Abstract
Plants trigger endoplasmic reticulum (ER) pathways to survive stresses, but the assistance of ER in plant tolerance still needs to be explored. Thus, we selected sensitive and tolerant contrasting abiotic stress sorghum varieties to test if they present a degree of tolerance to ER stress. Accordingly, this work evaluated crescent concentrations of tunicamycin (TM µg mL−1): control (0), lower (0.5), mild (1.5), and higher (2.5) on the initial establishment of sorghum seedlings CSF18 and CSF20. ER stress promoted growth and metabolism reductions, mainly in CSF18, from mild to higher TM. The lowest TM increased SbBiP and SbPDI chaperones, as well as SbbZIP60, and SbbIRE1 gene expressions, but mild and higher TM decreased it. However, CSF20 exhibited higher levels of SbBiP and SbbIRE1 transcripts. It corroborated different metabolic profiles among all TM treatments in CSF18 shoots and similarities between profiles of mild and higher TM in CSF18 roots. Conversely, TM profiles of both shoots and roots of CSF20 overlapped, although it was not complete under low TM treatment. Furthermore, ER stress induced an increase of carbohydrates (dihydroxyacetone in shoots, and cellobiose, maltose, ribose, and sucrose in roots), and organic acids (pyruvic acid in shoots, and butyric and succinic acids in roots) in CSF20, which exhibited a higher degree of ER stress tolerance compared to CSF18 with the root being the most affected plant tissue. Thus, our study provides new insights that may help to understand sorghum tolerance and the ER disturbance as significant contributor for stress adaptation and tolerance engineering.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12192-023-01382-5.
Keywords: Cell stress, Chaperones, Primary metabolites, Metabolic profile, GC-MS
Introduction
Forage sorghum (Sorghum bicolor (L.) Moench) stand out as a significant crop for human, particularly in Africa, Asia, and other semi-arid regions all over the world, as well as a source for ruminant diets used mainly in countries like Brazil and the United States (Afify et al. 2012; Taleon et al. 2012). The sorghum benefits are related to high levels of carbohydrates and proteins in some varieties associated with high digestibility, productivity, and adaptation to dry and hot environments (Stefoska-Needham and Tapsell 2020; Carvalho et al. 2020), including the potential for bioenergy production (Velmurugan et al. 2020). Sorghum is the fifth most produced cereal worldwide, behind only wheat, rice, maize, and barley (FAO 2019). In Brazil, the production is around 3.065 million tons per year, considering 2.985 kg/ha, and 1027.1 hectares of planted area (CONAB 2022).
Similar to all living organisms, sorghum can be affected by several stress types that compromise seedling establishment, plant development, and yield. The abiotic stresses include the ion excess (mainly sodium, and chloride) or toxic metals (aluminum, arsenate, and cadmium, for example) in the soil, drought or flooding (water stresses), low or high temperatures, and nutrient deficiency (Reddy 2019). In general, temperature, salinity, and drought are the main factors that can harm plant productivity (Fedoroff et al. 2010; Zhu 2016). On the other hand, considering plant diseases, the attack of fungi and bacteria, for example, induces biotic stress pathways (Schlemper et al. 2018). Due to their C4 metabolism, sorghum plants tolerate dry and high-temperature environments by a complex mechanism involving morphological, physiological, and biochemical alterations that manage to develop and grow in cultivated regions with irregular rainfall distribution (Taleon et al. 2012; McCormick et al. 2018). Moreover, some genotypes have shown differential tolerance to adverse environments, such as those presenting differential drought tolerance levels (Seyoum et al. 2019) and different salt stress tolerance levels (Silva et al. 2019). For instance, two sorghum varieties, CSF18 and CSF20, demonstrating different tolerance levels to salinity, are characterized as sensitive and tolerant to biotic stress, respectively (Vieira et al. 2005; Oliveira et al. 2020).
In general, plants display several responses to unfavorable conditions, such as morphophysiological and metabolic adjustments and adaptive cellular responses during development (Bai et al. 2018). Chaperones, heat shock proteins, and antioxidant enzymes are molecules related to most of these mechanisms, which can protect the plant from damage (Chi et al. 2019; Ul et al. 2019). In most cases, the Endoplasmic Reticulum (ER) synthesizes and exports these molecules to other organelles. Thus, it is noteworthy that this compartment became a crucial organelle for modulating plant responses to stresses, including primary metabolite adjustments in the cytosol (Park and Park 2019). Indeed, one general response of both biotic and abiotic stress is the induction of ER stress (Park and Park 2019; Pastor-Cantizano et al. 2020; Afrin et al. 2020). It decreases protein folding which causes an accumulation of unfolded or misfolded proteins in the ER lumen (Xiang et al. 2017; Manghwar and Li 2022). In Arabidopsis, at the first moment, it triggers the activation of a signaling pathway called unfolded protein response (UPR) which guarantees the maintenance of quality control of the secreted proteins (Wang et al. 2019). Also, in different crops, several other genes link ER stress response, multiple stresses, and development (Manghwar and Li 2022). If ER does not recover its homeostasis, apoptosis pathways are activated, leading to programmed cell death (Xiang et al. 2017), a conserved process among animals, yeasts, and plants (Simoni et al. 2022).
Several studies have exposed plant tissues or cells to tunicamycin (TM), a chemical agent widely used in experimental conditions that inhibit protein N-glycosylation disturbing ER homeostasis (Angelos and Brandizzi 2018; Yu et al. 2019). Although the increase of TM hardly impairs plant development, physiological and metabolic responses are not explored (Yang et al. 2014; Chakraborty et al. 2017). Otherwise, ER stress is often assessed by the upregulation of ER-resident proteins, mainly by transcriptome and proteome approaches (Feldeverd et al. 2020). Indeed, the cellular response is much broader and involves phytohormones, reactive oxygen species, membrane phospholipids, and primary metabolites (Lima et al. 2022; Kanehara et al. 2022). It results in a tight adjustment between cell survival and death, including a complex intracellular network among ER, chloroplast, mitochondria, and cytosol (Depaepe et al. 2021). Following these metabolic alterations that the plants use to face stresses, advanced tools have been employed, such as known omics. Among them, metabolomics stands out as a multidisciplinary science that offers unique possibilities to decode biochemistry at a cellular level, beyond transcriptome and proteome (Tugizimana et al. 2018; Hamany Djande et al. 2020). It provides a qualitative and quantitative analysis of metabolites, predicting discriminant ones and influencing plant growth and development under different environmental conditions (Razzaq et al. 2019; Lima et al. 2022; Raza 2022).
The ER efficiency in dealing with the accumulation of misfolded proteins is fundamental for adaptation and plant survival in front of any situation of stress. Nevertheless, cellular adjustments need to be explored and correlated to ER response. The ER efficiency in dealing with the accumulation of misfolded proteins is fundamental for adaptation and plant survival in front of any situation of stress. Nevertheless, cellular adjustments need to be explored and correlated to ER response. The influence of TM exposition over time shows amino acids are discriminants at 24 h, and components of the tricarboxylic cycle decrease to accumulate sugars at 96 h under high concentrations of TM (Lima et al. 2022). Hence, it is relevant to detect crop varieties that display different physiological, biochemical, and molecular responses regarding low, mild, and high-stress conditions by the increasing levels of TM.
Thus, we hypothesized that tolerance and sensibility are related to ER performance by different metabolite modulation that helps plants to acclimate to abiotic stress. Hence, the purpose of this research was to evaluate changes in the metabolic profiles in seedlings under endoplasmic reticulum stress induced by low, mild, and higher concentrations of tunicamycin (TM) in two contrasting sorghum varieties, CSF18 and CSF20 described in the literature as sensitive and tolerant to other abiotic stresses, respectively. The relationship between ER stress and metabolite modulation may provide new data to understand the differential responses between contrasting genotypes involved in stress adaptation, which may help future breeding programs to engineer stress tolerance.
Material and methods
Plant material and growth conditions
Sorghum seeds of two varieties (CSF18 and CSF20) from the Agronomic Institute of Pernambuco (IPA) were selected and surface sanitized with 0.04% sodium hypochlorite. The germination process was carried out on a germitest paper (28 × 38 cm), previously autoclaved at 120 °C, containing 20 seeds on each sheet of paper. The experiment was placed in a germination chamber (BOD) with average temperatures of 30 ºC during the day and 25 ºC at night, with a photoperiod of 12 h. After the third day after sowing, seedlings were carefully transferred to a new germitest sheet of paper moistened with the corresponding treatment solution. The control treatment (T1) consisted only of paper sheets moistened distilled water. Whereas T2, T3, and T4 treatments, the Tunicamycin (TM) solution, a specific ER stress inducer, was added to moist the sheet paper in the concentrations of 0.5 µg mL−1, 1.5 µg mL−1, and 2.5 µg mL−1, respectively.
Growth analysis: seedling length and dry matter determination
Seedlings were harvest from the germination chamber on the 4th day of treatment (7 days old) and photographed since it was the limit of root growth on germitest papers. Pictures were uploaded to the ImageJ software to measure the length of roots and shoots (Rasband 2016). Half of the plant material was divided by shoot tissues (leaf + stem) and root tissue and placed in an oven at 60 ºC for 72 h to obtain dry masses of shoots (SDM) and root (RDM) by weighing in an analytical balance. The other half was immediately frozen in liquid nitrogen and stored at -80 °C for further analyses.
Peroxidation of membrane lipids and hydrogen peroxide (H2O2) contents
Extracts were prepared by grinding 0.2 g of fresh vegetable tissue (shoots and roots) in 1.0 ml of trichloroacetic acid (TCA 5%) for 1 min. The macerate was transferred to a tube and centrifuged at 12.000 × g, for 15 min at 4.0 ºC. All procedures described here were conducted at 4.0 ºC, and the extracts were stored in an ultra-freezer at -80 ºC until the beginning of the analyses.
For peroxidation of membrane lipids, the content of malondialdehyde (MDA) was measured by substances reactive to the thiobarbituric acid method (Heath and Packer 1968). The reaction occurred from the addition of 250 µL of extract, 250 µL of distilled water, and 500 µL thiobarbituric acid solution (TBA 0.5%) in 20% trichloroacetic acid. The mixture was incubated in a water bath at 95 ºC, for 30 min, and the reaction was interrupted by cooling the tubes in an ice bath. Subsequently, it was centrifuged at 3.000 × g for 10 min at 4.0 ºC, and the supernatant was collected to determine readings at 532 nm and 600 nm. MDA contents were estimated by subtracting these readings, using their molar extinction coefficient (ɛ = 155 mM−1 cm−1) expressed as mmol g−1 FM.
