Abstract
Listeria monocytogenes is well known for its robust physiology, which permits growth at low temperatures under conditions of high osmolarity and low pH. Although studies have provided insight into the mechanisms used by L. monocytogenes to allay the physiological consequences of these adverse environments, little is known about how these responses are coordinated. In the studies presented here, we have cloned the sigB gene and several rsb genes from L. monocytogenes, encoding homologs of the alternative sigma factor ςB and the RsbUVWX proteins, which govern transcription of a general stress regulon in the related bacterium Bacillus subtilis. The L. monocytogenes and B. subtilis sigB and rsb genes are similar in sequence and physical organization; however, we observed that the activity of ςB in L. monocytogenes was uniquely responsive to osmotic upshifting, temperature downshifting, and the presence of EDTA in the growth medium. The magnitude of the response was greatest after an osmotic upshift, suggesting a role for ςB in coordinating osmotic responses in L. monocytogenes. A null mutation in the sigB gene led to substantial defects in the ability of L. monocytogenes to use betaine and carnitine as osmoprotectants. Subsequent measurements of betaine transport confirmed that the absence of ςB reduced the ability of the cells to accumulate betaine. Thus, ςB coordinates responses to a variety of physical and chemical signals, and its function facilitates the growth of L. monocytogenes under conditions of high osmotic strength.
Listeria monocytogenes is a ubiquitous gram-positive bacterium that occasionally causes outbreaks and sporadic cases of food-borne illness. It is estimated that more than 1,700 cases occur annually in the United States, with outcomes ranging from flu-like illness to meningitis, meningoencephalitis, septicemia, abortion, and death, particularly in pregnant women and immunocompromised individuals (24, 41).
As an intracellular parasite that is transmitted primarily by contaminated food, L. monocytogenes must reach and/or maintain a cell density in the food environment that is necessary for infection. This population must further mitigate the physiological consequences of passage through the gastrointestinal tract and entry into host cell phagosomes. Although genetic analysis of virulence has provided many of the details of how virulence genes allow L. monocytogenes to elude potentially lethal outcomes of entry into host cells (23, 40, 43), little is known about the role of adaptive physiological responses in promoting the growth and survival of this bacterium in the food, intestinal, and intracellular environments.
L. monocytogenes is well known for its robust physiological characteristics and is one of the few pathogenic bacteria capable of growth at refrigeration temperatures, at low pHs, and/or under conditions of high osmolarity (21, 22, 32, 36, 55). These characteristics play an integral role in its potential as a food-borne pathogen, since they permit the growth of L. monocytogenes under conditions that prohibit the growth of many commensal organisms. Physiological studies have previously demonstrated that specific transport systems which mediate the uptake of osmoprotectants and cryoprotectants facilitate growth under these environmental conditions (9, 31, 32). A further role for adaptive physiological responses in pathogenesis has also been proposed on the basis of genetic experiments demonstrating that genes participating in acid tolerance (35) and stress-induced proteolysis (42) are necessary for full virulence in the mouse model. Thus, significant evidence is beginning to mount for a central role of adaptive physiological responses of L. monocytogenes in pathogenesis as well as in growth and survival in the environment.
To further our understanding of the connection between physiology and the transmission of food-borne illness, we have begun an analysis of genetic pathways that modulate adaptive responses in L. monocytogenes. One candidate for mediating such responses in gram-positive organisms is ςB, a secondary subunit of RNA polymerase that is known to govern a large stress response regulon in the related bacterium Bacillus subtilis. The ςB regulon of B. subtilis comprises at least 40 genes (52) and includes the katE gene, encoding a catalase (19, 20), the opuE gene, encoding transport machinery for osmoprotectants (53), the clpC gene, which is similar to stress-induced ATPase subunits of ClpP-type proteases (33), the gtaB gene, encoding a UDP-glucose pyrophosphorylase believed to participate in trehalose biosynthesis (46), and several genes whose functions cannot be inferred from their sequences (3, 11, 47).
The activity of ςB is joined to several physical and chemical signals through a postranslational mechanism that partitions ςB between inactive complexes with an anti-sigma factor protein, RsbW (for regulator of sigma B), and free ςB, which is capable of forming holoenzyme complexes with core RNA polymerase (6). At least two independent pathways exist which can alter the affinity of RsbW for its antagonist, RsbV, or ςB. First, it has been proposed that ATP stimulates the formation of RsbW-ςB complexes and activates a serine kinase activity in RsbW that phosphorylates its antagonist, RsbV, into an inactive state (2, 18, 58). Physical signals such as temperature, pH, and osmolarity can also invoke ςB activity (7, 11–13, 51) by stimulating the function of the RsbV-phosphate phosphatase RsbU (29, 58). At least four other Rsb proteins (RsbR, RsbS, RsbT, and RsbX) modulate the activity of RsbU and serve as distinct points of signal input into the system (1, 29).
To examine how the ςB regulon contributes to adaptive responses of L. monocytogenes, we have cloned the genes encoding homologs of ςB and several of the Rsb proteins. In this report, we demonstrate that the L. monocytogenes rsbVW-sigB-rsbX transcription unit is structurally analogous to its counterpart in B. subtilis and that its activity in L. monocytogenes is responsive to several physical and environmental signals. Genetic and physiological studies indicate that ςB facilitates growth under conditions of high osmolarity when betaine and carnitine are supplied as the primary osmoprotectants. Thus, the ςB regulon in L. monocytogenes participates in responses to several physiological and chemical insults, and it may serve as a primary osmosensor in this organism.
MATERIALS AND METHODS
Strains and plasmids.
L. monocytogenes Scott A was obtained from R. Hutkins (University of Nebraska), and L. monocytogenes LO4035 was obtained from N. Freitag (Wayne State University). L. monocytogenes cells were grown in brain heart infusion (BHI) (Difco, Detroit, Mich.) at 30°C unless otherwise specified. Where specified, kanamycin (30 μg/ml) or chloramphenicol (6 μg/ml) was added. Escherichia coli DH5α and E. coli MC1061 were used as cloning hosts and were grown in Luria broth with 150 μg of ampicillin/ml or 40 μg of kanamycin/ml where appropriate.
Cloning of sigB from L. monocytogenes.
Preliminary Southern blot experiments using a 540-bp EcoRI-PstI fragment of the B. subtilis sigB gene (10) as a probe indicated that a single 3.8-kb EcoRI fragment of the L. monocytogenes Scott A or LO4035 chromosome hybridized under conditions of moderate stringency (with 5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate] at 56°C). A plasmid minilibrary was therefore constructed by cloning 3- to 4-kb EcoRI fragments of L. monocytogenes chromosomal DNA into EcoRI-digested pUC19. The resulting plasmid library was transformed into a DH5α strain, and approximately 200 colonies containing plasmids with inserts were patched onto replica plates and transferred to nylon membranes. The colonies were screened by colony hybridization with the 540-bp EcoRI-PstI B. subtilis sigB probe under the same hybridization conditions as those used for the Southern blots. Two positive clones with identical restriction patterns were obtained. One clone was designated pLM413 and was used for further studies.
DNA sequence analysis.
