterial strains and culture conditions
All bacterial strains and plasmids used in this study are shown in Tables S4 and S5. All Bacteroides were cultured in brain heart infusion (BHI) agar medium supplemented with 10% horse blood, Bacteroides TYG (tryptone-yeast extract-glucose) broth, or minimal medium (MM). All Clostridium strains were cultured in Clostridium TYG (3% wt/vol tryptone, 2% wt/vol yeast extract, 0.1% wt/vol sodium thioglycolate) broth or MM at 37°C in anaerobic chamber. Blautia hydrogenotrophica DSM 10507 was cultured in peptone yeast glucose (PYG) broth (Recipe #1139 on Recommended Media for Microorganisms list in DSMZ). Escherichia coli strains were aerobically cultured in Luria-Bertani (LB) broth. When appropriate, 100 µg/mL carbenicillin, 25 µg/mL erythromycin, 200 µg/mL gentamicin, and 200 µg/mL 5-fluoro-2’-deoxyuridine were supplemented into the medium.
Untargeted GC-MS analysis of human plasma samples
Untargeted gas chromatography mass spectrometry analyses of human plasma samples were performed as previously reported (58). ChromaTOF (Leco) software was used for data acquisition. Raw data files were processed using the metabolomics BinBase database (59). All database entries in BinBase were matched against UC Davis metabolomics center’s mass spectral library. p-Cresol was detected as trimethylsilyl (TMS) derivative (fatty acid methyl ester [FAME] retention index 280360) and m/z 165 was used as quantification ion.
Targeted LC-MS/MS analysis of human plasma samples
Targeted stable isotope-dilution liquid chromatography with on-line electrospray ionization tandem mass spectrometry (LC-MS/MS) was used for quantification of pCS and IS as previously described (60).
Gene disruption in B. thetaiotaomicron
All PCR amplifications were done with PrimeSTAR Max DNA polymerase (Takara Bio) according to the manufacturer’s instructions. Sequences of primers are shown in Table S6. To construct target gene deletion mutants, a previously described double-crossover recombination method was used (61).
For complementation of the BT0430 deletion, the BT0430 gene was amplified and assembled with pNBU2-bla-ermGb under control of promoter from the sigma 70 (BT1311) of B. thetaiotaomicron VPI 5482 (62) using Gibson Assembly Kit (New England Bio) to yield pMFT01. Escherichia coli S17-1 λ pir competent cells were transformed with the assembled plasmids by electroporation and transformants were confirmed by PCR. The positive clone harboring assembled plasmid was cultivated, prepared, and sequenced. The correct plasmid was introduced into B. thetaiotaomicron ∆tdk ∆BT0430 by conjugation, and erythromycin-resistant strains were selected followed by PCR confirmation.
Overexpression of BT1492 and hpdBCA in B. thetaiotaomicron
Vector maps used in this study for overexpression are shown in Fig. S2. BT1492 was amplified by PCR and assembled with pNBU2_bla_ermGb and pNBU2_bla_tetQb under the phage promoter from Bacteroides phage B124-14 using Gibson Assembly kit to make pMFT03 and pMFT04. The hpdBCA operon (gene ID: 640155897, 640155898, 640155899) from C. difficile JIR8094 was also amplified by PCR and assembled with pNBU2_bla_ermGb under the phage promoter to yield pMFT02. Escherichia coli S17-1 λ pir competent cells were transformed with these assembled plasmids by electroporation, and transformants were confirmed by PCR. The positive clone harboring assembled plasmid was cultivated, and plasmid was prepared and verified by sequencing. The plasmid was introduced and integrated into B. thetaiotaomicron ∆tdk or B. thetaiotaomicron ∆tdk ∆BT1492 by conjugation, and appropriate antibiotic-resistant strains were selected followed by PCR confirmation. For the strain expressing hpdBCA, pMFT02 was integrated into B. thetaiotaomicron ∆tdk ∆BT1492 by conjugation. For the strain expressing BT1492, pMFT03 and pMFT04 were integrated into B. thetaiotaomicron ∆tdk sequentially. For the strain expressing HpdBCA and BT1492, pMFTC02 and pMFTC04 were integrated into B. thetaiotaomicron ∆tdk sequentially.
Extraction of metabolites from bacterial culture medium
For metabolite analysis of Clostridium sp. D5 and B. hydrogenotrophica DSM 10507, they were cultured anaerobically in rich medium overnight, as described above. Cells were harvested and washed twice with MM. The pellets were re-suspended in MM with 0.1 mg/mL tyrosine or 0.1 mg/mL 4-HPA at a final optical density at 600 nm (OD600) of 1.0 and incubated for 24 h. Cell suspensions were extracted with equal amount of ethyl acetate and organic layers were analyzed by GC-MS.
For metabolite analysis of Bacteroides strains in mono-culture, strains were grown in rich medium overnight and the cells were harvested followed by MM wash. The cell pellet was re-suspended with MM containing 0.5 mg/mL tyrosine (for p-cresol production) or tryptophan (for indole production) to a final OD600 of 1.0 and incubated anaerobically for 24 h. For p-cresol and indole detection, the cultures were extracted with ethyl acetate and analyzed by GC-MS as outlined below. For 4-HPA detection, cells were extracted with acetone (20% in final) and supernatants after centrifugation were analyzed by LC/MS.
