Abstract
Membrane proteins determine the precise function of each membrane and, therefore, the function of each cell type. These proteins essential roles in cell physiology, participating in the maintenance of the cell metabolism, its homeostasis or promoting proper cell growth. Membrane proteins, as has long been described, are located both in the plasma membrane and in complex subcellular structures. However, they can also be released into the extracellular environment associated with extracellular vesicles (EVs). To date, most of the research have been focused on understanding the role of exosomal RNA in several processes. Recently, there has been increasing interest in studying the function of exosome membrane proteins for exosome-based therapy, but not much research has been done yet on the function of exosome membrane proteins. One of the major limitations of studying exosome membrane proteins and their application to translational research of exosome-based therapeutics is the low yield of exosome isolation. Here, we have introduced a new perspective on exosome membrane protein research by reviewing studies showing the important role of exosome membrane proteins in exosome-based therapies. Furthermore, we have proposed a new strategy to boost the yield of exosome isolation: hybridization of liposomes with exosome-derived membrane. Liposomes have already been reported to affect the cell excitation to increase exosome production in tumor cells. Therefore, increasing cellular uptake of these liposomes would enhance exosome release by increasing cellular excitation. This new perspective could be a breakthrough in exosome-based therapeutic research.
Keywords: Membrane proteins, Extracellular vesicles, Exosomes, Liposomes
Highlights
-
•
Introducing a novel perspective on exosome membrane proteins research.
-
•
Reviewing various research where exosome membrane proteins play an important role.
-
•
Advantages and challenges in exosome-based therapies.
-
•
Liposome as substrate material to boost isolation yield of exosome.
-
•
Fusing cells with exosome-derived membranes and liposomes enhance cell excitation.
Abbreviations
- ATP
Adenosine triphosphate
- DOPE
1,2-dioleoyl-sn-glycero-3-phosphoethanolamine.
- DOPE-PEG2000
1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-[amino(polyethylene glycol)-2000]
- DOPC
1,2-dioleoyl-sn-glycero-3-phosphocholine.
- DOPS
1,2-dioleoyl-sn-glycero-3-phospho-l-serine.
- DOTAP
1,2-dioleoyl-3-trimethylammonium-propane
- DSPE-PEG2000
1,2-Distearoyl-sn-glycero-3-phosphoethanolamine-N-[amino(polyethylene glycol)-2000]
- EVs
Extracellular vesicles
- ILVs
Intraluminal vesicles
- MBP
Myelin basic protein
- MSC
Mesenchymal stem cell
- MVBs
Multivesicular bodies
- PEG
Polyethylene glycol
- PLP
Proteolipid protein
- PrPc
Cellular prion protein
- PrPsc
Scrapie isoform of the prion protein
1. Introduction. Membrane proteins
One of the most important characteristics of a cell is the presence of a membrane, which covers not only the cell itself but also many complex subcellular structures [1,2]. The membrane is responsible for regulating the passage of substances [3]. The main structure of the membrane is formed by a phospholipid bilayer, but membrane-associated proteins are responsible for most of the specific functions of the membrane. In consequence, the functional characteristics of each type of cell membrane are determined by these proteins [4].
The function of these membrane proteins is to sustain the metabolism of the cell, its homeostasis and proper cell growth [5]. Some of these functions are mediated through the transport of ions, metabolites and larger molecules (such as RNA or proteins) across the membrane, propagation of electrical impulses, adhesion to adjacent cells or extracellular matrix and regulation of intracellular vesicular transport, among many others [6].
These membranes host a wide variety of proteins, such as receptors, ion channels, lipid domains lipid signals and scaffolding complexes. Some of these membrane proteins do not reach the hydrophobic interior of the lipid bilayer, instead they associate with the membrane by interacting with other membrane proteins or lipids through non-covalent binding. These proteins are known as peripheral membrane proteins. [1,4] In contrast, proteins that are permanently embedded in the lipid bilayer are called integral membrane proteins. In this case, proteins are held in the bilayer by lipid groups or in the membrane by tight junctions with other proteins [4,7].
Almost half of the proteins in our cells are in the membranes [5]. However, the quantity and type of proteins associated with the membrane are highly variable [4]. To illustrate this fact, we will now look at the case of myelin sheaths, modified cell membranes that wrap around the nerve axons several times to provide electrical insulation. The lipid-to-protein ratio of myelin sheaths differs from the composition of a typical cell membrane. Dry myelin sheaths are characterized by a high lipid content (75–80 %) and low protein content (20–25 %), whereas most other cell membranes have approximately equal proportions of lipid-to-protein (50-50 %). High lipid content reduces the ion permeability of the membrane, increasing its electrical insulation and resulting in rapid propagation of nerve impulses [[8], [9], [10]]. In addition, the myelin sheath is associated with certain proteins that contribute to its function, such as the proteolipid protein (PLP) or the myelin basic protein (MBP). The PLP plays an important role in maintaining the compact multilayered membrane structure by bringing the myelin membranes closer to each other. MBP participates in transmitting extracellular signals to the cytoskeleton and tight junctions [8].
As mentioned above, these membrane proteins are also found in the membrane of complex subcellular structures. Both the quantity and type of proteins are closely related to the function of these subcellular structures within the cell. In contrast, we will now review the case of the inner mitochondrial membrane. Mitochondria represent the main source of adenosine triphosphate (ATP), an energy-rich compound essential for fundamental cellular processes. The human body produces large amounts of ATP, up to 50 kg are synthesized per day in a healthy adult. The inner membrane of mitochondria presents multiple invaginations into the matrix, called cristae, where the protein complexes of the respiratory chain and mitochondrial ATP synthase reside. As a result, mitochondrial cristae is characterized by being among the most protein-rich membranes in the cell (more than 75 % of protein content) [[11], [12], [13]].
Membrane proteins play an important biological role in physiological mechanisms for cell survival, making them a major therapeutic target for drugs. Membrane proteins constitute a significant proportion of the human proteome (around 20–30 %) and therefore represent a vast source of therapeutic targets (more than 60 %). Some of the most common drug targets include enzymes, transporters, ion channels and receptors [14,15].
2. Extracellular vesicles: exosomes
As aforementioned, these membrane proteins are not only found in the plasma membrane, but also inside the cells in complex subcellular structures, such as the mitochondria. However, these proteins can also be released into the extracellular environment transported in the membrane of exosomes [16]. These exosomes contain a wide range of transmembrane proteins, lipid-anchored proteins, peripherally membrane-associated proteins and soluble proteins from the exosome lumen [17].
