Abstract
Objective
The classification of clinical note sections is a critical step before doing more fine-grained natural language processing tasks such as social determinants of health extraction and temporal information extraction. Often, clinical note section classification models that achieve high accuracy for 1 institution experience a large drop of accuracy when transferred to another institution. The objective of this study is to develop methods that classify clinical note sections under the SOAP (“Subjective,” “Object,” “Assessment,” and “Plan”) framework with improved transferability.
Materials and methods
We trained the baseline models by fine-tuning BERT-based models, and enhanced their transferability with continued pretraining, including domain-adaptive pretraining and task-adaptive pretraining. We added in-domain annotated samples during fine-tuning and observed model performance over a varying number of annotated sample size. Finally, we quantified the impact of continued pretraining in equivalence of the number of in-domain annotated samples added.
Results
We found continued pretraining improved models only when combined with in-domain annotated samples, improving the F1 score from 0.756 to 0.808, averaged across 3 datasets. This improvement was equivalent to adding 35 in-domain annotated samples.
Discussion
Although considered a straightforward task when performing in-domain, section classification is still a considerably difficult task when performing cross-domain, even using highly sophisticated neural network-based methods.
Conclusion
Continued pretraining improved model transferability for cross-domain clinical note section classification in the presence of a small amount of in-domain labeled samples.
Keywords: section classification, text classification, natural language processing, transfer learning, continued pretraining
Introduction and background
Electronic health record (EHR) systems contain important clinical information in unstructured text, and natural language processing (NLP) is an important tool for its secondary use. Clinical note section classification is a foundational NLP task, as it facilitates many downstream tasks, and section information has been found beneficial for a diversity of clinical NLP tasks including named entity recognition,1 abbreviation resolution,2 cohort retrieval,3 and temporal relation extraction.4
Existing work in section classification5,6 has shown that the task is solvable for a given dataset, but that performance drops substantially when applying a trained system to a new dataset. In clinical note section classification, researchers have found that statistical methods and modern pretrained transformers (e.g., BERT7) achieved high performance for single institution modeling.5,6 In a study for classifying emergency departments reports into SOAP (“Subjective,” “Objective,” “Assessment,” and “Plan”)8 sections, researchers built an SVM classifier with lexical syntactic, semantic, contextual, and heuristic features and the macro-F1 score was 0.85.9 In Rosenthal et al.,6 BERT achieved 0.99 and 0.9 F1 score for 2 section classification datasets with fine-grained section names. In Tepper et al.,5 researchers studied performing note segmentation and section classification together with fine-grained section names. Maximum entropy classifiers with fine-grained features (e.g., capital letters, numbers, blank lines, previous section names) achieved an F1 score of over 0.9 for 2 discharge summary datasets and 1 radiology report dataset. When transferring models learned from 1 dataset to another, the F1 score dropped to 0.6.
To address the gap in developing section classifiers that can perform accurately across domains, we developed domain adaptation methods in the context of the SOAP section classification task. In clinical practice, SOAP style notes are a widely used note-writing format taught for documenting the daily care of patients.8,10 In simplifying the section classification task to the SOAP classification task, we make it possible to perform more cross-domain experiments and simplify the task to examine the cross-domain performance loss in a setting where we can eliminate 1 variable—the differences in the output space between datasets.
Automatically classifying sections into SOAP categories is still beneficial for better understanding the sourcing of information extracted by other NLP systems. For example, social determinants of health information may be more likely to be found in the social history section of a clinical note which is a “Subjective” section in the SOAP framework. Medication mentions may have different interpretation if they are in an “Objective” section (e.g., treatments in a medication list) versus a “Subjective” section (e.g., medication misuse in a social history). In addition, state-of-the-art NLP models (pretrained transformers) have memory constraints that limit the number of words they can process,7 so processing only relevant sections may make these models more applicable.
Domain adaptation refers to the study of improving model’s transferability from a source dataset to a target dataset and is a common theme in clinical NLP. In this work, we used the methods of domain-adaptive pretraining (DAPT) and task-adapted pretraining (TAPT).11 These methods work by applying the masked language modeling pretraining objective to target domain data, before doing fine-tuning using labeled source domain data. We then experimented with using small amounts of labeled data in the target domain to quantify the interaction between unsupervised and supervised domain adaptation techniques.
Objective
The objective of this study is to develop methods that classify clinical note sections with SOAP labels. The secondary objective of this work is to examine the generalizability of existing datasets and methods by performing cross-domain validation, and to attempt to address any performance degradation with domain adaptation methods.
Methods
Datasets
We used 3 independent datasets across multiple health systems and different note types. The first dataset (discharge) consists of discharge summaries from the i2b2 2010 challenge from Partners Healthcare and Beth Israel Deaconess Medical Center.5 The second dataset (thyme) includes colorectal clinical notes of the THYME (temporal history of your medical events) corpus of Mayo Clinic data.12 The third dataset (progress) consists of MIMIC-III progress notes derived from providers across different specialty intensive care units13–15 We created classification instances for each dataset by extracting sections from all the notes. While all 3 datasets had available section label annotations, the section labels were different across datasets.
