Abstract
EnvZ, a membrane receptor kinase-phosphatase, modulates porin expression in Escherichia coli in response to medium osmolarity. It shares its basic scheme of signal transduction with many other sensor-kinases, passing information from the amino-terminal, periplasmic, sensory domain via the transmembrane helices to the carboxy-terminal, cytoplasmic, catalytic domain. The native receptor can exist in two active but opposed signaling states, the OmpR kinase-dominant state (K+ P−) and the OmpR-P phosphatase-dominant state (K− P+). The balance between the two states determines the level of intracellular OmpR-P, which in turn determines the level of porin gene transcription. To study the structural requirements for these two states of EnvZ, mutational analysis was performed. Mutations that preferentially affect either the kinase or phosphatase have been identified and characterized both in vivo and in vitro. Most of these mapped to previously identified structural motifs, suggesting an important function for each of these conserved regions. In addition, we identified a novel motif that is weakly conserved among two-component sensors. Mutations that alter this motif, which is termed the X region, alter the confirmation of EnvZ and significantly reduce the phosphatase activity.
To survive, bacteria must adapt to their rapidly changing environment, and two-component regulatory systems are key to this process. More than 250 two-component pairs have been discovered, and these systems sense and respond to a variety of environmental parameters and insults (5). A two-component system is generally composed of a sensor histidine kinase and a response regulator. The sensor kinase is often localized to the cytoplasmic membrane, where it senses external stimuli and transduces this information to the response regulator. The response regulator, which is normally a transcriptional regulatory protein that controls expression of a set of related genes (9, 18), mediates the proper cellular response to the stimuli.
A key feature of the information flow from the sensor to the regulator is protein phosphorylation and dephosphorylation (references 9 and 18 and references therein). The sensor can be autophosphorylated by ATP at a conserved histidine residue and subsequently transfer the phosphoryl group onto a conserved aspartic acid residue in the regulator. In many cases, the sensor kinase can dephosphorylate the phosphorylated response regulator. Thus, unlike their counterparts in eukaryotic signal transduction systems, three enzymatic activities are often associated with bacterial two-component sensor kinases: the autokinase, response regulator kinase, and response regulator phosphate phosphatase. How the catalytic domain of one protein catalyzes these three different reactions is a topic of considerable interest.
In this study, we approached this question by mutational analysis of EnvZ. EnvZ is a well-characterized sensor kinase that is involved in osmoregulation in Escherichia coli (6, 7). It shares basic architectural features with many other sensor kinases, having two transmembrane segments that divide the protein into an N-terminal sensory domain and a C-terminal catalytic domain. The C-terminal domain contains all of the conserved motifs common to the sensor family: H, N, D/F, G1, and G2 boxes (Fig. 1) (18, 25). Thus, mechanisms underlying the functions of EnvZ have implications for many other transmembrane sensors as well.
FIG. 1.
Schematic presentation of the structure of EnvZ. Conserved regions in the catalytic cytoplasmic domain are indicated with lettered boxes.
EnvZ forms a two-component pair with its cognate response regulator, OmpR. Together, EnvZ and OmpR enable cells to sense the external osmolarity and respond to it by regulating the transcription of two porin genes: ompF and ompC (4, 16, 19). Genetic evidence suggests that EnvZ can exist in two active but opposed signaling states: the OmpR kinase-dominant (K+ P−) state and the OmpR-P phosphatase-dominant (K− P+) state (19, 22, 24). High levels of OmpR-P activate ompC but repress ompF transcription; low levels of OmpR-P activate ompF transcription only. Neither ompC nor ompF is transcribed in the absence of OmpR-P. Thus, the level of ompF and ompC transcription reflects the level of OmpR-P, which is set by the sum of EnvZ kinase and phosphatase activities in vivo. By using lacZ fusions to the porin gene promoters, this can be easily monitored.
To investigate the structural components involved in the OmpR kinase or OmpR-P phosphatase activity of EnvZ, mutations that shift the balance of these enzymatic activities toward either the K− P+ or K+ P− state were identified and characterized both in vivo and in vitro. We found that most of the mutations altered a previously defined structural motif, demonstrating their critical importance. In addition, our studies revealed a novel motif that we have termed the X region, which is shared by other two-component sensors.
MATERIALS AND METHODS
Bacterial strains, plasmids, and phage.
The E. coli strains and plasmids used in this study are described in Table 1. Phage P1vir was used for transduction. Standard microbiological techniques were used for strain construction and bacterial growth (23). Cells were grown at 37 or 30°C with shaking in appropriate media.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Genotype | Reference or source |
|---|---|---|
| E. coli strains | ||
| FR247 | MC4100 Φ(ompC′-lacZ+)10-25 ompR101 [λpSG10(ompR+)] envZ247 zhf-37::Tn10 | 22 |
| FR250 | MC4100 Φ(ompC′-lacZ+)10-25 ompR101 [λpSG10(ompR+)] envZ250 zhf-37::Tn10 | 22 |
| FR1050 | MC4100 Φ(ompC′-lacZ+)10-25 ompR101 [λpSG10(ompR+)] envZ247 | This study |
| FR1070 | MC4100 Φ(ompC′-lacZ+)10-25 ompR101 [λpSG10(ompR+)] envZ250 | This study |
| MC4100 | F−araD139 Δ(argF-lac)U169 rpsL150 relA flb-5301 ptsF25 deoC1 thiA1 | 2 |
| SG480Δ900 | MC4100 Δ(envZ-malP)900 malP::neo | Lab stock |
| WH10 | MC4100 Φ(ompC′-lacZ+)10-25 araB Δ(envZ-malP)900 malP::neo | This study |
| WH20 | MC4100 Φ(ompF′-lacZ+)16-23 araB Δ(envZ-malP)900 malP::neo | 10 |
| WH30 | MC4100 Φ(ompC′-lacZ+)10-25 [λSG10(ompR+)] Δ(envZ-malP) malP::neo | This study |
| WH40 | MC4100 Φ(ompF′-lacZ+)16-23 [λpSG10(ompR+)] Δ(envZ-malP) malP::neo | This study |
| WH56 | MC4100 envZ::kan Φ(ompC′-lacZ+)10-25 | 10 |
| WH67 | MC4100 recA::cam envZ::kan Φ(ompF′-lacZ+)16-23 | 10 |
| Plasmids | ||
| pFR29 | ompR+ envZ+Apr | 22 |
| pFR32 | ompR+ envZ+Camr (Ts) | Lab stock |
| pEnvZ | envZ+ Apr | 10 |
| pMBP-OmpR | Φ(malE′-′ompR) Apr | 11 |
| pSG115 | envZ115 Apr | 12 |
Media and reagents.