For metabolite analysis of co-culture experiment, Bacteroides strains and Clostridium strains were cultivated anaerobically in rich medium overnight and cells were harvested and washed with MM. The cell pellets were re-suspended in MM containing 0.5 mg/mL tyrosine to OD600 of 1.0 and combined. After 24 h incubation, samples for GC-MS analysis were prepared as described above.
Analysis of bacterial culture metabolites and mouse urine
p-Cresol and indole were analyzed with a split ratio of 10:1 using a GC-MS (Agilent 7890 GC coupled to an Agilent 5,977 MSD) with an HP-5MS fused silica capillary column (30 m × 250 µm × 0.25 µm, Agilent) or DB-WAXetr capillary column (30 m × 250 µm × 0.25 µm, Agilent) with helium as carrier gas, injector temperature of 250°C with non-linear temperature gradient between 40°C and 320°C; m/z 107.1 and 117.1 were used as quantifying ions for p-cresol and indole, respectively. 4-HPA, pCS, and IS were analyzed by an LC-MS/MS system (Agilent 1260 coupled to an Agilent 6120 quadrupole) or an liquid chromatography time-of-flight (LC-TOF) system (Agilent 1290 LC system coupled to an Agilent 6530 QTOF) with a UK-C18 column (3 µm, 4.6 × 75 mm Unison, Imtakt). A discontinuous gradient employing a mixture of 10 mM ammonium acetate with 0.1% formic acid (solvent A) and acetonitrile with 0.1% formic acid (solvent B) was used for chromatographic separation; m/z 151.0401, 187.0071, and 212.0023 in negative ion mode were used for measuring 4-HPA, pCS, and IS, respectively. Concentration of creatinine was measured using Colorimetric Creatinine Assay Kit (Abcam) according to manufacturer’s instructions.
Gnotobiotic mouse colonization
Co-colonization of Bacteroides and Clostridium sp. D5
Stationary phase liquid cultures of Clostridium and Bacteroides spp. were combined at a 1:1 (vol/vol) ratio while maintained in an anaerobic chamber and sealed in airtight tubes until administration to mice. Germ-free C57Bl/6 mice (6 weeks old) from the Mazmanian Laboratory GF colony were gavaged with 100 µL of the combined culture and strictly maintained in autoclaved microisolator cages. One week following colonization, urine samples were collected passively during brief restraint of mice by hand and analyzed for metabolite levels as described above. A normalized mass of fecal material from each mouse was re-suspended in BHI medium and plated on brain heart infusion supplemented with yeast extract (BHIS) plates. Visual observation confirmed equivalent levels of colony morphologies of both the respective Bacteroides and Clostridium spp. across all plates and bacterial pairs. PCR of fecal pellets from these mice also showed accurate colonization. This is now included in the revised manuscript.
Mono-colonization with wild-type and engineered strains of B. thetaiotaomicron
B. thetaiotaomicron mutants were grown on tryptic soy blood agar plates (Anaerobe Systems) anaerobically for 48–72 h at 37°C. Single colonies were picked and used to inoculate Mega Medium (3 mL) in prepared hungate tubes. Cultures were grown anaerobically for 18–24 h at 37°C. At that time, an aliquot of culture (500 µL) was removed and the remaining bacterial culture was diluted 1:1 with glycerol (40%) in water (vol:vol) and stored at −80°C. Germ-free, C57BL/6, male, 8- to 10-week-old mice were mono-colonized by oral gavage with ~0.2 mL of bacterial culture inside the biological safety cabinet, using the indicated mutants. Mice were maintained on a sterilized diet and, 24 h prior to in vivo thrombosis, were injected with filter sterilized folic acid (250 mg/kg) to induce kidney injury and increase metabolites levels. At the time of sacrifice (~7 days post colonization), mice were subjected to carotid artery FeCl3 injury thrombosis assay and tissues were collected immediately after the assay, frozen, and stored at −80°C. Following colonization, the investigators were not blinded from treatment groups to avoid cross contamination.
To confirm colonization, DNA was isolated from flash frozen cecal contents of colonized mice with the NucleoSpin Tissue kit (Macherey-Nagel) according to manufacturer’s instructions for bacterial DNA isolation. Isolated DNA was used in a PCR reaction with GoTaq Green Master Mix and the 8F and 1492R 16S rRNA universal primers. PCR reactions were carried out in a 96-well plate with 20 µL final reaction volumes as follows: 95°C for 2 minutes; 95°C for 30 seconds, 51°C for 30 seconds, 72°C for 1 minute and 20 seconds (×30); 72°C for 5 minutes. Completed reactions were sent to Eurofins Genomics for PCR cleanup and 16S rRNA gene sequencing using their standard house primers (16F). Sequence identity was confirmed using NCBI BLAST (Basic Local Alignment Search Tool developed by the National Center for Biotechnology Information). To determine cecal bacterial load counting, a portion of the frozen ceca were weighed, thawed under anaerobic conditions, and suspended in PBS buffer. Dilutions of the cecal suspensions were then plated on BHI agar plates, incubated for 72 h at 37°C under anaerobic conditions for colony counting.
