Abstract
The species Pseudomonas syringae encompasses plant pathogens with differing host specificities and corresponding pathovar designations. P. syringae requires the Hrp (type III protein secretion) system, encoded by a 25-kb cluster of hrp and hrc genes, in order to elicit the hypersensitive response (HR) in nonhosts or to be pathogenic in hosts. DNA sequence analysis of the hrpC and hrpRS operons of P. syringae pv. syringae 61 (brown spot of beans), P. syringae pv. glycinea U1 (bacterial blight of soybeans), and P. syringae pv. tomato DC3000 (bacterial speck of tomatos) revealed that the 13 genes comprising the right half of the hrp cluster (including those in the previously sequenced hrpZ operon) are conserved and identically arranged. The hrpC operon is comprised of hrpF, hrpG, hrcC, hrpT, and hrpV. hrcC encodes a putative outer membrane protein that is conserved in all type III secretion systems. The other four genes appear to be characteristic of group I Hrp systems, such as those possessed by P. syringae and Erwinia amylovora. The predicted products of these four genes in P. syringae pv. syringae 61 are HrpF (8 kDa), HrpG (15.4 kDa), HrpT (7.5 kDa), and HrpV (13.4 kDa). HrpT is a putative outer membrane lipoprotein. HrpF, HrpG, and HrpV are all hydrophilic proteins lacking N-terminal signal peptides. The HrpG, HrcC, HrpT, and HrpV proteins of P. syringae pathovars syringae and tomato (the two most divergent pathovars) had at least 76% amino acid identity with each other, whereas the HrpF proteins of these two pathovars had only 36% amino acid identity. The HrpF proteins of P. syringae pathovars syringae and glycinea also showed significant similarity to the HrpA pilin protein of P. syringae pathovar tomato. Functionally nonpolar mutations were introduced into each of the genes in the hrpC operon of P. syringae pv. syringae 61 by insertion of an nptII cartridge lacking a transcription terminator. The mutants were assayed for their ability to elicit the HR in nonhost tobacco leaves or to multiply and cause disease in host bean leaves. Mutations in hrpF, hrcC, and hrpT abolished or greatly reduced the ability of P. syringae pv. syringae 61 to elicit the HR in tobacco. The hrpG mutant had only weakly reduced HR activity, and the activity of the hrpV mutant was indistinguishable from that of the wild type. Each of the mutations could be complemented, but surprisingly, the hrpV subclone caused a reduction in the HR elicitation ability of the ΔhrpV::nptII mutant. The hrpF and hrcC mutants caused no disease in beans, whereas the hrpG, hrpT, and hrpV mutants had reduced virulence. Similarly, the hrcC mutant grew little in beans, whereas the other mutants grew to intermediate levels in comparison with the wild type. These results indicate that HrpC and HrpF have essential functions in the Hrp system, that HrpG and HrpT contribute quantitatively but are not essential, and that HrpV is a candidate negative regulator of the Hrp system.
Many gram-negative plant-pathogenic bacteria elicit a rapid, localized necrosis in infiltrated tissues of plants that are outside their host range. This defense-associated apparent programmed cell death is known as the hypersensitive response (HR) (34). The ability of these bacteria to elicit the HR in nonhost plants, or to be pathogenic in their hosts, is dependent on hrp genes, which may be universal in plant-pathogenic Pseudomonas, Xanthomonas, Erwinia, and Ralstonia spp. (3, 34). The hrp genes are clustered, and many encode components of a type III protein secretion system that appears to be dedicated to the secretion of virulence proteins in both plant and animal pathogens. Nine of the hrp genes have homologs in animal-pathogenic Yersinia, Shigella, and Salmonella spp., and these have been renamed hrc (for HR and conserved) (8).
The species Pseudomonas syringae is divided into pathovars largely on the basis of host specificity (42). hrp genes have been studied in the P. syringae pathovars syringae (brown spot of beans), phaseolicola (halo blight of beans), tomato (bacterial speck of tomatoes), and glycinea (bacterial blight of soybeans) (3, 10). The hrp cluster of P. syringae pv. syringae 61, cloned on cosmid pHIR11, has been studied most extensively because it has the useful property of conferring on nonpathogenic bacteria, such as Pseudomonas fluorescens and Escherichia coli, the ability to elicit the HR in tobacco and several other plants (25). pHIR11 contains four major operons (hrpJ, hrpU, hrpC, and hrpZ), which encode all of the type III pathway components, one harpin (HrpZ), and one pilus subunit (HrpA) (3, 21, 22, 24, 26, 36, 46, 51). This cluster also contains hrmA, which is an apparent avr (avirulence) gene (4), and several hrp genes of unknown function that either have no homologs or have homologs only in the closely related hrp cluster of Erwinia amylovora (9, 24, 32, 45). pHIR11 also carries three regulatory genes that encode the positive regulators hrpR and hrpS and the hrp-activating alternate ς factor HrpL (51).
The sequence of the hrpZ operon, which encodes HrpA, HrpZ, and secretion pathway components such as HrcJ, has been analyzed in P. syringae pv. syringae 61, P. syringae pv. tomato DC3000, and P. syringae pv. glycinea U1 (45). The comparison suggests that the arrangement of hrp genes is conserved among P. syringae pathovars and that HrpZ does not directly control host range. The actual role of HrpZ in elicitation of the HR or pathogenesis remains uncertain (1, 43), and a primary function of the Hrp system may be the delivery of Avr effector proteins directly into plant cells (3, 18). Whether any components of the Hrp system itself affect host specificity is not known.
Eight of the Hrc proteins show similarity to a group of proteins involved in flagellar basal body biogenesis and flagellum-specific secretion (3, 8). HrcC (formerly known as HrpH), the remaining Hrc protein, is a member of the PulD-pIV superfamily of secretins, which are outer membrane proteins involved in macromolecular traffic across the bacterial outer membrane (22, 47). HrcC has been shown to be required for both HrpZ secretion and the delivery of Avr signals (18, 21). hrcC is carried in the hrpC operon and is preceded by hrpF and hrpG, two small open reading frames (ORFs) of unknown function that were found as a byproduct of our previous analysis of the hrpZ operons of P. syringae pv. syringae 61, P. syringae pv. tomato DC3000, and P. syringae pv. glycinea U1 (24, 45). Recently, Kim et al. (32) reported the presence in E. amylovora of ORFs similar to hrpF and hrpG upstream of hrcC and two new ORFs, hrpT and hrpV, downstream of hrcC, and they confirmed the products of all four ORFs by T7 expression.