Both strands of the 3.75-kb EcoRI fragment of pLM413 were subjected to DNA sequence analysis on an automated sequencer (LiCor, Inc.) by primer walking. Sequence alignments were performed with the PILEUP and BESTFIT programs from the Genetics Computer Group.
Preparation and analysis of RNA.
RNA samples were prepared from 200 ml of mid-logarithmic-phase L. monocytogenes cells (optical density at 600 nm [OD600], ∼0.4) before and 20 min after the addition of either 4% NaCl, 1 mM EDTA, 2% ethanol, 0.15% H2O2, or glacial acetic acid to a pH of 5.3. Additional samples were prepared after an aliquot of the cells was allowed to enter stationary phase (OD600, ∼3.0) and after mid-logarithmic-phase cells were shifted from 25 to 48°C for 20 min or to 4°C for 24 h. After treatment, the cells were harvested by centrifugation at 8,000 × g for 5 min at 4°C, and the cell pellets were frozen at −70°C overnight. The cells were subsequently resuspended in 5 ml of buffer D (4 M guanidinium thiocyanate, 25 mM sodium citrate, 0.5% Sarkosyl, 0.1 M 2-mercaptoethanol) and passed through a French pressure cell at 1,500 lb/in2. RNA was then extracted as described by Chomczynski and Sacchi (15). The final RNA pellet was dissolved in diethylpyrocarbonate-treated water, quantified spectrophotometerically, and stored frozen at −70°C. The integrity and relative concentrations of the RNA samples were also checked by agarose gel electrophoresis and ethidium bromide staining.
Primer extension analysis of sigB transcripts.
For each reaction, 10 pmol of the oligonucleotide VPROM2 (5′ CGCTGTATAAGCATCGATCAC 3′) was end labeled with 50 μCi of [γ-32P]ATP. The labeled primer was then mixed with 50 μg of RNA, heated to 70°C for 10 min, chilled on ice for 30 s, and incubated at 42°C with 3 U of SUPERSCRIPT II RNase H-reverse transcriptase (Life Technologies). After 50 min of extension, the products were ethanol precipitated, washed in 70% ethanol, and dried. The primer extension products were then dissolved in 90% formamide–10 mM EDTA and loaded onto a 6% denaturing polyacrylamide gel alongside a sequencing ladder prepared by using the VPROM2 primer and the pLM413 template DNA.
Construction of the sigB::km strain.
A null mutation in the L. monocytogenes sigB gene (sigB::km) was generated by cloning an aph3′5′′ gene encoding kanamycin resistance from pDG783 (26) into the unique StyI site of pLM413, which lies within the sigB coding region. The aph3′5′′ gene was removed from pDG783 as a HindIII fragment and was subsequently blunt ended with Klenow fragment prior to ligation to StyI-restricted pLM413 that had also been blunt ended with Klenow fragment. The resulting plasmid was designated pLM425 and was restriction mapped to confirm insertion of the aph3′5′′ gene into the appropriate position. The entire EcoRI fragment of pLM425, containing the rsbU, rsbV, rsbW, sigB::km, and rsbX genes, was then cloned into the EcoRI site of the temperature-sensitive integration vector pKSV7, which carries a chloramphenicol resistance gene (44). The resulting plasmid, designated pKSV7K8, was restriction mapped to ensure its integrity.
To place the sigB::km allele onto the L. monocytogenes chromosome by allelic exchange, we used pKSV7K8 in an integration-excision procedure outlined by Smith and Youngman (44). The plasmid was introduced into L. monocytogenes LO4035 cells by electroporation as described elsewhere (14). After 2 h of recovery in BHI, the transformants were plated onto BHI containing 6 μg of chloramphenicol/ml and were incubated at 30°C for 36 h. Two chloramphenicol-resistant transformants were then chosen and grown for three successive generations at the nonpermissive temperature for plasmid replication (42°C) in BHI supplemented with 6 μg of chloramphenicol/ml, followed by growth for three generations in BHI alone at the permissive temperature for plasmid replication (30°C). The cultures were then plated on BHI with 30 μg of kanamycin/ml and were incubated at 42°C. Kanamycin-resistant colonies were then patched to BHI-chloramphenicol plates to screen for loss of the plasmid-linked chloramphenicol resistance determinant. Kanamycin-resistant, chloramphenicol-sensitive colonies were subsequently analyzed by Southern blot analysis to confirm that plasmid excision had occurred, leaving the sigB::km allele on the chromosome.
Because insertion of the kanamycin resistance cassette into sigB is likely to be polar on downstream rsbX expression, we compared the phenotype of the sigB::km strain to that of an rsbX mutant in order to determine if the polarity could contribute to the phenotypic characteristics of the sigB::km strain. The rsbX gene was inactivated by integration of a pKSV7 derivative carrying an internal fragment of the L. monocytogenes rsbX gene. Transformation of L. monocytogenes LO4035 with this plasmid insertionally inactivates rsbX and subsequently gives rise to pinpoint colonies after 2 days of incubation. Transfer of the colonies to fresh medium consistently gave rise to flares of apparently faster-growing cells that maintained a large-colony phenotype upon subsequent passage. These observations are consistent with the phenotype of rsbX mutants of B. subtilis, which grow poorly, due to the derepression of ςB activity, and which acquire suppressor mutations that give rise to cells with normal growth rates (5, 11, 50). The fact that insertion of the kanamycin resistance gene into the sigB gene of L. monocytogenes gives rise to normal-sized colonies indicates that any polar effects on rsbX are epistatic to inactivation of sigB and are therefore negligible with regard to the phenotype of the sigB mutant.
Growth in DM.
To assess the ability of the wild-type and sigB::km strains to use betaine and carnitine as osmoprotectants, the cells were grown in a defined medium (DM) derived from formulations described by Pine et al. (39) and Beumer et al. (9). DM contains, per liter, 15 g of KH2PO4, 1 g of (NH4)2SO4, 0.2 g of MgSO4 · 7H2O, 0.02 g of CaCl2, 10 g of glucose, 0.088 g of ferric ammonium citrate, 0.1 g each of l-leucine, l-isoleucine, l-valine, l-methionine, and l-cysteine, 0.6 g of l-glutamine, 0.5 mg each of riboflavin and biotin, 1.0 mg of thiamine, and 0.005 mg of thioctic acid. After stock solutions of the components were combined, the pH was adjusted to 6.7 with potassium hydroxide and was filter sterilized. Cultures were grown in DM for 18 h at 30°C and were subsequently inoculated at 2% (vol/vol) in 250-ml baffled culture flasks containing DM with or without 3% NaCl, or DM with 3% NaCl that was supplemented either with 1 mM glycine betaine or with 1 mM carnitine. The cultures were incubated with shaking at 30°C, and growth was monitored by spectrophotometric measurements of OD600 over the course of several days.
Betaine uptake.
To measure the ability of LO4035 and LMA2B to accumulate betaine, log-phase cells grown in BHI at 30°C were harvested by centrifugation, washed twice, and resuspended in 50 mM potassium phosphate buffer (pH 6.8) to an OD600 of ca. 1.0. The cells were energized by the addition of glucose (final concentration, 5 mM), and where indicated, 3% NaCl (514 mM) was also added. After 20 min of incubation at room temperature, the assays were initiated by the addition of [14C]betaine (final concentration, 1 mM; 65 μCi/mmol; American Radiolabeled Chemicals, Inc., St. Louis, Mo.). The mixtures were incubated at room temperature, and 1-ml samples were removed and centrifuged through silicon oil as described previously (16). Radioactivity was measured by scintillation counting. The results are reported as the averages of duplicate samples. Independent experiments were conducted on successive days to confirm these results. The protein concentrations of cell suspensions were derived from a standard curve relating OD to protein concentration (54).