Exosomes are a subset of small extracellular vesicles (EVs), with diameters ranging from 30 to 150 nm, secreted from most of the cells including normal and diseased cells [[70], [71], [72], [73], [74]]. In addition to exosomes, two main types of EVs can be distinguished according to their size and origin. Apoptotic bodies (500 nm to several micrometers) are released by those cells undergoing programmed cell death, and microvesicles (100–1000 nm) are released during the budding and detachment of the plasma membrane [[18], [19], [20]]. The term “exosomes” was adopted to distinguish them from the rest of EVs. Although this term is widely used, it has been recommended to use “small EVs” following the International Society for Extracellular Vesicles’ 2018 guidelines, due to methodological difficulties in separating exosomes from other EVs [21]. These guidelines, known as MISEV2018, is an updated version of MISEV2014 which was published in 2014 with the purpose of establishing the minimum information necessary for EVs research. The MISEV2018 aims to guide the further standardization of studies on EVs. In the present review, we will use the term “exosome” for convenience.
In vivo, these vesicles are stable in a wide variety of biological fluids, such as amniotic fluid, blood, breast milk, urine and saliva among many others [22,23]. The stability of these EVs in the extracellular medium can be attributed to the proteins they harbor, which protect them from phagocytosis and complement-mediated lysis [24]. We can also find them in vitro in the conditioned medium of cell cultures [23].
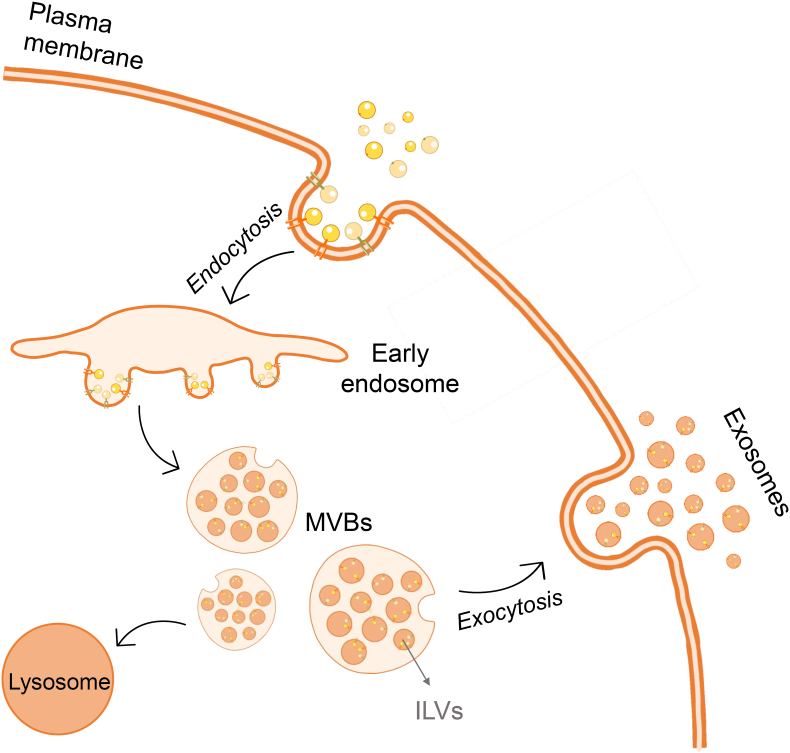
The formation of exosomes (Fig. 1) begins with the inward budding of the plasma membrane to form early endosomes, which is the first step on the endosomal transport pathway. Their function is to perform primary sorting and determine the fate of the endocytosed cargo. There are three different pathways that cargo can follow from the early endosome [75,76], however our focus would be the pathway that leads to exosome formation. After the early endosome is formed, the endocytosed cargo concentrates in the vacuolar regions of the early endosome and enters the endosomal maturation phase, eventually forming the late endosome. During the maturation, specific regions of the endosomal membrane partially invaginate, leading to the formation of intraluminal vesicles (ILVs) that are contained within the luminal interior of the endosome. These late endosomal structures containing dozens of ILVs are known as multivesicular bodies (MVBs). During this process, certain proteins are incorporated into the invaginating membrane, while cytosolic components are swollen and incorporated into the interior of the ILVs. Most of the MVBs will fuse with the plasma membrane of the cell, releasing the ILVs into the extracellular space. These vesicles are then called “exosomes”. Alternatively, these MVBs are directed to the lysosome for degradation [18,25,26].
Fig. 1.
Exosome biogenesis. After endocytosis of the cargo, the early endosome undergoes a maturation process until finally the late endosome is originated, also known as multivesicular bodies (MVBs). During the process of maturation, dozens of intraluminal vesicles (ILVs) will be formed and gathered inside the MVBs. Finally, the MVBs will fuse with the plasma membrane, releasing to the extracellular space the ILVs, receiving now the name of “exosomes”. Alternatively, this MVBs can be targeted to the lysosome for degradation. Illustration created with the support of SMART (Servier Medical Art).
These vesicles have numerous complex functions in intracellular communication and in exchanging substances [19]. All exosomes are equipped with surface molecules that allow them to target recipient cells. Once bound to the targeted cells, they can trigger signalization through receptor-ligand interaction and can be internalized by endocytosis and/or phagocytosis. Exosomes even can be fused with the recipient cell membrane, releasing their contents into the cytosol and modifying the physiological state of the cell [27]. Thus, exosomes secreted by a certain parent cell have a high potential to produce systemic effects on the behavior of neighboring cells, the cell microenvironment, and the phenotype of distant cells and tissues [28].
3. Exosome content
The RNA, proteins, and other cargoes of exosomes from different cell types reflect the phenotype of their parent cells. They may also carry cell- or tissue-specific factors that can be used to identify their origin [28]. For example, in the case of stem cell-derived exosomes, it has been reported that stem cells exert their reparative properties through paracrine effects rather than by direct cellular interaction [29]. In the study performed by Doeppner et al., in 2015, they observed that mesenchymal stem cell (MSC)-derived exosomes attenuated postischemic peripheral immunosuppression and promoted neuronal survival and angio-neurogenesis [30]. In contrast, exosomes released by cancer cells have been reported to inhibit the immune response against tumor cells, mainly by inducing T lymphocyte apoptosis [31]. In the study by Shen et al., in 2020, they demonstrated that exosomes derived from pancreatic cancer cells target T lymphocytes altering their gene expression profile, and thus impair their antitumor properties. It was found that these exosomes altered genes involved in apoptosis induction, such as p38 MAPK, PERK, ATF4, EIF2α and others [32].