We have focused our analysis of hrp genes on three P. syringae strains: P. syringae pv. syringae 61 (the source of pHIR11), P. syringae pv. tomato DC3000 (a model pathogen of Arabidopsis spp. as well as the tomato), and P. syringae pv. glycinea U1 (a strain in race 4, which is used extensively in avr gene studies). Here we report two results. One is the sequence of the hrpC and hrpRS operons of P. syringae pv. syringae 61, P. syringae pv. tomato DC3000, and P. syringae pv. glycinea U1, which reveals the complete conservation of hrp gene arrangement in the right half of the hrp clusters of these three pathovars and the relative variation among sets of homologous genes. The other is the construction, complementation, and phenotypic analysis of functionally nonpolar mutations in hrpF, hrpG, hrcC, hrpT, and hrpV in P. syringae pv. syringae 61, which reveals that these genes differ significantly in their contributions to plant reaction phenotypes.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli was routinely grown in Luria-Bertani medium or Terrific broth at 37°C (6). Pseudomonas strains were routinely grown in King’s B (KB) medium (33) at 28 to 30°C, but for certain experiments the hrp-derepressing minimal medium containing fructose (28), adjusted to pH 5.5, was used. Antibiotics were used in selective media at the following concentrations (micrograms per milliliter): ampicillin, 100; kanamycin, 50; tetracycline, 20; and nalidixic acid, 20.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| E. coli | ||
| HB101 | F′ hsd20 recA13 thr leu thi pro Smr | 6 |
| DH10B | endA1 hsdR17 recA1 relA Δ(argF-lacZYA)U169 f80d lacZΔM15 | Life Sciences Technologies (Gaithersburg, Md.) |
| P. syringae pv. syringae 61 | Wild type isolated from wheat; Nalr | 25 |
| 61-N393 | Derivative carrying ΔhrcC::nptII nonpolar mutation | 12 |
| 61-N402 | Derivative carrying ΔhrpT::nptII nonpolar mutation | This study |
| 61-N407 | Derivative carrying ΔhrpV::nptII nonpolar mutation | This study |
| 61-N491 | Derivative carrying ΔhrpF::nptII nonpolar mutation | This study |
| 61-N492 | Derivative carrying hrpG::nptII nonpolar mutation | This study |
| P. syringae pv. tomato DC3000 | Wild type; Rpr | D. E. Cuppels |
| P. syringae pv. glycinea Race 4 U1 | Wild type | C. J. Baker |
| Plasmids | ||
| pRK2013 | IncP Tra RK2+ ΔrepE1+ Kanr | 14 |
| pBluescript KS or SK | ColE1 Apr mcs-lacZ | Stratagene |
| pT7-6 | Apr, φ10 T7 RNA polymerase promoter | 48 |
| pCPP30 | IncP LacZ′ Tcr | D. W. Bauer (Cornell University) |
| pCPP2988 | pBluescript II SK(−) carrying 1.5-kb HindIII-SalI fragment from pRZ102 with nptII lacking terminator | 1 |
| pRK415 | Broad-host-range vector unstable in absence of selection; Tcr | 30 |
| pHIR11 | 25-kb hrp cluster fragment from P. syringae pv. syringae 61 in pLAFR3 | 25 |
| pNCHU7 | 5.4-kb P. syringae pv. syringae 61 EcoRI-BamHI fragment in pCPP30; contains hrpRS-hrpD | This study |
| pNCHU162 | 3.6-kb P. syringae pv. syringae 61 SmaI-HindIII fragment in pBluescript SK(−); contains hrpB, hrcJ, and hrpDEFG genes | This study |
| pNCHU169 | 6-kb P. syringae pv. syringae 61 BglII-EcoRI fragment in pBluescript SK(−); contains hrcRSTU and hrpVT genes | This study |
| pNCHU316 | prs1-prs2-generated 2.1-kb BamHI-HindIII fragment subcloned in pET29a; contains P. syringae pv. syringae 61 hrcC with N-terminal S · Taga fusion | This study |
| pNCHU329 | 1.8-kb SalI fragment containing P. syringae pv. syringae 61 hrpG subcloned in pT7-6 | This study |
| pNCHU366 | 0.4-kb NcoI-EcoRI fragment subcloned in pET29a; contains P. syringae pv. syringae 61 hrcQB with N-terminal S · Tag fusion | C.-J. Chang |
| pNCHU421 | prs1-prs2-generated 2.1-kb BamHI-HindIII fragment subcloned in pRK415; contains P. syringae pv. syringae 61 hrcC | This study |
| pNCHU451 | 250-bp EcoRV-HindIII fragment from pNCHU407 subcloned in pRK415; contains P. syringae pv. syringae 61 hrpT | This study |
| pNCHU513 | 750-bp KpnI-EcoRI fragment from pNCHU491 subcloned in pRK415; contains P. syringae pv. syringae 61 hrpG | This study |
| pNCHU515 | 400-bp PvuII-HindIII fragment from pNCHU329 subcloned in pRK415; contains P. syringae pv. syringae 61 hrpF | This study |
| pCPP2107 | a 7.2-kb P. syringae pv. syringae 61 KpnI fragment from pCPP2145 subcloned in pBluescript KS(−); contains hrpB, hrcJ, hrpD, hrpE, and complete hrpC operon | 22 |
| pCPP2145 | 10.6-kb BglII fragment from pHIR11 subcloned in pCPP30; contains complete hrpZ and hrpC operons and hrcRSTU genes | 22 |
| pCPP2200 | pUCP19 carrying 10-kb Sau3A1 partial fragment of P. syringae pv. glycinea U1 DNA containing hrpRS-hrcU | 45 |
| pCPP2201 | pUCP19 carrying 10-kb fragment of P. syringae pv. tomato DNA containing hrpRS-hrpV | 45 |
| pCPP2372 | 0.3-kb fragment from prs10-prs11-generated PCR product in pRK415; contains P. syringae pv. syringae 61 hrpV for complementation | This study |
S · Tag, a peptide encoded by the sequences of pET29a (Novagen Inc., Madison, Wis.).
Recombinant DNA techniques.
Restriction endonuclease digestion, agarose gel electrophoresis, DNA fragment preparation, plasmid extraction, DNA ligation, and transformation by CaCl2 followed standard procedures (6). Plasmids were introduced into bacteria by transformation, electroporation (Gene Pulsar; Bio-Rad, Richmond, Calif.), or triparental mating (14).
DNA sequencing and analysis.
The hrcC operon of P. syringae pv. tomato DC3000, carried on plasmid pCPP2201 (45), and the hrpF, hrpG, hrpT, and hrpV genes of P. syringae pv. glycinea U1, carried on plasmid pCPP2200 (45), were sequenced with the ABI 373A DNA sequencer at the Cornell Biotechnology Center DNA-sequencing facility, with specific primers synthesized by Integrated DNA Technologies (Coralville, Iowa). Nucleotide and derived amino acid sequences were analyzed with the Genetics Computer Group sequence analysis software package (13) and DNAStar (DNAStar Inc., Madison, Wis.). Homology searches against major sequence databases were done with the BLAST program (5). BESTFIT alignments were considered significantly similar if the score determined with default parameters was at least five times the standard deviation above the mean quality score of 100 randomized alignments (13, 15).
Construction of functionally nonpolar mutations in the hrpC operon.
