Abstract
Rapid diagnosis of urogenital schistosomiasis caused by Schistosoma haematobium requires an accurate and timely assay, especially for low-intensity S. haematobium infection cases and in non-endemic areas. The mitochondrial cytochrome c oxidase 1 (cox1) gene fragment of S. haematobium was selected as detection target as this short fragment, which can be rapidly sequenced and yet possess good diagnostic resolution. A pair of primers and a fluorescent probe were designed according to the principle of recombinase-aided amplification (RAA), which was subsequently optimized and applied as an S. haematobium-specific RAA assay. Its diagnostic performance was validated for sensitivity and specificity in comparison to microscopy-based egg counting after urine filtration. The RAA assay could detect as little as 10 copies/μL of S. haematobium recombinant plasmid, and no cross-reactions were observed with S. mansoni, S. japonicum, Ancylostoma duodenale, Clonorchis sinensis, Echinococcus granulosus, or Ascaris lumbricoides. This test can be conducted at 39 °C and the whole RAA reaction can be completed within 20 min. The validation of the RAA assay showed that it had 100 % consistency with urine-egg microscopy, as it does not require an elaborate reading tool, is simple to use, and should be useful for field diagnostics and point-of-care applications.
Keywords: Schistosoma haematobium, Field diagnosis, Recombinase-aided amplification (RAA), Cytochrome c oxidase subunit 1 (cox1), Urine
Highlights
-
•
Molecular assays have successfully detected the S. haematobium cox1 gene fragment in urine samples.
-
•
The newly established RAA assay offers an accurate diagnostic method for S. haematobium infection compared with microscopy.
-
•
The lyophilization process had minimal impact on the detection of the S. haematobium cox1 fragment in urine samples.
-
•
This RAA assay for S. haematobium detection holds great promise for field diagnostics and point-of-care applications.
1. Introduction
Schistosomiasis is a major neglected tropical disease that can lead to severe bladder, ureteral, kidney, and genital pathologies; approximately 258 million people are currently infected worldwide, and approximately 779 million people are at risk [[1], [2], [3], [4]]. It is estimated that more than 85 % schistosomiasis cases occur in sub-Saharan Africa, approximately two-thirds of which are caused by Schistosoma haematobium, the pathogenic agent of urogenital schistosomiasis [5,6]. This type of schistosomiasis is often misdiagnosed as a urinary tract infection, whereas other schistosomiasis cases, such as S. japonicum and S. mansoni infections, are often misdiagnosed as gastrointestinal tract infections. S. haematobium can cause infections via the bladder vein and pelvic venous plexus, leading to genitourinary tract diseases, such as terminal hematuria, bladder irritation, and urinary tract obstruction, which can lead to renal failure. Thus, the late stages can be life threatening [2]. S. haematobium eggs deposited in the bladder tissue can also lead to chronic infection and bladder squamous cell carcinoma [7,8].
Several countries with endemic S. haematobium infections have successfully reduced morbidity and, in some cases, interrupted transmission by regularly treating infected or at-risk individuals with schistosomiasis [9]. As the prevalence and intensity of infections have decreased, low-intensity infections have become more common and a high proportion of them are asymptomatic. Molecular diagnostic assays for S. haematobium in urine are more sensitive than immunological methods [10,11] and these infections are also more difficult to diagnose using techniques, such as reagent strip testing, to detect microhaematuria or microscopy [12,13] to detect the presence of S. haematobium eggs in urine [[14], [15], [16]]. In addition, cases of schistosomiasis in travellers and migrants, especially those with S. haematobium infection, still occur in various non-endemic countries. For example, imported schistosomiasis cases were reported in China, with 73.8 % (262/355) of all imported schistosomiasis cases in 1979–2019 being caused by S. haematobium [17,18]. Europe has also received imported schistosomiasis in the last decade, mainly from endemic countries in Africa and the Middle East; for example, up to 21.2 % of individuals in cohorts of newly arrived asylum seekers coming to Italy between April 2014 and June 2015 had schistosomiasis [19]. In Corsica (France), an outbreak of locally transmitted urogenital schistosomiasis occurred in 2013, infecting more than 120 people [20]. Owing to the lack of diagnosis and treatment experience among clinicians and specialized diagnostic assays, cases of imported schistosomiasis are prone to be misdiagnosed [[21], [22], [23]].
Efforts have been made to establish a diagnostic method to detect low-intensity and asymptomatic S. haematobium infections in endemic areas and to monitor cases of imported S. haematobium infections in non-endemic areas. Both polymerase chain reaction (PCR) and real-time fluorescence quantification are sensitive and specific [10,24,25]; however, they are relatively time-consuming and require a laboratory with specific equipment [12]. Loop-mediated isothermal amplification (LAMP) can detect S. haematobium DNA with a higher sensitivity than PCR [[26], [27], [28]].