Most media were prepared as described previously (23). ortho-Nitrophenyl-β-d-galactoside (ONPG) for β-galactosidase assay was purchased from Sigma. Restriction enzymes, β-agarase, and T4 DNA ligase were from New England BioLabs, Inc. Taq polymerase and reagents used for PCR amplification, T4 DNA polymerase, and Sequenase were from United States Biochemical Corp. [γ-33P]ATP (1,000 to 3,000 Ci/mmol; 10 mCi/ml) and [α-33P]ATP (2,000 Ci/mmol; 10 mCi/ml) were from NEN Life Science Products. ATP and ADP were from Boehringer Mannheim. The oligonucleotide primers used for PCR and DNA sequencing were provided by the Princeton University Department of Molecular Biology Synthesis/Sequencing Facility. NuSieve low-melting-point agarose was purchased from FMC BioProducts. Reagents for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) were purchased from National Diagnostics. The protein assay reagent was from Bio-Rad.
β-Galactosidase assay.
β-Galactosidase activities were determined by using a microtiter plate assay that has been previously described (24). Cells were grown overnight in appropriate medium at 37°C, subcultured (1:40) into 2 ml of the same medium, and then grown to mid-log phase at 37°C. Activities are expressed as (units/A600) × 103, where 1 U = 1 μmol of ortho-nitrophenol formed per min. A minimum of four independent assays was performed for each strain, and the results were averaged for display as bar graphs.
PCR amplification and DNA sequence analysis.
PCR amplification and DNA sequence analyses were performed essentially as previously described (22). DNA was amplified from either the bacterial cells or plasmids. For sequencing analysis, the template DNA was either from PCR amplification or from restriction digestion. The template was purified by gel electrophoresis in 1% NuSieve low-melting-point agarose, and the agarose was digested with β-agarase. Sequence analysis was then performed as indicated in the supplier’s instructions.
Screening for K− P+ mutations.
pEnvZ was first subjected to UV irradiation and then transformed into WH67. Transformants were plated onto lactose-MacConkey agar. Plasmids were isolated from the Lac− white transformants and retransformed into WH67. The plasmids that maintain the Lac− phenotype were sequenced by using primers throughout the entire length of envZ.
Isolation of intragenic suppressors for the K− P+ Mutants.
Several independent cultures of each strain (FR247 and FR250) were grown overnight in Luria-Bertani medium. They were washed in minimal salts, and an aliquot of each culture was plated on lactose minimal agar. Multiple isolated colonies appeared after 2 to 3 days of growth, of which two were picked from each plate and purified by restreaking on lactose minimal agar. Each isolate was characterized for its expression of the ompC′-lacZ+ fusion by streaking on lactose-MacConkey and lactose-tetrazolium agars to examine different ranges of β-galactosidase expression. If both isolates from any given plate were the same color on lactose-tetrazolium, they were considered siblings and only one was kept for further analysis.
Mapping of the suppressor mutations.
P1 transduction was performed to map the suppressor mutations. Each isolate was used as the donor. FR1050 and FR1070, which are identical to FR247 and FR250, respectively, except that they do not carry a Tn10 insertion, were used as recipients. In such a transduction, both the donor and recipient carry the same kinase deficiency mutation. The only difference among transductants is the additional presence or absence of a given suppressor mutation. If the suppressor is not linked to the Tn10, which is about 85% linked to envZ, then all transductants should be Lac−, but if the suppressor lies in envZ, Lac+ progeny will also result. After confirmation of their linkage to Tn10, the suppressors were moved into a clean ompC′-lacZ+ and ompF′-lacZ+ background by P1 transduction with WH30 and WH40 as recipients. The presence of suppressors in the ompC′-lacZ+ transductants was confirmed by the Lac+ phenotype. Marker rescue was used for the ompF′-lacZ+ transductants.
Separation of the suppressor mutation from the original kinase deficiency mutation.
The fragment of DNA that contains only the suppressor mutation was first subcloned onto pFR32, which has a temperature-sensitive origin of replication, and then recombined onto the chromosome as described by Hamilton et al. (8). WH56 was used as the recipient strain, and lactose-tetrozolium agar was used to facilitate the screening of chromosomal recombinants. The presence of each mutation on the chromosomal location was confirmed by PCR amplification and sequence analysis. Each envZ allele was then moved into clean strain backgrounds by P1 transduction with WH10 and WH20 as recipients.
In vitro phosphorylation and dephosphorylation analysis.
Membranes that were enriched for EnvZ through the multicopy plasmid pEnvZ were used for these assays. Procedures for protein phosphorylation and dephosphorylation were the same as previously described (10).
UV-cross-linking experiments.
Mutant envZ alleles were first subcloned onto pSG115 through in vitro DNA manipulation. Mutant EnvZ115 proteins were then purified from transformants carrying pSG115 with the mutant alleles and solubilized by using a previously described procedure (13, 14). Concentrations of purified EnvZ115 were quantified by use of the Bio-Rad protein assay reagent. The UV-cross-linking procedure was similar to that described by Ninfa et al. (17). The reaction mix contains 6 μl of EnvZ115 (0.3 mg/ml) in phosphate-buffered saline and 0.2% deoxycholate, 0.5 μl of [α-33P]ATP, and 4 μl of assay buffer (0.1 M Tris-HCl buffer [pH 8.0], 50 mM KCl, 5 mM CaCl2, 1 mM phenylmethylsulfonyl fluoride, and 10% glycerol). The cross-linking of ATP onto EnvZ115 was achieved by exposing the reaction mix to UV light for 10 min on ice. SDS sample buffer was then added to the reaction mix. After being boiled for 5 min, the sample was subjected to SDS-PAGE, and the amount of [α-33P]ATP cross-linked onto EnvZ115 was visualized by autoradiography.
RESULTS
In order to identify regions in EnvZ that are important for both the kinase and phosphatase activities, we have sought mutations that specifically alter only one of these activities. In particular, we have searched for K− P+ and K+ P− mutations.
Identification of K− P+ mutations.
To obtain information on the structural requirements for kinase activity, we performed a genetic screen to isolate mutations that render EnvZ kinase deficient but still maintain the phosphatase activity.
We know that in an envZ null background, ompF is expressed to a certain degree, because OmpR-P, which is formed by an acetyl phosphate-dependent mechanism, accumulates to significant levels in strains lacking EnvZ phosphatase activity (10). Thus, an envZ null strain with an ompF′-lacZ+ fusion is Lac+ and forms red colonies on lactose-MacConkey agar. Subsequent introduction of a plasmid carrying a K− P+ envZ allele will decrease the accumulated OmpR-P and confer a Lac− phenotype; therefore, the colony will appear white on this medium. This forms the basis for a screen for K− P+ mutations of envZ.