Thrombosis assay
Monocolonized or i.p. injected mice (vehicle [normal saline]; pCS [50 mg/kg]; IS [50 mg/kg]) were anesthetized with ketamine and xylazine and subjected to a carotid artery injury, and both the rate of thrombus generation and blood flow monitored in real time by fluorescence intravital microscopy with continuous image capturing as previously described (11). Briefly, rhodamine 6G (100 µL; 0.5 mg/mL) was injected directly into the right jugular vein to label platelets. The left carotid artery was exposed and injured by placing 1.5 × 1.5 mm2 Whatman filter paper soaked in 10% FeCl3 solution to the surface of the vessel for 1 minute. After removing the paper, the vessel was covered with saline. Thrombus formation was observed in real time using intravital fluorescence microscopy equipped with video recording. Time to cessation of blood flow through clot formation for all studies was determined by visual inspection by two independent investigators.
Metagenomic analyses of tryptophanase and hpdBCA gene abundances and prevalent ASCVD
We used the publicly available metagenomics data from a previously published ASCVD case/control cohort (41). ASCVD subjects (n = 218) were Han Chinese, 40–80 years old with stable angina, unstable angina, or acute myocardial infarction (AMI). ASCVD diagnosis was confirmed by coronary angiography and defined as ≥50% stenosis in single or multiple vessels were included. Han Chinese Controls (n = 187) comprised of subjects free of clinically evident ASCVD symptoms at the time of the medical examination.
Custom reference database creation
The Entrez Programming Utilities (E-utilities: https://www.ncbi.nlm.nih.gov/books/NBK25501/) implemented in the eutils package (https://github.com/biocommons/eutils) were used to retrieve all microbial protein-coding sequences (keywords: -db protein -query "(gene name) AND "bacteria"[porgn: txid2]" | efetch -format fasta_cds_na nucleotides) from NCBI. The protein-coding sequences (nucleotides) provided by NCBI’s protein database were compared to protein-coding sequences in the MGnify database (63) using blastn (64). Using a minimum identity and length cutoff of 90% and 80%, a tabular output was generated from blastn output using R (https://github.com/bioinfo-core-BGU/parse_blast). The MGnify database’s homologs were combined with NCBI sequences to generate a custom microbial protein-coding sequence database.
De novo metagenome assembly and screening
Shotgun microbiome data from the ASCVD cohort (41) were quality filtered and trimmed using the nesoni pipeline (https://github.com/Victorian-Bioinformatics-Consortium/nesoni). Quality-filtered reads were assembled into contigs using a megahit assembler (65). Metagenome assemblies were screened for possible mis-assembly events using metaMIC pipeline (66). Quality-filtered metagenome contigs were annotated using the prokka pipeline (67). Protein-coding genes from individual metagenome assemblies were compared against our customized reference database using blastn (minimum identity = 90% and minimum length = 80%) 2. To quantify the abundance of protein-coding genes, quality-filtered metagenome reads were mapped on the protein-coding genes that showed hits to our custom reference database using vsearch (68). Abundance (counts) patterns of hpdBCA and tryptophanase gene were used for differential abundance and metadata association analyses. Relative abundance of rplB, rplC, rpsS, rplD, rplE, and rpsQ genes was used to normalize the gene count matrix.
Statistical analyses
Student’s t-test (two tailed) or Wilcoxon’s rank-sum test for continuous variables and χ2 test for categorical variables were used to examine the differences between groups. Rank-based nonparametric Kruskal-Wallis test was used for non-normally distributed data. In the box-whisker plot, the upper and lower boundaries of the box represent the 25th and 75th percentiles, the median is marked by a horizontal line inside the box, and whiskers represent 10% and 90% of relative measured values. Categorical data are presented as n (%). HR for death at 5-year follow-up and corresponding 95% CI were estimated using both univariable (unadjusted) and multivariable (adjusted) Cox models. Kaplan-Meier analysis with Cox proportional hazards regression was used for time-to-event analysis to determine HR and 95% CI for 5 year mortality. Adjustments were made for individual traditional cardiac risk factors including age, sex, high-density lipoprotein, low-density lipoprotein, triglycerides, current smoking, diabetes mellitus, systolic blood pressure, and high-sensitivity C-reactive protein level (model 1) and all factors form model 1 and CKD (define as eGFR <60 mL/min/1.73 m2) (model 2). To further test for the relationship between tryptophanase and hpdBCA gene abundance and ASCVD, odds ratio for binary ASCVD and corresponding 95% CI were calculated using both univariable (unadjusted) and multivariable (adjusted) logistic regression models with cohort adjustments made for age, sex, and dyslipidemia. All data are presented as mean ± standard deviation or SEM or median with interquartile range. Statistical tests used to compare conditions are indicated in figure legends. GraphPad PRISM versions 8.0 and R 3.4.2 (Vienna, Austria, 2017) were used for generation of graphs and statistics.