For determination of H2O2 contents, 100 μL of the extract, 100 μL of 10 mM potassium phosphate buffer, pH 7.0, and 200 μL of 1 M KI (potassium iodide) were mixed (Sergiev et al. 1997). Samples were incubated in the dark for 15 min at room temperature, and then absorbance readings were taken at 390 nm. The H2O2 content was calculated from a standard curve reference with increasing concentrations of H2O2 expressed as mmol g−1 FM.
Gene expression
Total RNA was isolated from different treatments using TRizol (Sigma Aldrich's). The quantification and integrity were carried out in a NanoDrop 2000 spectrophotometer (Thermo Scientific TM, Waltham, USA), and electrophoresis in 1.5% (m/v) agarose gel electrophoresis system, at 50 mA, 100 V. cDNA libraries were built employing M-MLV reverse transcriptase. Reverse transcription-polymerase chain reaction (RT-PCR) was performed using RNase-free water and oligo(dT)s incubation at 70 ºC for 5 min and cooling at 4 ºC for 5 min. It was mixed with RNase inhibitor, Reverse Transcription Mix, oligo(dT)s, and submitted to 25 ºC annealing temperature for 5 min, followed by elongation at 42 ºC for 60 min, and enzyme denaturation at 70 ºC for 15 min. Synthesized cDNA was stored under -20 ºC until used. The qPCR amplification reactions were carried out in a real-time thermal cycler (Esco Swift, Esco), composed of initial denaturation at 95 °C for 2 min, and 40 thermal cycles of 15 s at 95 °C, followed by 15 s at a specific annealing temperature for each primer (Supplementary Table S1) and finally at 20 s at 60 °C in a total volume of 10 µL, according to the manufacturer’s instructions of GoTaq qPCR Master Mix (Promega) in biological triplicates. Relative expression levels of ER stress marker genes (to confirm the ER stress event) SbBiP1, SbPDI, SbIRE1, SbbZiP17/28, and housekeeping reference gene SbUBC18 of roots and shoots were performed based on previous studies in the same conditions (Queiroz et al. 2020; Lima et al. 2022). The relative quantification of transcripts was carried out using the mean Ct (Cycle threshold) and 2−ΔΔCT method (Livak and Schmittgen 2001).
Metabolic profile by GC-MS and data analysis
Approximately 50 mg of the samples were ground and mixed in methanol, chloroform, and ultrapure water (2: 1: 2, v/v) solution for polar metabolite extraction (Lisec et al. 2006). Then, 30 µL of the internal standard ribitol (0.2 mg ml−1) was added to the mixture, and 150 µl aliquot of the upper (polar) water-methanol phase was transferred to a new tube and dried in a vacuum concentrator at room temperature (SpeedVac Concentrator, Eppendorf, Hamburg, Germany). In the derivatization stage, the dried samples were treated with methoxylamine hydrochloride (10 mg / 0.5 mL in pyridine) with stirring at 37 °C for 2 h, followed by the addition of N-methyl-N- (trimethylsilyl) -trifluoro acetamide (MSTFA) with stirring at 37 °C for 30 min. Metabolite profiles were obtained by gas chromatography coupled with mass spectrometry (GC-MS, QP-PLUS 2010, Shimadzu, Japan). One microliter of each sample was injected in split mode (1:10 ratio). Helium gas was used as carrier gas with a flow rate of 1.0 mL min−1 and a capillary column RTX-5MS (30 m × 0.25 mm × 0.25 um) for the separation of the metabolites, being programmed with the initial temperature at 80 °C for 5 min, then increased from 8 °C per minute to 310 °C and maintained for 1 min at that temperature. The injection, ion source, and MS interface temperatures were 230 ºC, 200 °C, and 250 °C, respectively. The mass spectrometer was operated at 70 eV (Electron Ionization, EI) in a scan range of 80–700 (m/z), started after a solvent cut-off time of 3 min. Chromatograms and mass spectrum analysis were evaluated using the XcaliburTM 2.1 software, being compared with a mass spectrum library composed of a mix of standards previously identified (Batista et al. 2019; Lima et al. 2022). Files with the relative concentration of metabolites for the tissues of the leaves and roots of sorghum under treatments T1, T2, T3, and T4 were loaded on the server MetaboAnalyst 5.0 (http://www.metaboanalyst.ca) for further analysis. Then, metabolites were identified and classified using the Kyoto Gene and Genome Encyclopedia (KEGG) database.
Experimental design and statistical analysis
The experiment comprised of a completely randomized design (DIC), with a factorial scheme (4 × 2), two varieties of sorghum and four treatments previously explained (T1, T2, T3, and T4). Results were subjected to ANOVA analysis of variance and the data means were compared by the Tukey test (p ≤ 0.05), using the SISVAR statistical program. For metabolic profile statistics, data were normalized by log transformation and Pareto scaling. Mean values of the metabolites were compared using the T-tests to assess the effect within each treatment. Principal Component Analysis (PCA) was performed to identify the differences between the four treatments (T1, T2, T3, and T4) within each variety and tissue, and the most contributing metabolites were highlighted in the loading plots. To verify the effect of TM 2.5 µg mL−1 compared to the control on the metabolic profile, Orthogonal Partial Least Squares-Discriminant Analysis (OPLS-DA) was performed for leaves and roots separately of each variety, and main discriminant metabolites were provided.
Results
Effect of ER stress on sorghum seedling growth
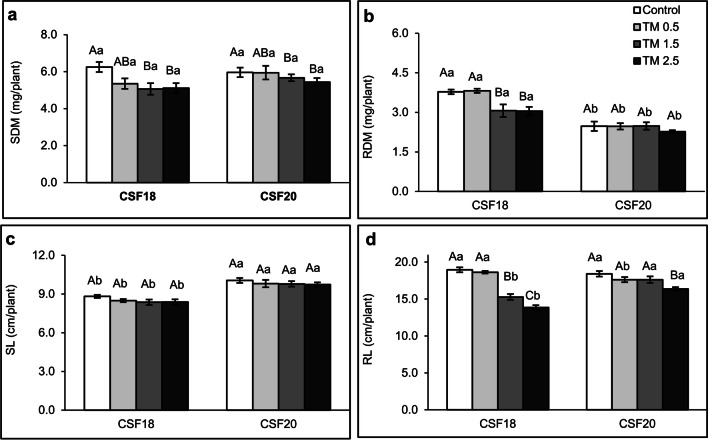
There was no difference between the control (0 µg mL−1 TM) and lower TM treatments (0.5 µg mL−1). Also, the dry mass weight of shoots in both CSF18 and CSF20 varieties were similar as TM increased (Fig. 1a). A significant shoot dry mass reduction occurred from the mild TM treatment (1.5 µg mL−1) compared to the control group. It remained constant in the highest TM concentration evaluated. Differently, there was a reduction in the root dry mass of CSF18 from mild TM treatment (Fig. 1b), which remained constant in the higher TM. Otherwise, there was no statistical significance among control and TM treatments of CSF20, despite this CSF18 had greater weights than CSF20. Comparing shoot lengths of all TM treatments within the same variety, the effect was the same for CSF18 and CSF20, and there were no significant differences among control and TM concentrations (Fig. 1c). However, when comparing the shoot length of both varieties, there were significant differences for this parameter in each TM treatment, with CSF20 showing the highest values regardless of TM concentration. CSF18 shoot length values ranged between 8.4 and 8.8 cm, whereas CSF20 ones were between 9.7 and 10.1 cm. Conversely, the root length of CSF18 and CSF20 from the control group are nearly the same size (Fig. 1d). Nevertheless, as the stress inducer was applied, CSF18 tended to be inhibited earlier than CSF20, especially at TM 1.5 µg mL−1. In which CSF20 maintained the root length similar to control treatment. At the highest TM concentration, CSF20 decreased the root length by only 10% (compared to the control group), whereas CSF18 exhibited a reduction of 27% in the same treatment. Clearly, the plant phenotypes showed a better performance of CSF20 than CSF18 (Supplementary Fig. S1). It is possible to perceive the morphological differences between both varieties, which may be influencing the differences in dry mass and length that exist between control and TM 2.5 µg mL−1, observing an increased volume of secondary roots in the CSF18 and a longer root length of CSF20 at the highest concentration of TM.
Fig. 1.
Growth parameters of two sorghum varieties under ER stress. SDM (a) and RDM (b) display the dry masses of shoots and roots, respectively. SL (c) and RL (d) indicate the length of shoots and roots, respectively. Three days old sorghum seedlings (CSF18 and CSF 20) were grown under control, TM 0.5 µg mL−1, 1.5 µg mL−1, and 2.5 µg mL−1 treatments until they reach seven days old. The values are the mean of 7 repetitions, and bars represent standard error. Data were submitted to analysis of variance (ANOVA), and all four treatments were compared by the Tukey test (p < 0.05) in each TM concentration. Lowercase letters compare the two sorghum varieties. Uppercase letters compare the treatments within each variety of sorghum
Lipid degradation and production of reactive oxygen species under ER stress
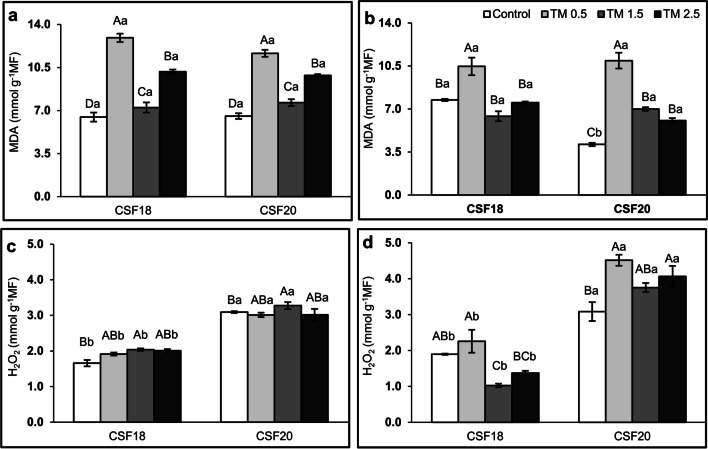
There was no significant difference in malondialdehyde (MDA) contents between shoots of both sorghum varieties comparing the same treatment (Fig. 2a). Additionally, CSF18 and CSF20 exhibited similar behavior as TM increased, in which MDA was higher than the control. Though, it was remarkably higher at the lowest TM concentration (0.5 µg mL−1). On the other hand, the H2O2 shoot was higher in CSF20 than in CSF18, comparing each treatment (Fig. 2c). Comparing the TM treatment, the levels of H2O2 in shoots were significantly higher at TM 1.5 µg mL−1 for both varieties but not different from other TM concentrations. In roots, there was no difference in MDA values between varieties under TM, except by control treatment (Fig. 2b). Likewise, H2O2 contents were different for the varieties in each treatment in which the values were higher in the CSF20 than CSF 18 (Fig. 2d). In CSF18 roots, the application of TM induced MDA content only at 0.5 µg mL−1 and reduced it at 1.5 µg mL−1, compared to the absence of TM. For CSF20 roots, all TM treatments induced MDA, but it was higher at 0.5 µg mL−1, and H2O2 increased at 0.5 and 2.5 µg mL−1 TM treatments.