In general, exosomes are enriched in a wide variety of proteins with different functions. Some of these proteins are considered as specific exosome markers and they are used to distinguish them from other EVs in exosome research. One of the major exosome proteins are tetraspanins, which are involved in cell penetration, invasion and fusion of exosomes [19,26]. A study by Escola et al., in 1998 have already demonstrated that the groups of membrane proteins are highly enriched in exosomes, while generic plasma membrane and lysosomal proteins do not reach these levels. Some of these tetraspanins are CD81, CD82, CD9 and CD63, with CD81 being the most abundant [33]. Other major proteins found in exosomes are heat shock proteins, such as HSP70 and HSP90, which are involved in stress response processes, and participate in the binding and presentation of antigens. Finally, we can find MVB formation proteins involved in the release of exosomes, such as Alix and TSG101, and proteins responsible for the transport and fusion to the membrane, such as annexins and Rab [26].
In addition to these proteins, exosomes have also been shown to carry genetic material, such as long non-coding RNAs, small nuclear RNA, microRNAs, mRNAs and DNA fragments [17,26,34]. Valadi et al. demonstrated that exosomes containing mRNA and microRNA can be transferred to target cells where they can be translated into proteins. It was observed that after transferring mouse exosomal RNA into human-derived mast cells, new mouse proteins were found in these cells, indicating that the transferred exosomal mRNA can be functional at its target location [35].
3.1. Exosome membrane proteins
Research on exosome membrane proteins represents a field in which much remains to be explored. With this review article, we aim to give some insight into this topic to encourage research in this field and to illustrate the importance of these proteins, both in the propagation of diseases and in their beneficial effects in exosome-based therapies.
Membrane proteins are generally responsible for the regulation of membrane-mediated cellular processes. Therefore, any dysregulation in their biological activity can have negative effects on cells, such as inappropriate cellular responses to exogenous infections, cancers, and genetic diseases. Focusing on the case of exosomes, most of the research analyzing the composition of exosomes to study the mechanism of pathologies, have focused on characterizing the functional changes of exosomal RNA. However, there is an increasing interest in exosome membrane proteins [16,28].
3.1.1. Role in disease propagation
A clear example of a membrane protein involved in disease is the cellular prion protein (PrPc). This protein is generally located in the outer layer of the plasma membrane anchored to glycosylphosphatidylinositol, and it is most abundantly expressed in the central nervous system [36]. The normal, non-pathogenic conformation of this protein is an alpha helix. However, its conversion to a beta-sheet would result a misfolded, pathogenic form, known as PrPsc (scrapie isoform of PrPc). This PrPsc acts as a template to facilitate the conversion of PrPc to PrPsc, resulting an accumulation of the pathogenic protein. This accumulation will lead to prion neurodegenerative diseases, such as Creutzfeldt-Jakob disease or Bovine Spongiform Encephalopathy [37]. Interestingly, both PrPc and its pathogenic isoform PrPsc can be found in EVs such as exosomes, underlying the contribution of exosomes in the propagation of diseases. Exosome-associated PrPsc has the ability to infect other cells [36,38].
Heisler et al. investigated the role of muskelin in the vesicular transport of PrPc. This protein binds to PrPc together with cytoplasmic dynein and KIF5C, and under normal conditions promotes lysosomal degradation over exosomal release of PrPc. In the absence of muskelin, PrPc is not properly targeted to the lysosome, but it is recycled to the plasma membrane and sorted as cargo for extracellular transport. Therefore, muskelin deficiency have an important role in the accelerated progression of prion diseases [39].
Further research on the role of muskelin in PrPc vesicle transport may be of great interest in understanding the propagation of neurodegenerative diseases. One of the main characteristics of neurodegenerative diseases is the presence of neuropathological protein aggregates, such as β-amyloid peptides in Alzheimer's disease or α-synuclein aggregates in Parkinson's disease. Numerous studies have demonstrated cell-to-cell transmission of these aggregates to other areas of the brain as the disease progresses. Exosomes have been presented as potential vehicles for the transmission of these neuropathological aggregates [40,41]. In the case of Alzheimer's disease, some studies have already demonstrated that PrPc plays an important role in the pathogenesis of this disease. For example, Takahashi et al. observed that PrPc deposits are often associated with β-amyloid plaques in Alzheimer's disease [42]. PrPc acts as a high-affinity receptor for amyloid β-42 oligomers on cells [43]. Because of the association between PrPc and Alzheimer's disease, it could be interesting to study the role of muskelin in the propagation of β-amyloid peptides through exosomes mediated by PrPc.
3.1.2. Role in exosome-based therapies
Over the past decade, the study of the therapeutic effects of exosomes for the treatment of diseases has become a growing field. The proteins contained in the exosome membrane have unique properties that qualify them as potential candidates for their use in therapies. Both ligands and receptors present on the membrane of exosomes will promote their interaction with target cells, conferring them a certain degree of cell specificity. In addition, they will also contribute to the low immunogenicity of the exosomes when exposed to the immune system [16,28].
The use of exosomes for the treatment of diseases is very versatile. Due to their numerous advantages, such as their enhanced biocompatibility, stability, or low toxicity compared to other nanocarriers, exosomes have been extensively studied for drug delivery [44]. However, many other research groups have focused on studying the effects caused by the contents of the exosomes themselves. A great example is the study by Otero-Ortega et al. in which they studied the effect of MSC-derived exosomes in an animal model of subcortical stroke in rats. They observed improved functional recovery, fiber tract integrity, axonal sprouting, and markers of white matter repair as well as proteomic analysis of these MSC-derived exosomes. Exosome proteins were identified to 2416 proteins involved in three different functions: molecular function regulator, catalytic activity, and binding. Furthermore, exosomes contain proteins involved in brain repair functions, such as synaptic transmission, neuronal differentiation from neural stem cells, angiogenesis, neuronal projections, neurite outgrowth and axonal growth [19,45].