ACKNOWLEDGMENTS
This work is supported by grants from the NIH (NHLBI and Office of Dietary Supplements): P01HL147823 (to S.L.H. and M.A.F.), R01HL103866 (to S.L.H.), R01HL160747 (to I.N.), and R01DK101674 (to M.A.F.); the Leducq Foundation (17CVD01 to S.L.H. and M.A.F.); and Chan Zuckerberg Biohub (to M.A.F.). K.A.R. was supported in part by NIH/NHLBI training grant HL134622. Mass spectrometry studies were performed on instrumentation housed in a facility supported in part through a Shimadzu Center of Excellence.
Contributor Information
Michael A. Fischbach, Email: fischbach@fischbachgroup.org.
Stanley L. Hazen, Email: hazens@ccf.org.
Edward G. Ruby, University of Hawaii at Manoa, Honolulu, Hawaii, USA
Joseph Zackular, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
DATA AVAILABILITY
All source data for figures included in the manuscript were deposited in Zenodo repository (https://zenodo.org/record/8338175). There are restrictions to the availability of some of the clinical data generated in the present study because we do not have permission in our informed consent from research subjects to share data outside our institution without their authorizations. Under these situations, data shared is in summary format.
ETHICS APPROVAL
This study was approved by the Cleveland Clinic Institutional Review Board, and all participants provided written informed consent. All animal model studies were approved by the Institutional Animal Care and Use Committee at the Cleveland Clinic, University of Wisconsin, and California Institute of Technology.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/mbio.01331-23.
Tables S1-S6; Fig. S1-S3.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Chiang C-E, Schwartz GG, Elbez Y, Szarek M, Bhatt DL, Bittner VA, Diaz R, Erglis A, Goodman SG, Hagström E, Jukema JW, Liberopoulos E, Loy M, Pordy R, White HD, Simon T, Steg PG, ODYSSEY OUTCOMES Investigators . 2022. Alirocumab and cardiovascular outcomes in patients with previous myocardial infarction: prespecified subanalysis from ODYSSEY OUTCOMES. Can J Cardiol 38:1542–1549. doi: 10.1016/j.cjca.2022.05.021 [DOI] [PubMed] [Google Scholar]
- 2. Cholesterol Treatment Trialists C, Baigent C, Blackwell L, Emberson J, Holland LE, Reith C, Bhala N, Peto R, Barnes EH, Keech A, Simes J, Collins R. 2010. Efficacy and safety of more intensive lowering of LDL cholesterol: a meta-analysis of data from 170,000 participants in 26 randomised trials. Lancet 376:1670–1681. doi: 10.1016/S0140-6736(10)61350-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cannon CP, Blazing MA, Giugliano RP, McCagg A, White JA, Theroux P, Darius H, Lewis BS, Ophuis TO, Jukema JW, De Ferrari GM, Ruzyllo W, De Lucca P, Im K, Bohula EA, Reist C, Wiviott SD, Tershakovec AM, Musliner TA, Braunwald E, Califf RM, IMPROVE-IT Investigators . 2015. Ezetimibe added to statin therapy after acute coronary syndromes. N Engl J Med 372:2387–2397. doi: 10.1056/NEJMoa1410489 [DOI] [PubMed] [Google Scholar]
- 4. Sabatine MS, Giugliano RP, Keech AC, Honarpour N, Wiviott SD, Murphy SA, Kuder JF, Wang H, Liu T, Wasserman SM, Sever PS, Pedersen TR, FOURIER Steering Committee and Investigators . 2017. Evolocumab and clinical outcomes in patients with cardiovascular disease. N Engl J Med 376:1713–1722. doi: 10.1056/NEJMoa1615664 [DOI] [PubMed] [Google Scholar]
- 5. Witkowski M, Weeks TL, Hazen SL. 2020. Gut microbiota and cardiovascular disease. Circ Res 127:553–570. doi: 10.1161/CIRCRESAHA.120.316242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fischbach MA. 2018. Microbiome: focus on causation and mechanism. Cell 174:785–790. doi: 10.1016/j.cell.2018.07.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung Y-M, Wu Y, Schauer P, Smith JD, Allayee H, Tang WHW, DiDonato JA, Lusis AJ, Hazen SL. 2011. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472:57–63. doi: 10.1038/nature09922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tang WHW, Bäckhed F, Landmesser U, Hazen SL. 2019. Intestinal microbiota in cardiovascular health and disease: JACC state-of-the-art review. J Am Coll Cardiol 73:2089–2105. doi: 10.1016/j.jacc.2019.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Koren O, Spor A, Felin J, Fåk F, Stombaugh J, Tremaroli V, Behre CJ, Knight R, Fagerberg B, Ley RE, Bäckhed F. 2011. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc Natl Acad Sci U S A 108 Suppl 1:4592–4598. doi: 10.1073/pnas.1011383107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fromentin S, Forslund SK, Chechi K, Aron-Wisnewsky J, Chakaroun R, Nielsen T, Tremaroli V, Ji B, Prifti E, Myridakis A, et al. 2022. Microbiome and metabolome features of the cardiometabolic disease spectrum. Nat Med 28:303–314. doi: 10.1038/s41591-022-01688-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhu W, Gregory JC, Org E, Buffa JA, Gupta N, Wang Z, Li L, Fu X, Wu Y, Mehrabian M, Sartor RB, McIntyre TM, Silverstein RL, Tang WHW, DiDonato JA, Brown JM, Lusis AJ, Hazen SL. 2016. Gut microbial metabolite TMAO enhances platelet hyperreactivity and thrombosis risk. Cell 165:111–124. doi: 10.1016/j.cell.2016.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nemet I, Saha PP, Gupta N, Zhu W, Romano KA, Skye SM, Cajka T, Mohan ML, Li L, Wu Y, Funabashi M, Ramer-Tait AE, Naga Prasad SV, Fiehn O, Rey FE, Tang WHW, Fischbach MA, DiDonato JA, Hazen SL. 2020. A cardiovascular disease-linked gut microbial metabolite acts via adrenergic receptors. Cell 180:862–877. doi: 10.1016/j.cell.2020.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pluznick JL. 2016. Gut microbiota in renal physiology: focus on short-chain fatty acids and their receptors. Kidney Int 90:1191–1198. doi: 10.1016/j.kint.2016.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Koh A, Molinaro A, Ståhlman M, Khan MT, Schmidt C, Mannerås-Holm L, Wu H, Carreras A, Jeong H, Olofsson LE, Bergh P-O, Gerdes V, Hartstra A, de Brauw M, Perkins R, Nieuwdorp M, Bergström G, Bäckhed F. 2018. Microbially produced imidazole propionate impairs insulin signaling through mTORC1. Cell 175:947–961. doi: 10.1016/j.cell.2018.09.055 [DOI] [PubMed] [Google Scholar]
- 15. Zhao M, Wei H, Li C, Zhan R, Liu C, Gao J, Yi Y, Cui X, Shan W, Ji L, Pan B, Cheng S, Song M, Sun H, Jiang H, Cai J, Garcia-Barrio MT, Chen YE, Meng X, Dong E, Wang DW, Zheng L. 2022. Gut microbiota production of trimethyl-5-aminovaleric acid reduces fatty acid oxidation and accelerates cardiac hypertrophy. Nat Commun 13:1757. doi: 10.1038/s41467-022-29060-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Needham BD, Funabashi M, Adame MD, Wang Z, Boktor JC, Haney J, Wu WL, Rabut C, Ladinsky MS, Hwang SJ, Guo Y, Zhu Q, Griffiths JA, Knight R, Bjorkman PJ, Shapiro MG, Geschwind DH, Holschneider DP, Fischbach MA, Mazmanian SK. 2022. A gut-derived metabolite alters brain activity and anxiety behaviour in mice. Nature 602:647–653. doi: 10.1038/s41586-022-04396-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lin CJ, Chuang CK, Jayakumar T, Liu HL, Pan CF, Wang TJ, Chen HH, Wu CJ. 2013. Serum p-cresyl sulfate predicts cardiovascular disease and mortality in elderly hemodialysis patients. Arch Med Sci 9:662–668. doi: 10.5114/aoms.2013.36901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Go AS, Chertow GM, Fan D, McCulloch CE, Hsu C. 2004. Chronic kidney disease and the risks of death, cardiovascular events, and hospitalization. N Engl J Med 351:1296–1305. doi: 10.1056/NEJMoa041031 [DOI] [PubMed] [Google Scholar]
- 19. Ramezani A, Raj DS. 2014. The gut microbiome, kidney disease, and targeted interventions. J Am Soc Nephrol 25:657–670. doi: 10.1681/ASN.2013080905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vanholder R, Schepers E, Pletinck A, Nagler EV, Glorieux G. 2014. The uremic toxicity of Indoxyl sulfate and p-cresyl sulfate: a systematic review. J Am Soc Nephrol 25:1897–1907. doi: 10.1681/ASN.2013101062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Devlin AS, Marcobal A, Dodd D, Nayfach S, Plummer N, Meyer T, Pollard KS, Sonnenburg JL, Fischbach MA. 2016. Modulation of a circulating uremic solute via rational genetic manipulation of the gut microbiota. Cell Host Microbe 20:709–715. doi: 10.1016/j.chom.2016.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brix LA, Duggleby RG, Gaedigk A, McManus ME. 1999. Structural characterization of human aryl sulphotransferases. Biochem J 337 (Pt 2):337–343. [PMC free article] [PubMed] [Google Scholar]
- 23. Wilborn TW, Comer KA, Dooley TP, Reardon IM, Heinrikson RL, Falany CN. 1993. Sequence analysis and expression of the cDNA for the phenol-sulfating form of human liver phenol sulfotransferase. Mol Pharmacol 43:70–77. [PubMed] [Google Scholar]