The progress dataset already contains mappings from section labels to SOAP.14 To facilitate cross-domain experiments and following the SOAP definition guideline,8 an expert physician informaticist (M.A.) mapped the thyme and discharge datasets’ section labels into SOAP labels.9,16 The expert was first provided with the section names, and for section names that could not be directly mapped, instances of sections were provided for additional context. The sections that did not fit into the SOAP (e.g., “Comments,” “Administrative”) were labeled as “Others.” This created a 5-way classification instance for each section. The complete list of section names and their SOAP categories are included in Supplementary Appendices.
Table 1 presents the size, average word count, label distribution, and train/test split ratio for each dataset. During SOAP mapping, we observed that the “Assessment and Plan” in the progress dataset covered both “Assessment” and “Plan” contents, and were not easily separable. We mapped such sections to the “Assessment” label. As a result, the progress dataset has a not applicable (N/A) for the “Plan” category in Table 1. When splitting the dataset into training and test set, for discharge, we randomly split the dataset with a 0.8/0.2 ratio. For thyme and progress, we followed the original train/test splits.12,14
Table 1.
Size, average section word count (with standard deviation), and label distribution of the discharge, thyme, and progress dataset.
| Dataset | Total section counts | Average word count | “Subjective” section count | “Objective” section count | “Assessment” section count | “Plan” section count | “Others” section count | Train/test split |
|---|---|---|---|---|---|---|---|---|
| Discharge | 1372 | 61 ± 112 | 376 (27.4%) | 628 (45.8%) | 243 (17.7%) | 103 (7.5%) | 22 (1.6%) | 0.8/0.2 |
| Thyme | 4223 | 74 ± 121 | 1878 (44.5%) | 1329 (31.5%) | 676 (16.0%) | 100 (2.4%) | 240 (5.7%) | 0.73/0.27 |
| Progress | 13 367 | 46 ± 97 | 4521 (33.8%) | 7039 (52.7%) | 787a (5.9%) | N/A | 1020 (7.6%) | 0.89/0.11 |
Assessment and plan combined.
Table 1 also suggests some potential challenge of transferring SOAP classifiers between domains, as the distribution of SOAP categories drastically differs. Although “Subjective” and “Objective” are always the 2 most prevalent categories, discharge and progress have “Objective” being the largest count while thyme has “Subjective” being the largest.
In-domain section classification
We used the pretrained transformer framework for section classification. We fine-tuned BioBERT17 for the thyme, discharge, and progress datasets. We used BioBERT as the BERT implementation because BioBERT was pretrained using biomedical texts and performed better than BERT on a variety of biomedical NLP tasks, including named entity recognition, relation extraction and question answering.17 BioBERT also performs well in medical concept/entity recognition18,19 and bleeding event relation extraction.20 Other domain-appropriate BERT variants (e.g., BioClinicalBERT21) are already pretrained on MIMIC-III, the source of our progress dataset, so we avoid those models for the initial fine-tuning experiments to avoid data leakage. The use of BioBERT as a baseline model, instead of other models such as BEHRT22 and GatorTron,23 also allows for making BioClinicalBERT an off-the-shelf DAPT version of BioBERT (see future sections for details).
We first measured the in-domain classification performance for the 3 datasets. These performance values represented the upper bounds for our subsequent experiments. We fine-tuned a model on the training set and applied the model to the same dataset’s test set. In the domain adaptation literature, the source domain refers to the domain of the dataset used for model training, and the target domain to the domain of the dataset used for model testing. The source and target domain are the same for in-domain experiments, so we denote the in-domain experiments as FTtarget, indicating that we fine-tuned directly on target data.
When fine-tuning BERT, we used a learning rate of 1e-5, epoch size of 40 and batch size of 10. These hyperparameters were tuned using the training set. The best model during model training (determined by the best F1 score on the held-out validation set) was saved and used for testing. We report a single run with the best model rather than averaging, which has the tradeoff of showing realistic amounts of noise due to small numbers of instances, while not giving a stable estimate of the expected change in performance. The micro-F1 score (referred to as F1 score in future sections) was used as the evaluation metric because SOAP categories in this study are highly imbalanced both within and across datasets. The micro-F1 score helps provide high-level insights given that we are comparing across multiple datasets and experimental settings. We implemented the Huggingface Transformers pipeline with AdamW optimizer for fine-tuning.24 Experiments in this study were done on a 24GB NVIDIA TITAN RTX GPU with FP16 precision.
Cross-domain section classification
We then measured the cross-domain classification performance for the 3 datasets. We measured the cross-domain classification performance by testing the fine-tuned model on the other 2 datasets’ test sets. For example, when we trained a model on discharge, we tested it on thyme and progress. We denote these experiments as FTsource because the models were fine-tuned on a source domain and tested on a different domain. We use the same model hyperparameter settings as the in-domain experiments, to simulate the realistic case where target domain resources are too limited to conduct hyperparameter search.