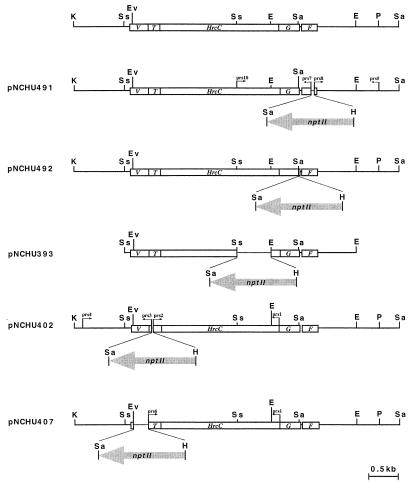
To create nonpolar mutations in the P. syringae pv. syringae 61 hrpC operon, a 1.5-kb nptII cassette lacking a rho-independent transcription terminator (1, 7) was used to disrupt hrpF, hrpG, hrcC, hrpT, and hrpV. The cassette marked deletions in hrpF, hrcC, hrpT, and hrpV and was used for insertional inactivation of hrpG. These recombinant constructions were cloned into vector pRK415 (30) (see Fig. 4). The DNA fragments used in the construction were amplified by PCR with Pfu polymerase (Stratagene, La Jolla, Calif.), and the corresponding primers are shown in Table 2. Inactivation of hrpF was achieved with two PCR-generated fragments by using pHIR11 as a template and prs5-prs8 and prs7-prs9 as primers. The amplified 1.1-kb DNA fragment of prs5 plus prs8 was treated with XbaI and BamHI and then cloned into pCPP2988 to generate a 2.6-kb XbaI-KpnI fragment. This 2.6-kb fragment was then subcloned into a pRK415 derivative, which had previously received the 1.3-kb prs7-prs9-generated fragment in the KpnI and SstI sites, to produce pNCHU491. To mutate hrpG, a 1.8-kb BamHI-HindIII fragment from pNCHU329 was cloned into pCPP2988 containing an insertion of the 4-kb SalI-KpnI fragment isolated from pCPP2107, and the total 7.3-kb fragment was subsequently cloned into pRK415 at the BamHI-KpnI sites to produce pNCHU492. The 5′ (ca. 2-kb)- and 3′ (ca. 1-kb)-flanking sequences of the hrpT gene were obtained from PCR-amplified DNA fragments by using prs1-prs2 and prs3-prs4, respectively, as primers. These DNA fragments were cloned into pCPP2988 at appropriate restriction sites. This construct resulted in a 34-bp deletion of hrpT that was replaced by the nptII gene. The 4.7-kb BamHI-KpnI fragment isolated as described above was cloned into pRK415 to produce pNCHU402. Primer prs1 was also used in the construction of the hrpV mutation. A 1,040-bp EcoRV-KpnI fragment and prs1-prs6-generated fragments were cloned in two steps into pCPP2988. The 4.7-kb BamHI-KpnI fragment containing nptII was subsequently ligated with pRK415 to produce pNCHU407. pNCHU393 (ΔhrcC::nptII), pNCHU402 (ΔhrpT::nptII), pNCHU407 (ΔhrpV::nptII), pNCHU491 (ΔhrpF::nptII), and pNCHU492 (hrpG::nptII) (see Fig. 4) were introduced into P. syringae pv. syringae 61 by triparental mating, using E. coli DH10B (carrying the constructed plasmids) as the donor and the helper strain E. coli HB101(pRK2013) (14). The mating mixtures were spotted on KB agar supplemented with nalidixic acid, tetracycline, and kanamycin at 30°C for 2 to 3 days. Cells from single transconjugant colonies were inoculated in 5 ml of KB broth supplemented with nalidixic acid and kanamycin. The bacteria were subcultured for 5 days, and then the final cultures were diluted and spread on KB agar plates containing nalidixic acid and kanamycin. Mutants were identified by screening on KB agar for kanamycin resistance and tetracycline sensitivity (23).
FIG. 4.
Construction of functionally nonpolar mutations in the P. syringae pv. syringae 61 hrpC operon. In each construction, the gray arrow represents the nptII cassette lacking a terminator and its orientation; the dotted line represents the internal deletion that is replaced by the nptII cassette. Primers involved with the PCR-amplified DNA fragments are indicated by small arrows. F, G, T, and V represent the hrpF, -G, -T, and -V genes, respectively. The restriction enzymes are abbreviated as follows: K, KpnI; E, EcoRI; Sa, SalI; EV, EcoRV; H, HindIII; P, PstI; Ss, SstI; Bg, BglII.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′-3′)a | Restriction enzymes |
|---|---|---|
| prs1 | 5′-TATGGGATCCATGCGCAAGGCCTTGATGTG-3′ | BamHI |
| prs2 | 5′-TAATAAGCTTCATCACGCATGTCCGGAGGCCA-3′ | HindIII |
| prs3 | 5′-ACTAGGTACCGTCGTGAAGT-3′ | KpnI |
| prs4 | 5′-GCGCGAATTCCATGCGAAAA-3′ | EcoRI |
| prs5 | 5′-CTGGATCTAGAGCAGCGCAT-3′ | XbaI |
| prs6 | 5′-TGAGAAGCTTCTCCTCGACCTCG-3′ | HindIII |
| prs7 | 5′-TGAGGTCGGTACCAACATCGC-3′ | KpnI |
| prs8 | 5′-ATTCGGATCCTCGATGTCGTCGGCGGT-3′ | BamHI |
| prs9 | 5′-AACGCATTGGTATCGAGG-3′ | |
| prs10 | 5′-ACATTCTAGATGATCGAGGTCGAG-3′ | XbaI |
| prs11 | 5′-TGCTGAATTCCTAAGCCAGATGA-3′ | EcoRI |
Restriction sites are indicated in boldface.
Complementation of mutations.
The hrpF, hrpG, hrcC, and hrpT genes from the P. syringae pv. syringae 61 hrpC operon were subcloned from available constructs (Table 1) or amplified by PCR as described above and were cloned individually into pRK415. The primers used are listed in Table 2. Resultant plasmids pNCHU515 (hrpFPss), pNCHU 513 (hrpGPss), pNCHU421 (hrcCPss), and pNCHU451 (hrpTPss) were then transformed into the corresponding mutants by triparental mating as described above.
Plant assays.
HR assays were performed in tobacco (Nicotiana tabacum L. cv. Xanthi) plants that were grown under greenhouse conditions at 23 to 25°C with a photoperiod of 16 to 24 h and transferred to the laboratory for the assays. Bacterial samples were prepared by suspending them in distilled water at a density of 108 to 109/ml. The cells were then grown for 24 h on KB agar plates. Inoculations were performed by pricking leaves with a dissecting needle and then infiltrating the bacterial suspension with a 1-ml syringe lacking a needle. The development of the HR at room temperature was scored within 24 h. Virulence assays were performed in bean (Phaseolus vulgaris cv. Eagle) plants that were grown under greenhouse conditions at 23 to 25°C with a photoperiod of 16 to 18 h. Bacteria were grown overnight on KB agar plates and suspended in 5 mM MES (morpholinoethanesulfonic acid), pH 5.5, at a density of 105 CFU/ml. Inoculations were performed by infiltration as described above. Plants were incubated at high humidity, and the appearance of disease symptoms was scored after 5 days. Multiplication assays were performed by grinding 0.6-cm-diameter leaf discs from infiltrated leaves in 1 ml of 5 mM MES (pH 5.5), followed by serial dilution and plating of the samples onto agar plates with 1 μg of cycloheximide/ml and appropriate antibiotics.
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this paper have been deposited in GenBank under accession no. AF051694 (P. syringae pv. syringae 61 hrpTV), AF061028 (P. syringae pv. tomato DC3000 hrpRS), AF061029 (P. syringae pv. tomato DC3000 hrpF to -V), AF069650 (P. syringae pv. glycinea U1 hrpRS), AF069651 (P. syringae pv. glycinea U1 hrpJ to -G), and AF069652 (P. syringae pv. glycinea U1 hrp TVU).
RESULTS
Sequence analysis of the hrpRS and hrpC operons of P. syringae pv. tomato DC3000 and P. syringae pv. glycinea U1 and the hrpT and hrpV genes of P. syringae pv. syringae 61.