- 24. Duranton F, Cohen G, De Smet R, Rodriguez M, Jankowski J, Vanholder R, Argiles A, European Uremic Toxin Work Group . 2012. Normal and pathologic concentrations of uremic toxins. J Am Soc Nephrol 23:1258–1270. doi: 10.1681/ASN.2011121175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Barreto FC, Barreto DV, Liabeuf S, Meert N, Glorieux G, Temmar M, Choukroun G, Vanholder R, Massy ZA, European Uremic Toxin Work G. 2009. Serum indoxyl sulfate is associated with vascular disease and mortality in chronic kidney disease patients. Clin J Am Soc Nephrol 4:1551–1558. doi: 10.2215/CJN.03980609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liabeuf S, Barreto DV, Barreto FC, Meert N, Glorieux G, Schepers E, Temmar M, Choukroun G, Vanholder R, Massy ZA, European Uraemic Toxin Work G. 2010. Free p-cresylsulphate is a predictor of mortality in patients at different stages of chronic kidney disease. Nephrol Dial Transplant 25:1183–1191. doi: 10.1093/ndt/gfp592 [DOI] [PubMed] [Google Scholar]
- 27. Rossi M, Campbell K, Johnson D, Stanton T, Pascoe E, Hawley C, Dimeski G, McWhinney B, Ungerer J, Isbel N. 2014. Uraemic toxins and cardiovascular disease across the chronic kidney disease spectrum: an observational study. Nutr Metab Cardiovasc Dis 24:1035–1042. doi: 10.1016/j.numecd.2014.04.006 [DOI] [PubMed] [Google Scholar]
- 28. Karbowska M, Kaminski TW, Marcinczyk N, Misztal T, Rusak T, Smyk L, Pawlak D. 2017. The uremic toxin Indoxyl sulfate accelerates thrombotic response after vascular injury in animal models. Toxins (Basel) 9:229. doi: 10.3390/toxins9070229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Smith EA, Macfarlane GT. 1996. Enumeration of human colonic bacteria producing phenolic and Indolic compounds: effects of pH, carbohydrate availability and retention time on dissimilatory aromatic amino acid metabolism. J Appl Bacteriol 81:288–302. doi: 10.1111/j.1365-2672.1996.tb04331.x [DOI] [PubMed] [Google Scholar]
- 30. Saito Y, Sato T, Nomoto K, Tsuji H. 2018. Identification of phenol- and p-cresol-producing intestinal bacteria by using media supplemented with tyrosine and its metabolites. FEMS Microbiol Ecol 94:fiy125. doi: 10.1093/femsec/fiy125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dawson LF, Donahue EH, Cartman ST, Barton RH, Bundy J, McNerney R, Minton NP, Wren BW. 2011. The analysis of para-cresol production and tolerance in clostridium difficile 027 and 012 strains. BMC Microbiol 11:86. doi: 10.1186/1471-2180-11-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Harrison MA, Kaur H, Wren BW, Dawson LF. 2021. Production of p-cresol by decarboxylation of p-HPA by all five lineages of clostridioides difficile provides a growth advantage. Front Cell Infect Microbiol 11:757599. doi: 10.3389/fcimb.2021.757599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sato Y, Atarashi K, Plichta DR, Arai Y, Sasajima S, Kearney SM, Suda W, Takeshita K, Sasaki T, Okamoto S, et al. 2021. Novel bile acid biosynthetic pathways are enriched in the microbiome of centenarians. Nature 599:458–464. doi: 10.1038/s41586-021-03832-5 [DOI] [PubMed] [Google Scholar]
- 34. Russell WR, Duncan SH, Scobbie L, Duncan G, Cantlay L, Calder AG, Anderson SE, Flint HJ. 2013. Major phenylpropanoid-derived metabolites in the human gut can arise from microbial fermentation of protein. Mol Nutr Food Res 57:523–535. doi: 10.1002/mnfr.201200594 [DOI] [PubMed] [Google Scholar]
- 35. Lal PB, Schneider BL, Vu K, Reitzer L. 2014. The redundant aminotransferases in lysine and arginine synthesis and the extent of aminotransferase redundancy in Escherichia coli. Mol Microbiol 94:843–856. doi: 10.1111/mmi.12801 [DOI] [PubMed] [Google Scholar]
- 36. Buffa JA, Romano KA, Copeland MF, Cody DB, Zhu W, Galvez R, Fu X, Ward K, Ferrell M, Dai HJ, et al. 2022. The microbial gbu gene cluster links cardiovascular disease risk associated with red meat consumption to microbiota L-carnitine catabolism. Nat Microbiol 7:73–86. doi: 10.1038/s41564-021-01010-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhu Y, Dwidar M, Nemet I, Buffa JA, Sangwan N, Li XS, Anderson JT, Romano KA, Fu X, Funabashi M, Wang Z, Keranahalli P, Battle S, Tittle AN, Hajjar AM, Gogonea V, Fischbach MA, DiDonato JA, Hazen SL. 2023. Two distinct gut microbial pathways contribute to meta-organismal production of phenylacetylglutamine with links to cardiovascular disease. Cell Host Microbe 31:18–32. doi: 10.1016/j.chom.2022.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Whitaker WR, Shepherd ES, Sonnenburg JL. 2017. Tunable expression tools enable single-cell strain distinction in the gut microbiome. Cell 169:538–546. doi: 10.1016/j.cell.2017.03.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gross P, Massy ZA, Henaut L, Boudot C, Cagnard J, March C, Kamel S, Drueke TB, Six I. 2015. Para-cresyl sulfate acutely impairs vascular reactivity and induces vascular remodeling. J Cell Physiol 230:2927–2935. doi: 10.1002/jcp.25018 [DOI] [PubMed] [Google Scholar]