Nucleotide sequence accession number.
The sequence of the 3.75-kb EcoRI fragment of pLM413 has been deposited in GenBank under accession no. AFO74855.
RESULTS
Cloning and sequence analysis of the sigB operon from L. monocytogenes.
DNA sequence analysis of the cloned fragment revealed that five putative coding regions were present, including one with homology to the 3′ end of rsbU, as well as the entire coding regions of the rsbV, rsbW, sigB, and rsbX homologs (Fig. 1). The contiguous arrangement of these genes is similar to that observed in B. subtilis and Staphylococcus aureus (28, 34, 56, 57).
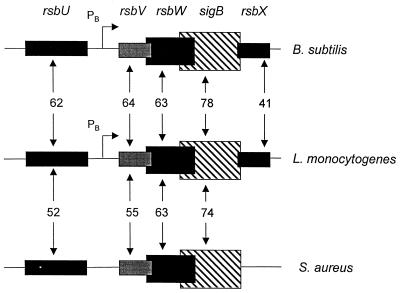
FIG. 1.
Similarity of the structure and organization of RsbU, RsbV, RsbW, ςB, and RsbX proteins with their B. subtilis and S. aureus homologs. The percent similarity scores of pairwise alignments of the primary amino acid sequences of the L. monocytogenes proteins with the B. subtilis and S. aureus proteins are shown between the blocks representing the respective genes. Coding regions that overlap are indicated by overlapping of the respective blocks representing each gene. The ςB-dependent promoters (PB) identified upstream of rsbV in B. subtilis (28) and in L. monocytogenes (this report) are indicated by arrows.
Detailed comparisons of the physical organization of the genes from L. monocytogenes to that of the genes in B. subtilis and S. aureus showed two highly conserved features. First, a significant spacer region of 153 bases is positioned between the 3′ end of rsbU and the 5′ end of rsbV in L. monocytogenes; although not conserved in sequence, this spacer region also exists in B. subtilis (59 bp) and S. aureus (110 bp) (28, 57). This region contains a known ςB-dependent promoter in B. subtilis that serves as an autocatalytic device for amplifying mRNA encoding ςB and the RsbVWX regulatory components in response to stress (7, 13, 28). Secondly, there appears to be translational coupling between rsbV and rsbW and between rsbW and sigB, as these coding regions overlap by at least 1 codon. The overlap is most striking between rsbW and sigB (13 codons) and appears to be an important feature of these genes, because it is conserved in B. subtilis (13 codons) and S. aureus (8 codons) (28, 34, 57). Translational coupling has been proposed for these genes (28), and experimental evidence supports this hypothesis in the case of rsbW and sigB (8). The conservation of the rsbW-sigB overlap further supports the importance of coupling as a device for ensuring equimolar synthesis of ςB and its primary regulator; however, the mechanism through which the coupling is mediated through this extensive overlap remains to be determined.
Homology of the Rsb and ςB proteins.
Alignment of the predicted amino acid sequences of RsbUVWX and ςB with their homologs from B. subtilis and S. aureus indicates that the L. monocytogenes homologs are more closely related to their counterparts in B. subtilis (Fig. 1 and 2). Overall, the putative RsbU, RsbV, RsbW, and ςB proteins display 60 to 75% similarity (40 to 50% identity) with their B. subtilis and S. aureus homologs. In contrast, however, the RsbX protein displays only limited homology (29% identity) to its B. subtilis counterpart.
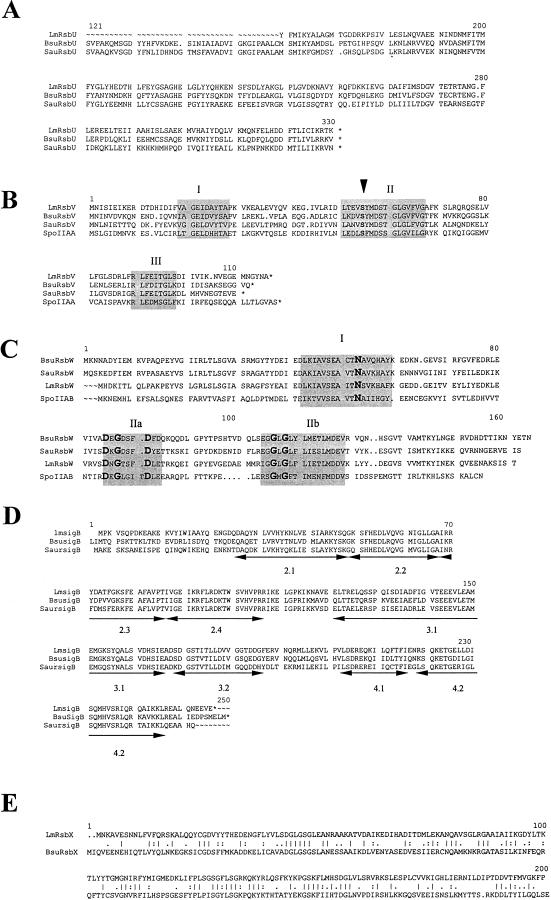
FIG. 2.
Multiple sequence alignments of the RsbU (A), RsbV (B), RsbW (C), ςB (D), and RsbX (E) proteins with their homologs from B. subtilis (Bsu) and S. aureus (Sau). Lm, L. monocytogenes. (A through D) Alignments were derived by using the PILEUP program of the Genetics Computer Group. (B) The serine residue of SpoIIAA that is phosphorylated by SpoIIAB and is believed to be the site of RsbW-dependent phosphorylation of RsbV is indicated by an arrowhead. Highly conserved domains are shaded. (C) Alignment of the RsbW proteins and the SpoIIAB homolog from B. subtilis. Domains showing homology with regions I and II of the histidine protein kinase family of two-component system proteins are shaded, and residues within these domains that are identical in every homolog are boldfaced. (D) The conserved subregions of sigma factors that were identified previously by Helman and Chamberlin (27) are underlined with arrows and numbered. (E) Pairwise alignment of the B. subtilis and L. monocytogenes RsbX proteins was performed with BESTFIT. Vertical lines indicate identical residues, and the periods and colons indicate semiconservative and conservative changes, respectively.
Multiple sequence alignments of the L. monocytogenes RsbV, RsbW, and ςB proteins with their counterparts from B. subtilis and S. aureus revealed highly conserved subregions within each (Fig. 2). Alignment of the RsbU homologs predicts that our cloned fragment contains only about half of the L. monocytogenes rsbU gene (Fig. 2A). Within RsbV, we identified three subregions, designated I through III, that appear to be highly conserved (Fig. 2B). Region II includes the conserved serine residue that can be aligned with the serine residue in the homolog SpoIIAA, which is the site of phosphorylation by the SpoIIAB homolog RsbW (17, 37, 38). How domains I and III contribute to RsbV structure, function, or both remains to be determined. As with RsbV, we also observed three highly conserved subregions, designated regions I, IIa, and IIb, within the L. monocytogenes RsbW homolog (Fig. 2C). These regions have previously been proposed to play a role in ATP binding of the RsbW homolog SpoIIAB by virtue of their similarities to domains I and II of histidine protein kinases (37). The ςB proteins, which demonstrated the highest identity scores, showed the highest degree of identity in regions 2.2, 2.4, and 4.2, which are believed to participate in core RNA polymerase binding and −10 and −35 recognition, respectively (27).