Currently, the impact of membrane proteins on the beneficial effects of exosome-based therapies remains an under-explored area. For this reason, these proteomics results could be a good starting point to find out which of them are located in the membrane of the exosome and to study their function in more detail.
4. Challenges in the use of exosomes in research
As a result of all the unique functions of exosomes, the use of exosome technology for clinical applications has become an increasingly popular field of research in recent years. Recent studies have proposed exosomes as innovative therapeutic tools for the treatment of a wide range of disease due to their ability to transport specific compounds and surface proteins [[46], [47], [48]]. However, despite the promising aspects of using exosomes for disease treatment, there are numerous challenges that have slowed their use in clinical trials. Some of these challenges include: no standardized protocol for exosome isolation, low purity of the exosomes obtained, poor characterization of exosomes and low production of exosomes by cells [21,47,49].
Regarding the latter point, the low yield of exosome isolation has been a limitation to the widespread application of exosomes in clinical research. Various strategies have been adopted to overcome this limitation, however, to the best of our knowledge, none of them has obtained successful results. Therefore, how to enrich the production level of exosomes is crucial for a better analysis of exosomes and their further application [47,50]. In 2018, a study by Emam et al. investigated the possibility of using liposomes to overcome this limitation. They evaluated the effect of incubating different liposome preparations in tumor cells in terms of exosome production. They observed that depending on the physicochemical properties of the liposomes, exosome production by tumor cells could be increased or even suppressed [50].
The use of nanomaterials has revolutionized the world of clinical research. Because of their size, they can interact directly with cells and affect their biological pathways. It has been shown that there is an active interaction between nanomaterials and organisms, which influences the physiology of the cell [51]. Liposomes are also known to interact with cell surfaces in a physicochemical dependent manner, resulting in cell stimulation [50]. In response to this stimulation, shedding vesicles are released by budding from the plasma membrane and released into the extracellular medium [52].
5. Liposomes: new insight
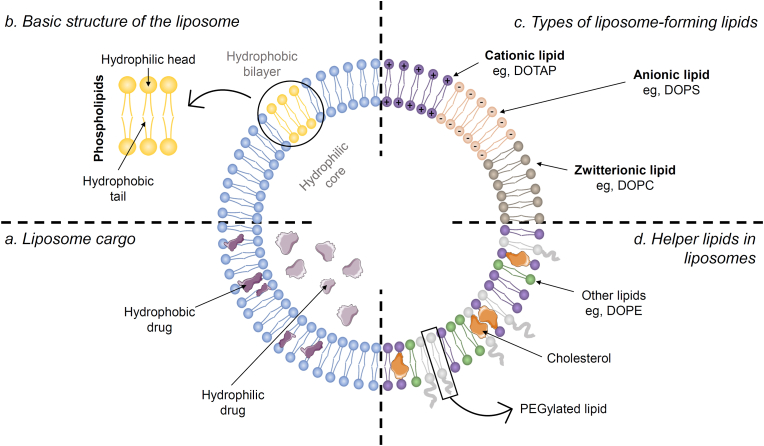
Within the wide variety of lipid nanoparticles, liposomes, along with solid lipid nanoparticles and nanostructured lipid carriers, are well known for their great clinical success in drug delivery [53]. This breakthrough is mainly due to their biocompatibility, low variability, ease of synthesis and scalability. Moreover, liposomes are highly versatile for drug delivery due to their ability to transport both hydrophilic compounds in their lumen and hydrophobic compounds in their lipid bilayer (Fig. 2a). This feature gives them the ability to release a wide variety of biologically active molecules [54,55].
Fig. 2.
Structure, composition, and cargo of liposomes. (a) Both hydrophilic (liposome lumen) and hydrophobic drugs (bilayer) can be loaded into the liposome. (b) Liposomes consist of a hydrophobic bilayer normally formed by phospholipids (hydrophilic head and hydrophobic tails) and a hydrophilic core. (c) The hydrophobic bilayer can be formed by cationic lipids (e.g., DOTAP), anionic lipids (e.g., DOPS) or zwitterionic lipids (e.g., DOPC). (d) Accompanying this bilayer, we can find some helper lipids like cholesterol, PEGylated lipids and other lipids like DOPE. Illustration created with the support of SMART (Servier Medical Art).
5.1. Liposome lipidic composition and structure
Liposomes are spherical vesicles (nanoparticles) built by the self-assembly of phospholipids into closed structures when hydrated in aqueous solutions. This assembly results a lipid bilayer composed of a hydrophilic (or polar) head and a hydrophilic (or non-polar) tail, which results in an amphipathic structure (Fig. 2b). Typically, the size of these nanoparticles ranges from 50 to 500 nm in diameter [55,56].
When formulating liposomes, either neutral (zwitterionic), positively (cationic) or negatively (anionic) charged lipids can be added to their composition. The polar heads of these lipids are the ones that will be neutral, positively or negatively charged (Fig. 2c) [55]. Each one will provide different properties to the liposome. For in vivo studies, neutrally charged liposomes are preferred to avoid non-specific uptake of negatively charged biomolecules [57]. An example of zwitterionic lipid is 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC). Among the cationic and anionic liposome formulations, the former has been the most studied [58]. On the one hand, cationic lipids will induce a positive charge on the surface of liposomes and are often used for the delivery of nucleic acids. It has been observed that the interaction between the anionic charge of the nucleic acid and the cationic lipid enhances its encapsulation [57,59]. In addition, the presence of these cationic lipids will increase their association with cell surfaces (negatively charged), leading to cellular uptake by endocytosis. Furthermore, after cellular uptake, the cationic lipid will promote destabilization of the endosomal membrane, facilitating the release of its cargo into the cytoplasm [60]. One of the most commonly used cationic lipids is 1,2-dioleoyl-3-trimethylammonium-propane, also known as DOTAP. On the other hand, anionic lipids will provide a negative charge to the liposome. Some of the advantages of these liposomes are: higher stability in solution and lower aggregation than neutral liposomes [58]. An example of anionic lipid is 1,2-dioleoyl-sn-glycero-3-phospho-l-serine (DOPS).