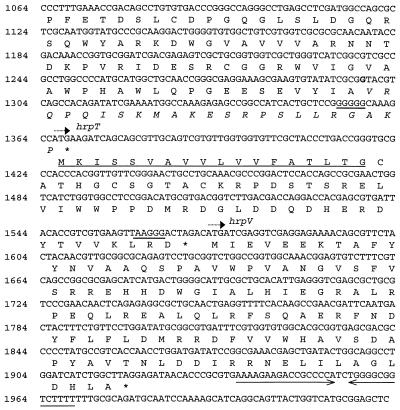
The complete sequence of the hrpRS operons of P. syringae pv. tomato DC3000 and P. syringae pv. glycinea U1, the hrpC operon of P. syringae pv. tomato DC3000, and portions of the hrpC operon of P. syringae pv. glycinea U1 were obtained by using a series of specific oligonucleotide primers to sequence pCPP2200 (P. syringae pv. glycinea U1) and pCPP2201 (P. syringae pv. tomato DC3000), each of which possesses an approximately 10-kb insert containing the hrpZ operon and flanking DNA (45). The sequenced DNA displayed the same organization as the equivalent region from P. syringae pv. syringae 61, as shown in Fig. 1, except for the hrpT and hrpV genes, which were not observed previously in the P. syringae pv. syringae 61 hrcC (hrpH) region (22). Upon resequencing this region in P. syringae pv. syringae 61, we discovered a missing nucleotide (G) (Fig. 2). The corrected hrcC sequence predicts a protein that is 47 amino acids smaller than that originally reported and has a different sequence for the last 23 amino acids, and the corrected hrcC is followed by the hrpT and hrpV genes.
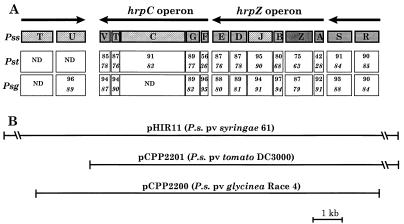
FIG. 1.
Organization of the right half of the hrp-hrc clusters of P. syringae pathovars syringae, tomato, and glycinea and primary clones used in this study. (A) Organization of the P. syringae hrp cluster from hrpRS to hrpT. Conserved type III secretion genes (hrc genes) are indicated by the crosshatched boxes, genes encoding proteins known to be secreted are shaded dark gray, and the remaining hrp genes are shown as light-gray boxes. The identity (italic) and similarity scores for the predicted proteins from P. syringae pv. tomato DC 3000 (Pst) and P. syringae pv. glycinea U1 (Psg), relative to homologous proteins in P. syringae pv. syringae 61 (Pss), are shown in the open boxes beneath the relevant genes, except for those not done (ND). (B) Primary clones are aligned beneath the operon maps. P.s., P. syringae.
FIG. 2.
Sequence of nucleotides 1064 to 2022 from the 2,022-bp SstI fragment containing the hrcC, hrpT, and hrpV genes of P. syringae pv. syringae 61. Potential ribosome-binding sites and the potential signal peptide of HrpT are underlined. The inverted repeat, marked by opposing arrows, forms the putative terminator for the hrpC operon. The italicized amino acids at the C terminus of HrcC represent the revised amino acid sequence predicted from the corrected sequence of hrcC, which has an additional G, indicated by boldface. The beginning of coding regions and stop codons are indicated by broken arrows and asterisks, respectively.
Each of the subclones from P. syringae pv. tomato DC3000 and P. syringae pv. glycinea U1 contained the entire hrpRS, hrpZ, and hrpC operons. In addition pCPP2201 contained 1 kb of DNA upstream of hrpR, which displayed no significant ORFs or similarity to sequences in the databases, while pCPP2200 contained very little DNA upstream of hrpR but contained all of hrcU and part of hrcT. A detailed analysis of the sequences of the hrpC and hrpRS operons is presented below (Fig. 3).
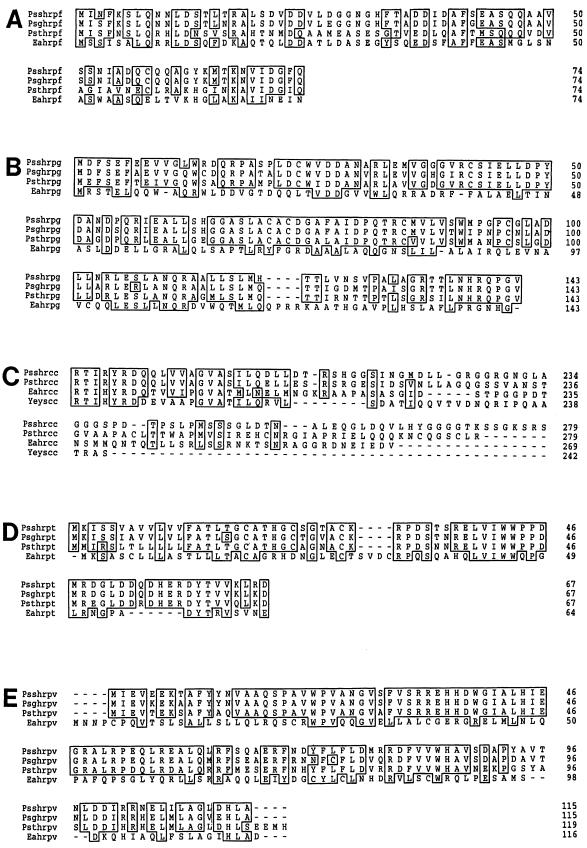
FIG. 3.
Alignments of the predicted amino acid sequences for HrpF, HrpG, HrcC, HrpT, and HrpV from P. syringae pv. syringae 61 (Pss) with those of homologous proteins from other bacteria possessing the type III secretion system. (A) HrpF; (B) HrpG; (C) an internal portion of HrcC containing amino acids 186 to 279; (D) HrpT; (E) HrpV. Sequences are from P. syringae pv. tomato DC 3000 (Pst), P. syringae pv. glycinea U1 (Psg), E. amylovora Ea321 (Ea), and Yersinia enterocolitica (Ye). Alignments were made by using the Genetics Computer Group PILEUP algorithm. The accession numbers are U56662 (E. amylovora HrpF to -V), U25813 (P. syringae pv. syringae 61 HrpF to -G), L01064 (P. syringae pv. syringae 61 HrcC), and M74011 (Y. enterocolitica YscC). Open boxes indicate identical amino acids.
HrpF and HrpG.
HrpF is a 74-amino-acid (8-kDa), putatively cytoplasmic protein which exhibited a high degree of divergence between P. syringae pathovars syringae and tomato. Sequence homology searches identified two extensive homologies. The first of these was between the HrpF proteins of P. syringae and E. amylovora (32). The HrpF proteins of P. syringae pv. syringae 61 and E. amylovora Ea321 possess 27% identity and 47% similarity and are homologous according to BESTFIT alignment randomization analysis (15). The search also identified similarity between HrpF from P. syringae pathovars syringae and glycinea and HrpA from P. syringae pathovar tomato. HrpA has been identified as a secreted protein in P. syringae pv. tomato DC3000 and is thought to be involved in the assembly of a pilus-like structure (46, 52). HrpFPss and HrpFPsg possessed 31% identity and 55% similarity, and 32% identity and 58% similarity, respectively, to HrpAPst. Both alignments passed the randomization test for significance but did incorporate gaps. The similarity was most marked within a region of 30 amino acids at the amino terminus of the protein.