Stress-dependent activation of ςB in L. monocytogenes.
The intergenic region between rsbU and rsbV is the location of a known ςB-dependent promoter in B. subtilis (28) that serves as an autocatalytic device for increasing the levels of mRNA encoding the RsbV, RsbW, RsbX, and ςB proteins under conditions of ςB activation (7, 13). We therefore examined this region by primer extension to determine if a ςB-dependent promoter is similarly positioned in L. monocytogenes and if its activity is stress dependent. A primer (VPROM2) that is complementary to positions +63 to +83 relative to the rsbV initiation codon was end labeled and used in primer extension analyses on mRNA extracted from logarithmically growing cells before and after environmental stress (4% NaCl or pH 5.3) or entrance into stationary phase. As shown in Fig. 3, no transcript from this region was detected in logarithmically growing cells; however, increasing the osmolarity of the medium, decreasing its pH, or permitting the cells to enter stationary phase led to the appearance of an extension product mapping to position 646 of the nucleotide sequence. The initiation site of this transcript lies immediately downstream of sequences that can be aligned with known ςB-dependent promoters (Fig. 4), indicating that this promoter may be utilized by ςB-RNA polymerase holoenzyme under stress conditions.
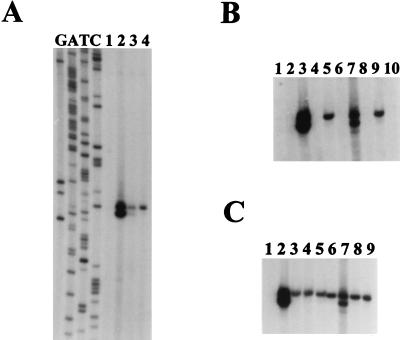
FIG. 3.
Primer extension analyses of transcripts originating upstream of rsbV in L. monocytogenes. In each panel, 50 μg of each RNA sample was used as a template for extension of the labeled VPROM2 primer. (A) RNA was derived from logarithmically growing cells before (lane 1) and after the addition of 4% NaCl (lane 2), entrance into stationary phase (lane 3), or acidification of the medium to pH 5.3 with glacial acetic acid (lane 4). The sequence ladder was derived from sequencing reactions using the VPROM2 primer. (B and C) Bands are shown at the same position relative to the sequencing ladder in panel A. (B) RNA samples were derived from logarithmically growing cells (25°C) of LO4035 (wild type) (lanes 1, 3, 5, 7, and 9) and the isogenic sigB::km mutant LMA2B (lanes 2, 4, 6, 8, and 10) before stress (lanes 1 and 2) and 20 min after the addition of 4% NaCl (lanes 3 and 4), acidification to pH 5.3 with glacial acetic acid (lanes 5 and 6), a temperature upshift to 48°C (lanes 7 and 8), and entrance into stationary phase (lanes 9 and 10). (C) RNA samples were derived from LO4035 (wild type) during logarithmic growth at 25°C (lane 1) and 20 min after the addition of 4% NaCl (lane 2), 2% ethanol (lane 3), 1 mM EDTA (lane 4), 0.15% H2O2 (lane 5), or glacial acetic acid to pH 5.3 (lane 6), a temperature upshift to 48°C (lane 7), a downshift to 4°C (lane 8), or entrance into stationary phase (lane 9).
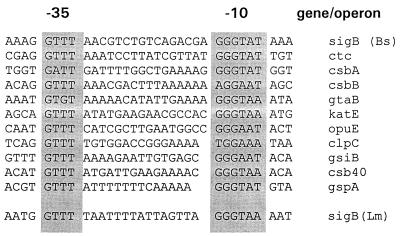
FIG. 4.
Alignment of known ςB-dependent promoters. The sequences immediately upstream of the ςB-dependent transcription start site of the L. monocytogenes rsbV gene are aligned with known ςB-dependent promoters from B. subtilis. The most highly conserved residues, centering around the −10 and −35 regions, are shaded. The alignment of B. subtilis promoters was derived from von Blohn et al. (53). Bs, B. subtilis; Lm, L. monocytogenes.
To confirm that the stress-induced transcript is ςB dependent, we performed primer extension assays on RNA extracted from both the wild type, LO4035, and its isogenic derivative, LMA2B, which carries an allele of sigB (sigB::km) that has been insertionally inactivated by the introduction of a kanamycin resistance gene (see Materials and Methods). As predicted, only RNA samples from the wild-type strain yielded primer extension products after osmotic upshifting, temperature upshifting, acidification, or entry into stationary phase (Fig. 3B). Thus, the stress-induced transcript relies on ςB and appears to be a conserved autocatalytic mechanism for increasing the abundance of the rsbVW-sigB-rsbX transcript upon the perception of stress.
Because of the autocatalytic nature of ςB, the level of the ςB-dependent transcript that originates upstream of rsbV is a convenient measure of ςB activity in the cell. We therefore used our primer extension assay to measure the relative effects of different environmental conditions on ςB activity in L. monocytogenes (Fig. 3C). As observed in the previous experiment, no transcript was detected in logarithmically growing cells; however, significant levels of the transcript were observed after cells had been exposed to 4% NaCl, 2% ethanol, acidification to a pH of 5.3, 1 mM EDTA, 0.15% H2O2, or a temperature upshift (from 25 to 48°C) or downshift (from 25 to 4°C). Each of these treatments led to the appearance of the ςB-dependent transcript, indicating that ςB activity was stimulated.
Although many of the conditions examined above are known to activate ςB in B. subtilis, we noted unique aspects of the L. monocytogenes responses. First, based on the intensity of the labeled bands, osmotic upshifting gave rise to the highest degree of stimulation relative to the other treatments. The magnitude of this response relative to the other conditions is striking and appears to be greater than what has been observed in B. subtilis after a similar upshift (13). We note, however, that the dose-response curve of ςB activity is complex and that the “doses” of each stress used in our experiments may not be optimal. Next, we observed two conditions, temperature downshifting and the addition of EDTA, which activated ςB only in L. monocytogenes. Experiments with B. subtilis cells grown under similar conditions failed to yield significant activation of ςB activity based on measurements of transcription from the ςB-dependent transcriptional fusion (ctc::lacZ) and on measurements of ςB by Western blotting (4).
Effects of the sigB::km mutation on osmotolerance in L. monocytogenes.