Plasmid pEnvZ was first mutagenized by UV light and then transformed into an envZ null ompF′-lacZ+ recA strain background. The transformants were plated onto lactose-MacConkey agar, and Lac− (white) colonies were picked and purified. After confirmation of the linkage of the Lac− phenotype to the plasmid by retransforming the plasmid into the original strain background, the entire envZ gene was sequenced. Two novel mutations were identified, envZ343 (N343K) and envZ390 (F390L), which have changed conserved residues in the N and D/F boxes, respectively (Fig. 1 and Table 2).
TABLE 2.
Mutations isolated in this study
| envZ allele | Residue change(s) | Location | Phenotypea | Isolationb |
|---|---|---|---|---|
| envZ982 | L23R | TM1 | Cc F− | envZ250 sup |
| envZ972 | L26R | TM1 | Cc F− | envZ250 sup |
| envZ977 | T30P | TM1 | Cc F− | envZ250 sup |
| envZ973 | L43P | TM1 | Cc F− | envZ250 sup |
| envZ976 | Q44P | TM1 | Cc F− | envZ250 sup |
| envZ981 | L230Q | H region | Cc F− | envZ250 sup |
| envZ975 | V241G | H region | Cc F− | envZ250 sup |
| envZ974 | Δ(A225-A239) | H region | Cc F− | envZ250 sup |
| envZ971 | D273Y | X region | Cc F− | envZ250 sup |
| envZ978 | Q283P | X region | Cc F− | envZ250, envZ247 sup |
| envZ959 | P248Q | H region | Cc F− | envZ247 sup |
| envZ954 | P248L | H region | Cc F− | envZ247 sup |
| envZ966 | Y287D | X region | Cc F− | envZ247 sup |
| envZ958 | Y287N | X region | Cc F− | envZ247 sup |
| envZ965 | Y287S | X region | Cc F− | envZ247 sup |
| envZ964 | L288P | X region | Cc F− | envZ247 sup |
| envZ962 | T402K | G2 region | Cc F− | envZ247 sup |
| envZ343 | N343K | N region | Cc F− | K− P+ screen |
| envZ390 | F390L | D/F region | Cc F− | K− P+ screen |
C and F, OmpC and OmpF, respectively; c, constitutive.
envZ250 sup mutations were isolated as the intragenic suppressors of envZ250; envZ247 sup mutations were isolated as the intragenic suppressors of envZ247. K− P+ screen, mutations were isolated by screening for mutants with no OmpR-P.
Characterization of the K− P+ mutations.
The two K− P+ mutations isolated as described above were subcloned onto a plasmid with a temperature-sensitive origin of replication, and subsequently recombined onto the chromosome at the normal location (see Materials and Methods). Neither of the mutations can activate ompC or ompF transcription as indicated by the Lac− phenotype in ompC′-lacZ+ or ompF′-lacZ+ fusion strains. The level of ompF transcription in either mutant envZ strain was similar to that in an envZ-ompR deletion mutant, and it was much lower than that in an envZ null (envZ::Kan) mutant (data not shown). We conclude that these two mutant EnvZ proteins are K− P+, since each prevents the accumulation of OmpR-P derived from acetyl phosphate.
Identification of K+ P− mutations.
As described above, the phosphatase activity of the K− P+ mutant EnvZ proteins diminishes OmpR-P in vivo, causing a porin-negative phenotype. The Lac− phenotype exhibited by ompC′-lacZ+ fusion strains carrying these mutations provided a means to isolate the other type of mutation that we are searching for, K+ P− mutations, through the analysis of intragenic suppressors that confer a Lac+ phenotype.
The nature of the original K− P+ mutation dictates the type of suppressor expected. Information flow within EnvZ occurs by a strictly ordered pathway that involves different domains of the receptor in turn. Environmental cues sensed by the periplasmic domain are transduced by the transmembrane helices to the catalytic cytoplasmic domain (Fig. 1). The defects caused by the original lesion should not be suppressed by mutations that affect steps upstream in this signaling pathway. To obtain a broader spectrum of mutations, we used two well-characterized K− P+ mutations for the suppressor analysis: envZ250 (22), which causes a P159S change at the junction between the periplasmic domain and the second transmembrane segment (TM2) of EnvZ, and envZ247 (22), which causes an A239T change in the conserved H box.
The two strains used to isolate intragenic suppressors were FR247 and FR250. Each carries an ompR101 null allele at the normal chromosomal locus, which is complemented in trans by the ompR+ prophage λpSG10 integrated at λatt. FR247 carries envZ247 linked to ompR101, while FR250 carries envZ250. Both strains harbor a Tn10 that is 85% linked to envZ. These features were chosen to facilitate suppressor mapping: suppressors in envZ must be linked to the Tn10. In addition, both strains carry the ompC-lacZ+ transcriptional fusion. In the presence of either envZ250 or envZ247, ompC is not transcribed, and the strains are phenotypically Lac−. Any suppressor mutation that restores expression from the ompC promoter will increase the production of β-galactosidase and allow the strain to grow on lactose. A fusion to the ompC promoter, as opposed to ompF, was chosen for two reasons. First, an envZ null mutation allows residual expression of the ompF-lacZ+ owing to OmpR-P produced from acetyl phosphate (10), and this residual expression is sufficient to support growth on lactose. The use of an ompC fusion excludes envZ null mutations from the selection. The second reason is that the phenotype of a known K+ P− mutation, envZ473, is OmpF− OmpC+. Such potentially interesting mutations would be excluded if the selection was performed in an ompF-lacZ+fusion background. Twelve suppressors for envZ250 and 10 suppressors for envZ247 were isolated. Linkage mapping showed that 10 suppressors of envZ250 and 9 suppressors of envZ247 were linked to envZ.
To determine the DNA sequence changes in the isolated suppressor strains, the entire envZ gene from each strain was amplified by PCR, and primers throughout the gene were used to sequence the products. All of these mutant envZ genes contain the original mutation plus a suppressor. All of the suppressor mutations are single-base-pair changes, except for one deletion, and most of them are transversions. As predicted, we obtained a different array of suppressors from each initial mutation. Suppressors of the envZ250 (P159S) mutation mapped within two domains (Table 2): five suppressors affect the TM1 region, and five affect the catalytic domain. Among the latter, two missense changes and one deletion fall immediately upstream of the conserved autophosphorylation site, histidine-243, and two additional mutations fall near each other in a region slightly downstream of the H box, which we have termed the X region. The suppressors of envZ247 (A239T) affect the catalytic domain only (Table 2). Two of these fall in the H box, six affect the X region (the Y287D mutation was isolated twice independently), and one affects a residue in the conserved G2 box. Q283P, which lies in the X region, was the only mutation isolated as a suppressor of both kinase deficiency alleles.