Fig. 2.
Malondialdehyde (MDA) and hydrogen peroxide (H2O2) in shoots (a, c) and roots (b, d), respectively. Three days old sorghum seedlings (CSF18 and CSF 20) were grown under control, TM 0.5 µg mL−1, 1.5 µg mL−1, and 2.5 µg mL−1 treatments until they reach seven days old. The values are the mean of 5 repetitions, and bars represent standard error. Data were submitted to analysis of variance (ANOVA) and all four treatments were compared by Tukey test (p < 0.05) in each TM concentration. Lowercase letters compare the two varieties of sorghum. Uppercase letters compare the treatments within each variety of sorghum
Gene expression analysis of chaperones and ER stress sensors
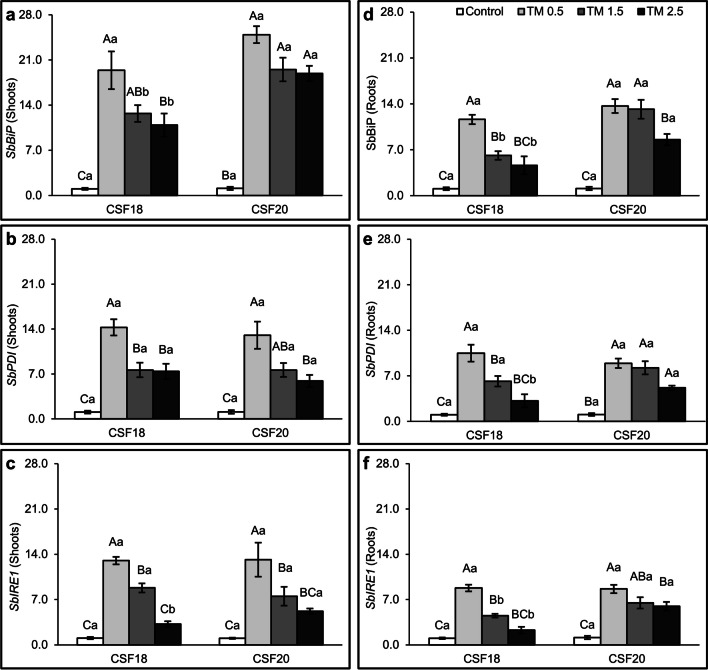
Sorghum seedlings varieties (CSF18 and CSF20) were treated with low, mild, and high concentrations of TM, 0 µg mL−1 (Control), 0.5 µg mL−1, 1.5 µg mL−1 and 2.5 µg mL−1 to evaluate the expression of SbBiP, SbPDI, SbIRE1, and SbbZIP17 genes as markers of ER stress (Fig. 3 and Supplementary Fig. S2). There was an increase in the expression of the three genes SbBiP, SbPDI, and SbIRE1 in the groups treated with different TM concentrations considering both varieties compared to the control group, as well as for both shoot and root. The lowest TM concentration (0.5 µg mL−1) induced the greatest changes in the relative expression values of genes in Sorghum. In general, at the lowest TM concentrations, shoots seemed to have higher values than roots, which was particularly evident in SbBiP. However, the expression of the genes started to decreased from mild TM (1.5 µg mL−1) to the higher TM concentration (2.5 µg mL−1). Although, the CFS20 presented high levels of SbBiP transcripts in roots and shoots, and and SbIRE1 in shoots under 2.5 µg mL−1 TM and roots from 1.5 µg mL−1 TM. This may indicate an efficient ER response of CFS20 than CFS18.
Fig. 3.
Relative gene expression of SbbBiP, SbPDI, and SbbIRE1. Sorghum bicolor seedlings were submitted to different concentrations of TM: 0 µg mL−1 (Control), 0.5 µg mL−1, 1.5 µg mL−1 and 2.5 µg mL−1. The expression profiles were determined in the shoot (a, b and c) and in the roots (d, e and f). Gene expression was normalized using SbUBC18 as a reference gene. Lowercase letters compare the two varieties of sorghum. Uppercase letters compare the treatments within each variety of sorghum. All four treatments were compared by Tukey test (p < 0.05) in each TM concentration. The columns indicate the means of three biological replicates and the bars indicate the standard deviation ( ±)
Metabolic profiling of sorghum shoots and roots under crescent TM concentrations
Using gas chromatography coupled with mass spectrometry (GC-MS), it was possible to separate and identify a total of 65 metabolites comprising all treatments and tissues (shoot and roots), including ribitol (internal standard). They were organized based on their retention time and KEGG ID classification (Supplementary Table S2). Out of this amount, the majority was represented by amino acids (34%), followed by carbohydrates (28%), organic acids (26%), and other metabolites, which included flavonoids, nitrogenous bases, and amines (12%).
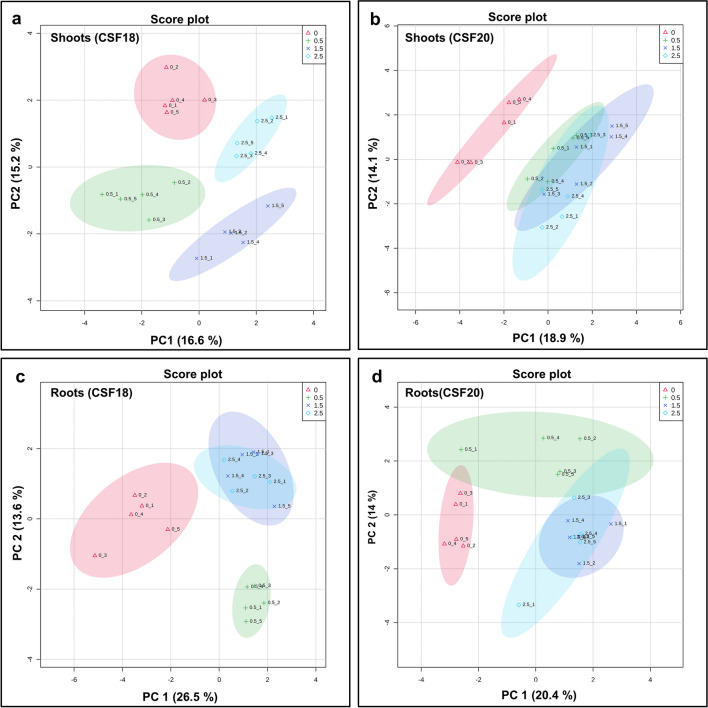
In order to demonstrate differences in the metabolic profiles among control and the three TM treatments, the principal component analysis (PCA) of shoots and roots in both sorghum varieties was carried out, separately. In CSF18 shoots, the score plot indicated a well-done separation of the four treatments in which PC1 and PC2 were responsible for 16.6% and 15.2%, respectively, of the total variance (Fig. 4a). The most positive contributing metabolites discrimination were uracil, dihydroxyacetone, threonine, ascorbic acid and ornithine for PC1, whereas maleic acid, talose, succinic acid, lactic acid, and, xylose were the most positive for PC2 (Supplementary Fig. S3 and Supplementary Table S3A). On the other hand, in CSF20 shoots, there were overlaps in treatments with TM (Fig. 4b). In this case, PC1 contributed 18.9% displaying dihydroxyacetone, pyruvic acid, cellobiose, uracil, and xylose as main metabolites and PC2 contributed 14.1% with the main contribution of hydroxylamine, butyric acid, ribose, fructose, and lactic acid (Fig. 4b, Supplementary Fig. S3 and Supplementary Table S3b).
Fig. 4.
Principal Component Analysis (PCA) of metabolic profiling of shoots (a, b) and roots (c, d) sorghum varieties. CSF 18 is on the left and CSF20 is on the right, they were grown under 0 (control), 0.5 µg mL−1, 1.5 µg mL−1, and 2.5 µg mL−1 tunicamycin (TM). The ellipses indicate the 95% confidence interval of the groups
Evaluating the PCA of roots, PC1 and PC2 contributed 26.5% and 13.6% in CSF18, respectively (Fig. 4c). Score plot results indicated a good separation between CSF18 control and TM treatments, mainly with TM 0.5 µg mL−1. However, there was a complete overlapping between 1.5 and 2.5 µg mL−1 TM treatments. The most positive metabolites observed in PC1 were glyceric acid, fumaric acid, talose, pyroglutamic acid, and malonic acid, whereas most of the contribution for PC2 was maleic acid, glycerol-3-phosphate, N-acetyl-serine, butyric acid, and tyrosine (Supplementary Fig. S3 and Supplementary Table S3c). Then, for CSF20 roots, the main contribution of PC1 was 20.4%, and PC2 was 14%. There was a separation of control and TM, although there was a slight overlapping between control and 0.5 µg mL−1 TM confidence intervals. Also, there was overlapping between 1.5 and 2.5 µg mL−1 TM, followed by a small overlap between 0.5 and 2.5 µg mL−1 TM confidence intervals. The main contributions for PC1 were quercetin, cellobiose, tryptophan, maltose, and sucrose (Fig. 4d). For PC2, the most positive contributing metabolites were galactose, mannose, ribose, sorbitol and arabinose (Supplementary Fig. S3 and Supplementary Table S3d).