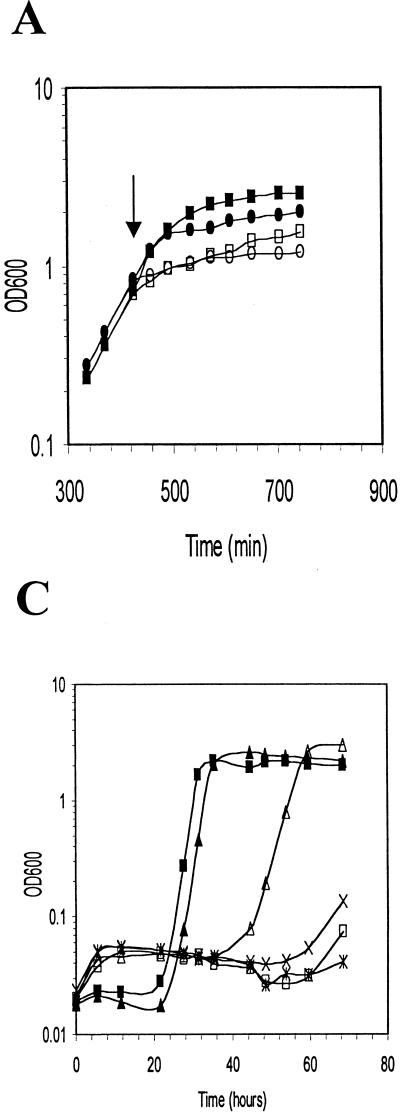
Because we observed the highest level of ςB activity after an osmotic upshift, we next examined whether the absence of sigB would have any effect on the ability of L. monocytogenes to grow under conditions of high osmotic strength. We initially evaluated the growth of LO4035 and the isogenic LMA2B derivative (sigB::km) in BHI after logarithmically growing cells were exposed to an osmotic upshift by the addition of 6% NaCl. As illustrated in Fig. 5A, the addition of NaCl to the cultures led to an abrupt decrease in the growth rate. Relative to the parental strain, however, the sigB::km mutant showed a slightly reduced, but reproducible, rate after upshifting of the culture during the latter stages of logarithmic growth, suggesting that the absence of ςB only slightly impaired adaptation to the osmotic upshift in this complex growth medium.
FIG. 5.
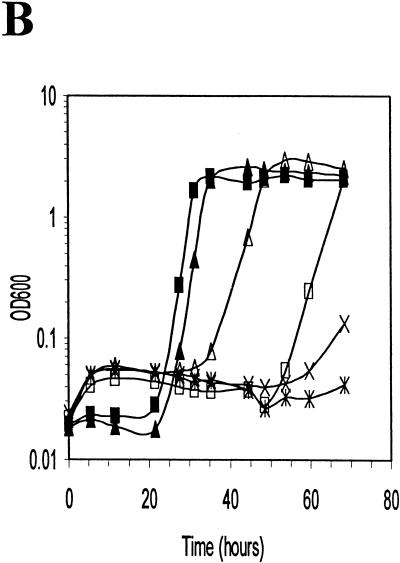
Growth of wild-type and sigB mutant strains of L. monocytogenes after osmotic upshift in BHI (A) or DM (B and C). (A) LO4035 (wild type) and LMA2B (sigB::km) were grown in BHI, and the cultures were divided into two equal portions at 425 min (arrow). Sodium chloride was added to one portion to a final concentration of 6%, and both portions were further incubated. Symbols: ■, LO4035 without NaCl; □, LO4035 with 6% NaCl; •, LMA2B without NaCl; ○, LMA2B with 6% NaCl. (B) Equivalent volumes of 18-h cultures of LO4035 and LMA2B in DM were inoculated into DM without NaCl (▴, LO4035; ■, LMA2B), DM with 3% NaCl (✕⃒, LO4035; ×, LMA2B), and DM with 3% NaCl supplemented with 1 mM betaine (▵, LO4035; □, LMA2B. (C) Equivalent volumes of 18-h cultures of LO4035 and LMA2B in DM were inoculated into fresh DM (▴, LO4035; ■, LMA2B), DM with 3% NaCl (✕⃒, LO4035; ×, LMA2B), and DM with 3% NaCl supplemented with 1 mM carnitine (▵, LO4035; □, LMA2B).
Since the growth medium used in the above experiment provides several amino acids and the quaternary ammonium compounds betaine and carnitine, which can be used by L. monocytogenes as osmoprotectants (9, 31, 32), we next tested the ability of the isogenic strains to grow in DM under conditions of high osmolarity when only a single osmoprotectant was provided (Fig. 5B and C). When overnight cultures were inoculated into fresh DM of low osmolarity, only a brief lag was observed. Inoculation of the same cells into DM containing 3% NaCl (514 mM), however, prevented growth of both strains during the course of the experiments. When the cells were inoculated into DM containing 3% NaCl and 1 mM betaine (Fig. 5B) or 1 mM carnitine (Fig. 5C), the wild-type strain was capable of growth after a prolonged lag period (30 h for betaine and 40 h for carnitine). In contrast, LMA2B demonstrated a substantially longer lag period in the presence of betaine (Fig. 5B) and failed to grow in the presence of carnitine within the time frame of the experiment (Fig. 5C). These results indicate that the absence of ςB impairs the ability of L. monocytogenes to use betaine and carnitine as osmoprotectants, most likely due to defects in the transport of the osmoprotectants.
Betaine accumulation is defective in the sigB::km strain.
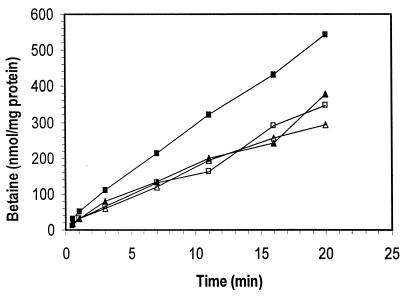
To confirm that the absence of ςB impairs the accumulation of betaine, we measured the ability of the isogenic strains to accumulate [14C]betaine under conditions of high and low osmolarity. Cells from both LO4035 and LMA2B were grown to mid-logarithmic phase in BHI (OD600, ∼0.4) and then were harvested and washed at room temperature to prevent temperature-dependent induction of ςB activity. As shown in Fig. 6, the two strains accumulated similar amounts of labeled betaine when incubated in phosphate buffer alone, confirming the presence of a constitutive betaine transport system that has been described previously (49). When 3% NaCl was added to the cells, betaine accumulation in the wild-type strain LO4035 was stimulated considerably. In contrast, inactivation of the sigB gene in LMA2B prohibited osmotic stimulation of betaine accumulation. These results are consistent with a model in which ςB mediates a sodium-inducible or osmotically inducible component of betaine transport. Thus, ςB appears to play an integral role in coordinating the physiological adaptation of L. monocytogenes to osmotic upshifting.
FIG. 6.
Betaine accumulation in LO4035 and LMA2B. Cells in the mid-logarithmic growth phase (OD600, ∼0.4) were harvested, washed, and resuspended in potassium phosphate buffer. The washed cell suspension from each strain was energized by the addition of glucose and then divided into two equal volumes, and sodium chloride to a final concentration of 3% was added to one of the samples. After 20 min of induction, 14C-labeled betaine was added to each sample, and aliquots were removed over time, harvested by centrifugation through oil, and counted by scintillation counting. Symbols: □, LO4035; ■, LO4035 plus 3% NaCl; ▴, LMA2B; ▵, LMA2B plus 3% NaCl.