Cross-domain section classification with continued pretraining
Recent work has provided evidence that continued pretraining of pretrained language models on a target domain allows for better adaptability of the model.11 Domain-adaptive pretraining is an unsupervised domain adaptation technique where a pretrained model is trained for additional steps, using the same pretraining task of masked language modeling objective, on a large collection of unlabeled data from the target domain. Task-adaptive pretraining is similar, but uses a smaller amount of target domain data—only that portion that was labeled for the task of interest. For example, for the progress dataset, the DAPT used the entire MIMIC-III dataset, and the TAPT considered the training set of progress. In previous work on general domain datasets,11 both DAPT and TAPT improved better cross-domain performance, and combining them sequentially (i.e., DAPT + TAPT) obtained the best performance. We thus experimented with pretrained transformer models that have been adapted either with DAPT or DAPT + TAPT. In these experiments, the DAPT, TAPT, or DAPT + TAPT training is done on top of a base language model (BioBERT), followed by fine-tuning BERT on labeled examples in a source and/or target domain (as in the FTsource experiments in the last section). We denote these experiments as DAPT + FTsource and DAPT + TAPT + FTsource in the remainder of the article.
We note that existing work in the clinical domain could be interpreted as DAPT. For example, BioClinicalBERT21 was created by doing continued pretraining on MIMIC-III13 using BioBERT17 as a starting point. From the perspective of downstream tasks that use MIMIC-III as a target domain (e.g., the progress dataset), comparing a BioBERT that has been fine-tuned on a source domain to BioClinicalBERT that has been fine-tuned on a source domain is essentially testing DAPT. Since BioClinicalBERT has already been shown to perform well on multiple tasks, in this work, we use the existing BioClinicalBERT checkpoint as our DAPT model when progress is the target domain. When thyme is the target domain, we used an unreleased section of additional unlabeled notes for the patients in the THYME labeled corpus12 to perform the continued pretraining for DAPT. For discharge, no additional unlabeled data are available. As a proxy, we again used MIMIC-III and used BioClinicalBERT as the DAPT model for progress.
In DAPT pretraining for thyme, we followed the setup of the BioClinicalBERT paper21 and used a maximum training step count of 15 000 and a learning rate of 5e-5. For TAPT, we followed the continued pretraining paper11 and trained the model on the labeled data from the target domain (with the masked language modeling task, so it is still unsupervised) for 100 epochs with other settings being the same.
Our TAPT experiments used only the training splits of the discharge, progress, and thyme datasets.
To summarize our experimental settings, Table 2 presents the configuration details of experiments for when the thyme dataset is the target domain. The corresponding tables for discharge and progress datasets are included in Supplementary Appendices.
Table 2.
Summarization of experiment configurations with thyme being the target domain.
| Method | Experiment | Source domain | Target domain | Number of target domain labeled samples added to fine-tuning | DAPT corpus | TAPT corpus |
|---|---|---|---|---|---|---|
| In-domain section classification | FTtarget | Thyme | Thyme | All | Unlabeled notes in THYME corpus | Thyme training set |
| Cross-domain section classification | FTsource | Discharge or progress | 0 | |||
| Cross-domain section classification with continued pretraining | DAPT + FTsource | |||||
| DAPT + TAPT + FTsource | ||||||
| Cross-domain section classification with continued pretraining and target domain labeled data | FTsource + target | 10, 20, 30, 40, 50 | ||||
| DAPT + FTsource + target | ||||||
| DAPT + TAPT + FTsource + target | ||||||
| Quantifying the value of domain adaptation methods | FTtarget | – | 10, 20, …, 190, 200 | – | – |
Cross-domain section classification with continued pretraining and target domain labeled data
In the DAPT and DAPT + TAPT experiments, we used only the source domain data for BERT fine-tuning, simulating the realistic setting where no annotation is possible at the target site (i.e., unsupervised domain adaptation). We next performed experiments that simulate the possibility that a small amount of labeled data is available at the target site, by including small numbers of labeled samples from the target domain during BERT fine-tuning (i.e., supervised domain adaptation). We also explore how the addition of labeled target domain data interacts with DAPT and TAPT. We varied the number of target domain samples from 10, 20, 30, 40 to 50. We denote these experiments as FTsource + target, DAPT + FTsource + target, and DAPT + TAPT + FTsource + target.
Quantifying the value of domain adaptation methods
Annotation can be expensive, and domain adaptation methods can reduce annotation costs. However, the exact relationship between the domain adaption method and the number of annotations it can save is unknown for section classification. We next take a step to address this question.
We do this by estimating the number of in-domain annotations needed for an in-domain model to achieve the F1 score of the domain adapted model. For example, if a cross-domain model with 10 target domain samples (FTsource + target) can achieve an F1 score of 0.66, and that F1 score will take an in-domain model (FTtarget) 61 samples to achieve, we can say the domain adaption method (transfer learning) saved 51 annotations.