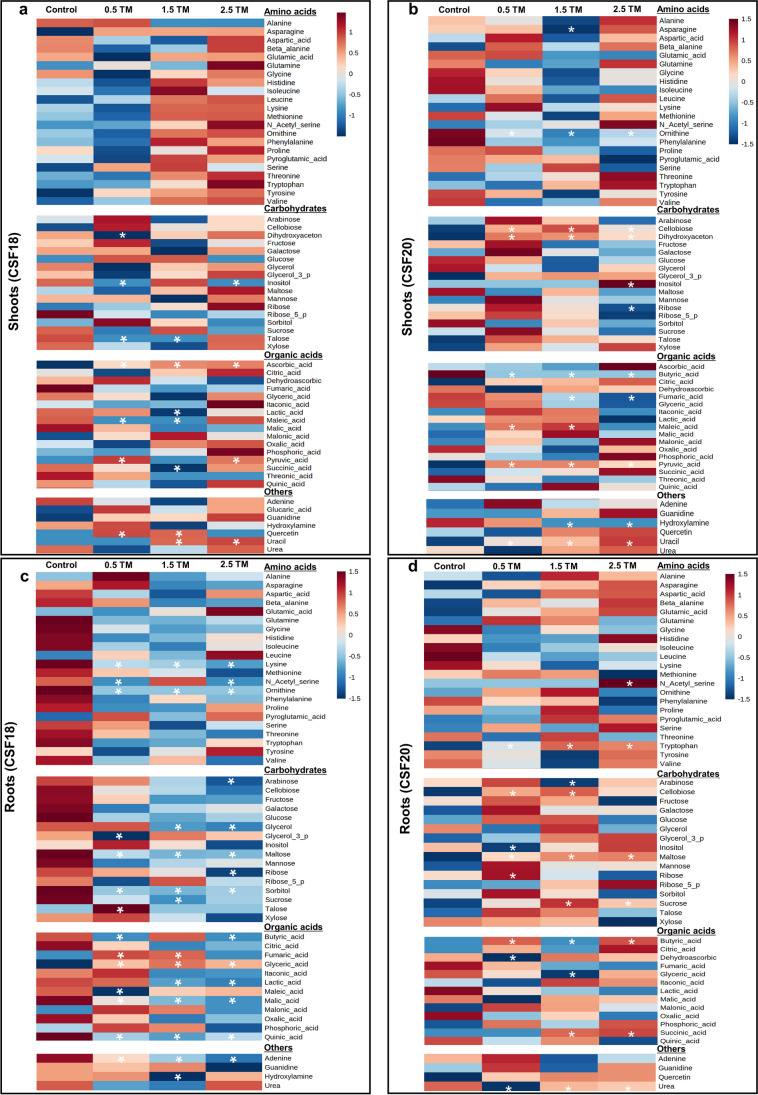
A heatmap of the relative abundance of metabolites in shoots and roots showed an overview of the modulation promoted by TM at different concentrations (Fig. 5). There is no standard of shoot heatmaps since crescent TM-induced stress modulated distinct metabolites in both cultivars. Otherwise, there was a prevailing modulation of metabolites in CFS18 roots, while CSF20 exhibited a general increase of metabolites. Differentially significant metabolites were indicated by asterisks in the heatmap, and statistical differences were detailed in supplementary Table S4. There were 10 and 12 differentially modulated metabolites identified in shoots of CSF 18 and CSF20, respectively (Fig. 5). In CSF18, the differential metabolites were five organic acids (ascorbic acid, lactic acid, maleic acid, pyruvic acid, and succinic acid), which seems to be decreased under 1.5 µg mL−1 TM, followed by three carbohydrates (dihydroxyacetone, inositol and talose) that were decreased mainly under 0.5 µg mL−1 TM, and two others (quercetin and uracil) increased in different concentrations (Fig. 5a). In CSF 20, there were three carbohydrates (dihydroxyacetone, inositol, and ribose), four organic acids (butyric acid, fumaric acid, maleic acid and pyruvic acid), two amino acids (asparagine and ornithine), and two others (hidroxilamine and uracil) statistically different. In general, these metabolites were increased, particularly under 0.5 and 1.5 µg mL−1 TM (Fig. 5b).
Fig. 5.
Heat map representation of the relative abundance metabolites in shoots (a, c) and roots (b, d). Sorghum variety CSF 18 is on the left and CSF20 is on the right. They were grown in the absence or presence of ER stress inducer TM (0.5 µg mL−1, 1.5 µg mL−1, and 2.5 µg mL−1). Each square represents the log2 mean of five replicates. The color map shows an increasing (red scale) or decreasing (blue scale) of each metabolite in comparison to control plants. Asterisks indicated statistically differential metabolites by Anova p < 0,05
In roots, there were 20 and 13 differently significant metabolites in the CSF18 and CSF20, respectively (Fig. 5). In CSF18, the main differential metabolites were eight carbohydrates (arabinose, glycerol, glyceraldehyde-3-phosphate, maltose, ribose, sorbitol, sucrose, and talose), seven organic acids (butyric acid, fumaric acid, glyceric acid, maleic acid, malic acid, quinic acid), three amino acids (lysine, n-acetyl-serine, and ornithine), and two others (adenine and hydroxylamine). Although a few of them had increase under the lowest TM, there is a tendency to decrease it in the concentrations of 1.5 and 2.5 µg mL−1 TM (Fig. 5c). In CSF 20, the metabolites showing statistically differences between control and TM treatments were six carbohydrates (arabinose, cellobiose, inositol, maltose, ribose, and sucrose) followed by for organic acids (butyric acid dehydroascorbic acid, glyceric acid, and succinic acid) and quercetin. In general, these metabolites were increased from 0.5 to 2.5 µg mL−1 TM (Fig. 5d).
Comparing the metabolic profiles of CSF18 and CSF20 sorghum varieties and treatments
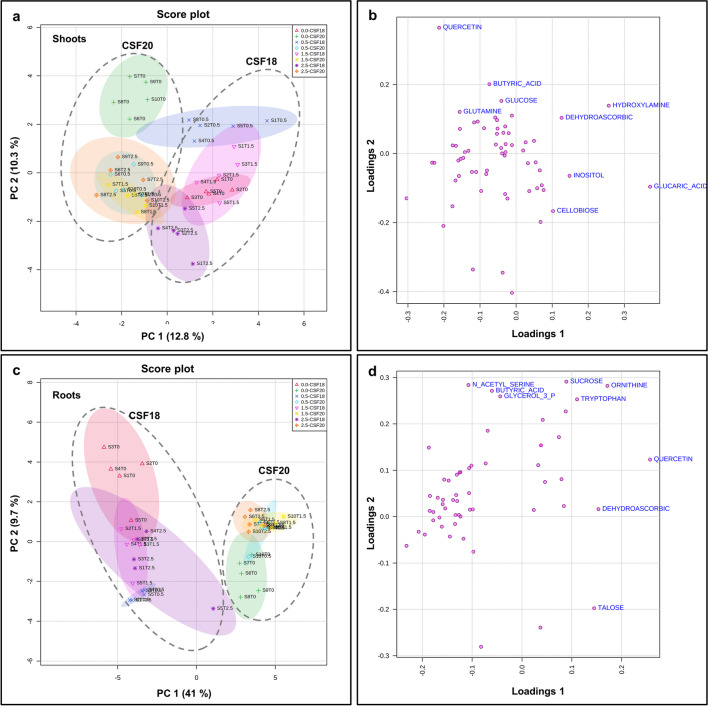
Combining all variables of two sorghum varieties and their treatments (Control and TM concentrations), the PCA of shoots and roots showed a pattern of results since there was mainly the separation of sorghum varieties (Fig. 6). However, shoot samples exhibited some overlapping between CSF18 under 2.5 µg mL−1 TM and other CSF 20 TM treatments (Fig. 6a). Notably, CSF20 root profiles were closer to each other than CSF18 root profiles (Fig. 6c). The main contributions for PC1 and PC2 for shoots profiles were 12.8% and 10.3%, respectively. In roots, PC1 was 41%, and PC2 was 9.7%. The loading plots demonstrate the contribution of each metabolite in PC1 and PC2. In shoot profiles, the top five contributions for PC1 were glucaric acid, hydroxylamine, inositol, dehydroascorbic, and cellobiose, and for PC2 were quercetin, butyric acid, glucose, hydroxylamine, and glutamine (Fig. 6b and Supplementary Table S5a). In roots, the top five contributions for PC1 were quercetin, ornithine, dehydroascorbic, talose, and tryptophan, and for PC2 were sucrose, N- acetyl-serine, ornithine, butyric acid, and glycerol-3-phosphate (Fig. 6d and Supplementary Table S5b).
Fig. 6.
Principal Component Analysis (PCA) of metabolic profiling in two varieties of sorghum, analyzing shoots CSF18 and CSF20 (a) and roots CSF18 and CSF20 (c). The loading plot for shoot (b) and root (d) indicate the positive and negative contributions of each metabolite in two varieties of sorghum: CSF 18 e CSF20. The ellipses indicate the 95% confidence interval of the groups
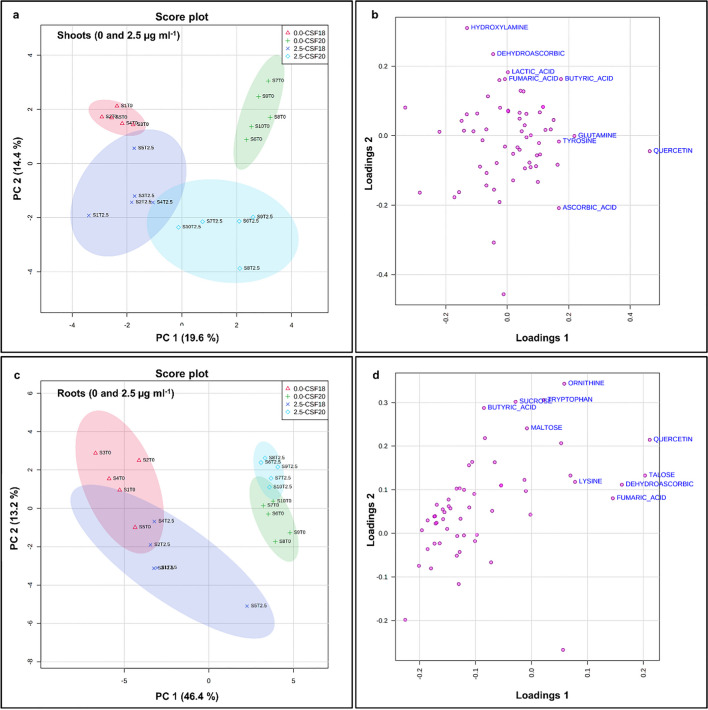
Due to the overlap of TM treatments within the varieties showed above, it was decided to perform the PCAs comprising only control and 2.5 µg mL−1 TM profiles which are the distant treatments (Fig. 7a and c). In shoot profiles, PC1 contributed with 19.6% and PC2 14.4% of distribution, which showed a slight overlapping between TM treatments of CSF 18 and CSF 20. In root profiles, the separation between sorghum varieties was much higher than in shoots. The contribution of PC1 and PC2 were 46.4% and 13.2%, respectively. In shoot profiles, the top five contributions for PC1 were quercetin, glutamine, butyric acid, tyrosine, and ascorbic acid, and for PC2 were hydroxylamine, dehydroascorbic, lactic acid, butyric acid and fumaric acid (Fig. 7b and Supplementary Table S6). In roots, the top five contributions for PC1 were quercetin, talose, dehydroascorbic, fumaric acid, and lysine, and for PC2 were ornithine, tryptophan, sucrose, butyric acid, and maltose (Fig. 7d and Supplementary Table S6).
Fig. 7.