Recombinase-aided amplification (RAA), based on the principle of recombinase-mediated isothermal amplification, can rapidly detect specific nucleic acid fragments, such as S. japonicum, S. mansoni, SARS-CoV-2, and Orf virus [[29], [30], [31], [32]]. The recombinant enzyme can combine with the primer DNA at room temperature to form an aggregate of enzymes and primers. When primers search for complementary sequences that exactly match the template DNA, the double-stranded structure of the template DNA can be unrolled with the help of a single-stranded DNA-binding protein, resulting in the formation of new complementary DNA strands under the action of DNA polymerase, and the amplification product grows exponentially. It is simple to conduct and allows real-time monitoring at 37–42 °C [29,30,33]. Here, we aimed to establish and assess an RAA assay to detect a S. haematobium cytochrome c oxidase subunit 1 (cox1) gene fragment in 5 min and validate it using urine samples from S. haematobium infection. This assay represents a new tool to detect S. haematobium and may play an important role in point-of-care applications and in the diagnosis of imported cases of urogenital schistosomiasis.
2. Methods
2.1. Urine sample collection and processing
Urine samples were collected from 57 pupils from a primary school in Zanzibar (United Republic of Tanzania) maintained by the China-Zanzibar-World Health Organization (WHO) Schistosomiasis Control and Elimination Project [[34], [35], [36]]. All urine samples were collected between 10:00 and 14:00 to determine the optimum egg output [14]. All urine samples had been previously screened for S. haematobium infection by urine filtration (10 mL) and examined under a microscope by skilled local technicians [35,37]. They were classified according to the WHO recommendations [38] as no egg load (considered as negative samples), having a light (<50 eggs/10 mL urine) or heavy (≥50 eggs/10 mL urine) egg load. The samples were then lyophilized, stored at −80 °C and transported to our laboratory at the Jiangsu Institute of Parasitic Diseases in Wuxi, China. The Ethics Review Committee of Zanzibar approved sample collection (ZAMREC/002/MAY/014).
2.2. Extraction of nucleic acids
Nucleic acids were extracted from urine samples using an AxyPrep™ Body Fluid Viral DNA/RNA Miniprep Kit (Axygen Scientific, Union City, CA, USA). Thereafter, the DNA concentration was determined using a NanoDrop 2000 system (Thermo Fisher Scientific, Waltham, MA, USA), and the nucleic acid was stored at −80 °C.
According to the principle of recombinase-aided isothermal amplification and with reference to previous research [29,30], primers were designed using an S. haematobium cox1 gene fragment as the target sequence (GenBank accession no. MT579449.1) (Table 1). Primers and probes were synthesized by Sangon Biotech (Shanghai, China).
Table 1.
Primers for S. haematobium cox1 gene fragment and internal probe for RAA assay.
| Primer/probe | Sequence (5′–3′) |
|---|---|
| Forward primer | CTATGATTATAGGGATTCCTACAGGTATAAAG |
| Reverse primer | TAATATATCTAATGAAGAAGCTGATAAAGC |
| Probea | CTGTGGGTCTCGTGTATGAGATCCTATAGTTTGATGATTGGTTGGTTTTA |
The 31st base modifies the fluorescence reporter group, the 35th base modifies the quenching group, and the 33rd base modifies the tetrahydrofuran residue.
2.3. RAA assay
RAA reactions were performed in a 50-μL reaction volume using RAA Nucleic Acid Amplification Kit (Lot. FEZFB0; Jiangsu Qitian Gene Biotechnology Co., Ltd., China). The reaction mixtures contained 5 μL template DNA, 25 μL reaction buffer, 4.2 μL primers (forward and reverse; 10 μM), 0.6 μL probe (10 μM), 12.7 μL double-distilled water (ddH2O), and 2.5 μL 280 mM magnesium acetate. The reaction system was mixed and incubated for 4 min in a 0.2-mL PCR tube in a constant-temperature oscillation mixer (RAA B6100; Jiangsu Qitian Gene Biotechnology Co., Ltd., Jiangsu, China). Thereafter, the fluorescence values were assessed every 20 s for the whole 20 min using a real-time fluorometer (RAA F1620, Jiangsu Qitian Gene Biotechnology Co., Ltd., China) at 39 °C.