To do this, we created a function that estimates the number of in-domain samples needed for FTtarget to achieve a specific F1 score, using a measure-then-interpolate approach. We measured the F1 score of FTtarget (i.e., in-domain training) when given 10, 20, … 200 (with an interval of 10) in-domain annotated samples. With the measured F1 scores from this setup we fit a power law function, , over these data points, with nt being the target domain annotated sample size. The power law function has previously been used for predicting model performance from sample size.25–27 We invert that function to obtain a function that, given an F1 score, can estimate . We used this function to convert the F1 score of multiple domain adapted models to the equivalent FTtarget sample sizes, rounding to the closest integer. To improve the generalizability of reporting, the results of this analysis are all based on the 3 datasets’ average.
Results
In-domain section classification
In Table 3, the shaded cells of the FT columns show the results of the in-domain section classification experiments, with F1 scores greater than 0.95 for all 3 datasets. The high accuracy of in-domain section classification findings are consistent with other studies such as Tepper et al.5
Table 3.
Overall F1 scores of in-domain and cross-domain models, with DAPT and TAPT when applicable.
| Source domain (→) |
Discharge |
Thyme |
Progress |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Target domain (↓) | FT | DAPT + FT | DAPT + TAPT + FT | FT | DAPT + FT | DAPT + TAPT + FT | FT | DAPT + FT | DAPT + TAPT + FT |
| Discharge | 0.972 | – | – | 0.572 | 0.6 | 0.675 | 0.541 | 0.5 | 0.501 |
| Thyme | 0.601 | 0.469 | 0.53 | 0.99 | – | – | 0.646 | 0.632 | 0.544 |
| Progress | 0.656 | 0.67 | 0.749 | 0.717 | 0.58 | 0.528 | 0.973 | – | – |
The best F1 score for each combination of source and target domain is in bold.
Cross-domain section classification
In Table 3, the unshaded cells in the FT columns show the results of the cross-domain section classification experiments, with significantly worse performance than the in-domain setting. When moving from in-domain to cross-domain, the F1 scores dropped from 0.97-0.99 to 0.541-0.717 range. The average in-domain (FTtarget) F1 score is 0.977 while the average cross-domain (FTsource) F1 score is 0.618.
Cross-domain section classification with continued pretraining
Table 3 also shows that continued pretraining (DAPT + FT and DAPT + TAPT + FT columns) led to decreased performance when thyme was the target domain. The effect of continued pretraining was mixed for progress and discharge. No significant performance improvement was observed when continued pretraining (DAPT or DAPT + TAPT) was applied directly on cross-domain section classification.
Cross-domain section classification with continued pretraining and target domain labeled data
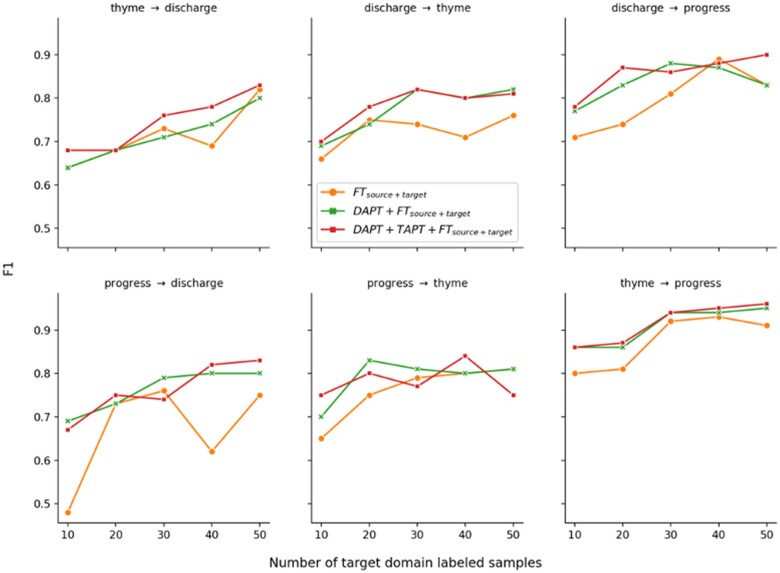
Figure 1 shows learning curves when some target-domain labeled data were provided for fine-tuning. When comparing before and after continued pretraining (DAPT or DAPT + TAPT), we found continued pretraining generally improved model performance when combined with small numbers of target domain instances.
Figure 1.
F1 scores of FTsource + target, DAPT + FTsource + target, and DAPT + TAPT + FTsource + target with 10, 20, 30, 40, and 50 target domain samples for different source and target domain experiments. For example, thyme → discharge represents the experiment with thyme being the source domain and discharge being the target domain.
The F1 scores of FTsource + target exhibit substantial noise when the source domain is progress and the target domain is discharge (Figure 1, bottom left). This is likely due to noise that can occur with small amounts of labeled target data, especially given the large difference in section count and section ratio between the 2 datasets. In discharge, almost all of its section categories are 10 times fewer than that of progress. In terms of section ratio, “Assessment” accounts for 17.7% sections in discharge, but only 5.9% in progress. “Others” accounts only for 1.6% sections in discharge, but 7.6% for progress.