The X region of EnvZ has not generally been recognized as a conserved motif. However, scattered but significant homology within this region can be found in all two-component sensors (5). The sequence alignment in this region of some representative sensors is shown in Fig. 2.
FIG. 2.
Sequence alignment of the X regions from some E. coli sensor kinases. Sequences of the sensor kinases were obtained from Swissbank. A multiple sequence alignment program, PIMA 1.4 (Pattern-Induced [local] Multiple Alignment) was used. The previously identified H, N, D/F, and G boxes were identified as conserved motifs through this program. Highly conserved residues are shaded. Mutational changes within the X region are shown in boldface.
Characterization of the K+ P− mutations.
All of the suppressors restore ompC expression in the original K− P+ mutant background, suggesting that they all restore kinase activity. To quantitate the strength of suppression, we measured the levels of porin transcription under both high- and low-osmolarity conditions by using ompF′-lacZ+ and ompC′-lacZ+ fusion strains. Our results showed that despite the presence of the original K− P+ mutation, most of the suppressors activated ompC and repressed ompF at both low and high osmolarity (data not shown).
As noted in the introduction, levels of porin gene transcription are determined by the amount of OmpR-P, which is set by the sum of the kinase and phosphatase activities of EnvZ. At high osmolarity, EnvZ is mainly K+ P−, resulting in high levels of OmpR-P, which in turn will activate ompC and repress ompF. At low osmolarity, however, EnvZ is shifted toward K− P+, resulting in low levels of OmpR-P, which can activate ompF only. The constitutive activation of ompC and repression of ompF in most of the double envZ mutants indicate high levels of OmpR-P even under low-osmolarity conditions. This suggests that these suppressor mutations shifted the balance of enzymatic activities from the original K− P+ state toward K+ P−. The balance was reset such that even at low osmolarity, the level of OmpR-P was still higher than that in the wild type at high osmolarity.
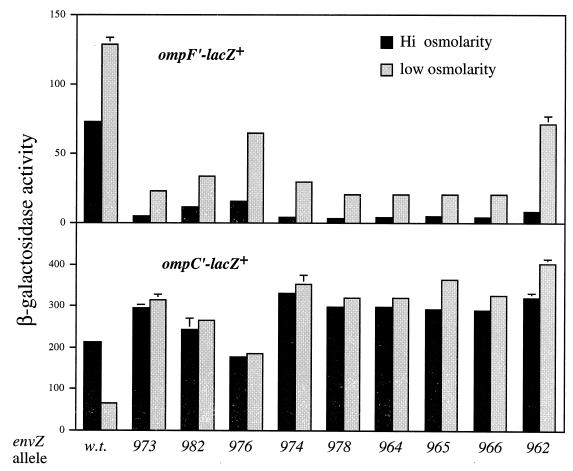
To determine if the increased kinase activity caused by the intragenic suppressors depends on the original K− P+ mutation (envZ250 or envZ247), the suppressors were isolated by in vitro DNA manipulation and recombined in single copy at the chromosomal location (see Materials and Methods). The kinase and phosphatase activities conferred by the resulting single mutant envZ alleles were then assessed in vivo by assaying the β-galactosidase activities in ompF′-lacZ+ and ompC′-lacZ+ fusion strains following growth at different osmolarities. As illustrated in Fig. 3, all of the isolated suppressor mutations tested activate ompC and repress ompF constitutively, suggesting that these suppressor mutations alone reset the balance of the enzymatic activity toward K+ P−: the restoration of kinase activity does not depend on the original K− P+ mutation. This is consistent with the locations of the suppressor mutations at or downstream from the original mutations in the intramolecular signal transduction pathway (Fig. 1). All further analyses were performed with these single mutant (K+ P−) genes or their products.
FIG. 3.
Osmoregulation of porin gene expression in strains carrying the isolated envZ intragenic suppressor mutations. Transcription of ompF and ompC was monitored by using ompF′-lacZ+ and ompC′-lacZ+ fusions. β-Galactosidase assays were performed with log-phase cells grown under either high (A medium plus 15% sucrose)- or low (A medium)-osmolarity conditions. Cells of wild-type (w.t.) and suppressor strains were tested. Note the difference in scale for the β-galactosidase activities from the ompF and ompC fusion strains. Error bars indicate standard deviations.
Membrane localization and stability of the mutant EnvZ proteins.
The K+ P− mutations in or near TM1 introduce either an arginine or a proline. Accordingly, they could affect the localization of mutant EnvZ proteins to the membrane. To test this, fractionation experiments were performed with cells carrying the mutant genes on the plasmid pEnvZ. We found that although the TM1 mutant EnvZ proteins fractionated with the cell membranes, their levels were generally about one-fourth that of the wild type (data not shown). We suspect that most of the TM1 mutations reduce the efficiency of membrane targeting and that mislocalized molecules are degraded. All of the K+ P− mutations in the H region caused similar reductions in the yield of mutant EnvZ protein. We suspect that these mutant proteins are unstable. Low yields and mutant protein instability complicate meaningful interpretation of the in vitro activity assays we employ. Accordingly, we limited biochemical analysis to those mutant proteins that are membrane localized at levels equivalent to the wild type (see below). This subset includes those with the X-region mutations (envZ964 and envZ966), the two K− P+ mutations in the N and D/F boxes (envZ343 and envZ390), and the K+ P− mutations in TM1 (envZ976) and the G2 box (envZ962).
Enzymatic activities of the mutant EnvZ proteins.
To confirm and extend the predictions based on genetic analysis with the lacZ fusion strains, autokinase, OmpR kinase, and OmpR-P phosphatase assays were performed with cell membranes that were enriched for the mutant EnvZ proteins. This was done by using strains carrying the mutant envZ genes on the multicopy plasmid pEnvZ (see Materials and Methods).
(i) The N-box (envZ343) and D/F-box (envZ390) mutations.