- 40. Pletinck A, Glorieux G, Schepers E, Cohen G, Gondouin B, Van Landschoot M, Eloot S, Rops A, Van de Voorde J, De Vriese A, van der Vlag J, Brunet P, Van Biesen W, Vanholder R. 2013. Protein-bound uremic toxins stimulate crosstalk between leukocytes and vessel wall. J Am Soc Nephrol 24:1981–1994. doi: 10.1681/ASN.2012030281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jie Z, Xia H, Zhong S-L, Feng Q, Li S, Liang S, Zhong H, Liu Z, Gao Y, Zhao H, et al. 2017. The gut microbiome in atherosclerotic cardiovascular disease. Nat Commun 8:845. doi: 10.1038/s41467-017-00900-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Meijers BKI, Claes K, Bammens B, de Loor H, Viaene L, Verbeke K, Kuypers D, Vanrenterghem Y, Evenepoel P. 2010. p-cresol and cardiovascular risk in mild-to-moderate kidney disease. Clin J Am Soc Nephrol 5:1182–1189. doi: 10.2215/CJN.07971109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Attaye I, Lassen PB, Adriouch S, Steinbach E, Patiño-Navarrete R, Davids M, Alili R, Jacques F, Benzeguir S, Belda E, et al. 2023. Protein supplementation changes gut microbial diversity and derived metabolites in subjects with type 2 diabetes. iScience 26:107471. doi: 10.1016/j.isci.2023.107471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang K, Du C, Wang X, Li F, Xu Y, Wang S, Chen S, Chen F, Shen M, Chen M, Hu M, He T, Su Y, Wang J, Zhao J. 2017. Indoxyl sulfate induces platelet hyperactivity and contributes to chronic kidney disease-associated thrombosis in mice. Blood 129:2667–2679. doi: 10.1182/blood-2016-10-744060 [DOI] [PubMed] [Google Scholar]
- 45. Chitalia VC, Shivanna S, Martorell J, Balcells M, Bosch I, Kolandaivelu K, Edelman ER. 2013. Uremic serum and solutes increase post-vascular interventional thrombotic risk through altered stability of smooth muscle cell tissue factor. Circulation 127:365–376. doi: 10.1161/CIRCULATIONAHA.112.118174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Skye SM, Zhu W, Romano KA, Guo CJ, Wang Z, Jia X, Kirsop J, Haag B, Lang JM, DiDonato JA, Tang WHW, Lusis AJ, Rey FE, Fischbach MA, Hazen SL. 2018. Microbial transplantation with human gut commensals containing CutC is sufficient to transmit enhanced platelet reactivity and thrombosis potential. Circ Res 123:1164–1176. doi: 10.1161/CIRCRESAHA.118.313142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Craciun S, Balskus EP. 2012. Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proc Natl Acad Sci U S A 109:21307–21312. doi: 10.1073/pnas.1215689109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang Z, Roberts AB, Buffa JA, Levison BS, Zhu W, Org E, Gu X, Huang Y, Zamanian-Daryoush M, Culley MK, DiDonato AJ, Fu X, Hazen JE, Krajcik D, DiDonato JA, Lusis AJ, Hazen SL. 2015. Non-lethal inhibition of gut microbial trimethylamine production for the treatment of atherosclerosis. Cell 163:1585–1595. doi: 10.1016/j.cell.2015.11.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Roberts AB, Gu X, Buffa JA, Hurd AG, Wang Z, Zhu W, Gupta N, Skye SM, Cody DB, Levison BS, Barrington WT, Russell MW, Reed JM, Duzan A, Lang JM, Fu X, Li L, Myers AJ, Rachakonda S, DiDonato JA, Brown JM, Gogonea V, Lusis AJ, Garcia-Garcia JC, Hazen SL. 2018. Development of a gut microbe-targeted nonlethal therapeutic to inhibit thrombosis potential. Nat Med 24:1407–1417. doi: 10.1038/s41591-018-0128-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Meijers B, Evenepoel P, Anders HJ. 2019. Intestinal microbiome and fitness in kidney disease. Nat Rev Nephrol 15:531–545. doi: 10.1038/s41581-019-0172-1 [DOI] [PubMed] [Google Scholar]
- 51. Hobby GP, Karaduta O, Dusio GF, Singh M, Zybailov BL, Arthur JM. 2019. Chronic kidney disease and the gut microbiome. Am J Physiol Renal Physiol 316:F1211–F1217. doi: 10.1152/ajprenal.00298.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dodd D, Spitzer MH, Van Treuren W, Merrill BD, Hryckowian AJ, Higginbottom SK, Le A, Cowan TM, Nolan GP, Fischbach MA, Sonnenburg JL. 2017. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 551:648–652. doi: 10.1038/nature24661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhu W, Romano KA, Li L, Buffa JA, Sangwan N, Prakash P, Tittle AN, Li XS, Fu X, Androjna C, DiDonato AJ, Brinson K, Trapp BD, Fischbach MA, Rey FE, Hajjar AM, DiDonato JA, Hazen SL. 2021. Gut Microbes impact stroke severity via the trimethylamine N-oxide pathway. Cell Host Microbe 29:1199–1208. doi: 10.1016/j.chom.2021.