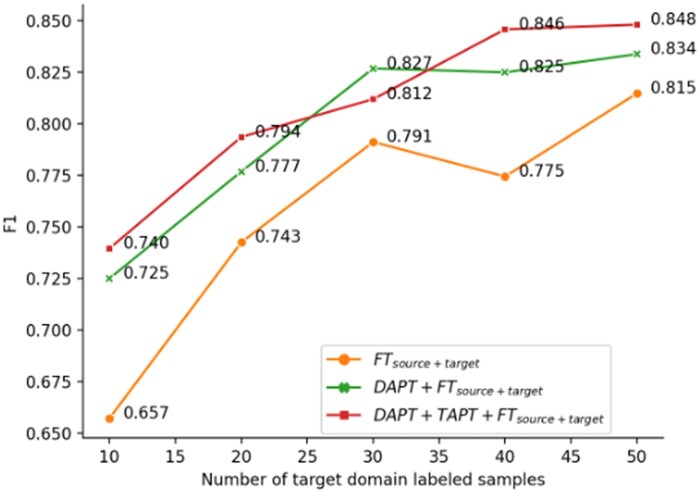
Figure 2 shows the F1 score curves (from Figure 1) averaged over the 6 source and target domain dataset combinations. FTsource + target has a drop at sample size 40, which can be a result of the more fluctuating progress → discharge model in Figure 1. On average, continued pretraining (DAPT or DAPT + TAPT) improved over the model without it (FTsource + target) consistently. When comparing within continued pretraining models (DAPT + FTsource + target and DAPT + TAPT + FTsource + target), we found applying TAPT after DAPT further increased the F1 score for 4 out of 5 sample sizes. DAPT + TAPT on averaged increased the F1 score from 0.756 to 0.808 (+0.052).
Figure 2.
Dataset averaged F1 scores of FTsource + target, DAPT + FTsource + target, and DAPT + TAPT + FTsource + target with 10, 20, 30, 40, and 50 target domain samples included in fine-tuning.
Quantifying the value of domain adaptation methods
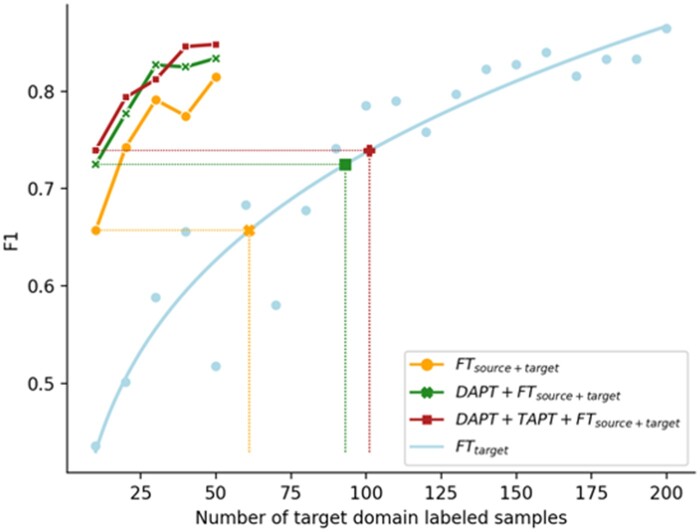
Figure 3 shows the F1 scores (averaged over datasets) of the in-domain model (FTtarget) with sample size varying from 10 to 200, and the power law curve. The results in Figure 2 were overlaid on Figure 3 to help illustrate the quantification process. With nt being the target domain annotated sample size, the fit power law function is, , and the corresponding inverse function is, .
Figure 3.
Dataset averaged F1 scores of FTtarget when trained with 10-200 samples. Results from Figure 2 are overlaid for illustration.
By applying the function to the domain adapted models’ F1 scores, we can convert them to the training sample size needed for FTtarget to achieve the same F1. We can therefore quantify domain-adapted models in the unit of target domain annotations. As an example and visualized in Figure 3 (full visualization included in Supplementary Appendices), FTsource + target, when trained with 10 target domain samples, achieved an F1 score equivalent to an in-domain model (FTtarget) trained using 61 target domain samples. Similarly, the equivalent target domain sample size is 93 for DAPT + FTsource + target and 101 for TAPT + DAPT + FTsource + target. The complete result is included in Table 4. We observed that, compared to FTtarget, FTsource + target on average saved 85.2 annotations, DAPT + FTsource + target saved 112.4, and DAPT + TAPT + FTsource + target saved 120.2. Continued pretraining (DAPT + TAPT) saved 120.2 − 85.2 = 35 annotations.
Table 4.
The number of samples FTtarget needs to reach to the same F1 score as a domain adapted model.
| Domain adaptation method (→) Target domain (↓) labeled sample size |
FTsource + target | DAPT + FTsource + target | DAPT + TAPT + FTsource + target |
|---|---|---|---|
| 10 | 61 (51) | 93 (83) | 101 (91) |
| 20 | 103 (83) | 125 (105) | 137 (117) |
| 30 | 135 (105) | 163 (133) | 151 (121) |
| 40 | 124 (84) | 162 (122) | 180 (140) |
| 50 | 153 (103) | 169 (119) | 182 (132) |
| Average | 115.2 (85.2) | 142.4 (112.4) | 150.2 (120.2) |
The F1 scores of domain adapted models varied with the number of in-domain samples provided (10-50). Numbers in parenthesis show the number of annotations saved by the domain adaptation methods.