In addition to the lipids mentioned above, the liposome formulation also contains other lipids, commonly known as helper lipids (Fig. 2d). These lipids are used to improve the properties of the liposome, contributing to its stability and delivery efficiency [61,62]. For example, cholesterol, an important component of animal cells, does not form the cell membrane by itself. However, cholesterol plays a very important role in maintaining the structure of the bilayer, provides higher fluidity and stability, and limits the permeability of water-soluble molecules across the membrane [63,64]. Another commonly used helper lipid is DOPE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine), which has a conical geometry that promotes the formation of the hexagonal H(II) phase, destabilizing endosomal membranes and facilitating endosomal escape of the liposome [62]. This endosomal escape is essential for the delivery of the liposome cargo into the cytosol [65]. Last but not least, the use of PEGylated lipids (PEG as an abbreviation for Polyethylene glycol) offers several advantages, such as contributing to particle stability and reducing particle aggregation. This aggregation occurs mainly because the attraction between them is stronger than the attraction for the solvent. Therefore, coating the surface of the liposome with PEG would reduce this attraction by increasing the steric distance between them [66]. Some PEGylated lipids are: DSPE-PEG2000 (1,2-Distearoyl-sn-glycero-3-phosphoethanolamine-N-[amino(polyethylene glycol)-2000]) or DOPE-PEG2000 (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-[amino(polyethylene glycol)-2000]).
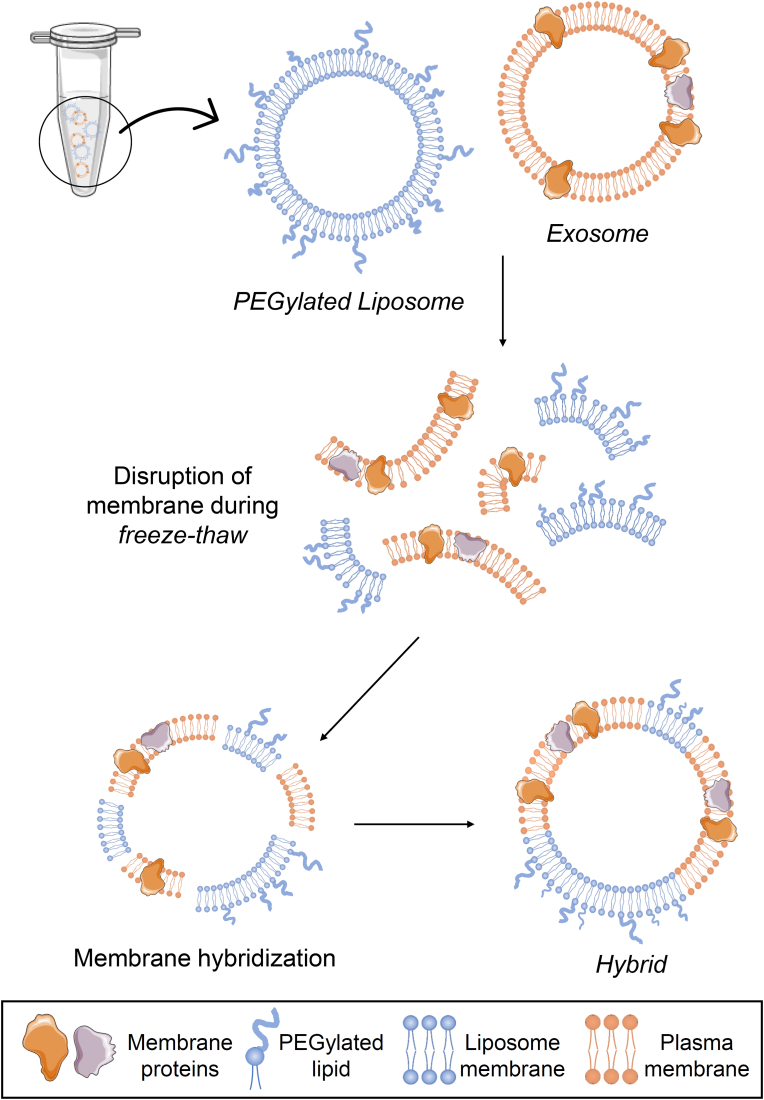
To enhance the properties of standard liposomes, several approaches have been pursued to modify them. One of them is the formulation of liposomes in combination with well-designed polymers, such as PEG [67]. Another approach taken by Sato et al. was to develop hybrid nanoparticles by fusing the exosome membrane to liposomes. This research demonstrated that the delivery of exosome can be modified by changing the lipid composition of the nanoparticles or the properties of the membrane by fusion with exosomes, in the view of cellular uptake [68]. The freeze-thaw method was used to hybridize the exosome membrane with the liposomes (Fig. 3). The freeze-thaw method has been used to incorporate water-soluble molecules into the internal aqueous phase of liposomes. This may suggest that the freeze-thaw disrupts the plasma membrane by temporarily forming ice crystals [68,69].
Fig. 3.
Scheme to illustrate the formation of hybrids. In the freeze-thaw process, the membrane of both liposomes and exosomes is disrupted and rejoined with each other, forming hybrids. Illustration created with the support of SMART (Servier Medical Art).
6. Conclusions and perspectives
Membrane proteins play a critical role in numerous physiological processes. Despite the importance of exosome proteins and the growing relevance of exosomes in translational research, there is a lack of literature that integrates these two areas of study. The main goal of this review is to provide a new perspective on the study of exosomes by highlighting the importance of exosome membrane proteins and the potential therapeutic target they may represent for the development of novel therapies.
Exosomes face numerous limitations that restrict their widespread application in research, and one of the major challenges is the low production level of exosomes. This review proposes a novel strategy to overcome this obstacle by introducing the hybridization concept of liposomes. These lipidic nanoparticles are widely known for their clinical success in drug delivery. It was hypothesized that by increasing the cellular uptake of liposomes, we could enhance cell stimulation and exosome production by cells. The approach we propose to enhance cellular uptake is the fusion of liposomes with exosome membrane.
Fundings
This work has been supported by grant no. 0420230480 from the Seoul National University Hospital Research Fund.
CRediT authorship contribution statement
Nuria Palomar-Alonso: Data curation, Investigation, Methodology, Writing – original draft. Mijung Lee: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. Manho Kim: Conceptualization, Resources, Supervision, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data availability
Data will be made available on request.