DISCUSSION
The ability of L. monocytogenes to grow under conditions of suboptimal temperature, osmolarity, and pH may favor its growth in contaminated food products and promote survival during transit through the gastrointestinal tract and entry into host cell phagosomes. In this report, we have used genetic analyses and physiological studies to examine how L. monocytogenes coordinates such physiological responses through the activity of an alternative sigma subunit of RNA polymerase, ςB.
Analysis of the physical organization of the L. monocytogenes rsbU, rsbV, rsbW, sigB, and rsbX genes demonstrated that the genes are organized in much the same manner as they are in the related gram-positive organisms B. subtilis and S. aureus, with the exception of rsbX, which is absent downstream in S. aureus. These conserved features include an extensive overlap of the rsbW and sigB coding regions, further supporting an important role for translational coupling of ςB to its primary regulator, RsbW. Multiple sequence alignments of the putative ςB and Rsb proteins further revealed highly conserved subregions within the anti-anti-sigma factors RsbV and SpoIIAA, which may facilitate further genetic and biochemical analysis of the structure-function relationships among these proteins.
Given the high degree of conservation among the RsbUVW and ςB proteins, we are intrigued by the low degree of identity between the L. monocytogenes and B. subtilis RsbX proteins. In B. subtilis, RsbX is the farthest-upstream member of the Rsb signal transduction cascade (29, 50, 58). This divergence may therefore reflect adaptation of rsbX to sensing functions that are attuned to unique aspects of the physiology and ecology of L. monocytogenes. We are currently identifying the other rsb genes in L. monocytogenes to determine how differences in structure might correspond to differences in function.
In addition to the divergence in the primary sequence of RsbX, we also observed unique characteristics of the physical and chemical conditions that elicit ςB activity in L. monocytogenes, including responses to temperature downshifting and to the presence of EDTA, and the magnitude of the response to osmotic upshifting. Induction of ςB activity subsequent to a temperature downshift suggests that ςB may contribute to the unique psychrotrophic characteristics of this organism. Since betaine has been demonstrated to act as a cryoprotectant as well as an osmoprotectant in L. monocytogenes (31), the participation of ςB in betaine accumulation suggests that low-temperature induction of ςB activity may also stimulate betaine accumulation after a temperature downshift. We are currently testing this hypothesis.
Induction of ςB activity in response to EDTA is also notable. The response may be due to decreased availability of divalent cations within the cytoplasm. Alternatively, it could be an indirect response to destabilization of teichoic acids or effects on membrane proteins that require divalent ions for structure or function. Given that the Rsb cascade responds to many types of physical signals (pH, temperature, and osmolarity), it may be that compromising the integrity of the envelope is sufficient to elicit ςB activity. In support of this idea is the fact that the S. aureus sigB gene was originally discovered as a transposon insertion that led to sensitivity of the cells to methicillin (57), suggesting a role for ςB in cell wall-associated functions in this organism. We are currently determining which cation limitation invokes ςB activity and whether it is due to its declining concentration inside or outside the cell.
Role of ςB in osmotolerance in L. monocytogenes.
The magnitude of ςB induction that was observed after an osmotic upshift (Fig. 3) initially suggested that ςB function might be important in mediating osmotolerance. Experiments with the isogenic sigB+ and sigB mutant strains subsequently demonstrated that the absence of ςB impaired the ability of L. monocytogenes to use betaine and carnitine as osmoprotectants (Fig. 5B and C). This defect was further shown to result from the inability of the sigB mutant strain to stimulate betaine accumulation after an osmotic upshift (Fig. 6).
Physiological studies with L. monocytogenes have previously demonstrated the existence of independent systems for betaine and carnitine uptake (25, 48). Betaine is transported by a sodium-driven transporter (25) and is the principal osmoprotectant in the presence of other osmoprotectants, while carnitine transport appears to be ATP dependent and subject to inhibition by betaine (25, 48). We did not observe defects in constitutive betaine accumulation in the sigB mutant but instead found that this strain failed to stimulate accumulation after an osmotic upshift (Fig. 6). Previous studies have demonstrated that betaine uptake in L. monocytogenes is stimulated by osmotic upshifting (31). These results are consistent with ςB mediating a sodium-inducible or osmotically inducible component of betaine transport. This function could be a consequence of ςB directing the transcription of one or more genes encoding a betaine transporter or, alternatively, a regulatory protein that modulates the synthesis or activity of a betaine transport system. Since betaine is the osmoprotectant of choice, when available (48), the ςB-dependent component of betaine transport in L. monocytogenes may therefore be the primary system for osmotic stimulation. Consistent with this hypothesis, we observed significant induction of ςB activity in response to osmotic upshifting.
The use of ςB as a primary means for coordinating osmotic responses in L. monocytogenes is apparently unique. B. subtilis sigB mutants do not show obvious defects in osmotolerance (53), likely due to the existence of both multiple regulatory systems and redundant transporters in this organism. Indeed, three independent transporters can be used for proline and betaine uptake in B. subtilis (30, 53). Moreover, in the case of the opuE gene, which encodes a proline transporter, its ςB-dependent promoter is one of two osmotically inducible promoters (53). The adoption of redundant genes encoding transport systems in this organism may have led to dissemination of the role of coordinating their expression among other regulatory systems in order to bring expression into harmony with the availability of different substrates. Given that several sophisticated adaptive responses such as sporulation and competence are also available to B. subtilis, the role of the ςB regulon may have been further diminished to accommodate these pathways. Thus, in organisms, such as L. monocytogenes, that do not possess such sophisticated adaptive responses, ςB may serve a primary role in coordinating responses to physical changes in the environment. Further comparative studies of the ςB regulon from related gram-positive organisms would therefore provide important information about the role of adaptive responses in the physiology and ecology of these organisms.
ACKNOWLEDGMENTS
This work was supported by grants from Li-Cor, Inc., and from the USDA Midwest Advanced Food Manufacturing Alliance to A.K.B. L.A.B. is a recipient of the Widaman Trust Distinguished Graduate Assistant Fellowship.
We thank Abraham Oommen and Margaret Esser for performing the DNA sequence analysis and Mark Morrison and Jeff Cirillo for critical reading of the manuscript.
L. A. Becker and M. S. Çetin contributed equally to this work.
Footnotes
Journal Series paper 12219 of the Nebraska Agricultural Experimental Station.
REFERENCES
- 1.Akbar S, Kang C M, Gaidenko T A, Price C W. Modulator protein RsbR regulates environmental signalling in the general stress pathway of Bacillus subtilis. Mol Microbiol. 1997;24:567–578. doi: 10.1046/j.1365-2958.1997.3631732.x. [DOI] [PubMed] [Google Scholar]
- 2.Alper S, Duncan L, Losick R. An adenosine nucleotide switch controlling the activity of a cell type-specific transcription factor in Bacillus subtilis. Cell. 1994;77:195–205. doi: 10.1016/0092-8674(94)90312-3. [DOI] [PubMed] [Google Scholar]
- 3.Antelmann H, Bernhardt J, Schmid R, Hecker M. A gene at 333° on the Bacillus subtilis chromosome encodes the newly identified ςB-dependent general stress protein GspA. J Bacteriol. 1995;177:3540–3545. doi: 10.1128/jb.177.12.3540-3545.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker, L. A., and A. K. Benson. 1998. Unpublished data.