Discussion
Challenges of transferring SOAP section classification
Our results show that, while SOAP section classification is a straightforward task for humans, and one that can be effectively solved for individual datasets, current state of the art supervised methods did not solve the task in a generalizable way. Part of the challenge may be attributable to different institutions having different documentation practices by providers, different note types in the EHR, and changes in label distribution. Many tasks are not adequately tested in out-of-sample environments across different domains and we provided a rigorous approach across multiple centers and note types to show that even “simple” tasks are difficult to generalize. The results also follow a similar finding in a finer-grained version of the task,5 as well as other clinical NLP tasks,28 but is perhaps more surprising here due to the relative simplicity of the task and the degree to which it is solved within each dataset. The attempts to leverage pretrained language models, and multiple fine-tuning and continual training approaches, still did not completely overcome the cross-domain challenges.
Transferability difference by SOAP category
To understand which SOAP categories are more transferable, we analyzed the results from the “Cross-domain section classification” section by SOAP categories, averaged across the 3 datasets. The by-category F1 scores are 0.68 for “Subjective,” 0.73 for “Objective,” 0.15 for “Assessment,” 0.13 for “Plan,” and 0.05 for “Others.”
“Subjective” and “Objective” are the 2 most prevalent categories, and are more transferable than the others. The 2 combined always account for more than 70% sections. In comparison, the other 3 categories are rare and their prevalence differ greatly between datasets. For example, the prevalence of “Plan” is 7.5% for discharge, 2.4% for thyme, and N/A for progress.
We also noted that “Subjective” and “Objective” categories usually have more homogenous fine-grained section names across datasets. Section names like “Chief complaint,” “Social History,” and “Vital Sign” are common across datasets. In contrast, the other 3 categories often have section names unique to certain datasets, such as “Special Instructions” which only occurs in thyme’s “Plan.”
Benefits of domain adaptation methods
The experiments between different combinations of training sets and training methods highlight trade-offs between different ways of mitigating the performance drop-offs when crossing domains. Unsupervised adaptation methods like DAPT and TAPT show benefits that are equivalent to dozens of target-domain training samples, but only when some target samples are already annotated. We also noted minimal performance gain from TAPT over DAPT, unlike prior work.11 The small benefit from TAPT could be due to the fact that transfer learning already brought knowledge to the model in a similar form as pretraining. One important direction moving forward is to regularly report quantification of this type of information across tasks so that different NLP tasks can be situated amongst each other in terms of the relative benefit they receive from unsupervised adaptation versus labeling additional instances.
The modest performance gain of continued pretraining in this study is similar to other papers that reported domain adaptation methods in clinical NLP tasks. In a study, researchers developed BioClincalBERT21 by continued pretraining BioBERT17 on clinical notes and reported that the performance gain is usually less than 2%. Similarly, in a study experimenting continued pretraining of T5 on clinical notes, only 2%-4% performance gain was observed.29 In a study on negation classification,28 it was found that even with a supervised domain adaption method, the performance gain is minimal.
The value of unsupervised domain adaptation of pretrained transformers when paired with small amounts of in-domain data is an encouraging result of this work. We caution, however, that it does not tell a complete story. Target domain annotation and continued pretraining, our 2 adaptation methods, both can be challenging and require resources at a target site. So, while the improvements of DAPT and TAPT are large in some cases, for this task they do seem to require some small amount of target-domain labeling. It could be the case that annotating a few hundred more instances is actually a more efficient decision than setting up continued pretraining infrastructure. In summary, even for the straightforward SOAP section classification task, these questions around adapting NLP systems are complex.
Limitations and future work
Each of the individual datasets we used were derived from single centers, which may be a contributing factor to the lack of generalizability. Future work in this task should explore the benefits of incorporating more variability in the types of notes and health systems used as source training data, to see whether combinations of datasets generalize better.
Future work should also extend to the segmentation version of the task, to see whether the same conclusions apply in that setting. Finally, future work should study whether the same findings may also be applicable to the more fine-grained section classification task, where the problem is more challenging due to lack of label standardization and sparsity of different section labels.
Conclusion
Our primary conclusion is that SOAP section classification is challenging in the cross-domain setting, even despite recent advances in modeling and the simplification of the task from full section classification. Our experiments with domain adaptation showed that straightforward unsupervised methods were not helpful on their own, but when combined with small amounts of supervision in the target domain had a larger impact.
Supplementary Material
Contributor Information
Weipeng Zhou, Department of Biomedical Informatics and Medical Education, School of Medicine, University of Washington-Seattle, Seattle, WA, United States.
Meliha Yetisgen, Department of Biomedical Informatics and Medical Education, School of Medicine, University of Washington-Seattle, Seattle, WA, United States.
Majid Afshar, Department of Medicine, School of Medicine and Public Health, University of Wisconsin-Madison, Madison, WI, United States.
Yanjun Gao, Department of Medicine, School of Medicine and Public Health, University of Wisconsin-Madison, Madison, WI, United States.