Principal Component Analysis (PCA) of metabolic profiling in two varieties of sorghum, analyzing shoots CSF18 and CSF20 (a) and roots CSF18 and CSF20 (c). The loading plot for shoot (b) and root (d) indicate the positive and negative contributions of each metabolite in two varieties of sorghum: CSF 18 e CSF20 under control and TM 2.5 µg mL−1. The ellipses indicate the 95% confidence interval of the groups
Identification of potential biomarkers for ER stress in sorghum varieties
Orthogonal Projections to Latent Structures Discriminant Analysis (OPLS-DA) was performed to predict positive and negative metabolites influenced by TM. The discriminant metabolites were identified and recognized as potential biomarkers of the treatment (Figs. 8, S4, S5, and Supplementary Table S7). Shoots and roots were analyzed separately in each variety of sorghum in this test, comparing each TM concentration against control.
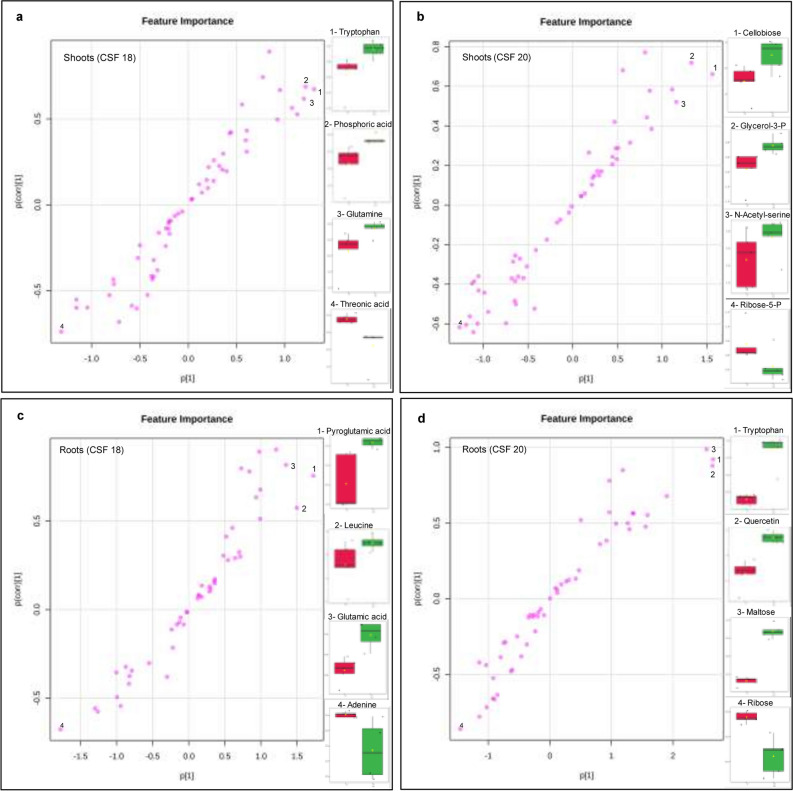
Fig. 8.
Orthogonal Projections to Latent Structures Discriminant Analysis (OPLS-DA) in shoots (a, b) and roots (c, d) of two sorghum varieties grown under 2.5 µg mL−1 TM. The top 3 significant metabolites that had a more contribution to the stressed seedlings (top right) and the top one for the control (bottom left) were highlighted. The names and the amounts were plotted in the right side of each plot
For lower TM concentration (0.5 µg mL−1), the top three discriminant metabolites were pyruvic acid, quercetin, and ascorbic acid for CSF18 shoots, and dihydroxyacetone was the most negative (Fig. S4). Likewise, the top discriminants for CSF20 shoots were dihydroxyacetone, pyruvic acid, and uracil, while butyric acid had the most negative contribution. For roots, CSF18 exhibited fumaric acid, talose, and glyceric acid as the top three discriminants, followed by glycerol-3-phosphate as the most negative contribution. Accordingly, CSF20 showed butyric acid, ribose, and mannose as the principal contributions, and dehydroascorbic acid as the most negative contribution.
For mild TM concentration (1.5 µg mL−1), the top three discriminant metabolites were ascorbic acid, uracil, and quercetin for CSF18 shoots, and lactic acid was the most negative (Fig. S5). Likewise, the top discriminants for CSF20 shoots were uracil, cellobiose, and pyruvic acid, while butyric acid had the most negative contribution. For roots, CSF18 exhibited fumaric acid, glyceric acid, and malonic acid as the top three discriminants and hydroxilamine as the most negative contribution. Hence, CSF20 showed succinic acid, ribose, and tryptophan as the principal contributions and glyceric acid as the most negative contribution.
In shoots treated with 2.5 µg mL−1 TM, the three major discriminant compounds that positively influenced CSF18 metabolome under ER stress were pyruvic acid, ascorbic acid and uracil (Fig. 8a). In contrast, inositol was the lowest discriminant. In CSF20 shoots (Fig. 8b), the three potential metabolites were uracil, inositol and pyruvic acid that positively discriminate TM treatment, and hydrixilamine was the discriminant metabolite. In CSF18 roots (Fig. 8c), three amino acids represented by glyceric acid, pyroglutamic acid, and leucine, were most positive discriminants to TM treatment, and adenine was the most negative discriminant. For roots of CSF20 variety (Fig. 8d), the three majors positive discriminant compounds induced by ER stress were tryptophan, quercetin, and maltose, and the most negative was ribose.
Discussion
The sorghum varieties present contrasting sensibility to ER stress
Plants sense environmental stresses and trigger cellular processes to maintain their homeostasis. In most cases, the healthy endoplasmic reticulum plays a crucial role in keeping a regular protein folding and protein secretory pathway (Chi et al. 2019; Ul et al. 2019). Accordingly, the ER efficiency in dealing with the accumulation of misfolded proteins is fundamental for adaptation and plant survival in stressful situations (Irsigler et al. 2007; Carvalho et al. 2014). Additionally, some genotypes exhibit a degree of tolerance to these environmental stresses, and the relation with ER is unclear. For instance, the sorghum genotypes CSF18 and CSF20 are characterized as sensible and tolerant to salinity, respectively (Vieira et al. 2005; Oliveira et al. 2020). Thus, we investigated two sorghum genotypes under low, medium, and high TM concentrations and evaluated the modulation of primary metabolites to acclimate to crescent ER stress.
The increase of TM concentration triggered ER stress in a tissue-specific way since it affected shoots and roots of sorghum seedlings differently (Fig. 1). The growth of CSF18 was impacted by ER stress at the mild TM, while CSF20 was slightly affected only at high TM. Losses of dry masses of organs and root length were more noticeable in CSF18 than in CSF20, accentuating it at the highest concentration. Even though morphologically, CSF18 had a higher abundance of lateral roots (Fig. S1), it did not increase the root length. Indeed, TM inhibits mainly the primary root growth of Arabidopsis plants, and it became more highlighted by the days of TM exposition (Lai et al. 2018; Ruberti et al. 2018; Hong et al. 2019), similar results occur during sorghum seedling establishment under ER stress overtime (Lima et al. 2022). Here, there was no change in sorghum shoot length, but TM effects were noticeable by the dry mass decrease. Likewise, the harmful effects on shoots occur in Arabidopsis as the impairment of leaf development and fresh weight decreases (Lai et al. 2018; Ruberti et al. 2018; Hirata et al. 2019; Nguyen et al. 2019).
Overall, the multivariate analysis of the main component corroborated the contrasting effects between root and shoots of CSF18 and CSF20 and the differences in TM concentrations. It showed different patterns in which metabolic profiles varied for each plant tissue and sorghum variety, whereas TM treatment profiles of CSF20 were relatively similar (Fig. 4). Although tunicamycin was applied directly to roots, it can be vascularly translocated to shoot without compromising the results (Irsigler et al. 2007). Roots are crucial organs that respond to environmental changes, including ER stress triggered by TM. There is more impairment of morphological growth during the long-term TM exposition in Arabidopsis and molecular processes induced to maintain cellular homeostasis are quick, intense in root tissues, and spread to leaves (Cho and Kanehara 2017). It was also exhibited here since the notable metabolic modulation occurs in the roots, and there is a tendency of decrease it in CSF18 and increase it in CSF20 (Fig. 5). In agreement, 397 transcripts of grapevine roots were influenced by TM (5.0 µg mL−1), it was related to several biological processes such as signal transduction, amino acid metabolism, protein processing, carbohydrate metabolism, and apoptosis (Aydemir and Ergül 2021). It is already observed that during stressful situations, Arabdopisis young seedlings encode the BiP3 gene (Noh et al. 2003). Certainly, the ER is the main organelle required, as it acts in the modulation of responses to stressful situations through an Unfolded Protein Response (UPR), increasing chaperones and ER proteins to assist in the correct folding of these proteins, such as BiP (Zhang et al. 2017; Park and Park 2019). Indeed, the relative expression of endoplasmic reticulum genes SbBiP, SbPDI, SbbZIP60, and SbbIRE1 in sorghum seedlings is induced by 2.5 μg ml−1 TM demonstrating the ER stress (Queiroz et al. 2020; Lima et al. 2022). The higher levels of SbBiP and SbIRE1 transcripts indicate that CSF20 variety preserved an efficient ER response at mild and higher TM concentrations than CFS18 (Fig. 3). It promoted a reduction of CSF18 seedling growth from mild TM and metabolic adjustments for acclimation occurred from lower TM. It led us to investigate the contrasting effects of both varieties.
It has also shown that environmental pressures induce oxidative stress in plant tissues. It occurs mainly marked by the production of MDA originating from lipid peroxidation (Nxele et al. 2017). Similarly, ER stress also induces this damage in the cellular membrane (Nawkar et al. 2017; Yu et al. 2019; Beaugelin et al. 2020), leading to an increase in MDA, an important leaf senescence hallmark (Melo et al. 2018). MDA results followed this feature, and more differences were observed in the shoots than roots comparing the control group and the TM treatments (Fig. 2), which was reported by ER stressors (Lima et al. 2022). Interestingly, the lowest concentration of TM (0.5 μg ml−1) promoted the highest lipid peroxidation. One possible explanation links it to a signaling mechanism that seems to be unrelated to an enzymatic anti-oxidant system in sorghum (Queiroz et al. 2020). Additionally, it may be related to a non-enzymatic system and an efficient ER response observed here by expressing high levels of SbBIP that help to keep the protein processing. However, it suggests new insights to be explored in the future. Moreover, hydrogen peroxide levels did not appear to be linked to the formation of MDA observed in this study, but they may also act as a signaling effector. Indeed, studies reported the role of ER transducers to induce IRE1, and genes responsible for membrane lipid metabolism via a decrease of myo-inositol levels (Kanehara et al. 2022). Hence, the MDA and H2O2 levels management contributes to intact UPR signaling and ER stress survival in plants (Angelos and Brandizzi 2018; Uzilday et al. 2018). It is already known that H2O2 is produced mainly in organelles such as mitochondria, chloroplasts, and peroxisomes. It has been reported that this oxygen species can also be generated in the endoplasmic reticulum (Farooq et al. 2019). Thus, we assume that there is a fine-tuning of H2O2 signaling for acclimatization to ER stress, mainly in the roots of the CSF20 variety, which has the highest accumulation of hydrogen peroxide (Fig. 2). In addition, the accumulation of H2O2 may be linked to the increase of ER genes, SbBiP, SbPDI, and SbIRE, particularly in the CSF20 (Fig. 3) that help to deal with stress decreasing misfolded protein accumulation (Ozgur et al. 2015). Even so, further studies are needed to understand the H2O2 signaling mechanism and the acclimation to ER stress, especially the CSF 20 variety that presented a better performance in this situation.