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gupta N, Buffa JA, Roberts AB, Sangwan N, Skye SM, Li L, Ho KJ, Varga J, DiDonato JA, Tang WHW, Hazen SL. 2020. Targeted inhibition of gut microbial trimethylamine N-oxide production reduces renal tubulointerstitial fibrosis and functional impairment in a murine model of chronic kidney disease. Arterioscler Thromb Vasc Biol 40:1239–1255. doi: 10.1161/ATVBAHA.120.314139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Stewart Campbell A, Needham BD, Meyer CR, Tan J, Conrad M, Preston GM, Bolognani F, Rao SG, Heussler H, Griffith R, Guastella AJ, Janes AC, Frederick B, Donabedian DH, Mazmanian SK. 2022. Safety and target engagement of an oral small-molecule sequestrant in adolescents with autism spectrum disorder: an open-label phase 1b/2a trial. Nat Med 28:528–534. doi: 10.1038/s41591-022-01683-9 [DOI] [PubMed] [Google Scholar]
- 56. Organ CL, Li Z, Sharp TE, Polhemus DJ, Gupta N, Goodchild TT, Tang WHW, Hazen SL, Lefer DJ. 2020. Nonlethal inhibition of gut microbial trimethylamine N-oxide production improves cardiac function and remodeling in a murine model of heart failure. J Am Heart Assoc 9:e016223. doi: 10.1161/JAHA.119.016223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Benson TW, Conrad KA, Li XS, Wang Z, Helsley RN, Schugar RC, Coughlin TM, Wadding-Lee C, Fleifil S, Russell HM, et al. 2023. Gut microbiota-derived trimethylamine N-oxide contributes to abdominal aortic aneurysm through inflammatory and apoptotic mechanisms. Circulation 147:1079–1096. doi: 10.1161/CIRCULATIONAHA.122.060573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fiehn O. 2016. Metabolomics by gas chromatography-mass spectrometry: combined targeted and untargeted profiling. Curr Protoc Mol Biol 114:30. doi: 10.1002/0471142727.mb3004s114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Fiehn O, Scholz M, Wohlgemuth G. 2005. Data integration in the life sciences, p 224–239. In Setup and annotation of Metabolomic experiments by integrating biological and mass spectrometric Metadata. Springer Berlin Heidelberg, Berlin, Heidelberg. [Google Scholar]
- 60. Nemet I, Li XS, Haghikia A, Li L, Wilcox J, Romano KA, Buffa JA, Witkowski M, Demuth I, König M, Steinhagen-Thiessen E, Bäckhed F, Fischbach MA, Tang WHW, Landmesser U, Hazen SL. 2023. Atlas of gut microbe-derived products from aromatic amino acids and risk of cardiovascular morbidity and mortality. Eur Heart J 44:3085–3096. doi: 10.1093/eurheartj/ehad333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Koropatkin NM, Martens EC, Gordon JI, Smith TJ. 2008. Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices. Structure 16:1105–1115. doi: 10.1016/j.str.2008.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. García-Bayona L, Comstock LE. 2019. Streamlined genetic manipulation of diverse bacteroides and parabacteroides isolates from the human gut microbiota. mBio 10:e01762-19. doi: 10.1128/mBio.01762-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mitchell AL, Almeida A, Beracochea M, Boland M, Burgin J, Cochrane G, Crusoe MR, Kale V, Potter SC, Richardson LJ, Sakharova E, Scheremetjew M, Korobeynikov A, Shlemov A, Kunyavskaya O, Lapidus A, Finn RD. 2020. MGnify: the microbiome analysis resource in 2020. Nucleic Acids Res 48:D570–D578. doi: 10.1093/nar/gkz1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Li D, Liu CM, Luo R, Sadakane K, Lam TW. 2015. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph . Bioinformatics 31:1674–1676. doi: 10.1093/bioinformatics/btv033 [DOI] [PubMed] [Google Scholar]
- 66. Lai S, Pan S, Sun C, Coelho LP, Chen WH, Zhao XM. 2022. metaMIC: reference-free misassembly identification and correction of de novo metagenomic assemblies. Genome Biol 23:242. doi: 10.1186/s13059-022-02810-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- 68. Rognes T, Flouri T, Nichols B, Quince C, Mahé F. 2016. VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. doi: 10.7717/peerj.2584 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1-S6; Fig. S1-S3.
Data Availability Statement
All source data for figures included in the manuscript were deposited in Zenodo repository (https://zenodo.org/record/8338175). There are restrictions to the availability of some of the clinical data generated in the present study because we do not have permission in our informed consent from research subjects to share data outside our institution without their authorizations. Under these situations, data shared is in summary format.