Guergana Savova, Computational Health Informatics Program, Boston Children’s Hospital, Department of Pediatrics, Harvard Medical School, Boston, MA, United States.
Timothy A Miller, Computational Health Informatics Program, Boston Children’s Hospital, Department of Pediatrics, Harvard Medical School, Boston, MA, United States.
Author contributions
W.Z. and T.M. designed the study, with feedback from all coauthors. W.Z. carried out all of the experiments in the study under the guidance of T.M. M.A. created the mappings from section labels to SOAP categories. M.Y., M.A., Y.G., and G.S. contributed to understanding the datasets and participated in article editing.
Supplementary material
Supplementary material is available at Journal of the American Medical Informatics Association online.
Funding
This work was supported by the National Library of Medicine of the National Institutes of Health under Award Number R01LM012973, R01LM012918, and R01LM013486. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of Interest
None declared.
Data availability
The labeled datasets used in this manuscript were obtained from external resources, and data use agreements prevent us from redistributing them. The progress dataset is available at https://physionet.org/content/task-1-3-soap-note-tag/1.0.0/. The thyme dataset is available at http://center.healthnlp.org/. The discharge dataset text can be found at https://portal.dbmi.hms.harvard.edu/projects/n2c2-nlp/, and the section annotations can be obtained at a GitHub repository that we have created: https://github.com/2533245542/SectionChunker_i2b2_2010_data. We also used 2 unlabeled datasets. MIMIC-III can be obtained at: https://physionet.org/content/mimiciii/1.4/. The THYME unlabeled dataset cannot be redistributed because this data was made available to Boston Children’s Hospital researchers with a Data Use Agreement that does not allow redistribution.
References
- 1. Lei J, Tang B, Lu X, Gao K, Jiang M, Xu H.. A comprehensive study of named entity recognition in Chinese clinical text. J Am Med Inform Assoc. 2014;21(5):808-814. 10.1136/amiajnl-2013-002381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zweigenbaum P, Deléger L, Lavergne T, Névéol A, Bodnari A. A supervised abbreviation resolution system for medical text. Presented at the Conference and Labs of the Evaluation Forum, 2013. Accessed February 20, 2023. https://www.semanticscholar.org/paper/A-Supervised-Abbreviation-Resolution-System-for-Zweigenbaum-Del%C3%A9ger/b3ba1306d0afb9f69412df1ca35ee1c49cf27a13
- 3. Edinger T, Demner-Fushman D, Cohen AM, Bedrick S, Hersh W.. Evaluation of clinical text segmentation to facilitate cohort retrieval. AMIA Annu Symp Proc. 2018;2017:660-669. [PMC free article] [PubMed] [Google Scholar]
- 4. Kropf S, Krücken P, Mueller W, Denecke K.. Structuring legacy pathology reports by openEHR archetypes to enable semantic querying. Methods Inf Med. 2017;56(3):230-237. 10.3414/ME16-01-0073 [DOI] [PubMed] [Google Scholar]
- 5. Tepper M, Capurro D, Xia F, Vanderwende L, Yetisgen-Yildiz M. Statistical section segmentation in free-text clinical records. In Proceedings of the Eighth International Conference on Language Resources and Evaluation (LREC’12), Istanbul, Turkey: European Language Resources Association (ELRA), May 2012, pp. 2001-2008. Accessed October 31, 2022. http://www.lrec-conf.org/proceedings/lrec2012/pdf/1016_Paper.pdf [Google Scholar]
- 6. Rosenthal S, Barker K, Liang Z. Leveraging medical literature for section prediction in electronic health records. In Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing (EMNLP-IJCNLP), Hong Kong, China: Association for Computational Linguistics, November 2019, pp. 4864–4873. 10.18653/v1/D19-1492 [DOI]
- 7. Devlin J, Chang M-W, Lee K, Toutanova K. BERT: pre-training of deep bidirectional transformers for language understanding. In Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long and Short Papers), Minneapolis, Minnesota: Association for Computational Linguistics, June 2019, pp. 4171–4186. 10.18653/v1/N19-1423 [DOI]
- 8. Podder V, Lew V, Ghassemzadeh S.. SOAP Notes in StatPearls. StatPearls Publishing; 2022. Accessed January 4, 2023. http://www.ncbi.nlm.nih.gov/books/NBK482263/ [PubMed] [Google Scholar]
- 9. Mowery D, Wiebe J, Visweswaran S, Harkema H, Chapman WW.. Building an automated SOAP classifier for emergency department reports. J Biomed Inform. 2012;45(1):71-81. 10.1016/j.jbi.2011.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wright A, Sittig DF, McGowan J, Ash JS, Weed LL.. Bringing science to medicine: an interview with Larry Weed, inventor of the problem-oriented medical record. J Am Med Inform Assoc. 2014;21(6):964-968. 10.1136/amiajnl-2014-002776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gururangan S, Marasović A, Swayamdipta S, et al. Don’t stop pretraining: adapt language models to domains and tasks. In Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics, Seattle, WA: Association for Computational Linguistics, July 2020, pp. 8342–8360. 10.18653/v1/2020.acl-main.740 [DOI]