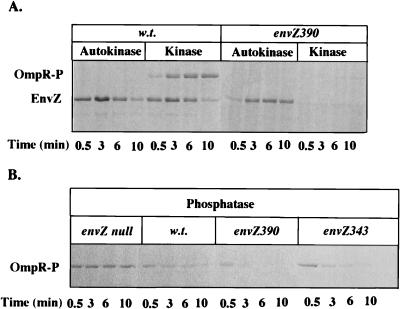
The envZ343 and envZ390 mutations confer the K− P+ phenotype when assayed in vivo (see previous section). Under our assay conditions, the EnvZ390 mutant protein exhibited reduced autokinase activity and barely detectable kinase activity (Fig. 4A). The EnvZ343 mutant protein could not be autophosphorylated by ATP (data not shown). Thus, neither protein could phosphorylate OmpR to any significant level. However, both proteins exhibited wild-type levels of OmpR-P phosphatase activity (Fig. 4B). Indeed, EnvZ390 may have elevated phosphatase activity. These results are consistent with the observed K− P+ phenotype conferred by these mutant proteins in vivo.
FIG. 4.
Enzymatic assays of the K− P+ mutants in vitro. (A) The autokinase and OmpR kinase activities were measured by incubating cell membranes that were enriched for EnvZ with [γ-33P]ATP in the absence (autokinase) or presence (kinase) of purified MBP-OmpR. Aliquots were removed at the indicated time points. Proteins were separated by SDS-PAGE and subjected to autoradiography. The EnvZ proteins enriched in cell membranes were either wild type (w.t.) or EnvZ390 (F390L). (B) The OmpR-P phosphatase activities were measured by using [33P]MBP-OmpR as the substrate (see Materials and Methods). Membranes isolated from strains that were enriched for wild-type, EnvZ390, or EnvZ343 were used for the assay. Proteins were displayed as described above. As a control, the phosphatase activity of membranes from the vector control strain (envZ null) was also measured.
Note that the total 33P labeling in the EnvZ390 kinase reaction (Fig. 4B) is much lower than that in the EnvZ390 autokinase reaction (Fig. 4A). One possible explanation is that in the kinase reaction, the 33P label on EnvZ390 is efficiently transferred to OmpR but subsequently undergoes rapid hydrolysis due to an elevated phosphatase activity of EnvZ390. Such a hypothesis predicts an increased yield of 33Pi in the EnvZ390 kinase reaction compared to the wild-type EnvZ kinase reaction. To test this, we examined the levels of ATP and Pi in these kinase reactions by thin-layer chromotography. Our results showed much higher concentrations of 33Pi in the wild-type EnvZ kinase reaction than in the EnvZ390 reaction (data not shown). Thus, we favor an alternative explanation: OmpR, the substrate for the kinase reaction, may inhibit the autophosphorylation of EnvZ390 by ATP (see Discussion).
(ii) The TM1 (envZ976), X-region (envZ966), and G2-box (envZ962) mutations.
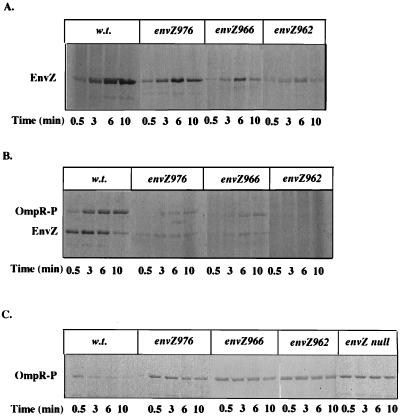
As shown in Fig. 3, the envZ976, envZ966, and envZ962 mutations confer a K+ P− phenotype. These mutant proteins were also subjected to biochemical analysis, and the results are presented in Fig. 5. Each of the mutant proteins retained the ability to autophosphorylate, although they had lower autokinase activity than the wild type (Fig. 5A). The observed OmpR kinase activities of these mutant proteins were significantly lower than that of the wild type, which may be due to their lower autokinase activities. Nonetheless, each of them could phosphorylate OmpR (Fig. 5B). In contrast, none of these mutant proteins exhibited OmpR-P phosphatase activity under conditions in which wild-type EnvZ could dephosphorylate OmpR-P completely in about 10 min. (Fig. 5C). Results similar to those for EnvZ966 were also obtained with proteins altered by two other X-region mutations, envZ964 and envZ965 (data not shown). These results are consistent with the observed K+ P− phenotype conferred by these mutant proteins in vivo. Although these mutations affected the autokinase and OmpR kinase activities to various degrees, they all severely decreased the phosphatase activity. This will shift the balance between the kinase and phosphatase reactions such that the formation of OmpR-P is strongly favored.
FIG. 5.
Enzymatic assays of the K+ P− mutations in vitro. Cell membranes that were enriched for wild-type or mutant EnvZ proteins were used for the in vitro phosphorylation and dephosphorylation assays. Mutant EnvZ proteins tested include suppressors near TM1 (envZ976), in the X region (envZ966), and in the G2 box (envZ962). (A) Autokinase assay. Cell membranes were incubated with [γ-33P]ATP in assay buffer, and the reaction was stopped at different time points. Proteins were separated by SDS-PAGE and subjected to autoradiography. (B) OmpR kinase assay. The procedure was the same as for panel A except that purified MBP-OmpR was added to the reaction mixture. (C) OmpR-P phosphatase assay. MBP–OmpR-33P was used as the substrate for the assay. Reactions were stopped at different time points after incubation of the substrate, cell membranes, and 1 mM cold ATP in the assay buffer. The remaining MBP–OmpR-P was visualized through autoradiography after separation by SDS-PAGE. Membranes from an envZ null strain were also tested as a control.
Interestingly, similar to the case for EnvZ390, the total 33P labeling in the EnvZ962 kinase reaction (Fig. 5B) is much lower than that in the EnvZ962 autokinase reaction (Fig. 5A). Again, no increased ATP cleavage to yield 33Pi by EnvZ962 during the kinase reaction can be detected by using the thin-layer chromatography assay (data not shown). Thus, we suspect that OmpR may inhibit the autophosphorylation of EnvZ962 by ATP as well (see Discussion).
ATP binding by the mutant EnvZ proteins.
The conserved N, D/F, and G boxes of two-component sensor kinases have been implicated in nucleotide binding because of sequence homologies to eukaryotic kinases (25). ATP cross-linking was performed to determine if the mutations that alter these boxes affect ATP binding. The mutant proteins to be tested were purified as a truncated form, EnvZ115, in which the amino terminus of the molecule, including TM1, has been removed. Following purification, the aggregated EnvZ115 proteins are solubilized and renatured (12–14) prior to the cross-linking experiment.