Metabolic profiles of sorghum varieties under lower, mild, and higher TM
We also investigated metabolic profiles of the contrasting sorghum varieties (CSF18 and CSF20) to ER stress (0.5, 1.5, and 2.5 μg ml-1 TM) in shoots and roots. TM treatments displayed completely distinct metabolic profiles between control and TM treatments (Fig. 4) that were evident in shoots and roots of CSF18, although mild and higher TM profiles in roots overlapped. There are distinct changes in the modulation of amino acids, carbohydrates, organic acids, and other metabolites. This differential modulation in the classes of compounds for each sorghum variety and organ analyzed was possible to perceive through the heatmap overviewing (Fig. 5). Changes in primary metabolism are common responses to stressful situations, whether due to biotic or abiotic factors (Tugizimana et al. 2018), and it is directly involved in the growth and development of plants (Hamany Djande et al. 2020). Indeed, the roots of both varieties showed the highest number of metabolites with differential modulation. Interestingly, there was a decrease in the relative abundance of most compounds in CSF18 due to the increase in TM that supports the root length decrease. In contrast, CSF20 showed increases in all groups of compounds as TM concentration increased. The overlapping of profiles also corroborates the maintenance of SDM, RDM, SL, and RL close to control measurements, including ER gene expressions, particularly under lower and mild TM (Figs. 1, 2, and 3). Thus, the metabolic modulation may reflect the better performance of CSF20 than CSF18 supporting our hypothesis that CSF20 tolerance from mild ER stress. It led us to explore these data to understand the influence of different levels of ER stress on the distinct metabolite groups and cellular metabolism of different sorghum seedlings varieties.
Carbohydrates contributed to mild and higher TM tolerance in CSF20 cultivar
The analysis revealed that carbohydrates were the group that had the most significant differences in the roots and shoots of both varieties. It is a common fact since these compounds are strictly related to plant growth and development, providing carbon skeletons being a source of energy (Lastdrager et al. 2014). They act as signaling molecules in response to stress, helping the plant to survive (Eveland and Jackson 2012). Sugars are the key molecules in UPR signaling, and their relationship with ROS is essential to a satisfactory response to ER stress (Depaepe et al. 2021). Indeed, the punctual increases of H2O2 may be linked to the maintenance of sugar levels in shoots and roots of CSF20 (Figs. 2 and 5), otherwise it was decreased in CSF18. It was observed that in sorghum varieties, CSF18 and CSF20 (the same as in the present study) subjected to saline stress, also presented differential modulation mainly of carbohydrates as an adaptative strategy (Oliveira et al. 2020). In sorghum seedlings under severe ER stress, there are inhibition of glycolysis and tricarboxylic acid cycle that promote sugars accumulation (Lima et al. 2022). It can further stabilize proteins and protect cellular structures, such as membranes, when stress becomes severe or persists for prolonged periods (Thalmann et al. 2016).
As mentioned earlier, mainly in roots, the vast majority of compounds were negatively modulated in CSF18, including carbohydrates. Therefore, it will be focused on carbohydrates and other groups that had their positive modulation and that in some way could contribute to greater stress tolerance of ER (for instance, maltose, sucrose, and cellobiose), especially CSF20. Maltose was a metabolite increased since the lowest concentration of TM (0.5 μg ml−1), probably due to the breakdown of starch for energy release (Kötting et al. 2010). There is an accumulation of maltose in wheat plants under osmotic (Darko et al. 2019) and saline stresses (Shelden et al. 2016), contributing to a significant increase in all growth parameters (Ibrahim and Abdellatif 2016). Besides, there is an evident maltose increase in the heatmap, it is also one of the most discriminant metabolites by the Ortho-PLSDA model for CSF 20 roots (Fig. 8d). Then, sucrose, the mainly transported carbohydrate in higher plants, is related to the maintenance of root elongation (Shelden et al. 2016), a fact observed in CSF20. It may act as a signal for pathways that lead to gene expression and physiological adaptation (Wind et al. 2010). Sucrose content increased mainly from the concentration of mild TM (1.5 μg ml−1) in the CSF20 roots. Finally, further studies are needed to understand the role of cellobiose in stress tolerance of CSF20, a discriminant metabolite for shoots under lower and mild TM (Figs. S4 and S5), which was increased from TM 0.5 μg ml−1 in booth shoots of CSF20. The maintenance of energy, defense metabolism, and antioxidant metabolism were reported under drought conditions, although they are still not well understood (Li et al. 2017). In fact, the earlier maintenance of other sugars act as the key for plant to survive under ER stress, for instance fructose, maltose, and kestose (Lima et al. 2022).
Organic acids are consumed in the CSF18 under ER stress
It is worth mentioning differential modulations of organic acids, in which they were decreased in the CFS18 roots, although punctual increases are perceived. Organic acids are important sources of carbon skeletons for various biochemical processes, able to participate in the TCA cycle for the production of energy that could be used in the metabolism of plants (Igamberdiev and Eprintsev 2016; Quan et al. 2016; Panchal et al. 2021). The mainly reported function of maleic acid is the tolerance to chromium and its role in the positive regulation of non-enzymatic and enzymatic antioxidant systems (Mahmud et al. 2017), requiring studies on the involvement of this metabolite in tolerance to other types of stress, including the ER stress. Here, it was increased in the CSF20 shoots under 0.5 and 1.5 μg ml−1 TM. Among these organic acid stands outs the pyruvic acid that was increased in shoots under TM concentrations, and it was a discriminant for higher TM (Fig. 8b). Pyruvic acid is considered a precursor to organic acids, linking the glycolysis pathway to the tricarboxylic acid cycle (TCA) (Jardine et al. 2010). Through the accumulation of this metabolite, there is a greater supply of material for the TCA cycle, an important defense system in situations of stress, verified by the increase in the tolerance to salt in barley (Wu et al. 2013). Possibly, in CSF18, ER stress acted by interrupting TCA cycle and decreasing the synthesis of organic acids, leading to a decline in the metabolic function and the electron transport chain, thus affecting the normal development of the plant (Zhong et al. 2016). On the other hand, in roots of CSF20, succinic acid is increased from 1.5 μg ml−1 TM being a discriminant for mild TM (Fig. S5d). Succinic acid acts as an intermediary in the TCA cycle and plays an important role in energy production and regulation of the TCA cycle (Fernie et al. 2004). The accumulation of this metabolite demonstrates a better tolerance to water stress, the result of greater efficiency of the TCA cycle in producing more energy to deal with a stressful situation (Khan et al. 2019a) supporting our results. It is worth remembering that these organic acids mentioned can participate in the TCA cycle or be diverted to the synthesis of amino acids (Igamberdiev and Eprintsev 2016).
Amino acids are consumed in the CSF18 roots under ER stress
A few amino acids had significant differences, mainly centered on asparagine (in shoots) and tryptophan (in roots) of CSF20. Tryptophan stands out at all TM concentrations and it is highlighted as a discriminant marker in the OPLS-DA models (Figs. 8, S4d, and S5d). Asparagine is acts in nitrogen storage and transport. It is reported that at high concentrations it is related to the response to a variety of abiotic and biotic stresses (Lea et al. 2007), such as water and nutrition stress (Curtis et al. 2018). Tryptophan is a precursor to secondary metabolites such as indoleacetic acid, a plant hormone necessary for cell expansion (Zemanová et al. 2014), and plays a role in tolerating abiotic stresses such as drought and reducing reactive oxygen species (Khan et al. 2019b). Studies demonstrate that the negative (Kamauchi et al. 2005) and positive (Okada et al. 2002; Çakır Aydemir et al. 2020) modulation of certain genes related to the metabolism of amino acids are crucial to the recovery from ER stress as well as in the protection against the oxidative stress of ER (Zeeshan et al. 2016). High levels of BiP (luminal binding protein) have also been reported to induce an increase in the concentration of different amino acids (Kawakatsu et al. 2010).
Furthermore, there are a remarkable positive modulation of quercetin in roots of CSF20 (Figs. 5d and 8d) which became a target for further studies since it was a flavonoid molecule that reduces ER stress in mammalian (Eisvand et al. 2022), and seems to be also an important metabolite for stress tolerance in plants.
Conclusions
Tunicamycin induced ER stress followed by the expression of several ER genes, including the chaperones SbBiP and SbPDI, as well as the sensors SbbZIP60, and SbbIRE1. The more significant damage occurred in the root development of the CSF18 variety under the high concentrations. The metabolic responses of shoots and roots were quite distinct; CSF18 showed a reduction in the relative concentration of metabolites, being more evident in the root part. The CSF20 roots displayed the greatest positive modulation of carbohydrates (sucrose, maltose, and cellobiose) and organic acids (maleic, pyruvic, and succinic acid) due to their importance in providing energy and intermediates of metabolic pathways necessary for plant growth, and quercetin a metabolite reported to decrease ER stress. Few significant changes were observed in the amino acids. The highest concentration of TM (2.5 μg ml−1), demonstrated the greatest effects, although it is observed that small concentrations of TM (from 0.5 μg ml−1) can induce ER stress, secondary oxidative stress by greater production of H2O2 to act as a possible signal to stresses. In general, CSF20 showed a higher degree of ER stress tolerance compared to CSF18. Thus, this work contributes to the understanding of primary metabolism alterations involved in the regulation of ER stress tolerance in plants and a significant contributor to stress adaptation and potential tolerance engineering.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary Fig. S1 Sorghum plants CSF 18 e CSF20 grown in the absence (A) e (C) or presence of ER stress inducer TM (B) e (D) in the TM 2,5 µg mL-1 concentration. (TIF 4973 KB)
Supplementary Fig. S2 Relative gene expression of SbbZIP17. Sorghum bicolor seedlings were submitted to different concentrations of TM: 0 µg mL-1 (Control), 0.5 µg mL-1, 1.5 µg mL-1 and 2.5 µg mL-1. The expression profiles were determined in the shoot (A) and in the roots (B). Gene expression was normalized using SbUBC18 as a reference gene (Queiroz et al. 2020; Lima et al. 2022). Lowercase letters compare the two varieties of sorghum. Uppercase letters compare the treatments within each variety of sorghum. All four treatments were compared by Tukey test (p<0.05) in each TM concentration. The columns indicate the means of three biological replicates and the bars indicate the standard deviation (±). (TIF 912 KB)
Supplementary Fig. S3 The loading plot for shoot (A, B) and root (C, D) indicate the positive and negative contributions of each metabolite in two varieties of sorghum: CSF 18 on the left and CSF20 on the right. The top five most positive metabolites for each component are highlighted. (TIF 373 KB)
Supplementary Fig. S4 Orthogonal Projections to Latent Structures Discriminant Analysis (OPLS-DA) in shoots (a, b) and roots (c, d) of two sorghum varieties grown under 0.5 µg mL-1 TM. (TIF 2354 KB)
Supplementary Fig. S5 Orthogonal Projections to Latent Structures Discriminant Analysis (OPLS-DA) in shoots (a, b) and roots (c, d) of two sorghum varieties grown under 1.5 µg mL-1 TM. (TIF 2452 KB)
Supplementary Table S2 List of detected metabolites in shoots and roots of sorghum with their classification in compound types, their retention time, respectively, and with their kyoto encyclopedia of genes and genomes identifier number (KEGG ID). (XLS 12 KB)
Supplementary Table S3 Loading score of primary metabolites found 0 (control), 0.5 µg mL-1, 1.5 µg mL-1, and 2.5 µg mL-1 tunicamycin (TM) treated seedlings ranked by principal component analyses values. (CSV 10 KB)
Acknowledgements
We would like to thank Instituto Agronômico de Pernambuco (IPA) to provide sorghum seeds and to Cryogenics Center’s Physics Department of Federal University of Ceará for nitrogen suppling. Thanks to Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Fundação Cearense de Apoio ao Desenvolvimento Científico e Tecnológico (FUNCAP), and Conselho Nacional de desenvolvimento Científico e Tecnológico (CNPq) for student’s scholarship.