- 5.Benson A K, Haldenwang W G. Characterization of a regulatory network that controls ςB expression in Bacillus subtilis. J Bacteriol. 1992;174:749–757. doi: 10.1128/jb.174.3.749-757.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benson A K, Haldenwang W G. Bacillus subtilis ςB is regulated by a binding protein (RsbW) that blocks its association with core RNA polymerase. Proc Natl Acad Sci USA. 1993;90:2330–2334. doi: 10.1073/pnas.90.6.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benson A K, Haldenwang W G. The ςB-dependent promoter of the Bacillus subtilis sigB operon is induced by heat shock. J Bacteriol. 1993;175:1929–1935. doi: 10.1128/jb.175.7.1929-1935.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benson A K, Haldenwang W G. Regulation of ςB levels and activity in Bacillus subtilis. J Bacteriol. 1993;175:2347–2356. doi: 10.1128/jb.175.8.2347-2356.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beumer R R, Te Giffel M C, Cox L J, Rombouts F M, Abee T. Effect of exogenous proline, betaine, and carnitine on growth of Listeria monocytogenes in a minimal medium. Appl Environ Microbiol. 1994;60:1359–1363. doi: 10.1128/aem.60.4.1359-1363.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Binne C, Lampe M, Losick R. Gene encoding the ς37 species of RNA polymerase ς factor from Bacillus subtilis. Proc Natl Acad Sci USA. 1986;83:5943–5947. doi: 10.1073/pnas.83.16.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boylan S A, Redfield A R, Price C W. Transcription factor ςB of Bacillus subtilis controls a large stationary-phase regulon. J Bacteriol. 1993;175:3957–3963. doi: 10.1128/jb.175.13.3957-3963.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boylan S A, Rutherford A, Thomas S M, Price C W. Activation of Bacillus subtilis transcription factor ςB by a regulatory pathway responsive to stationary-phase signals. J Bacteriol. 1992;174:3695–3706. doi: 10.1128/jb.174.11.3695-3706.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boylan S A, Rutherford A, Thomas S M, Price C W. Stress-induced activation of the ςB transcription factor of Bacillus subtilis. J Bacteriol. 1993;175:7931–7937. doi: 10.1128/jb.175.24.7931-7937.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Camilli A, Portnoy D A, Youngman P. Insertional mutagenesis of Listeria monocytogenes with a novel Tn917 derivative that allows direct cloning of DNA flanking transposon insertions. J Bacteriol. 1990;172:3738–3744. doi: 10.1128/jb.172.7.3738-3744.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 16.Christensen D P, Hutkins R W. Glucose uptake by Listeria monocytogenes Scott A and inhibition by pediocin JD. Appl Environ Microbiol. 1994;60:3870–3873. doi: 10.1128/aem.60.10.3870-3873.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diederich B, Wilkinson J F, Magnin T, Najafi M, Errington J, Yudkin M D. Role of interactions between SpoIIAA and SpoIIAB in regulating cell-specific transcription factor sigma F of Bacillus subtilis. Genes Dev. 1994;8:2653–2663. doi: 10.1101/gad.8.21.2653. [DOI] [PubMed] [Google Scholar]
- 18.Dufour A, Haldenwang W G. Interactions between a Bacillus subtilis anti-sigma factor (RsbW) and its antagonist (RsbV) J Bacteriol. 1994;176:1813–1820. doi: 10.1128/jb.176.7.1813-1820.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Engelmann S, Hecker M. Impaired oxidative stress resistance of Bacillus subtilis sigB mutants and the role of katA and katE. FEMS Microbiol Lett. 1996;145:63–69. doi: 10.1111/j.1574-6968.1996.tb08557.x. [DOI] [PubMed] [Google Scholar]
- 20.Englemann S, Linder C, Hecker M. Cloning, nucleotide sequence, and regulation of katE encoding a ςB-dependent catalase in Bacillus subtilis. J Bacteriol. 1995;177:5598–5605. doi: 10.1128/jb.177.19.5598-5605.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farber J M, Brown B E. Effect of prior heat shock on heat resistance of Listeria monocytogenes in meat. Appl Environ Microbiol. 1990;56:1584–1587. doi: 10.1128/aem.56.6.1584-1587.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farber J M, Peterkin P I. Listeria monocytogenes, a food-borne pathogen. Microbiol Rev. 1991;55:476–511. doi: 10.1128/mr.55.3.476-511.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finlay B B, Cossart P. Exploitation of mammalian host cell functions by bacterial pathogens. Science. 1997;276:718–725. doi: 10.1126/science.276.5313.718. [DOI] [PubMed] [Google Scholar]
- 24.Gellin B, Broome C. Listeriosis. JAMA. 1989;261:1313–1320. [PubMed] [Google Scholar]
- 25.Gerhardt P N, Smith L T, Smith G M. Sodium-driven, osmotically activated glycine betaine transport in Listeria monocytogenes membrane vesicles. J Bacteriol. 1996;178:6105–6109. doi: 10.1128/jb.178.21.6105-6109.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guerout-Fleury A-M, Shazand K, Frandsen N, Stragier P. Antibiotic-resistance cassettes for Bacillus subtilis. Gene. 1995;167:335–337. doi: 10.1016/0378-1119(95)00652-4. [DOI] [PubMed] [Google Scholar]
- 27.Helman J D, Chamberlin M J. Structure and function of bacterial sigma factors. Annu Rev Biochem. 1988;57:839–872. doi: 10.1146/annurev.bi.57.070188.004203. [DOI] [PubMed] [Google Scholar]
- 28.Kalman S, Duncan M L, Thomas S M, Price C W. Similar organization of the sigB and spoIIA operons encoding alternate sigma factors of Bacillus subtilis. J Bacteriol. 1990;172:5575–5585. doi: 10.1128/jb.172.10.5575-5585.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang C M, Brody M S, Akbar S, Yang X, Price C W. Homologous pairs of regulatory proteins control activity of Bacillus subtilis transcription factor ςB in response to environmental stress. J Bacteriol. 1996;178:3846–3853. doi: 10.1128/jb.178.13.3846-3853.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kappes R M, Kempf B, Bremer E. Three transport systems for the osmoprotectant glycine betaine operate in Bacillus subtilis: characterization of OpuD. J Bacteriol. 1996;178:5071–5079. doi: 10.1128/jb.178.17.5071-5079.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ko R, Smith L T, Smith G M. Glycine betaine confers enhanced osmotolerance and cryotolerance on Listeria monocytogenes. J Bacteriol. 1994;176:426–431. doi: 10.1128/jb.176.2.426-431.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kroll R G, Patchett P A. Induced acid tolerance in Listeria monocytogenes. Lett Appl Microbiol. 1992;14:224–227. [Google Scholar]
- 33.Krüger E, Msadek T, Hecker M. Alternate promoters direct stress-induced transcription of the Bacillus subtilis clpC operon. Mol Microbiol. 1996;20:713–723. doi: 10.1111/j.1365-2958.1996.tb02511.x. [DOI] [PubMed] [Google Scholar]