2.4. RAA assay sensitivity
An amplified and purified S. haematobium mitochondrial cox1 gene fragment (approximately 176 bp) was synthesized into a plasmid vector by Sangon Biotech (Shanghai, China), and its initial concentration was determined using a NanoDrop 2000 system (Thermo Fisher Scientific, Waltham, MA, USA). Recombinant plasmid samples with concentrations of 1.0 × 106, 1.0 × 105, 1.0 × 104, 1.0 × 103, 1.0 × 102, and 10 copies/μL were subjected to the RAA assay, involving detection of fluorescence signals. The negative control was ddH2O.
2.5. RAA assay specificity
The DNA of S. haematobium, S. mansoni, S. japonicum, Ancylostoma duodenale, Clonorchis sinensi, Echinococcus granulosus, and Ascaris lumbricoides was obtained from the Key Laboratory on Technology for Parasitic Disease Prevention and Control (Wuxi, China). DNA concentrations were determined using the NanoDrop 2000 system (Thermo Fisher Scientific, Waltham, MA, USA), and 2 ng of each DNA was used as a template and then subjected to the RAA assay, involving the detection of fluorescence signals. The negative control was ddH2O, which was used as a template.
2.6. RAA validation
Among the 57 urine samples, 9 were found positive and 48 were found negative via microscopy-based egg counting. Nine egg-positive urine samples with light or heavy S. haematobium egg loads (five with a light load [U1,U82,U89,U171, and U48] and four with a heavy load [U137, U79, U96, and U240]) and 48 egg-negative urine samples were used to validate the RAA assay. Two-hundred microliters of each urine DNA was extracted using an AxyPrep™ Body Fluid Viral DNA/RNA Miniprep Kit (Axygen Scientific, Union City, CA, USA). Lastly, 5 μL extracted DNA from each sample was subjected to the RAA assay, involving detection of fluorescence signals. In addition, 100 copies of the recombinant plasmid were used as a positive control.
3. Results
3.1. RAA assay sensitivity
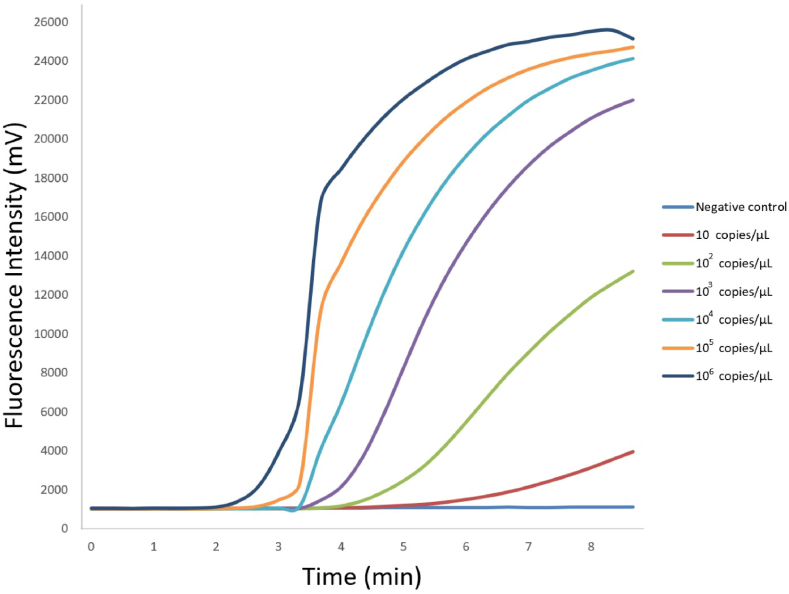
The S. haematobium cox1 recombinant plasmid samples at various concentrations (1.0 × 106, 1.0 × 105, 1.0 × 104, 1.0 × 103, 1.0 × 102, and 10 copies/μL) were amplified via RAA. The fluorescence values of all samples (but not of the negative control) increased within 5 min at the beginning of the detection period. The concentration of 1.0 × 106 copies/μL led to the earliest peak (Fig. 1).
Fig. 1.
Sensitivity of the RAA assay in detecting S. haematobium cox1 recombinant plasmid at various concentrations
The sensitivity of the RAA assay was demonstrated by its ability to detect and amplify S. haematobium cox1 recombinant plasmid at various concentrations. All concentrations of S. haematobium cox1 recombinant plasmid were successfully detected and amplified, highlighting the effectiveness of the assay. Notably, the negative control (ddH2O) did not show any amplification, confirming the specificity of the assay.