Author contributions
FLPC and SJS carried out the all research, data acquisition and wrote the manuscript. MIFG helped qPCR data acquisition, LSL contributed to MDA and hydrogen peroxide experiment; EG-F revised the manuscript, HHC designed the research concept, data acquisition and wrote the manuscript. All authors discussed the results and contributed to the final manuscript.
Funding
This work was supported by Fundação Cearense de Apoio ao Desenvolvimento Científico e Tecnológico. (FUNCAP)- DEP-0164-00152.01.00/19, and Conselho Nacional de desenvolvimento Científico e Tecnológico (CNPq), processo 408118/2021-0.
Data availability
Data will be made available on request.
Declarations
Conflicts of interest
The authors declare that there is no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Afify AEMMR, El-Beltagi HS, El-Salam SM, Omran AA. Protein solubility, digestibility and fractionation after germination of sorghum varieties. PLoS ONE. 2012;7:1–6. doi: 10.1371/journal.pone.0031154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afrin T, Diwan D, Sahawneh K, Pajerowska-Mukhtar K. Multilevel regulation of endoplasmic reticulum stress responses in plants: where old roads and new paths meet. J Exp Bot. 2020;71:1659–1667. doi: 10.1093/jxb/erz487. [DOI] [PubMed] [Google Scholar]
- Angelos E, Brandizzi F. NADPH oxidase activity is required for ER stress survival in plants. Plant J. 2018;96:1106–1120. doi: 10.1111/tpj.14091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydemir BÇ, Ergül A. Transcriptomic analysis of endoplasmic reticulum stress in roots of grapevine rootstock. Plant Biotechnol Rep. 2021;15:683–706. doi: 10.1007/s11816-021-00707-z. [DOI] [Google Scholar]
- Bai Y, Sunarti S, Kissoudis C, et al. The role of tomato WRKY genes in plant responses to combined abiotic and biotic stresses. Front Plant Sci. 2018;9:1–7. doi: 10.3389/fpls.2018.00801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista VCV, Pereira IMC, de Paula-Marinho S, O, , et al. Salicylic acid modulates primary and volatile metabolites to alleviate salt stress-induced photosynthesis impairment on medicinal plant Egletes viscosa. Environ Exp Bot. 2019;167:103870. doi: 10.1016/j.envexpbot.2019.103870. [DOI] [Google Scholar]
- Beaugelin I, Chevalier A, D’Alessandro S, et al. Endoplasmic reticulum-mediated unfolded protein response is an integral part of singlet oxygen signalling in plants. Plant J. 2020 doi: 10.1111/tpj.14700. [DOI] [PubMed] [Google Scholar]
- ÇakırAydemir B, Yüksel Özmen C, Kibar U, et al. Salt stress induces endoplasmic reticulum stress-responsive genes in a grapevine rootstock. PLoS One. 2020;15:e0236424. doi: 10.1371/journal.pone.0236424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Carvalho AP, Behling Neto A, de Moraes KNC, et al. Chemical composition, kinetics of degradation, and digestibility of forage of different purpose sorghum cultivars. Semin Ciênc Agrár. 2020;41:607–620. doi: 10.5433/1679-0359.2020v41n2p607. [DOI] [Google Scholar]
- Carvalho HH, Silva PA, Mendes GC, et al. The endoplasmic reticulum binding protein bip displays dual function in modulating cell death events. Plant Physiol. 2014;164:654–670. doi: 10.1104/pp.113.231928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty R, Macoy DM, Lee SY, et al. Tunicamycin-induced endoplasmic reticulum stress suppresses plant immunity. Appl Biol Chem. 2017;60:623–630. doi: 10.1007/s13765-017-0319-3. [DOI] [Google Scholar]
- Chi YH, Koo SS, Oh HT, et al. The physiological functions of universal stress proteins and their molecular mechanism to protect plants from environmental stresses. Front Plant Sci. 2019;10:1–13. doi: 10.3389/fpls.2019.00750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y, Kanehara K. Endoplasmic reticulum stress response in arabidopsis roots. Front Plant Sci. 2017;8:1–10. doi: 10.3389/fpls.2017.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CONAB (2022) Companhia Nacional de Abastecimento. http://www.conab.gov.br. Accessed 17 May 2022
- Curtis TY, Bo V, Tucker A, Halford NG. Construction of a network describing asparagine metabolism in plants and its application to the identification of genes affecting asparagine metabolism in wheat under drought and nutritional stress. Food Energy Secur. 2018;7:1–16. doi: 10.1002/fes3.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darko E, Végh B, Khalil R, et al. Metabolic responses of wheat seedlings to osmotic stress induced by various osmolytes under iso-osmotic conditions. PLoS ONE. 2019;14:1–19. doi: 10.1371/journal.pone.0226151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depaepe T, Hendrix S, Janse van Rensburg HC, et al. At the crossroads of survival and death: the reactive oxygen species–ethylene–sugar triad and the unfolded protein response. Trends Plant Sci. 2021;26:338–351. doi: 10.1016/j.tplants.2020.12.007. [DOI] [PubMed] [Google Scholar]
- Eisvand F, Tajbakhsh A, Seidel V, et al. Quercetin and its role in modulating endoplasmic reticulum stress: A review. Phyther Res. 2022;36:73–84. doi: 10.1002/ptr.7283. [DOI] [PubMed] [Google Scholar]
- Eveland AL, Jackson DP. Sugars, signalling, and plant development. J Exp Bot. 2012;63:3367–3377. doi: 10.1093/jxb/err379. [DOI] [PubMed] [Google Scholar]
- FAO (2019) Statistics - Food and Agriculture Organization of the United Nations. In: Statistics -. http://www.fao.org/statistics/en. Accessed 12 Apr 2022
- Farooq MA, Niazi AK, Akhtar J, et al. Acquiring control: the evolution of ROS-Induced oxidative stress and redox signaling pathways in plant stress responses. Plant Physiol Biochem. 2019;141:353–369. doi: 10.1016/j.plaphy.2019.04.039. [DOI] [PubMed] [Google Scholar]
- Fedoroff N V., Battisti DS, Beachy RN et al (2010) Radically rethinking agriculture for the 21st century. Science (80- ) 327:833–834. 10.1126/science.1186834 [DOI] [PMC free article] [PubMed]
- Feldeverd E, Porter BW, Yuen CYL et al (2020) The arabidopsis protein disulfide isomerase subfamily M isoform, PDI9, Localizes to the endoplasmic reticulum and influences pollen viability and proper formation of the pollen exine during heat stress. Front Plant Sci 11:610052. 10.3389/fpls.2020.610052 [DOI] [PMC free article] [PubMed]
- Fernie AR, Carrari F, Sweetlove LJ. Respiratory metabolism: glycolysis, the TCA cycle and mitochondrial electron transport. Curr Opin Plant Biol. 2004;7:254–261. doi: 10.1016/j.pbi.2004.03.007. [DOI] [PubMed] [Google Scholar]
- HamanyDjande CY, Pretorius C, Tugizimana F, et al. Metabolomics: a tool for cultivar phenotyping and investigation of grain crops. Agronomy. 2020;10:1–30. doi: 10.3390/agronomy10060831. [DOI] [Google Scholar]
- Heath RL, Packer L. Photoperoxidation in isolated chloroplasts. Arch Biochem Biophys. 1968 doi: 10.1016/0003-9861(68)90654-1. [DOI] [PubMed] [Google Scholar]
- Hirata R, Mishiba KI, Koizumi N, Iwata Y. Deficiency in the double-stranded RNA binding protein HYPONASTIC LEAVES1 increases sensitivity to the endoplasmic reticulum stress inducer tunicamycin in Arabidopsis. BMC Res Notes. 2019;12:1–6. doi: 10.1186/s13104-019-4623-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong ZH, Qing T, Schubert D, et al. BLISTER-regulated vegetative growth is dependent on the protein kinase domain of ER stress modulator IRE1A in Arabidopsis thaliana. PLoS Genet. 2019;15:1–19. doi: 10.1371/journal.pgen.1008563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim HA, Abdellatif YMR. Effect of maltose and trehalose on growth, yield and some biochemical components of wheat plant under water stress. Ann Agric Sci. 2016;61:267–274. doi: 10.1016/j.aoas.2016.05.002. [DOI] [Google Scholar]
- Igamberdiev AU, Eprintsev AT. Organic acids: the pools of fixed carbon involved in redox regulation and energy balance in higher plants. Front Plant Sci. 2016;7:1–15. doi: 10.3389/fpls.2016.01042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irsigler AST, Costa MDL, Zhang P, et al. Expression profiling on soybean leaves reveals integration of ER- and osmotic-stress pathways. BMC Genomics. 2007;8:1–15. doi: 10.1186/1471-2164-8-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine KJ, Sommer ED, Saleska SR, et al. Gas phase measurements of pyruvic acid and its volatile metabolites. Environ Sci Technol. 2010;44:2454–2460. doi: 10.1021/es903544p. [DOI] [PubMed] [Google Scholar]
- Kamauchi S, Nakatani H, Nakano C, Urade R. Gene expression in response to endoplasmic reticulum stress in Arabidopsis thaliana. FEBS J. 2005;272:3461–3476. doi: 10.1111/j.1742-4658.2005.04770.x. [DOI] [PubMed] [Google Scholar]
- Kanehara K, Cho Y, Yu C-Y. A lipid viewpoint on the plant endoplasmic reticulum stress response. J Exp Bot. 2022;73:2835–2847. doi: 10.1093/jxb/erac063. [DOI] [PubMed] [Google Scholar]
- Kawakatsu T, Wang S, Wakasa Y, Takaiwa F. Increased lysine content in rice grains by over-accumulation of BiP in the endosperm. Biosci Biotechnol Biochem. 2010;74:2529–2531. doi: 10.1271/bbb.100619. [DOI] [PubMed] [Google Scholar]