References
- 1.Nastou K.C., Tsaousis G.N., Iconomidou V.A. PerMemDB: a database for eukaryotic peripheral membrane proteins. Biochim Biophys Acta - Biomembr. 2020;1862(2) doi: 10.1016/j.bbamem.2019.183076. [DOI] [PubMed] [Google Scholar]
- 2.Sadowski P.G., Groen A.J., Dupree P., Lilley K.S. Sub-cellular localization of membrane proteins. Proteomics. 2008;8(19):3991–4011. doi: 10.1002/pmic.200800217. [DOI] [PubMed] [Google Scholar]
- 3.Stevens T.J., Arkin I.T. Do more complex organisms have a greater proportion of membrane proteins in their genomes? Proteins. 2000;39(4):417–420. doi: 10.1002/(sici)1097-0134(20000601)39:4<417::aid-prot140>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 4.Alberts B., Johnson A., Lewis J., et al. fourth ed. Garland Science; New York: 2002. Molecular Biology of the Cell. [Google Scholar]
- 5.Stahelin R.V. Lipid binding domains: more than simple lipid effectors. J. Lipid Res. 2009;50(Suppl):S299–S304. doi: 10.1194/jlr.R800078-JLR200. Suppl. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Von Heijne G. The membrane protein universe: what's out there and why bother? J. Intern. Med. 2007;261(6):543–557. doi: 10.1111/J.1365-2796.2007.01792.X. [DOI] [PubMed] [Google Scholar]
- 7.Escribá P.V., González-Ros J.M., Goñi F.M., et al. Membranes: a meeting point for lipids, proteins and therapies. J. Cell Mol. Med. 2008;12(3):829–875. doi: 10.1111/J.1582-4934.2008.00281.X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kister A., Kister I. Overview of myelin, major myelin lipids, and myelin-associated proteins. Front. Chem. 2023;10 doi: 10.3389/fchem.2022.1041961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams K.A., Deber C.M. The structure and function of central nervous system myelin. Crit. Rev. Clin. Lab Sci. 1993;30(1):29–64. doi: 10.3109/10408369309084665. [DOI] [PubMed] [Google Scholar]
- 10.Poitelon Y., Kopec A.M., Belin S. Myelin fat facts: an overview of lipids and fatty acid metabolism. Cells. 2020;9(4):812. doi: 10.3390/cells9040812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kühlbrandt W. Structure and function of mitochondrial membrane protein complexes. BMC Biol. 2015;13:89. doi: 10.1186/s12915-015-0201-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schlame M. Protein crowding in the inner mitochondrial membrane. Biochim Biophys Acta - Bioenerg. 2021;1862(1) doi: 10.1016/j.bbabio.2020.148305. [DOI] [PubMed] [Google Scholar]
- 13.Hill J., Olson E. Muscle: Fundamental Biology and Mechanisms of Disease. 2012. Mitochondrial morphology and function. [Google Scholar]
- 14.Aguayo-Ortiz R., Creech J., Jiménez-Vázquez E.N., et al. A multiscale approach for bridging the gap between potency, efficacy, and safety of small molecules directed at membrane proteins. Sci. Rep. 2021;11(1) doi: 10.1038/s41598-021-96217-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gong J., Chen Y., Pu F., et al. Understanding membrane protein drug targets in computational perspective. Curr. Drug Targets. 2019;20(5):551–564. doi: 10.2174/1389450120666181204164721. [DOI] [PubMed] [Google Scholar]
- 16.Yang Y., Hong Y., Cho E., Kim G.B., Kim I.S. Extracellular vesicles as a platform for membrane-associated therapeutic protein delivery. J. Extracell. Vesicles. 2018;7(1) doi: 10.1080/20013078.2018.1440131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pegtel D.M., Gould S.J. Exosomes. Annu Rev Biochem. 2019;88:487–514. doi: 10.1146/annurev-biochem-013118-111902. [DOI] [PubMed] [Google Scholar]
- 18.Yue B., Yang H., Wang J., et al. Exosome biogenesis, secretion and function of exosomal miRNAs in skeletal muscle myogenesis. Cell Prolif. 2020;53(7) doi: 10.1111/cpr.12857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Otero-Ortega L., Laso-García F., Gómez-de Frutos M.C., et al. Role of exosomes as a treatment and potential biomarker for stroke. Transl Stroke Res. 2019;10(3):241–249. doi: 10.1007/s12975-018-0654-7. [DOI] [PubMed] [Google Scholar]
- 20.Gurung S., Perocheau D., Touramanidou L., Baruteau J. The exosome journey: from biogenesis to uptake and intracellular signalling. Cell Commun. Signal. 2021;19(1):47. doi: 10.1186/s12964-021-00730-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y., Bi J., Huang J., Tang Y., Du S., Li P. Exosome: a review of its classification, isolation techniques, storage, diagnostic and targeted therapy applications. Int. J. Nanomed. 2020;15:6917–6934. doi: 10.2147/IJN.S264498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Z., Wang Y., Xiao K., Xiang S., Li Z., Weng X. Emerging role of exosomes in the joint diseases. Cell. Physiol. Biochem. 2018;47(5):2008–2017. doi: 10.1159/000491469. [DOI] [PubMed] [Google Scholar]
- 23.Purushothaman A. Exosomes from cell culture-conditioned medium: isolation by ultracentrifugation and characterization. Methods Mol. Biol. 2019;1952:233–244. doi: 10.1007/978-1-4939-9133-4_19. [DOI] [PubMed] [Google Scholar]
- 24.Kugeratski F.G., Kalluri R. Exosomes as mediators of immune regulation and immunotherapy in cancer. FEBS J. 2021;288(1):10–35. doi: 10.1111/febs.15558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krylova S.V., Feng D. The machinery of exosomes: biogenesis, release, and uptake. Int. J. Mol. Sci. 2023;24(2):1337. doi: 10.3390/ijms24021337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y., Liu Y., Liu H., Tang W.H. Exosomes: biogenesis, biologic function and clinical potential. Cell Biosci. 2019;9:19. doi: 10.1186/s13578-019-0282-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tkach M., Théry C. Communication by extracellular vesicles: where we are and where we need to go. Cell. 2016;164(6):1226–1232. doi: 10.1016/j.cell.2016.01.043. [DOI] [PubMed] [Google Scholar]