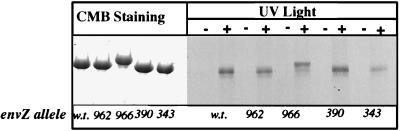
The mutant EnvZ115 proteins were incubated with [α-33P]ATP, and UV light was used to cross-link radioactive label onto the protein (17). As shown in Fig. 6, under our assay conditions with ATP concentrations of approximately 0.25 μM, cross-linking was observed with all of the mutant EnvZ115 proteins tested, including those containing lesions in the X region (envZ966) and the N (envZ343), D/F (envZ390), and G2 (envZ962) boxes. Cross-linking to the N-box mutant (envZ343) is noticeably less efficient than that to the others, and we suggest that this mutation decreases affinity for ATP. However, the other mutants behave in a manner indistinguishable from that of the wild type.
FIG. 6.
UV-cross-linking assay. Various mutant alleles were first subcloned onto pSG115, a plasmid that overproduces a truncated version of EnvZ, EnvZ115. After solubilization with deoxycholate, mutant proteins were incubated with [α-33P]ATP on ice in the presence (+) or absence (−) of UV light for 10 min. The amount of ATP cross-linked onto the protein was visualized through autoradiography after SDS-PAGE. The autoradiograph is shown on the right. A similar amount of each mutant EnvZ115 protein was used in the assay, as shown by the Coomassie blue-stained SDS-polyacrylamide gel on the left. envZ alleles tested included the wild type (w.t.), envZ962 (G2 box), envZ966 (X region), envZ390 (D/F box), and envZ343 (N box).
The use of truncated EnvZ115 mutant proteins complicates biochemical analysis because the truncation itself may alter activity. We have performed the UV-cross-linking assay with cell membranes enriched for the full-length mutant EnvZ proteins. The results showed that all of the mutant EnvZ proteins can bind ATP. However, the strength of binding varied from assay to assay and was less reproducible.
The X-region mutations alter the conformation of EnvZ.
Figure 6 shows that envZ966 slows the migration of EnvZ in SDS-polyacrylamide gels. Similar gel migration patterns were also observed with other X-region mutant proteins. We have also observed that the X-region mutant EnvZ proteins have proteolytic patterns different from those of the wild type after limited trypsin proteolysis (data not shown). Thus, we conclude that the X-region mutations alter the conformation of EnvZ.
DISCUSSION
We sought to identify the structural components of EnvZ that are involved in either the OmpR kinase or the OmpR-P phosphatase activity by mutational analysis. Two striking generalities emerge from this analysis. First, although the mutations identified decreased one of these activities more than the other, most affected both simultaneously. This supports and strengthens our previous proposal that these active sites overlap substantially (10). Second, all of the mutations lie in the conserved structural motifs previously identified by sequence homologies. Even the newly discovered X region is weakly conserved among all the sensors (5). Clearly these conserved structural motifs are critically important for function.
The structural motifs highlighted by these envZ mutations will be discussed in turn.
The newly identified X region.
The majority of the K+ P− mutations that alter the cytoplasmic domain of EnvZ fall within a region approximately 40 residues downstream of histidine-243. We have termed this the X region, and as noted above, this region is weakly conserved among all sensors. A representation of the alignment in this region is illustrated in Fig. 2. One codon in particular, that for tyrosine-287, is the site of mutations causing three different amino acid substitutions, one of which (Y287D) was independently isolated twice. Interestingly, neither this position nor other positions altered by X mutations in this study are highly conserved. Tyrosine-287 is located at the end of a fairly conserved sequence that could form an amphipathic α-helix, and it is followed by a conserved positive charge (R289). A mutation at this conserved site (R289C) confers a weak K+ P− phenotype that decreases but does not completely abolish the phosphatase activity when assayed in vivo and in vitro (data not shown).
At least one mutation in the X region in another sensor kinase has been characterized. A P188Q mutation in NtrB, the sensor protein involved in nitrogen utilization, also causes a K+ P− phenotype (1). Strikingly, P188 is located at a position in NtrB that corresponds to Y287 in EnvZ (Fig. 2). In addition, the dual sensors for nitrate and nitrite, NarX and NarQ, share a stretch of 11 identical residues in this region (3, 20), and it has been proposed that this stretch is important in conferring specificity on sensor-response regulator interaction (21).
Mutations within the X region of EnvZ have clearly defined properties. In vivo these mutations abolish the phosphatase activity and thus confer an OmpC+ OmpF− phenotype. In vitro, these mutations cause detectable defects in both the autokinase and kinase activities, but they abolish the phosphatase activity. Clearly the X region is important for the dephosphorylation of OmpR-P. Finally, unlike mutations in all of the other structural motifs, the X-region mutations cause a significant alteration in EnvZ conformation (Fig. 6).
Since the X-region mutations alter EnvZ conformation, it seems likely that their effect on the phosphatase is indirect. They may alter EnvZ structure in a way that prevents interaction with OmpR-P. Alternatively, and perhaps more likely, the mutations may prevent productive interaction with OmpR-P. We believe that the kinase and phosphatase active sites overlap substantially (10), and we know that ATP and the H box are required for both reactions. Accordingly, subtle changes in EnvZ conformation may shift the balance of these reactions as noted below. The indirect effect of mutations in the X region on the phosphatase activity is consistent with the conservation of an X region in all sensors, including those do not appear to have phosphatase activity.
The vicinity of the phosphorylated histidine.
A number of suppressors of the K− P+ mutations envZ247 and envZ250 fall into the conserved H region, which contains the autophosphorylation site, histidine-243. After they were separated from the original K− P+ mutations, these H-region mutations conferred an OmpC+ OmpF− phenotype, suggesting that they have shifted the balance of the kinase and phosphatase reactions toward the kinase reaction (i.e., K+ P−), resulting in increased OmpR-P levels in vivo.
Previous work indicates that the H box is directly involved in both OmpR kinase and OmpR-P phosphatase activities, and we have proposed a common transition state with histidine-243 in close contact with aspartate-55 of OmpR for both reactions. Phosphotransfer occurs from histidine-243-P to aspartate-55, but water replaces the phosphorylated histidine side chain, leading to hydrolysis (10). Thus, mutations in the H region could affect the kinase activity, the phosphatase activity, or both activities.
Perhaps the most striking H-region mutation reported here is the deletion caused by envZ974. This deletion removes 15 amino acids immediately upstream of the crucial histidine-243. One explanation for the K+ P− phenotype conferred by this mutation is that the deletion simply removes a structure that blocks access of ATP to this histidine. According to this view, autophosphorylation occurs when histidine-243 is accessible to ATP, and EnvZ-P will subsequently phosphorylate OmpR (the kinase reaction). However, when ATP is inaccessible to histidine-243, EnvZ will function as an OmpR-P phosphatase.