- 12. Styler WF IV, Bethard S, Finan S, et al. Temporal annotation in the clinical domain. Trans Assoc Comput Linguist. 2014;2:143–154. 10.1162/tacl_a_00172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Johnson AEW, Pollard TJ, Shen L, et al. MIMIC-III, a freely accessible critical care database. Sci Data. 2016;3(1):160035. 10.1038/sdata.2016.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gao Y, Dligach D, Miller T, et al. Hierarchical annotation for building a suite of clinical natural language processing tasks: progress note understanding. In Proceedings of the Thirteenth Language Resources and Evaluation Conference, Marseille, France: European Language Resources Association, June 2022, pp. 5184-5493. 10.48550/arXiv.2204.03035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gao Y, Caskey J, Miller T, et al. Tasks 1 and 3 from progress note understanding suite of tasks: SOAP note tagging and problem list summarization (version 1.0.0). PhysioNet. 2022. 10.13026/WKS0-W041 [DOI] [Google Scholar]
- 16. Häyrinen K, Saranto K, Nykänen P.. Definition, structure, content, use and impacts of electronic health records: A review of the research literature. Int J Med Inform. 2008;77(5):291-304. 10.1016/j.ijmedinf.2007.09.001 [DOI] [PubMed] [Google Scholar]
- 17. Lee J, Yoon W, Kim S, et al. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics. 2020;36(4):1234-1240. 10.1093/bioinformatics/btz682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yu X, Hu W, Lu S, Sun X, Yuan Z. BioBERT based named entity recognition in electronic medical record. In 2019 10th International Conference on Information Technology in Medicine and Education (ITME), Qingdao, China: IEEE, August 2019, pp. 49–52. 10.1109/ITME.2019.00022 [DOI]
- 19. Turchin A, Masharsky S, Zitnik M.. Comparison of BERT implementations for natural language processing of narrative medical documents. Inform Med Unlocked. 2023;36:101139. 10.1016/j.imu.2022.101139 [DOI] [Google Scholar]
- 20. Mitra A, Rawat BPS, McManus DD, Yu H.. Relation classification for bleeding events from electronic health records using deep learning systems: an empirical study. JMIR Med Inform. 2021;9(7):e27527. 10.2196/27527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Alsentzer E, et al. Publicly available clinical BERT embeddings. In Proceedings of the 2nd Clinical Natural Language Processing Workshop, Minneapolis, MN: Association for Computational Linguistics, June 2019, pp. 72–78. 10.18653/v1/W19-1909 [DOI]
- 22. Li Y, Rao S, Solares JRA, et al. BEHRT: transformer for electronic health records. Sci Rep. 2020;10(1):7155. 10.1038/s41598-020-62922-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yang X, Chen A, PourNejatian N, et al. A large language model for electronic health records. NPJ Digit Med. 2022;5(1):194. 10.1038/s41746-022-00742-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wolf T, et al. 2020. HuggingFace’s transformers: state-of-the-art natural language processing, arXiv, July 13, preprint: not peer reviewed. 10.48550/arXiv.1910.03771 [DOI]
- 25. Figueroa RL, Zeng-Treitler Q, Kandula S, Ngo LH.. Predicting sample size required for classification performance. BMC Med Inform Decis Mak. 2012;12(1):8. 10.1186/1472-6947-12-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Partin A, Brettin T, Evrard YA, et al. Learning curves for drug response prediction in cancer cell lines. BMC Bioinformatics. 2021;22(1):252. 10.1186/s12859-021-04163-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Larracy R, Phinyomark A, Scheme E.. Machine learning model validation for early stage studies with small sample sizes. Ann Int Conf IEEE Eng Med Biol Soc. 2021;2021:2314-2319. 10.1109/EMBC46164.2021.9629697 [DOI] [PubMed] [Google Scholar]
- 28. Wu S, Miller T, Masanz J, et al. Negation’s not solved: generalizability versus optimizability in clinical natural language processing. PLoS One. 2014;9(11):e112774. 10.1371/journal.pone.0112774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lehman E, et al. 2023. Do we still need clinical language models?, arXiv. Feb.16, preprint: not peer reviewed. 10.48550/arXiv.2302.08091 [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The labeled datasets used in this manuscript were obtained from external resources, and data use agreements prevent us from redistributing them. The progress dataset is available at https://physionet.org/content/task-1-3-soap-note-tag/1.0.0/. The thyme dataset is available at http://center.healthnlp.org/. The discharge dataset text can be found at https://portal.dbmi.hms.harvard.edu/projects/n2c2-nlp/, and the section annotations can be obtained at a GitHub repository that we have created: https://github.com/2533245542/SectionChunker_i2b2_2010_data. We also used 2 unlabeled datasets. MIMIC-III can be obtained at: https://physionet.org/content/mimiciii/1.4/. The THYME unlabeled dataset cannot be redistributed because this data was made available to Boston Children’s Hospital researchers with a Data Use Agreement that does not allow redistribution.