- 28.Hu Q., Su H., Li J., et al. Clinical applications of exosome membrane proteins. Precis Clin Med. 2020;3(1):54–66. doi: 10.1093/pcmedi/pbaa007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Timmers L., Lim S.K., Arslan F., et al. Reduction of myocardial infarct size by human mesenchymal stem cell conditioned medium. Stem Cell Res. 2007;1(2):129–137. doi: 10.1016/j.scr.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 30.Doeppner T.R., Herz J., Görgens A., et al. Extracellular vesicles improve post-stroke neuroregeneration and prevent postischemic immunosuppression. Stem Cells Transl Med. 2015;4(10):1131–1143. doi: 10.5966/sctm.2015-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huber V., Fais S., Iero M., et al. Human colorectal cancer cells induce T-cell death through release of proapoptotic microvesicles: role in immune escape. Gastroenterology. 2005;128(7):1796–1804. doi: 10.1053/j.gastro.2005.03.045. [DOI] [PubMed] [Google Scholar]
- 32.Shen T., Huang Z., Shi C., et al. Pancreatic cancer-derived exosomes induce apoptosis of T lymphocytes through the p38 MAPK-mediated endoplasmic reticulum stress. Faseb. J. 2020;34(6):8442–8458. doi: 10.1096/fj.201902186R. [DOI] [PubMed] [Google Scholar]
- 33.Escola J.M., Kleijmeer M.J., Stoorvogel W., Griffith J.M., Yoshie O., Geuze H.J. Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J. Biol. Chem. 1998;273(32):20121–20127. doi: 10.1074/jbc.273.32.20121. [DOI] [PubMed] [Google Scholar]
- 34.Raposo G., Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J. Cell Biol. 2013;200(4):373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J.J., Lötvall J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 36.Mahabadi H.M., Taghibiglou C. Cellular prion protein (PrPc): putative interacting partners and consequences of the interaction. Int. J. Mol. Sci. 2020;21(19):7058. doi: 10.3390/ijms21197058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Atkinson C.J., Zhang K., Munn A.L., Wiegmans A., Wei M.Q. Prion protein scrapie and the normal cellular prion protein. Prion. 2016;10(1):63–82. doi: 10.1080/19336896.2015.1110293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fevrier B., Vilette D., Archer F., et al. Cells release prions in association with exosomes. Proc. Natl. Acad. Sci. U.S.A. 2004;101(26):9683–9688. doi: 10.1073/pnas.0308413101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heisler F.F., Pechmann Y., Wieser I., et al. Muskelin coordinates PrPC lysosome versus exosome targeting and impacts prion disease progression. Neuron. 2018;99(6):1155–1169.e9. doi: 10.1016/j.neuron.2018.08.010. [DOI] [PubMed] [Google Scholar]
- 40.Lim Y.J., Lee S.J. Are exosomes the vehicle for protein aggregate propagation in neurodegenerative diseases? Acta Neuropathol Commun. 2017;5(1):64. doi: 10.1186/s40478-017-0467-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soria F.N., Pampliega O., Bourdenx M., Meissner W.G., Bezard E., Dehay B. Exosomes, an unmasked culprit in neurodegenerative diseases. Front. Neurosci. 2017;11:26. doi: 10.3389/fnins.2017.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takahashi R.H., Tobiume M., Sato Y., Sata T., Gouras G.K., Takahashi H. Accumulation of cellular prion protein within dystrophic neurites of amyloid plaques in the Alzheimer's disease brain. Neuropathology. 2011;31(3):208–214. doi: 10.1111/j.1440-1789.2010.01158.x. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Y., Zhao Y., Zhang L., Yu W., Wang Y., Chang W. Cellular prion protein as a receptor of toxic amyloid-β42 oligomers is important for Alzheimer's disease. Front. Cell. Neurosci. 2019;13:339. doi: 10.3389/fncel.2019.00339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cano A., Muñoz-Morales Á., Sánchez-López E., et al. Exosomes-based nanomedicine for neurodegenerative diseases: current insights and future challenges. Pharmaceutics. 2023;15(1):298. doi: 10.3390/pharmaceutics15010298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Otero-Ortega L., Laso-García F., Del Carmen Gómez-De Frutos M., et al. White matter repair after extracellular vesicles administration in an experimental animal model of subcortical stroke. Sci. Rep. 2017;7 doi: 10.1038/srep44433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li X., Corbett A.L., Taatizadeh E., et al. Challenges and opportunities in exosome research-Perspectives from biology, engineering, and cancer therapy. APL Bioeng. 2019;3(1) doi: 10.1063/1.5087122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hussen B.M., Faraj G.S.H., Rasul M.F., et al. Strategies to overcome the main challenges of the use of exosomes as drug carrier for cancer therapy. Cancer Cell Int. 2022;22(1):323. doi: 10.1186/s12935-022-02743-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perocheau D., Touramanidou L., Gurung S., Gissen P., Baruteau J. Clinical applications for exosomes: are we there yet? Br. J. Pharmacol. 2021;178(12):2375–2392. doi: 10.1111/bph.15432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ludwig N., Whiteside T.L., Reichert T.E. Challenges in exosome isolation and analysis in health and disease. Int. J. Mol. Sci. 2019;20(19):4684. doi: 10.3390/ijms20194684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Emam S.E., Ando H., Lila A.S.A., et al. A novel strategy to increase the yield of exosomes (extracellular vesicles) for an expansion of basic research. Biol. Pharm. Bull. 2018;41(5):733–742. doi: 10.1248/bpb.b17-00919. [DOI] [PubMed] [Google Scholar]
- 51.Villanueva-Flores F., Castro-Lugo A., Ramírez O.T., Palomares L.A. Understanding cellular interactions with nanomaterials: towards a rational design of medical nanodevices. Nanotechnology. 2020;31(13) doi: 10.1088/1361-6528/ab5bc8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Camussi G., Deregibus M.C., Bruno S., Cantaluppi V., Biancone L. Exosomes/microvesicles as a mechanism of cell-to-cell communication. Kidney Int. 2010;78(9):838–848. doi: 10.1038/ki.2010.278. [DOI] [PubMed] [Google Scholar]
- 53.Xu L., Wang, Xing Y.L., Yang G., Falconer R.J., Zhao Chun-Xia. Lipid nanoparticles for drug. Adv NanoBiomed Res. 2022;2 doi: 10.1002/anbr.202100109. [DOI] [Google Scholar]