The other conserved motifs in the cytoplasmic domain.
In addition to the H region, previous sequence alignment revealed three more conserved motifs among the bacterial two-component sensor-kinases: the N, D/F, and G boxes. It is thought that these elements form a nucleotide-binding surface within the active site (18, 25). We obtained mutations in all three boxes.
The N-box mutation, envZ343, changed the second-most-conserved asparagine residue in this box to a lysine residue. The properties of this mutation are similar to those of previously characterized mutations that changed the most-conserved asparagine residue in this region to several other amino acids (28). All of these N-box mutations confer a porin-negative phenotype, and as expected, the mutant proteins are K− P+ in vitro. Clearly the N region is critical for the kinase activity.
As shown by UV-cross-linking experiments, the N-region mutant does not bind ATP as well as wild-type EnvZ, consistent with the prediction that the N box is involved in nucleotide binding. Recall that ATP binding is required not only for the autokinase activity but also for the OmpR-P phosphatase activity. Indeed, ATP or ADP is required for the phosphatase activity of EnvZ343 (data not shown). To account for the differential effects of the N-box mutations on these two activities, we propose that these mutations affect ATP binding in such a way that the γ-phosphate of ATP is misaligned with histidine-243 for phosphorylation.
envZ390 is the first reported mutation in the conserved D/F box of sensor kinases. It confers a porin-negative phenotype, and the mutant protein is K− P+ in vitro. Thus, the D/F box is important for kinase activity. In eukaryotic kinases a conserved DFG motif is believed to be involved in chelating the Mg2+ that bridges the β- and γ-phosphates of ATP, thus helping to orient the phosphate moiety that will be transferred (26). By analogy, the D/F region of sensor kinases is thought to be involved in nucleotide binding as well (25). Interestingly, EnvZ390, in which the conserved phenylalanine-390 is changed to a leucine, does not appear to be defective in ATP binding (Fig. 6). Furthermore, this mutation does not affect the phosphatase activity or the autokinase activity severely, both of which directly require ATP binding. Instead, it most strongly affects the kinase activity, a reaction that does not require ATP when phosphorylated EnvZ is utilized. In fact, the addition of maltose-binding protein–OmpR (MBP-OmpR) to the autokinase reaction mixture appears to inhibit the autophosphorylation of EnvZ390. It is difficult to explain the effects of these D/F mutations solely in terms of nucleotide binding. Directly or indirectly, the D/F mutations appear to affect the presentation of OmpR to phosphorylated EnvZ.
An intragenic suppressor of envZ247, envZ962 (T402K), was isolated in the conserved G2 region. Previous attempts to alter all of the conserved glycine residues in this region simultaneously resulted in a mutant protein that was too unstable to be characterized (28). Thus, envZ962 is the first envZ mutation in this region to be characterized. Strains carrying this mutation are phenotypically OmpC+ OmpF−. When assayed in vitro, the mutant protein exhibits decreased autokinase, very low kinase, and no phosphatase activity, demonstrating the importance of this region for all three reactions, especially the phosphatase activity.
It is interesting to compare EnvZ962 with EnvZ390, since the G and D/F boxes are both thought to be important for positioning the β- and γ-phosphates of ATP. Although the two mutations confer opposite phenotypes in vivo, reflecting their negative effects on opposing activities in vitro, the mutants have several common features: neither appears to be defective in nucleotide binding, and for both, purified MBP-OmpR appears to inhibit the autophosphorylation reaction. Thus, mutations in both boxes can affect the interaction between EnvZ and OmpR.
The first transmembrane segment.
Five of the intragenic suppressors of the periplasmic mutant protein EnvZ250 (P159S), but none of the suppressors of the cytoplasmic mutant protein EnvZ247 (A239T), fall into or near the first transmembrane segment (TM1) of EnvZ (Table 2). Based on hydrophobicity, this segment is predicted to include residues from arginine-14 to serine-42 (27). All of these TM1 mutant proteins are localized to the inner membranes as shown by membrane fractionation experiments. Thus, it does not appear that these mutations work by causing the mislocalization of the mutant EnvZ proteins. These suppressors cause constitutive activation of ompC in the presence or absence of envZ250, and when assayed in vitro, these mutant EnvZ proteins were phosphatase defective. Interestingly, they were still capable of sensing and responding to osmolarity as indicated by the stronger repression of ompF at high osmolarity (Fig. 3). Apparently these functions are not critically dependent on TM1 (but see reference 15).
A helical-wheel representation of the TM1 α-helix shows that four suppressors (L23R, S26R, T30P, and Q44P) lie on the same face of this helix. In addition, two other previously identified K+ P− mutations, P41S and P41L, also map to this face, and a suppressor for these two TM1 mutations was found in TM2 (R180W) (27). These data suggest that a specific face of the TM1 helix, which contains residues I19 L23, S26, T30, F37, and P41, is critical to maintain the proper balance between the kinase and phosphatase activities. Point mutations in this face shift this balance toward K+ P−.
The model.
The external stimulus (15) modulates the relative amount of time that the external domain spends in either of two distinct conformations. The two conformations affect the transmembrane signaling differently. A specific face on TM1 is critical for this signaling process. In response to the transmembrane signaling, the region surrounding the critical histidine-243 is positioned into one of two different states by movements in which the X region is critically involved. In one state this histidine is properly aligned with ATP, which is bound at a surface composed of the N, D/F, and G boxes, for autophosphorylation; in the other histidine-243 is inaccessible to this bound ATP. EnvZ-P always functions as an OmpR kinase (K+ P−), and EnvZ is a phosphatase for OmpR-P (K− P+). The sum of these opposing enzymatic activities determines the amount of intracellular OmpR-P, and this in turn determines the relative levels of ompF and ompC transcription (22). We believe that this theme, a balance between two extreme states by regulation of the accessibility of the critical histidine residue to ATP, explains the function of EnvZ and probably applies to homologous sensors as well.
ACKNOWLEDGMENTS
We thank J. Stock for his helpful discussions and C. Harris for his critical reading of the manuscript. We also thank Liya Shi for her excellent lab assistance.
This work was supported by a Damon Runyon-Walter Winchell Cancer Research Postdoctoral Fellowship to W.H. and an NIGMS grant (GM35791) to T.J.S.