Curiously, HrpF and HrpA did not show significant similarity to each other when compared within each of the three pathovars. Although there was limited similarity between the amino-terminal amino acids, the overall scores for the proteins did not pass the randomization test. This reflects the fact that the HrpA proteins of P. syringae pv. tomato DC3000 and P. syringae pv. syringae 61 are only 27% identical and 43% similar and the HrpF proteins of P. syringae pv. tomato DC3000 and P. syringae pv. syringae 61 are only 36% identical and 56% similar. In contrast the HrpA and HrpF proteins of P. syringae pv. syringae 61 are highly similar to their homologs in P. syringae pv. glycinea U1. Therefore, HrpA and HrpF share the attribute of being highly divergent in P. syringae pathovar tomato relative to the other two pathovars, in addition to being small hydrophilic proteins encoded by the first ORFs of polycistronic operons.
HrpG is predicted to be a 143-amino-acid (15.4-kDa) cytoplasmic protein. It is highly conserved among P. syringae pathovars syringae, tomato, and glycinea. However, HrpG of P. syringae does not show significant homology to its counterpart in E. amylovora or to any other proteins in the database.
HrcC.
Many of the features and homologies of HrcC and related proteins have been described previously (17, 27). Here we will focus mostly on the new information derived from the corrected sequence of P. syringae pv. syringae 61 hrcC and from the comparison of HrcC in P. syringae pathovars syringae and tomato. Homology searches show that HrcC is a member of the PulD-pIV superfamily of secretins, which are outer membrane proteins involved in macromolecular traffic across the bacterial outer membrane (47). The superfamily includes the type III-specific proteins HrcC (E. amylovora, Xanthomonas campestris, and Ralstonia solanacearum), YscC (Yersinia spp.), PscC (Pseudomonas aeruginosa), SpiA and InvG (Salmonella typhimurium), MxiD (Shigella flexneri), and SepC (E. coli) (27). The N-terminal portion of HrcC also exhibits homology to the NolW protein of Rhizobium fredii, which is involved in host specificity (40). Comparing HrcC proteins from P. syringae pathovars syringae and tomato with each other and with homologs from other bacteria revealed that there is a region of approximately 70 amino acids, which begins about 230 amino acids into the 700-amino-acid HrcC protein, that is highly divergent between the two pathovars. This region is also divergent between HrcC in Erwinia, Xanthomonas, and Ralstonia and is largely absent in YscC from Yersinia. Moreover, although the C-terminal end of HrcC is highly conserved between P. syringae pathovars syringae and tomato, and moderately conserved between P. syringae and E. amylovora, the P. syringae HrcC is significantly longer than the corresponding proteins from Yersinia, Xanthomonas, and Ralstonia, and the C-terminal ends of the proteins from these different species are not highly conserved.
HrpT and HrpV.
The predicted product of P. syringae pv. syringae 61 hrpT is a 67-amino-acid, 7.5-kDa outer membrane lipoprotein and that of hrpV is a 115-amino-acid, 13.4-kDa hydrophilic protein. The predicted product of P. syringae pv. tomato DC 3000 hrpV is a slightly larger protein of 119 amino acids (13.9 kDa). HrpT and HrpV both have homologs with a significant degree of homology in E. amylovora but show no homology to other proteins in the database.
HrpR and HrpS.
The sequence of the operon encoding HrpR and HrpS has previously been reported for P. syringae pathovars phaseolicola and syringae (19, 20, 51). The hrpR and hrpS genes from P. syringae pv. tomato DC 3000 and P. syringae pv. glycinea U1 are more highly conserved than those of the other two pathovars, and as in P. syringae pathovars syringae and phaseolicola, they are also highly similar to each other (data not shown). The similarity between HrpR and HrpS in each of the P. syringae pathovars glycinea, tomato, and phaseolicola is significantly stronger than the similarity between the reported sequences of HrpR and HrpS from P. syringae pv. syringae 61. Furthermore, reexamination of the HrpR sequence from P. syringae pv. syringae 61 following comparison with those of the other pathovars suggested that there is a frameshift resulting from a sequencing error between amino acids 235 and 249. Resequencing of this region revealed an additional nucleotide corresponding to amino acid 239. After correction, the deduced amino acid sequences show even greater identity between HrpRPss and HrpSPss (data not shown).
In P. syringae pv. tomato DC 3000 and P. syringae pv. syringae 61 there is a region of approximately 1 kb of AT-rich, noncoding DNA upstream of hrpRS, which forms the “right” boundary of the conserved hrp cluster. The flanking DNA beyond this noncoding region encodes putative virulence-associated proteins, including AvrE, and other proteins involved in the Hrp system, such as HrpW (11, 38).
Construction of functionally nonpolar mutations in the P. syringae pv. syringae 61 hrpC operon.
To investigate the role of each gene in the hrpC operon, individual ORFs were disrupted by insertion of a 1.5-kb nptII (neomycin phosphotransferase II) cassette lacking a rho-independent transcription terminator, followed by marker exchange recombination with the P. syringae pv. syringae 61 chromosome. The construction of the individual mutations is outlined in Fig. 4, and the plasmids and primers used are listed in Tables 1 and 2. In brief, recombinant plasmids pNCHU402, pNCHU407, pNCHU491, and pNCHU492 were each transformed into P. syringae pv. syringae 61 by triparental mating, and kanamycin-resistant transformants were screened for loss of tetracycline resistance. The mutations were confirmed by DNA gel blotting and hybridization, using the nptII gene as a probe (data not shown).
Altered abilities of P. syringae pv. syringae 61 hrpF, hrpG, hrcC, hrpT, and hrpV mutants and complemented strains to elicit HR in tobacco leaves.
To evaluate the effect of mutations on the ability of P. syringae pv. syringae 61 to elicit an HR, the mutants—hrpF, hrpG, hrcC, hrpT, and hrpV—were infiltrated individually into tobacco leaves at a range of inoculum levels. The HR was evaluated for rapid tissue collapse at 24 h postinoculation. Nonpolar mutations in hrpF, hrcC, and hrpT abolished or greatly reduced the ability of P. syringae pv. syringae 61 to elicit an HR in tobacco. The hrpG mutant retained significant HR-eliciting ability, but the HR observed was weaker than that caused by the wild-type strain. However, mutation of the hrpV gene had no observable effect on the timing or intensity of the HR. The results of the HR elicitation experiments are summarized in Table 3. The ability of hrpF, hrpG, hrcC, and hrpT mutants to elicit the HR in tobacco plants was restored to the wild-type phenotype by complementation with the corresponding subclones (Table 3). Curiously, complementation of the hrpV mutation, which lacked any obvious HR phenotype, resulted in a significant reduction in HR-eliciting activity when relative levels of activity were assessed by serial dilution.
TABLE 3.
Phenotypes of P. syringae pv. syringae 61 hrpF, hrpG, hrcC, hrpT, and hrpV mutants in planta
| P. syringae strain | Genotype | HR in tobaccoa | Disease in beansb | Multiplication in beansc | Complementing plasmid | HR in tobacco (complemented strain) |
|---|---|---|---|---|---|---|
| 61 | Wild type | I | I | 21,005A | pNCHU515NA | NAd |
| 61-N491 | ΔhrpF::nptII | IV | III | 367C | pNCHU515 | I |
| 61-N492 | hrpG::nptII | II | II | 1,411B | pNCHU513 | I |
| 61-N393 | ΔhrcC::nptII | IV | III | 4D | pNCHU421 | I |
| 61-N402 | ΔhrpT::nptII | III | II | 1,020B | pNCHU451 | I |
| 61-N407 | ΔhrpV::nptII | I | II | 320C | pCPP2372 | II |
Phenotypic classes: I, wild-type HR; II, HR slightly reduced, with difference observable mostly at lower inoculum levels; III, HR very weak and spotty, with some inoculations producing no response; IV, no HR.