3.2. RAA assay specificity
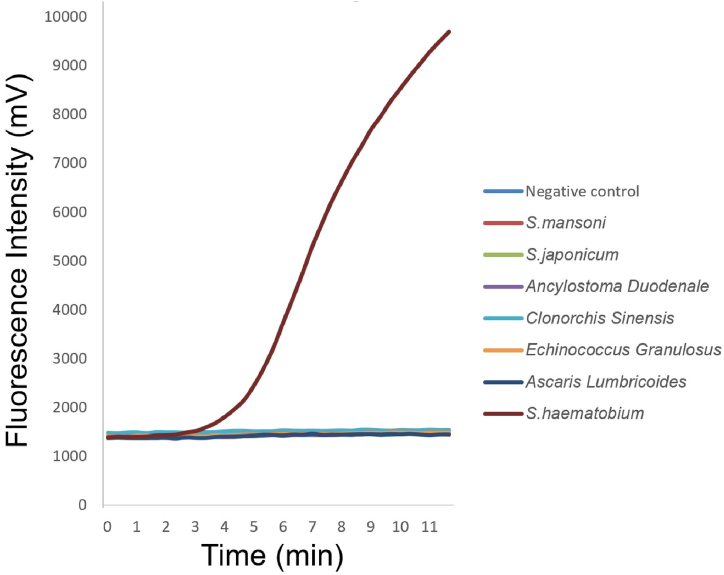
No fluorescence was observed for S. japonicum, S. mansoni, Ancylostoma duodenale, Clonorchis sinensis, Echinococcus granulosus, or Ascaris lumbricoides, whereas increased fluorescence (starting within 5 min of the detection period) was observed for S. haematobium (Fig. 2).
Fig. 2.
Specificity of the RAA assay in detecting various parasitic species
This figure illustrates the specificity of the RAA assay in detecting different parasite species. Notably, no fluorescence was observed in Schistosoma japonicum, Schistosoma mansoni, Ancylostoma duodenale, Clonorchis sinensis, Echinococcus granulosus, or Ascaris lumbricoides. In contrast, a noticeable increase in fluorescence that began within 5 min of the detection period was observed in S. haematobium. This highlights the ability of the assay to accurately differentiate and detect S. haematobium from other parasitic species.
3.3. RAA assay performance
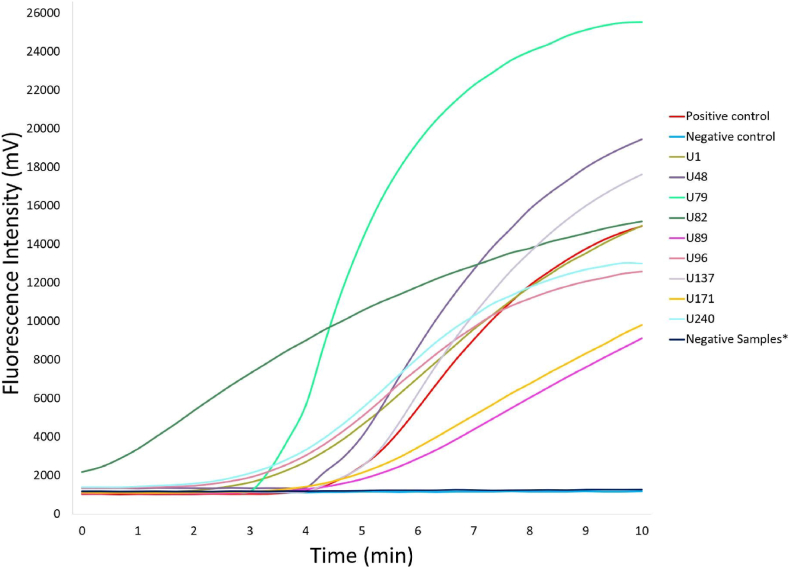
Nine urine samples with light or heavy S. haematobium egg loads (five with light loads [U1,U82,U89,U171, and U48] and four with heavy loads [U137, U79, U96, and U240]) and 48 egg-negative urine samples were amplified using RAA assay (Fig. 3, Supplementary Table 1). The fluorescence signal of all the nine of the samples (U1, U48, U79, U82, U89, U96, U137, U171, and U240) increased rapidly within 5 min of the beginning of the detection period (Fig. 3). No fluorescence was observed in any of the negative samples. Thus, among the nine egg-positive urine samples, none led to a false-negative result based on RAA (Fig. 3, Table 2), while none led to a false-positive result among the 48 egg-negative urine samples. The consistency between the RAA assay and the microscopy-based egg counting was 100 % (kappa value = 1). The positive and negative predictive values were 100 %, and nine egg-positive urine samples (from patients with S. haematobium infection) and 48 egg-negative urine samples were examined using microscopy-based egg counts in Zanzibar and RAA assay in China.
Fig. 3.