REFERENCES
- 1.Atkinson M R, Ninfa A J. Characterization of Escherichia coli glnL mutations affecting nitrogen regulation. J Bacteriol. 1992;174:4538–4548. doi: 10.1128/jb.174.14.4538-4548.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casadaban M J. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage Lambda and Mu. J Mol Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 3.Chiang R C, Cavicchioli R, Gunsalus R P. Identification and characterization of narQ, a second nitrate sensor for nitrate-dependent gene regulation in Escherichia coli. Mol Microbiol. 1992;6:1913–1923. doi: 10.1111/j.1365-2958.1992.tb01364.x. [DOI] [PubMed] [Google Scholar]
- 4.Forst S, Inouye M. Environmentally regulated gene expression for membrane proteins in Escherichia coli. Annu Rev Cell Biol. 1988;4:21–42. doi: 10.1146/annurev.cb.04.110188.000321. [DOI] [PubMed] [Google Scholar]
- 5.Grebe, T., and J. B. Stock. Personal communication.
- 6.Hall M N, Silhavy T J. Transcriptional regulation of Escherichia coli K-12 major outer membrane protein 1b. J Bacteriol. 1979;140:342–350. doi: 10.1128/jb.140.2.342-350.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hall M N, Silhavy T J. The ompB locus and the regulation of the major outer membrane porin proteins of Escherichia coli K12. J Mol Biol. 1981;146:23–43. doi: 10.1016/0022-2836(81)90364-8. [DOI] [PubMed] [Google Scholar]
- 8.Hamilton C M, Aldea M, Washburn B K, Babitzke P, Kushner S R. New method for generating deletions and gene replacements in Escherichia coli. J Bacteriol. 1989;171:4617–4622. doi: 10.1128/jb.171.9.4617-4622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoch J A, Silhavy T J. Two-component signal transduction. Washington, D.C: American Society for Microbiology; 1995. [Google Scholar]
- 10.Hsing W, Silhavy T J. Function of conserved histidine-243 in phosphatase activity of EnvZ, the sensor for porin osmoregulation in Escherichia coli. J Bacteriol. 1997;179:3729–3735. doi: 10.1128/jb.179.11.3729-3735.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang K, Igo M M. Identification of the bases in the ompF regulatory region, which interact with the transcription factor OmpR. J Mol Biol. 1996;262:615–628. doi: 10.1006/jmbi.1996.0540. [DOI] [PubMed] [Google Scholar]
- 12.Igo M M, Silhavy T J. EnvZ, a transmembrane environmental sensor of Escherichia coli K-12, is phosphorylated in vitro. J Bacteriol. 1988;170:5971–5973. doi: 10.1128/jb.170.12.5971-5973.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Igo M M, Ninfa A J, Silhavy T J. A bacterial environmental sensor that functions as a protein kinase and stimulates transcriptional activation. Genes Dev. 1989;3:598–605. doi: 10.1101/gad.3.5.598. [DOI] [PubMed] [Google Scholar]
- 14.Igo M M, Ninfa A J, Stock J B, Silhavy T J. Phosphorylation and dephosphorylation of a bacterial transcription activator by a transmembrane receptor. Genes Dev. 1989;3:1725–1734. doi: 10.1101/gad.3.11.1725. [DOI] [PubMed] [Google Scholar]
- 15.Leonardo M R, Forst S. Re-examination of the role of the periplasmic domain of EnvZ in sensing of osmolarity signal in Escherichia coli. Mol Microbiol. 1996;22:405–415. doi: 10.1046/j.1365-2958.1996.1271487.x. [DOI] [PubMed] [Google Scholar]
- 16.Mizuno T, Mizushima S. Signal transduction and gene regulation through the phosphorylation of two regulatory components: the molecular basis for the osmotic regulation of the porin genes. Mol Microbiol. 1990;4:1077–1082. doi: 10.1111/j.1365-2958.1990.tb00681.x. [DOI] [PubMed] [Google Scholar]
- 17.Ninfa E G, Atkinson M R, Kamberov E S, Ninfa A J. Mechanism of autophosphorylation of Escherichia coli nitrogen regulator II (NRII or NtrB): transphosphorylation between subunits. J Bacteriol. 1993;175:7024–7031. doi: 10.1128/jb.175.21.7024-7032.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parkinson J S, Kofoid E C. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 19.Pratt L A, Silhavy T J. Porin regulon of Escherichia coli. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: American Society for Microbiology; 1995. pp. 105–127. [Google Scholar]
- 20.Rabin R S, Stewart V. Either of two functionally redundant sensor proteins, NarX and NarQ, is sufficient for nitrate regulation in Escherichia coli K-12. Proc Natl Acad Sci USA. 1992;89:8419–8423. doi: 10.1073/pnas.89.18.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rabin R S, Stewart V. Dual sensors and dual response regulators interact to control nitrate- and nitrite-responsive gene expression in Escherichia coli. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: American Society for Microbiology; 1995. pp. 233–252. [Google Scholar]
- 22.Russo F D, Silhavy T J. EnvZ controls the concentration of phosphorylated OmpR to mediate osmoregulation of the porin genes. J Mol Biol. 1991;222:567–580. doi: 10.1016/0022-2836(91)90497-t. [DOI] [PubMed] [Google Scholar]
- 23.Silhavy T J, Berman M, Enquist L. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1984. [Google Scholar]
- 24.Slauch J M, Silhavy T J. Genetic analysis of the switch that controls porin gene expression in Escherichia coli K-12. J Mol Biol. 1989;210:281–292. doi: 10.1016/0022-2836(89)90330-6. [DOI] [PubMed] [Google Scholar]
- 25.Stock J B, Surette M G, Levit M, Park P. Two-component signal transduction systems: structure-function relationships and mechanisms of catalysis. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: American Society for Microbiology; 1995. pp. 25–51. [Google Scholar]
- 26.Taylor S S, Knighton D R, Zheng J, Ten Eyck L F, Sowadski J M. Structural framework for the protein kinase family. Annu Rev Cell Biol. 1992;8:429–462. doi: 10.1146/annurev.cb.08.110192.002241. [DOI] [PubMed] [Google Scholar]
- 27.Tokishita S, Mizuno T. Transmembrane signal transduction by the Escherichia coli osmotic sensor, EnvZ: intermolecular complementation of transmembrane signaling. Mol Microbiol. 1994;13:435–444. doi: 10.1111/j.1365-2958.1994.tb00438.x. [DOI] [PubMed] [Google Scholar]
- 28.Yang Y, Inouye M. Requirement of both kinase and phosphatase activities of an Escherichia coli receptor (Taz1) for ligand-dependent signal transduction. J Mol Biol. 1993;231:335–342. doi: 10.1006/jmbi.1993.1286. [DOI] [PubMed] [Google Scholar]