Phenotypic classes: I, wild-type symptoms of spreading lesion and chlorotic halo; II, symptoms reduced; III, no symptoms.
Fold increase in bacterial population (CFU) in 1 cm2 of leaf tissue 2 days after inoculation; mean of four tests. The superscript capital letters indicate that the values are significantly different from each other (P = 0.05) by the analysis of variables test (JMP version 2.0.5 [1989]; SAS Institute, Carry, N.C.).
NA, not applicable.
Altered abilities of P. syringae pv. syringae 61, hrpF, hrpG, hrcC, hrpT, and hrpV mutants and complemented strains to multiply and produce disease symptoms in bean leaves.
The ability of hrpC operon mutants to cause disease was assessed by infiltrating bacteria into bean leaves at 105 CFU/ml and determining bacterial multiplication after 2 days and symptom expression after 5 days (Table 3). Conditions of high humidity were used to favor disease development. The strains tested fell into three classes with regard to their ability to multiply: (i) the hrcC mutant multiplied the least; (ii) P. syringae pv. syringae 61 multiplied the most; and (iii) the hrpF, hrpG, hrpT, and hrpV mutants multiplied to an intermediate level. The strains could be divided into three different classes with regard to the production of symptoms on bean leaves: (i) the hrpF and hrcC mutants were symptomless; (ii) P. syringae pv. syringae 61 produced necrotic, water-soaked lesions; and (iii) the hrpG, hrpT, and hrpV mutants produced significantly smaller lesions. Thus, although the ability of the hrpV mutant to elicit the HR was indistinguishable from that of wild-type P. syringae pv. syringae 61, its ability to multiply and produce disease symptoms was impaired.
DISCUSSION
We have characterized the hrpC operons of three P. syringae pathovars and compared the effects on bacterium-plant interactions of mutations in the P. syringae pv. syringae 61, hrpF, hrpG, hrcC, hrpT, and hrpV genes. Our findings reveal that the structure of the hrpC operon is conserved in these P. syringae pathovars and in E. amylovora and that each of the five genes in the hrpC operon contributes differently to the ability of P. syringae pv. syringae 61 to interact with plants. The structure of the hrpC operon and the differential phenotypes of the genes within it are significant for several reasons.
The P. syringae HrcC protein is a member of a superfamily of outer membrane proteins that are thought to multimerize and form a channel for translocation of proteins or filamentous phages across the outer membrane (17, 22, 29). The HrcC (formerly HrpH) protein of P. syringae pv. syringae 61 is essential for secretion of the HrpZ harpin (21). HrcC (formerly HrpA1) of X. campestris pv. vesicatoria has been shown to be localized to the outer membrane, and the protein also induces the psp operon when produced in E. coli (50), which is indicative of multimerization in the outer membrane (37). P. syringae pv. syringae 61 hrcC mutants accumulate HrpZ in the periplasm, which provides further evidence for a role of HrcC in protein translocation across the outer membrane (12). HrcC and its homologs are conserved components of all known type III secretion systems, but the flanking genes differ widely in many of those systems. For example, hrcC and yscC are flanked by different genes in R. solanacearum, X. campestris pv. vesicatoria, and Y. enterocolitica, respectively (41, 49, 50). In contrast, the hrcQ, -R, and -S homologs are present in the same order in all of these bacteria, as are their flagellar biogenesis homologs (49).
The hrp clusters of plant-pathogenic bacteria have been divided into two groups based on their regulatory components and hrp gene compositions (3). Group I contains P. syringae and E. amylovora, and group II contains R. solanacearum and X. campestris. Conservation of the hrpC operon appears to be characteristic of group I hrp clusters, a notion that is further supported by the recent finding that Erwinia chrysanthemi also carries the hrpC operon (31). Since the hrpF-hrpG-hrcC-hrpT-hrpV arrangement is not widely conserved (in contrast, for example, to the hrcQ-hrcR-hrcS arrangement), the existence of the hrpC operon suggests a close relationship among the group I hrp clusters, probably as a result of horizontal transfer to these diverse bacteria.
Further evidence for conservation of the hrp gene clusters among P. syringae pathovars was found by comparing the hrpRS, hrpZ, and hrpC operons of P. syringae pv. syringae 61, P. syringae pv. tomato DC 3000, and P. syringae pv. glycinea U1. Three taxonomic groups have been identified among the P. syringae pathovars on the basis of PCR-restriction fragment length polymorphism analysis of rRNA operons, and P. syringae pathovars syringae, tomato, and glycinea each belong to a different group (39). The divergence of P. syringae pathovars tomato and syringae is further supported by DNA-DNA hybridization studies (16). The finding of an identical gene arrangement for the 13 hrp and hrc genes comprising the right half of the hrp-hrc gene clusters in these divergent pathovars supports the hypothesis that the type III pathways of the different P. syringae pathovars are similar in function and that the differing pathogenic properties of these bacteria are determined by proteins that travel the pathway and are encoded in more variable regions (2).
The functions of the four Hrp proteins encoded by the hrpC operon are unclear, although mutations affecting each of them alter interactions of the bacterium with diagnostic plants. It should be noted that these genes are all designated hrp, even if they do not have a typical Hrp phenotype, because they are in a hrp operon (8) and mutations in them have at least some effect on the Hrp system. As expected, the hrcC mutation completely abolished all plant reaction phenotypes tested. The hrpF mutation had a similarly strong effect. The possibility that HrpF is secreted is suggested by the significant similarity between the P. syringae pv. syringae 61 and P. syringae pv. glycinea U1 HrpF proteins and the P. syringae pv. tomato DC 3000 HrpA protein and by the sequence divergence of HrpF in the three pathovars. HrpA is a Hrp-secreted pilin (46), and the degree of divergence has been reported to be higher for extracellular components of the type III secretion system (35). In contrast, HrpG and HrpT were more conserved and the corresponding mutations had intermediate effects in each of the plant reaction assays. The phenotypes of the hrpF, hrpG, hrcC, and hrpT mutants were all consistent with the hypothesis that these genes encode components of the Hrp pathway, but the hrpV mutation was puzzling in two ways. First, it had a strong effect on multiplication in beans but not on the HR in tobacco. Second, when complemented with a hrpV subclone in trans, the hrpV mutant acquired a reduced HR phenotype. One explanation for this is that HrpV is a negative regulator of the Hrp system. In the accompanying paper (44), we further explore this issue and the hrpF, hrpG, hrpT, and hrpV mutations by examining the effects of these mutations on the expression of the Hrp regulon and on the secretion of the HrpZ harpin and we further discuss the role of each of these proteins in the Hrp system.
ACKNOWLEDGMENTS
We thank Jihyun F. Kim and Steven V. Beer for helpful discussions and for sharing data before publication.
This work was supported by NSF grant MCB-9631530 and NSC grant 85-2311-B-005-37 from Taiwan.