Evaluation of the RAA assay's performance using field-collected urine samples
The performance of the RAA assay was assessed using field-collected urine samples. A total of 57 urine samples were included in the study, 9 of them tested positive and 48 tested negative based on microscopy-based egg counting. Specifically, among the 57 samples:Nine urine samples were positive for S. haematobium eggs with varying egg loads (five with a light load [U1, U82, U89, and U171, and U48] and four with a heavy load [U137, U79, U96, and U240]). All 48 egg-negative urine samples were used as negative controls. This validation process helped confirm the accuracy and reliability of the RAA assay in diagnosing S. haematobium infections in a field setting.
Table 2.
Diagnostic performance of the RAA assay.
| Reference test (microscopy-based egg counting*) | |||||
|---|---|---|---|---|---|
| + | – | Total | |||
| RAA assay | + | 9 | 0 | 9 | PPV: 100 % |
| – | 0 | 48 | 48 | NPV: 100 % |
|
| Total | 9 | 48 | 57 | ||
| Sensitivity: 100 % (9/9) |
Specificity: 100 % (48/48) |
||||
PPV, positive predictive value; NPV, negative predictive value. Fifty seven egg-positive urine samples (from patients with S. haematobium infection) were examined using microscopy-based egg counting in Zanzibar and RAA assays in China. *Test conducted in duplicate; positive: ≥1 egg was observed; negative: 0 eggs were observed in either reading.
4. Discussion
Sensitive and specific diagnostic methods are critical for the elimination of schistosomiasis as they enable the timely and accurate identification and treatment of cases. In addition, better diagnostics can enable accurate surveillance of cases (including imported cases) in low- and non-endemic areas [39,40]. Molecular detection of S. haematobium DNA in urine is considered a highly sensitive indicator of infection [12,13] and RAA assay is a highly sensitive method that does not require costly tools or a sophisticated laboratory and can deliver results within 5–15 min.
The results of the RAA assay of the recombinant plasmids harboring the S. haematobium mitochondrial cox1 gene fragment showed that the higher the concentration, the earlier the fluorescent signal appeared; conversely, the lower the concentration, the later the fluorescent signal appeared (Fig. 1). This shows that the RAA assay has the potential to be used as a semi-quantitative assay for the detection S. haematobium in urine. In the present study, the RAA assay could detect 10 copies/μL of recombinant plasmids harboring the S. haematobium mitochondrial cox1 gene fragment, which was the lowest concentration assessed (Fig. 1). In theory, a single copy could be detected using the RAA assay if the assay is optimized to increase sensitivity (focusing on the primer and probe combination, target gene region, and concentration) in future research.
No cross-reactivity was observed with the other two major schistosome species or with other common parasitic helminths (Fig. 2), indicating that targeting the S. haematobium cox1 gene fragment led to high specificity for detecting S. haematobium infection using the RAA assay. The mitochondrial S. haematobium cox1 gene fragment was selected as the specific target to design the primers because it is conserved (despite the natural intra-species genetic variation observed across Africa) and can be used for species identification [[41], [42], [43]].
To validate the RAA assay, 57 lyophilized urine samples collected from a primary school were subjected to the RAA assay, including 9 egg-positive urine samples (five with a light infection and four with a heavy infection) and 48 egg-negative urine samples. The RAA assay correctly categorized all 9 (100 %) egg-positive samples and all 48 egg-negative samples (Table 2), indicating that the RAA assay can detect S. haematobium DNA in urine when patients have mild infections. For the nine egg-positive urine samples (five samples with a light load [U1,U82,U89,U171, and U48] and all four with a heavy load [U137, U79, U96, and U240]), fluorescence values were rapidly observed within 5 min, and the signals were strong (Fig. 3) and observed at the very beginning of the detection period.
Regarding the fluorescence values of all urine samples, the onset of fluorescence values varied and were not related to the egg load. Because the fluorescence values varied despite the samples having a low infection load, the difference may have been due to the original sample characteristics or sample degradation. As all egg-positive urine samples were collected from patients with S. haematobium infection in Zanzibar, lyophilized, transported to China, and then subjected to the RAA assay, differences among samples in the processes of sample collection, lyophilization, and transportation (which involve factors that can lead to DNA degradation) could be the reason for the difference in fluorescence values. However, although the urine samples were lyophilized and reconstituted in normal physiological saline prior to DNA extraction, the strong fluorescence values detected in the RAA assay (Fig. 3) showed that these processes had little effect on the detection of the S. haematobium cox1 fragment in egg-positive urine samples. The AxyPrep™ Body Fluid Viral DNA/RNA Miniprep Kit was used for DNA extraction because it is simple and does not require complex equipment. It can extract low quantities of nucleic acids at a low cost ($2.45/sample). The source of DNA in the RAA assay still needs to be determined; however, it has been reported that cell-free parasite DNA or S. haematobium eggs could be the source when DNA is detected using molecular methods [44].