- 34.Kullik I, Giachino P. The alternative sigma factor ςB in Staphylococcus aureus: regulation of the sigB operon in response to growth phase and heat shock. Arch Microbiol. 1997;167:151–159. doi: 10.1007/s002030050428. [DOI] [PubMed] [Google Scholar]
- 35.Marron L, Emerson N, Gahan C G, Hill C. A mutant of Listeria monocytogenes LO28 unable to induce an acid tolerance response displays diminished virulence in a murine model. Appl Environ Microbiol. 1997;63:4945–4947. doi: 10.1128/aem.63.12.4945-4947.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller A J. Combined water activity and solute effects on growth and survival of Listeria monocytogenes Scott A. J Food Prot. 1992;55:414–418. doi: 10.4315/0362-028X-55.6.414. [DOI] [PubMed] [Google Scholar]
- 37.Min K-T, Hilditch C M, Diedrich B, Errington J, Yudkin M D. ςF, the first compartment-specific transcription factor of Bacillus subtilis, is regulated by an anti-sigma factor that is also a protein kinase. Cell. 1993;74:735–742. doi: 10.1016/0092-8674(93)90520-z. [DOI] [PubMed] [Google Scholar]
- 38.Najafi S M, Willis A C, Yudkin M D. Site of phosphorylation of SpoIIAA, the anti-anti-sigma factor for sporulation-specific sigma F of Bacillus subtilis. J Bacteriol. 1995;177:2912–2913. doi: 10.1128/jb.177.10.2912-2913.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pine L, Malcom G B, Brooks J B, Daneshvar M I. Physiological studies on the growth and utilization of sugars by Listeria species. Can J Microbiol. 1989;35:245–254. doi: 10.1139/m89-037. [DOI] [PubMed] [Google Scholar]
- 40.Portnoy D A, Chakraborty T, Goebel W, Cossart P. Molecular determinants of Listeria monocytogenes pathogenesis. Infect Immun. 1992;60:1263–1267. doi: 10.1128/iai.60.4.1263-1267.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roberts T, Pinner R. Economic impact of disease caused by Listeria monocytogenes. In: Miller A J, Smith J L, Somkuti G A, editors. Foodborne listeriosis. Amsterdam, The Netherlands: Elsevier Science Publishing Co., Inc.; 1990. pp. 137–149. [Google Scholar]
- 42.Rouquette C, Ripio M T, Pellegrini E, Bolla J M, Tascon R I, Vázquez-Boland J A, Berche P. Identification of a ClpC ATPase required for stress tolerance and in vivo survival of Listeria monocytogenes. Mol Microbiol. 1996;21:977–987. doi: 10.1046/j.1365-2958.1996.641432.x. [DOI] [PubMed] [Google Scholar]
- 43.Sheehan B, Kocks C, Dramsi S, Gouin E, Klarsfield A D, Mengaud J, Cossart P. Molecular and genetic determinants of the Listeria monocytogenes infectious process. Curr Top Microbiol Immunol. 1994;192:187–216. doi: 10.1007/978-3-642-78624-2_9. [DOI] [PubMed] [Google Scholar]
- 44.Smith K, Youngman P. Use of a new integrational vector to investigate compartment-specific expression of the Bacillus subtilis spoIIM gene. Biochimie. 1992;74:705–711. doi: 10.1016/0300-9084(92)90143-3. [DOI] [PubMed] [Google Scholar]
- 45.Smith L T. Role of osmolytes in adaptation of osmotically stressed and chill-stressed Listeria monocytogenes grown in liquid media and on processed meat surfaces. Appl Environ Microbiol. 1996;62:3088–3093. doi: 10.1128/aem.62.9.3088-3093.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Varón D, Boylan S A, Okamoto K, Price C W. Bacillus subtilis gtaB encodes UDP-glucose pyrophosphorylase and is controlled by stationary-phase transcription factor ςB. J Bacteriol. 1993;175:3964–3971. doi: 10.1128/jb.175.13.3964-3971.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Varón D, Brody M S, Price C W. Bacillus subtilis operon under the dual control of the general stress transcription factor ςB and the sporulation transcription factor ςH. Mol Microbiol. 1996;20:339–350. doi: 10.1111/j.1365-2958.1996.tb02621.x. [DOI] [PubMed] [Google Scholar]
- 48.Verheul A, Glaasker E, Poolman B, Abee T. Betaine and l-carnitine transport by Listeria monocytogenes Scott A in response to osmotic signals. J Bacteriol. 1997;179:6979–6985. doi: 10.1128/jb.179.22.6979-6985.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verheul A, Rombouts F M, Beumer R R, Abee T. An ATP-dependent l-carnitine transporter in Listeria monocytogenes is involved in osmoprotection. J Bacteriol. 1995;177:3205–3212. doi: 10.1128/jb.177.11.3205-3212.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Voelker U, Dufour A, Haldenwang W G. The Bacillus subtilis rsbU gene product is necessary for RsbX-dependent regulation of ςB. J Bacteriol. 1995;177:114–122. doi: 10.1128/jb.177.1.114-122.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Voelker U, Voelker A, Maul B, Hecker M, Dufour A, Haldenwang W G. Separate mechanisms activate ςB of Bacillus subtilis in response to environmental and metabolic stress. J Bacteriol. 1995;177:3771–3780. doi: 10.1128/jb.177.13.3771-3780.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Völker U, Engelmann S, Maul B, Riethdorf S, Völker A, Schmid R, Mach H, Hecker M. Analysis of the induction of general stress proteins of Bacillus subtilis. Microbiology. 1994;140:741–752. doi: 10.1099/00221287-140-4-741. [DOI] [PubMed] [Google Scholar]
- 53.Von Blohn C, Kempf B, Kappes R M, Bremer E. Osmostress response in Bacillus subtilis: characterization of a proline uptake system (OpuE) regulated by high osmolarity and the alternative transcription factor sigma B. Mol Microbiol. 1997;25:175–187. doi: 10.1046/j.1365-2958.1997.4441809.x. [DOI] [PubMed] [Google Scholar]
- 54.Waite B L, Siragusa G R, Hutkins R W. Bacteriocin inhibition of two glucose transport systems in Listeria monocytogenes. J Appl Microbiol. 1998;84:715–721. doi: 10.1046/j.1365-2672.1998.00401.x. [DOI] [PubMed] [Google Scholar]
- 55.Wilkins P O, Bourgeois R, Murray R G E. Psychrotrophic properties of Listeria monocytogenes. Can J Microbiol. 1972;18:543–551. doi: 10.1139/m72-087. [DOI] [PubMed] [Google Scholar]
- 56.Wise A A, Price C W. Four additional genes in the sigB operon of Bacillus subtilis that control activity of the general stress factor ςB in response to environmental signals. J Bacteriol. 1995;177:123–133. doi: 10.1128/jb.177.1.123-133.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu S, de Lencastre H, Tomasz A. Sigma-B, a putative operon encoding alternate sigma factor of Staphylococcus aureus RNA polymerase: molecular cloning and DNA sequencing. J Bacteriol. 1996;178:6036–6042. doi: 10.1128/jb.178.20.6036-6042.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang X, Kang C M, Brody M S, Price C W. Opposing pairs of serine protein kinases and phosphatases transmit signals of environmental stress to activate a bacterial transcription factor. Genes Dev. 1996;10:2265–2275. doi: 10.1101/gad.10.18.2265. [DOI] [PubMed] [Google Scholar]