REFERENCES
- 1.Alfano J R, Bauer D W, Milos T M, Collmer A. Analysis of the role of the Pseudomonas syringae pv. syringae HrpZ harpin in elicitation of the hypersensitive response in tobacco using functionally nonpolar deletion mutations, truncated HrpZ fragments, and hrmA mutations. Mol Microbiol. 1996;19:715–728. doi: 10.1046/j.1365-2958.1996.415946.x. [DOI] [PubMed] [Google Scholar]
- 2.Alfano J R, Collmer A. Bacterial pathogens in plants: life up against the wall. Plant Cell. 1996;8:1683–1698. doi: 10.1105/tpc.8.10.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alfano J R, Collmer A. The type III (Hrp) secretion pathway of plant pathogenic bacteria: trafficking harpins, Avr proteins, and death. J Bacteriol. 1997;179:5655–5662. doi: 10.1128/jb.179.18.5655-5662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alfano J R, Kim H-S, Delaney T P, Collmer A. Evidence that the Pseudomonas syringae pv. syringae hrp-linked hrmA gene encodes an Avr-like protein that acts in a hrp-dependent manner within tobacco cells. Mol Plant-Microbe Interact. 1997;10:580–588. doi: 10.1094/MPMI.1997.10.5.580. [DOI] [PubMed] [Google Scholar]
- 5.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 6.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K E. Short protocols in molecular biology. 3rd ed. New York, N.Y: John Wiley & Sons; 1995. [Google Scholar]
- 7.Beck E, Ludwig G, Auerswald E A, Reiss B, Schaller H. Nucleotide sequence and exact location of the neomycin phosphotransferase gene from transposon Tn5. Gene. 1982;19:327–336. doi: 10.1016/0378-1119(82)90023-3. [DOI] [PubMed] [Google Scholar]
- 8.Bogdanove A J, Beer S V, Bonas U, Boucher C A, Collmer A, Coplin D L, Cornelis G R, Huang H-C, Hutcheson S W, Panopoulos N J, Van Gijsegem F. Unified nomenclature for broadly conserved hrp genes of phytopathogenic bacteria. Mol Microbiol. 1996;20:681–683. doi: 10.1046/j.1365-2958.1996.5731077.x. [DOI] [PubMed] [Google Scholar]
- 9.Bogdanove A J, Wei Z-M, Zhao L, Beer S V. Erwinia amylovora secretes harpin via a type III pathway and contains a homolog of yopN of Yersinia spp. J Bacteriol. 1996;178:1720–1730. doi: 10.1128/jb.178.6.1720-1730.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonas U. hrp genes of phytopathogenic bacteria. In: Dangl J L, editor. Current topics in microbiology and immunology. 192. Bacterial pathogenesis of plants and animals—molecular and cellular mechanisms. Berlin, Germany: Springer-Verlag; 1994. pp. 79–98. [DOI] [PubMed] [Google Scholar]
- 11.Charkowski A O, Conlin A, He S-Y, Collmer A. HopPtoA, a Pseudomonas syringae pv. tomato Hrp-secreted protein with homology to pectate lyases. Phytopathology. 1997;87:S17. [Google Scholar]
- 12.Charkowski A O, Huang H-C, Collmer A. Altered localization of HrpZ in Pseudomonas syringae pv. syringae hrp mutants suggests that different components of the type III secretion pathway control protein translocation across the inner and outer membranes of gram-negative bacteria. J Bacteriol. 1997;179:3866–3874. doi: 10.1128/jb.179.12.3866-3874.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Devereaux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Gene. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ditta G, Stanfield S, Corbin D, Helinski D R. Broad host range DNA cloning system for Gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci USA. 1980;77:7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doolittle R F. Of Urfs and Orfs: a primer on how to analyze derived amino acid sequences. Mill Valley, Calif: University Science Books; 1986. [Google Scholar]
- 16.Gardan L, Shafif H, Grimont P. DNA relatedness among pathovars of P. syringae and related bacteria. In: Rudolph K, Burr T J, Mansfield J W, Stead D, Vivian A, von Kietzell J, editors. Developments in plant pathology. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1997. pp. 445–448. [Google Scholar]
- 17.Genin S, Boucher C A. A superfamily of proteins involved in different secretion pathways in gram-negative bacteria: modular structure and specificity of the N-terminal domain. Mol Gen Genet. 1994;243:112–118. doi: 10.1007/BF00283883. [DOI] [PubMed] [Google Scholar]
- 18.Gopalan S, Bauer D W, Alfano J R, Loniello A O, He S Y, Collmer A. Expression of the Pseudomonas syringae avirulence protein AvrB in plant cells alleviates its dependence on the hypersensitive response and pathogenicity (Hrp) secretion system in eliciting genotype-specific hypersensitive cell death. Plant Cell. 1996;8:1095–1105. doi: 10.1105/tpc.8.7.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grimm C, Aufsatz W, Panopoulos N J. The hrpRS locus of Pseudomonas syringae pv. phaseolicola constitutes a complex regulatory unit. Mol Microbiol. 1995;15:155–165. doi: 10.1111/j.1365-2958.1995.tb02230.x. [DOI] [PubMed] [Google Scholar]
- 20.Grimm C, Panopoulos N J. The predicted protein product of a pathogenicity locus from Pseudomonas syringae pv. phaseolicola is homologous to a highly conserved domain of several procaryotic regulatory proteins. J Bacteriol. 1989;171:5031–5038. doi: 10.1128/jb.171.9.5031-5038.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He S Y, Huang H-C, Collmer A. Pseudomonas syringae pv. syringae harpinPss: a protein that is secreted via the Hrp pathway and elicits the hypersensitive response in plants. Cell. 1993;73:1255–1266. doi: 10.1016/0092-8674(93)90354-s. [DOI] [PubMed] [Google Scholar]
- 22.Huang H-C, He S Y, Bauer D W, Collmer A. The Pseudomonas syringae pv. syringae 61 hrpH product, an envelope protein required for elicitation of the hypersensitive response in plants. J Bacteriol. 1992;174:6878–6885. doi: 10.1128/jb.174.21.6878-6885.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang H-C, Hutcheson S W, Collmer A. Characterization of the hrp cluster from Pseudomonas syringae pv. syringae 61 and TnphoA tagging of genes encoding exported or membrane-spanning Hrp proteins. Mol Plant-Microbe Interact. 1991;4:469–476. [Google Scholar]
- 24.Huang H-C, Lin R-W, Chang C-J, Collmer A, Deng W-L. The complete hrp gene cluster of Pseudomonas syringae pv. syringae 61 includes two blocks of genes required for harpinPss secretion that are arranged colinearly with Yersinia ysc homologs. Mol Plant-Microbe Interact. 1995;8:733–746. doi: 10.1094/mpmi-8-0733. [DOI] [PubMed] [Google Scholar]
- 25.Huang H-C, Schuurink R, Denny T P, Atkinson M M, Baker C J, Yucel I, Hutcheson S W, Collmer A. Molecular cloning of a Pseudomonas syringae pv. syringae gene cluster that enables Pseudomonas fluorescens to elicit the hypersensitive response in tobacco. J Bacteriol. 1988;170:4748–4756. doi: 10.1128/jb.170.10.4748-4756.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang H-C, Xiao Y, Lin R-H, Lu Y, Hutcheson S W, Collmer A. Characterization of the Pseudomonas syringae pv. syringae hrpJ and hrpl genes: homology of HrpI to a superfamily of proteins associated with protein translocation. Mol Plant-Microbe Interact. 1993;6:515–520. doi: 10.1094/mpmi-6-515. [DOI] [PubMed] [Google Scholar]