The RAA assay requires a short reaction time (∼20 min), whereas the LAMP assay requires approximately 35–40 min to amplify DNA [45](Supplementary Table 2). Moreover, unlike the reference test (microscopy-based egg counting), the RAA assay is not time-consuming, which implies that the RAA assay has huge potential to rapidly detect low-intensity S. haematobium infections. RAA and recombinase polymerase amplification (RPA), which rely on the principle of isothermal nucleic acid amplification, have recently been developed as molecular diagnostic methods. The difference between these two methods is the source of the recombinase, with RPA using UvsX from the T4 phage and RAA using recombinases from an extensive range of sources, such as bacteria and fungi [46]. In addition, the reaction sample (urine) was lyophilized into a single unit, which simplified the sample pretreatment process, facilitated transportation, and allowed long-term preservation at room temperature. The limitation of our study is that the shortage of urine samples and the RAA cross-reaction test did not involve other pathogens that could cause urinary tract infection due to the shortage of samples.
5. Conclusion
The RAA assay presented here is a rapid and sensitive approach for the diagnosis of S. haematobium infection and can be used to assess schistosomiasis control measures and diagnose S. haematobium infection in low-endemic and non-endemic areas with imported cases of schistosomiasis.
Funding
This study was supported by the National Natural Science Foundation of China (grant number 82173586) and the Jiangsu Provincial Department of Science and Technology (grant number BZ2020003).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Ethics approval and consent to participate
All aspects of the study were performed in accordance with the national ethics regulations and approved by the Ethics Review Committee of Zanzibar (ZAMREC/002/MAY/014).
Consent for publication
Not applicable.
Data availability statement
Data included in article/supp. material/referenced in article.
CRediT authorship contribution statement
Song Zhao: Writing – review & editing, Writing – original draft, Methodology, Conceptualization. Qiaoqiao Zhang: Writing – original draft, Validation, Project administration, Data curation. Xinyao Wang: Validation. Wei Li: Validation, Resources. Saleh Juma: Validation, Resources, Data curation. Robert Berquist: Writing – review & editing. Jianfeng Zhang: Validation, Resources. Kun Yang: Writing – review & editing, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:Kun Yang reports financial support was provided by National Natural Science Foundation of China. Kun Yang reports financial support was provided by Jiangsu Provincial Department of Science and Technology.
Acknowledgements
No applicable.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e23031.
Contributor Information
Jianfeng Zhang, Email: zhangjianfeng@jipd.com.
Kun Yang, Email: yangkun@jipd.com.
Abbreviations
- RAA
Recombinase-aided amplification
- cox1
cytochrome c oxidase subunit I
- RPA
Recombinase polymerase amplification
- LAMP
Loop-mediated isothermal amplification
- PCR
Polymerase chain reaction
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.McManus D.P., et al. Schistosomiasis. Nat Rev Dis Primers. 2018;4:13. doi: 10.1038/s41572-018-0013-8. [DOI] [PubMed] [Google Scholar]
- 2.Colley D.G., et al. Human schistosomiasis. Lancet. 2014;383:2253–2264. doi: 10.1016/S0140-6736(13)61949-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanchard T.J. Schistosomiasis. Travel Med Infect Dis. 2004;2:5–11. doi: 10.1016/j.tmaid.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 4.Molehin A.J. Schistosomiasis vaccine development: update on human clinical trials. J. Biomed. Sci. 2020;27:28. doi: 10.1186/s12929-020-0621-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brindley P.J., Hotez P.J. Break Out: urogenital schistosomiasis and Schistosoma haematobium infection in the post-genomic era. PLoS Neglected Trop. Dis. 2013;7 doi: 10.1371/journal.pntd.0001961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hotez P.J., Kamath A. Neglected tropical diseases in sub-saharan Africa: review of their prevalence, distribution, and disease burden. PLoS Neglected Trop. Dis. 2009;3:e412. doi: 10.1371/journal.pntd.0000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adebayo A.S., et al. The microbiome in urogenital schistosomiasis and induced bladder pathologies. PLoS Neglected Trop. Dis. 2017;11 doi: 10.1371/journal.pntd.0005826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwartz D.A. Helminths in the induction of cancer II. Schistosoma haematobium and bladder cancer. Trop. Geogr. Med. 1981;33:1–7. [PubMed] [Google Scholar]