- 54.Noble G.T., Stefanick J.F., Ashley J.D., Kiziltepe T., Bilgicer B. Ligand-targeted liposome design: challenges and fundamental considerations. Trends Biotechnol. 2014;32(1):32–45. doi: 10.1016/j.tibtech.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 55.Nsairat H., Khater D., Sayed U., Odeh F., Al Bawab A., Alshaer W. Liposomes: structure, composition, types, and clinical applications. Heliyon. 2022;8(5) doi: 10.1016/j.heliyon.2022.e09394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Akbarzadeh A., Rezaei-Sadabady R., Davaran S., et al. Liposome: classification, preparation, and applications. Nanoscale Res. Lett. 2013;8(1):102. doi: 10.1186/1556-276X-8-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Evers M.J.W., Kulkarni J.A., van der Meel R., Cullis P.R., Vader P., Schiffelers R.M. State-of-the-Art design and rapid-mixing production techniques of lipid nanoparticles for nucleic acid delivery. Small Methods. 2018;2(9) doi: 10.1002/smtd.201700375. [DOI] [Google Scholar]
- 58.Neves L.F., Duan J., Voelker A., et al. Preparation and optimisation of anionic liposomes for delivery of small peptides and cDNA to human corneal epithelial cells. J. Microencapsul. 2016;33(4):391–399. doi: 10.1080/02652048.2016.1202343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu C., Zhang L., Zhu W., et al. Barriers and strategies of cationic liposomes for cancer gene therapy. Mol Ther - Methods Clin Dev. 2020;18:751–764. doi: 10.1016/j.omtm.2020.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hafez I.M., Maurer N., Cullis P.R. On the mechanism whereby cationic lipids promote intracellular delivery of polynucleic acids. Gene Ther. 2001;8(15):1188–1196. doi: 10.1038/sj.gt.3301506. [DOI] [PubMed] [Google Scholar]
- 61.Cheng X., Lee R.J. The role of helper lipids in lipid nanoparticles (LNPs) designed for oligonucleotide delivery. Adv. Drug Deliv. Rev. 2016;99(Pt A):129–137. doi: 10.1016/j.addr.2016.01.022. [DOI] [PubMed] [Google Scholar]
- 62.Hou X., Zaks T., Langer R., Dong Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 2021;6(12):1078–1094. doi: 10.1038/s41578-021-00358-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Laouini A., Jaafar-Maalej C., Limayem-Blouza I., Sfar S., Charcosset C., Fessi H. Preparation, characterization and applications of liposomes: state of the Art. J Colloid Sci Biotechnol. 2012;1(2):147–168. doi: 10.1166/jcsb.2012.1020. [DOI] [Google Scholar]
- 64.Koĉiŝová E., Antalik A., Prochazka M. Drop coating deposition Raman spectroscopy of liposomes: role of cholesterol. Chem. Phys. Lipids. 2013;172–173:1–5. doi: 10.1016/j.chemphyslip.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 65.Kondow-Mcconaghy H.M., Muthukrishnan N., Erazo-Oliveras A., Najjar K., Juliano R.L., Pellois J.P. Impact of the endosomal escape activity of cell-penetrating peptides on the endocytic pathway. ACS Chem. Biol. 2020;15(9):2355–2363. doi: 10.1021/acschembio.0c00319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jokerst J.V., Lobovkina T., Zare R.N., Gambhir S.S. Nanoparticle PEGylation for imaging and therapy. Nanomedicine. 2011;6(4):715–728. doi: 10.2217/nnm.11.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cao Y., Dong X., Chen X. Polymer-modified liposomes for drug delivery: from fundamentals to applications. Pharmaceutics. 2022;14(4):778. doi: 10.3390/pharmaceutics14040778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sato Y.T., Umezaki K., Sawada S., et al. Engineering hybrid exosomes by membrane fusion with liposomes. Sci. Rep. 2016;6 doi: 10.1038/srep21933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Talsma H., Van Steenbergen M.J., Crommelin D.J.A. The cryopreservation of liposomes: 3. Almost complete retention of a water-soluble marker in small liposomes in a cryoprotectant containing dispersion after a freezing/thawing cycle. Int. J. Pharm. 1991;77(2–3):119–126. doi: 10.1016/0378-5173(91)90309-C. [DOI] [Google Scholar]
- 70.Almohammai A., Rahbarghazi R., Keyhanmanesh R., Rezaie J., Ahmadi M. Asthmatic condition induced the activity of exosome secretory pathway in rat pulmonary tissues. J. Inflamm. 2021;18(1):14. doi: 10.1186/s12950-021-00275-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Feghhi M., Rezaie J., Akbari A., et al. Effect of multi-functional polyhydroxylated polyhedral oligomeric silsesquioxane (POSS) nanoparticles on the angiogenesis and exosome biogenesis in human umbilical vein endothelial cells (HUVECs) Mater. Des. 2021;197 doi: 10.1016/j.matdes.2020.109227. [DOI] [Google Scholar]
- 72.Hassanpour M., Rezaie J., Darabi M., Hiradfar A., Rahbarghazi R., Nouri M. Autophagy modulation altered differentiation capacity of CD146(+) cells toward endothelial cells, pericytes, and cardiomyocytes. Stem Cell Res. Ther. 2020;11(1):139. doi: 10.1186/s13287-020-01656-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hedayat M., Ahmadi M., Shoaran M., Rezaie J. Therapeutic application of mesenchymal stem cells derived exosomes in neurodegenerative diseases: a focus on non-coding RNAs cargo, drug delivery potential, perspective. Life Sci. 2023;320 doi: 10.1016/j.lfs.2023.121566. [DOI] [PubMed] [Google Scholar]
- 74.Rezaie J., Nejati V., Mahmoodi M., Ahmadi M. Mesenchymal stem cells derived extracellular vesicles: a promising nanomedicine for drug delivery system. Biochem. Pharmacol. 2022;203 doi: 10.1016/j.bcp.2022.115167. [DOI] [PubMed] [Google Scholar]
- 75.Shaban S.A., Rezaie J., Nejati V. Exosomes derived from senescent endothelial cells contain distinct pro-angiogenic miRNAs and proteins. Cardiovasc. Toxicol. 2022;22(6):592–601. doi: 10.1007/s12012-022-09740-y. [DOI] [PubMed] [Google Scholar]
- 76.Mahbubfam S., Rezaie J., Nejati V. Crosstalk between exosomes signaling pathway and autophagy flux in senescent human endothelial cells. Tissue Cell. 2022;76 doi: 10.1016/j.tice.2022.101803. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.