- 27.Hueck C J. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huynh T V, Dahlbeck D, Staskawicz B J. Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity. Science. 1989;245:1374–1377. doi: 10.1126/science.2781284. [DOI] [PubMed] [Google Scholar]
- 29.Kazmierczak B I, Mielke D L, Russel M, Model P. pIV, a filamentous phage protein that mediates phage export across the bacterial cell envelope, forms a multimer. J Mol Biol. 1994;238:187–198. doi: 10.1006/jmbi.1994.1280. [DOI] [PubMed] [Google Scholar]
- 30.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 31.Kim J F, Ham J H, Bauer D W, Collmer A, Beer S V. The hrpC and hrpN operons of Erwinia chrysanthemi EC16 are flanked by plcA homologs of hemolysin/adhesin genes and accompanying activator/transporter genes. Mol Plant-Microbe Interact. 1998;11:563–567. doi: 10.1094/MPMI.1998.11.6.563. [DOI] [PubMed] [Google Scholar]
- 32.Kim J F, Wei Z-M, Beer S V. The hrpA and hrpC operons of Erwinia amylovora encode components of a type III pathway that secretes harpin. J Bacteriol. 1997;179:1690–1697. doi: 10.1128/jb.179.5.1690-1697.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.King E O, Ward M K, Raney D E. Two simple media for the demonstration of pyocyanine and fluorescein. J Lab Clin Med. 1954;22:301–307. [PubMed] [Google Scholar]
- 34.Klement Z. Hypersensitivity. In: Mount M S, Lacy G H, editors. Phytopathogenic prokaryotes. Vol. 2. New York, N.Y: Academic Press; 1982. pp. 149–177. [Google Scholar]
- 35.Li J, Ochman H, Groisman E A, Boyd E F, Solomon F, Nelson K, Selander R K. Relationship between evolutionary rate and cellular location among the Inv/Spa invasion proteins of Salmonella enterica. Proc Natl Acad Sci USA. 1995;92:7252–7256. doi: 10.1073/pnas.92.16.7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lidell M C, Hutcheson S W. Characterization of the hrpJ and hrpU operons of Pseudomonas syringae pv. syringae Pss61: similarity with components of enteric bacteria involved in flagellar biogenesis and demonstration of their role in harpinPss secretion. Mol Plant-Microbe Interact. 1994;7:488–497. doi: 10.1094/mpmi-7-0488. [DOI] [PubMed] [Google Scholar]
- 37.Linderoth N A, Model P, Russel M. Essential role of a sodium dodecyl sulfate-resistant protein IV multimer in assembly-export of filamentous phage. J Bacteriol. 1996;178:1962–1970. doi: 10.1128/jb.178.7.1962-1970.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lorang J M, Keen N T. Characterization of avrE from Pseudomonas syringae pv. tomato: a hrp-linked avirulence locus consisting of at least two transcriptional units. Mol Plant-Microbe Interact. 1995;8:49–57. doi: 10.1094/mpmi-8-0049. [DOI] [PubMed] [Google Scholar]
- 39.Manceau C, Horvais A. Assessment of genetic diversity among strains of Pseudomonas syringae by PCR-restriction fragment length polymorphism analysis of rRNA operons with special emphasis on P. syringae pv. tomato. Appl Environ Microbiol. 1997;63:498–505. doi: 10.1128/aem.63.2.498-505.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meinhardt L W, Krishnan H B, Balatti P A, Pueppke S G. Molecular cloning and characterization of a sym plasmid locus that regulates cultivar-specific nodulation of soybean by Rhizobium fredii USDA257. Mol Microbiol. 1993;9:17–29. doi: 10.1111/j.1365-2958.1993.tb01665.x. [DOI] [PubMed] [Google Scholar]
- 41.Michiels T, Vanooteghem J-C, de Rouvroit C L, China B, Gustin A, Boudry P, Cornelis G R. Analysis of virC, an operon involved in the secretion of Yop proteins by Yersinia enterocolitica. J Bacteriol. 1991;173:4994–5009. doi: 10.1128/jb.173.16.4994-5009.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Palleroni N J. Genus 1. Pseudomonas Migula 1894, 237AL. In: Krieg N R, Holt J G, editors. Bergey’s manual of systematic bacteriology. Baltimore, Md: Williams and Wilkins; 1984. pp. 141–199. [Google Scholar]
- 43.Pirhonen M U, Lidell M C, Rowley D L, Lee S W, Jin S, Liang Y, Silverstone S, Keen N T, Hutcheson S W. Phenotypic expression of Pseudomonas syringae avr genes in E. coli is linked to the activities of the hrp-encoded secretion system. Mol Plant-Microbe Interact. 1996;9:252–260. doi: 10.1094/mpmi-9-0252. [DOI] [PubMed] [Google Scholar]
- 44.Preston G, Deng W-L, Huang H-C, Collmer A. Negative regulation of hrp genes in Pseudomonas syringae by HrpV. J Bacteriol. 1998;180:4532–4537. doi: 10.1128/jb.180.17.4532-4537.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Preston G, Huang H-C, He S Y, Collmer A. The HrpZ proteins of Pseudomonas syringae pvs. syringae, glycinea, and tomato are encoded by an operon containing Yersinia ysc homologs and elicit the hypersensitive response in tomato but not soybean. Mol Plant-Microbe Interact. 1995;8:717–732. doi: 10.1094/mpmi-8-0717. [DOI] [PubMed] [Google Scholar]
- 46.Roine E, Wei W, Yuan J, Nurmiaho-Lassila E-L, Kalkkinen N, Romantschuk M, He S Y. Hrp pilus: an hrp-dependent bacterial surface appendage produced by Pseudomonas syringae pv. tomato DC3000. Proc Natl Acad Sci USA. 1997;94:3459–3464. doi: 10.1073/pnas.94.7.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Russel M. Moving through membranes with filamentous phages. Trends Microbiol. 1995;3:223–228. doi: 10.1016/s0966-842x(00)88929-5. [DOI] [PubMed] [Google Scholar]
- 48.Tabor S, Richardson C C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Gijsegem F, Gough C, Zischek C, Niqueux E, Arlat M, Genin S, Barberis P, German S, Castello P, Boucher C. The hrp gene locus of Pseudomonas solanacearum, which controls the production of a type III secretion system, encodes eight proteins related to components of the bacterial flagellar biogenesis complex. Mol Microbiol. 1995;15:1095–1114. doi: 10.1111/j.1365-2958.1995.tb02284.x. [DOI] [PubMed] [Google Scholar]
- 50.Wengelnik K, Marie C, Russel M, Bonas U. Expression and localization of HrpA1, a protein of Xanthomonas campestris pv. vesicatoria essential for pathogenicity and induction of the hypersensitive response. J Bacteriol. 1996;178:1061–1069. doi: 10.1128/jb.178.4.1061-1069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiao Y, Heu S, Yi J, Lu Y, Hutcheson S W. Identification of a putative alternate sigma factor and characterization of a multicomponent regulatory cascade controlling the expression of Pseudomonas syringae pv. syringae Pss61 hrp and hrmA genes. J Bacteriol. 1994;176:1025–1036. doi: 10.1128/jb.176.4.1025-1036.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yuan J, He S Y. The Pseudomonas syringae Hrp regulation and secretion system controls the production and secretion of multiple extracellular proteins. J Bacteriol. 1996;178:6399–6402. doi: 10.1128/jb.178.21.6399-6402.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]