- 9.Rollinson D., et al. Time to set the agenda for schistosomiasis elimination. Acta Trop. 2013;128:423–440. doi: 10.1016/j.actatropica.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 10.Frickmann H., et al. Evaluation of a duplex real-time PCR in human serum for simultaneous detection and differentiation of Schistosoma mansoni and Schistosoma haematobium infections - cross-sectional study. Trav. Med. Infect. Dis. 2021;41 doi: 10.1016/j.tmaid.2021.102035. [DOI] [PubMed] [Google Scholar]
- 11.Obeng B.B., et al. Application of a circulating-cathodic-antigen (CCA) strip test and real-time PCR, in comparison with microscopy, for the detection of Schistosoma haematobium in urine samples from Ghana. Ann. Trop. Med. Parasitol. 2008;102:625–633. doi: 10.1179/136485908X337490. [DOI] [PubMed] [Google Scholar]
- 12.Weerakoon K.G., Gordon C.A., McManus D.P. DNA diagnostics for schistosomiasis control. Trav. Med. Infect. Dis. 2018;3 doi: 10.3390/tropicalmed3030081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vinkeles Melchers N.V., et al. Diagnostic performance of Schistosoma real-time PCR in urine samples from Kenyan children infected with Schistosoma haematobium: day-to-day variation and follow-up after praziquantel treatment. PLoS Neglected Trop. Dis. 2014;8 doi: 10.1371/journal.pntd.0002807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le L., Hsieh M.H. Diagnosing urogenital schistosomiasis: dealing with diminishing returns. Trends Parasitol. 2017;33:378–387. doi: 10.1016/j.pt.2016.12.009. [DOI] [PubMed] [Google Scholar]
- 15.Knopp S., et al. Urogenital schistosomiasis elimination in Zanzibar: accuracy of urine filtration and haematuria reagent strips for diagnosing light intensity Schistosoma haematobium infections. Parasites Vectors. 2018;11:552. doi: 10.1186/s13071-018-3136-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.King C.H., Bertsch D. Meta-analysis of urine heme dipstick diagnosis of Schistosoma haematobium infection, including low-prevalence and previously-treated populations. PLoS Neglected Trop. Dis. 2013;7 doi: 10.1371/journal.pntd.0002431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang L., et al. Imported schistosomiasis: a new public Health challenge for China. Front. Med. 2020;7 doi: 10.3389/fmed.2020.553487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang J., et al. Current status and transmission risks of oversea imported schistosomiasis in China. Chinese journal of schistosomiasis control. 2019;31:26–32. doi: 10.16250/j.32.1374.2019021. [DOI] [PubMed] [Google Scholar]
- 19.Buonfrate D., et al. Extended screening for infectious diseases among newly-arrived asylum seekers from Africa and Asia, Verona province, Italy, April 2014 to June 2015. Euro Surveill. 2018;23 doi: 10.2807/1560-7917.ES.2018.23.16.17-00527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boissier J., et al. Outbreak of urogenital schistosomiasis in Corsica (France): an epidemiological case study. Lancet Infect. Dis. 2016;16:971–979. doi: 10.1016/S1473-3099(16)00175-4. [DOI] [PubMed] [Google Scholar]
- 21.Shiff C. The importance of definitive diagnosis in chronic schistosomiasis, with reference to Schistosoma haematobium. J Parasitol Res. 2012;2012 doi: 10.1155/2012/761269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang W., et al. African schistosomiasis in mainland China: risk of transmission and countermeasures to tackle the risk. Parasites Vectors. 2013;6:249. doi: 10.1186/1756-3305-6-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gomes L.I., Enk M.J., Rabello A. Diagnosing schistosomiasis: where are we? Rev. Soc. Bras. Med. Trop. 2014;47:3–11. doi: 10.1590/0037-8682-0231-2013. [DOI] [PubMed] [Google Scholar]
- 24.Cnops L., et al. A Schistosoma haematobium-specific real-time PCR for diagnosis of urogenital schistosomiasis in serum samples of international travelers and migrants. PLoS Neglected Trop. Dis. 2013;7:e2413. doi: 10.1371/journal.pntd.0002413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keller D., et al. Performance of a real-time PCR approach for diagnosing Schistosoma haematobium infections of different intensity in urine samples from Zanzibar. Infect Dis Poverty. 2020;9:128. doi: 10.1186/s40249-020-00726-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bayoumi A., et al. Loop-mediated isothermal amplification(LAMP): sensitive and rapid detection of Schistosoma Haematobium DNA in urine samples of Egyptian suspected cases. J. Egypt. Soc. Parasitol. 2016;46:299–308. [PubMed] [Google Scholar]
- 27.Gandasegui J., et al. The rapid-heat LAMPellet method: a potential diagnostic method for human urogenital schistosomiasis. PLoS Neglected Trop. Dis. 2015;9 doi: 10.1371/journal.pntd.0003963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang C., et al. Application of DNA-based diagnostics in detection of schistosomal DNA in early infection and after drug treatment. Parasites Vectors. 2011;4:164. doi: 10.1186/1756-3305-4-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao S., et al. Establishment of a recombinase-aided isothermal amplification technique to detect Schistosoma japonicum specific gene fragments. Chinese journal of schistosomiasis control. 2018;30:273–277. doi: 10.16250/j.32.1374.2018120. [DOI] [PubMed] [Google Scholar]
- 30.Zhao S., et al. Establishment of the gene detection method of Schistosoma mansoni based on the recombinase-aided isothermal amplification assay. Chinese journal of schistosomiasis control. 2020;32:335–339. doi: 10.16250/j.32.1374.2020058. [DOI] [PubMed] [Google Scholar]
- 31.Xue G., et al. Reverse-transcription recombinase-aided amplification assay for rapid detection of the 2019 novel coronavirus (SARS-CoV-2) Anal. Chem. 2020;92:9699–9705. doi: 10.1021/acs.analchem.0c01032. [DOI] [PubMed] [Google Scholar]
- 32.Pang F., Long Q. Recent advances in diagnostic approaches for orf virus. Appl. Microbiol. Biotechnol. 2023;107:1515–1523. doi: 10.1007/s00253-023-12412-8. [DOI] [PubMed] [Google Scholar]
- 33.Li J., Macdonald J. Advances in isothermal amplification: novel strategies inspired by biological processes. Biosens. Bioelectron. 2015;64:196–211. doi: 10.1016/j.bios.2014.08.069. [DOI] [PubMed] [Google Scholar]
- 34.He M.Z., et al. A Google Earth-based database management for schistosomiasis control in Zanzibar. Geospat Health. 2019;14 doi: 10.4081/gh.2019.740. [DOI] [PubMed] [Google Scholar]
- 35.Wang X.Y., et al. Efficacy of China-made praziquantel for treatment of Schistosomiasis haematobium in Africa: a randomized controlled trial. PLoS Neglected Trop. Dis. 2019;13 doi: 10.1371/journal.pntd.0007238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang K., Mehlhorn H., editors. Sino-African Cooperation for Schistosomiasis Control in Zanzibar : a Blueprint for Combating Other Parasitic Diseases. Springer; Cham: 2021. [Google Scholar]
- 37.Muhsin M.A., et al. The indispensability of snail control for accelerating schistosomiasis elimination: evidence from Zanzibar. Tropical Medicine and Infectious Disease. 2022;7 doi: 10.3390/tropicalmed7110347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.World Health O. World Health Organization; Geneva: 2013. Schistosomiasis: Progress Report 2001 - 2011, Strategic Plan 2012 - 2020. [Google Scholar]
- 39.Bergquist R., Johansen M.V., Utzinger J. Diagnostic dilemmas in helminthology: what tools to use and when? Trends Parasitol. 2009;25:151–156. doi: 10.1016/j.pt.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 40.Cavalcanti M.G., et al. Schistosomiasis in areas of low endemicity: a new era in diagnosis. Trends Parasitol. 2013;29:75–82. doi: 10.1016/j.pt.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 41.Jia W.Z, et al. Research progress on complete mitochondrial genome sequences and their application for trematodes. Chin. J. Vet. Sci. 2011;31:926–932. [Google Scholar]
- 42.Webster B.L., et al. Rapid diagnostic multiplex PCR (RD-PCR) to discriminate Schistosoma haematobium and S. bovis. J. Helminthol. 2010;84:107–114. doi: 10.1017/S0022149X09990447. [DOI] [PubMed] [Google Scholar]
- 43.Webster B.L., et al. Genetic diversity within Schistosoma haematobium: DNA barcoding reveals two distinct groups. PLoS Neglected Trop. Dis. 2012;6 doi: 10.1371/journal.pntd.0001882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weerakoon K.G., McManus D.P. Cell-free DNA as a diagnostic tool for human parasitic infections. Trends Parasitol. 2016;32:378–391. doi: 10.1016/j.pt.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 45.Soroka M., Wasowicz B., Rymaszewska A. Loop-mediated isothermal amplification (LAMP): the better sibling of PCR? Cells. 2021;10 doi: 10.3390/cells10081931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fan X., et al. Clinical validation of two recombinase-based isothermal amplification assays (RPA/RAA) for the rapid detection of African swine fever virus. Front. Microbiol. 2020;11:1696. doi: 10.3389/fmicb.2020.01696. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.
Data included in article/supp. material/referenced in article.