- Khan N, Bano A, Babar MA. Metabolic and physiological changes induced by plant growth regulators and plant growth promoting rhizobacteria and their impact on drought tolerance in Cicer arietinum L. PLoS ONE. 2019;14:1–21. doi: 10.1371/journal.pone.0213040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan N, Bano A, Rahman MA, et al. UPLC-HRMS-based untargeted metabolic profiling reveals changes in chickpea (Cicer arietinum) metabolome following long-term drought stress. Plant Cell Environ. 2019;42:115–132. doi: 10.1111/pce.13195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kötting O, Kossmann J, Zeeman SC, Lloyd JR. Regulation of starch metabolism: the age of enlightenment? Curr Opin Plant Biol. 2010;13:320–328. doi: 10.1016/j.pbi.2010.01.003. [DOI] [PubMed] [Google Scholar]
- Lai YS, Renna L, Yarema J, et al. Salicylic acid-independent role of NPR1 is required for protection from proteotoxic stress in the plant endoplasmic reticulum. Proc Natl Acad Sci U S A. 2018;115:E5203–E5212. doi: 10.1073/pnas.1802254115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lastdrager J, Hanson J, Smeekens S. Sugar signals and the control of plant growth and development. J Exp Bot. 2014;65:799–807. doi: 10.1093/jxb/ert474. [DOI] [PubMed] [Google Scholar]
- Lea PJ, Sodek L, Parry MAJ, et al. Asparagine in plants. Ann Appl Biol. 2007;150:1–26. doi: 10.1111/j.1744-7348.2006.00104.x. [DOI] [Google Scholar]
- Li Z, Yu J, Peng Y, Huang B. Metabolic pathways regulated by abscisic acid, salicylic acid and γ-aminobutyric acid in association with improved drought tolerance in creeping bentgrass (Agrostis stolonifera) Physiol Plant. 2017;159:42–58. doi: 10.1111/ppl.12483. [DOI] [PubMed] [Google Scholar]
- Lima KRP, Cavalcante FLP, de Paula-Marinho S, O, , et al. Metabolomic profiles exhibit the influence of endoplasmic reticulum stress on sorghum seedling growth over time. Plant Physiol Biochem. 2022;170:192–205. doi: 10.1016/j.plaphy.2021.11.041. [DOI] [PubMed] [Google Scholar]
- Lisec J, Schauer N, Kopka J, et al. Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc. 2006;1:387–396. doi: 10.1038/nprot.2006.59. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Mahmud JA, Hasanuzzaman M, Nahar K, et al. Maleic acid assisted improvement of metal chelation and antioxidant metabolism confers chromium tolerance in Brassica juncea L. Ecotoxicol Environ Saf. 2017;144:216–226. doi: 10.1016/j.ecoenv.2017.06.010. [DOI] [PubMed] [Google Scholar]
- Manghwar H, Li J. Endoplasmic reticulum stress and unfolded protein response signaling in plants. Int J Mol Sci. 2022;23:828. doi: 10.3390/ijms23020828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick RF, Truong SK, Sreedasyam A, et al. The Sorghum bicolor reference genome: improved assembly, gene annotations, a transcriptome atlas, and signatures of genome organization. Plant J. 2018;93:338–354. doi: 10.1111/tpj.13781. [DOI] [PubMed] [Google Scholar]
- Melo BP, Fraga OT, Silva JCF, et al. Revisiting the soybean GmNAC superfamily. Front Plant Sci. 2018;9:1–22. doi: 10.3389/fpls.2018.01864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawkar GM, Kang CH, Maibam P, et al. HY5, a positive regulator of light signaling, negatively controls the unfolded protein response in Arabidopsis. Proc Natl Acad Sci. 2017;114:2084–2089. doi: 10.1073/pnas.1609844114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen VC, Nakamura Y, Kanehara K. Membrane lipid polyunsaturation mediated by FATTY ACID DESATURASE 2 (FAD2) is involved in endoplasmic reticulum stress tolerance in Arabidopsis thaliana. Plant J. 2019;99:478–493. doi: 10.1111/tpj.14338. [DOI] [PubMed] [Google Scholar]
- Noh SJ, Kwon CS, Oh DH, et al. Expression of an evolutionarily distinct novel BiP gene during the unfolded protein response in Arabidopsis thaliana. Gene. 2003;311:81–91. doi: 10.1016/S0378-1119(03)00559-6. [DOI] [PubMed] [Google Scholar]
- Nxele X, Klein A, Ndimba BK. Drought and salinity stress alters ROS accumulation, water retention, and osmolyte content in sorghum plants. South African J Bot. 2017;108:261–266. doi: 10.1016/j.sajb.2016.11.003. [DOI] [Google Scholar]
- Okada T, Yoshida H, Akazawa R, et al. Distinct roles of activating transcription factor 6 (ATF6) and double-stranded RNA-activated protein kinase-like endoplasmic reticulum kinase (PERK) in transcription during the mammalian unfolded protein response. Biochem J. 2002;366:585–594. doi: 10.1042/bj20020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira DF, de Sousa LL, Gomes-Filho E. Metabolic changes associated with differential salt tolerance in sorghum genotypes. Planta. 2020;252:34. doi: 10.1007/s00425-020-03437-8. [DOI] [PubMed] [Google Scholar]
- Ozgur R, Uzilday B, Sekmen AH, Turkan I. The effects of induced production of reactive oxygen species in organelles on endoplasmic reticulum stress and on the unfolded protein response in arabidopsis. Ann Bot. 2015;116:541–553. doi: 10.1093/aob/mcv072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchal P, Miller AJ, Giri J. Organic acids: versatile stress-response roles in plants. J Exp Bot. 2021 doi: 10.1093/jxb/erab019. [DOI] [PubMed] [Google Scholar]
- Park CJ, Park JM (2019) Endoplasmic reticulum plays a critical role in integrating signals generated by both biotic and abiotic stress in plants. Front Plant Sci 10. 10.3389/fpls.2019.00399 [DOI] [PMC free article] [PubMed]
- Pastor-Cantizano N, Ko DK, Angelos E, et al. Functional diversification of ER stress responses in Arabidopsis. Trends Biochem Sci. 2020;45:123–136. doi: 10.1016/j.tibs.2019.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan X, Qian Q, Ye Z, et al. Metabolic analysis of two contrasting wild barley genotypes grown hydroponically reveals adaptive strategies in response to low nitrogen stress. J Plant Physiol. 2016;206:59–67. doi: 10.1016/j.jplph.2016.07.020. [DOI] [PubMed] [Google Scholar]
- de Queiroz CS, Pereira IMC, Lima KRP, et al. Combined NaCl and DTT diminish harmful ER-stress effects in the sorghum seedlings CSF 20 variety. Plant Physiol Biochem. 2020;147:223–234. doi: 10.1016/j.plaphy.2019.12.013. [DOI] [PubMed] [Google Scholar]
- Rasband W. ImageJ. U. S. Natl. Bethesda: Institutes Heal; 2016. [Google Scholar]
- Raza A. Metabolomics: a systems biology approach for enhancing heat stress tolerance in plants. Plant Cell Rep. 2022;41:741–763. doi: 10.1007/s00299-020-02635-8. [DOI] [PubMed] [Google Scholar]
- Razzaq A, Sadia B, Raza A, et al. Metabolomics: a way forward for crop improvement. Metabolites. 2019;9:303. doi: 10.3390/metabo9120303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy PS (2019) Breeding for abiotic stress resistance in sorghum. In: Breeding sorghum for diverse end uses. Elsevier, pp 325–340. 10.1016/B978-0-08-101879-8.00020-6
- Ruberti C, Lai YS, Brandizzi F. Recovery from temporary endoplasmic reticulum stress in plants relies on the tissue-specific and largely independent roles of bZIP28 and bZIP60, as well as an antagonizing function of BAX-Inhibitor 1 upon the pro-adaptive signaling mediated by bZIP28. Plant J. 2018;93:155–165. doi: 10.1111/tpj.13768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlemper TR, van Veen JA, Kuramae EE. Co-variation of bacterial and fungal communities in different sorghum cultivars and growth stages is soil dependent. Microb Ecol. 2018;76:205–214. doi: 10.1007/s00248-017-1108-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergiev I, Alexieva V, Karanov E. Effect of spermine, atrazine and combination between them on some endogenous protective systems and stress markers in plants. Proc Bulg Acad Sci. 1997;51:121–124. [Google Scholar]
- Seyoum A, Gebreyohannes A, Nega A et al (2019) Performance evaluation of sorghum (Sorghum bicolor (L.) Moench) genotypes for grain yield and yield related traits in drought prone areas of ethiopia. Adv Crop Sci Tehnol 7. 10.4172/2329-8863.1000439
- Shelden MC, Dias DA, Jayasinghe NS, et al. Root spatial metabolite profiling of two genotypes of barley (Hordeum vulgare L.) reveals differences in response to short-term salt stress. J Exp Bot. 2016;67:3731–3745. doi: 10.1093/jxb/erw059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva MLDS, de Sousa HG, Silva MLDS, et al. Growth and photosynthetic parameters of saccharine sorghum plants subjected to salinity. Acta Sci Agron. 2019;41:42607. doi: 10.4025/actasciagron.v41i1.42607. [DOI] [Google Scholar]
- Simoni EB, Oliveira CC, Fraga OT et al (2022) Cell death signaling from endoplasmic reticulum stress: plant-specific and conserved features. Front Plant Sci 13. 10.3389/fpls.2022.835738 [DOI] [PMC free article] [PubMed]
- Stefoska-Needham A, Tapsell L. Considerations for progressing a mainstream position for sorghum, a potentially sustainable cereal crop, for food product innovation pipelines. Trends Food Sci Technol. 2020;97:249–253. doi: 10.1016/j.tifs.2020.01.012. [DOI] [Google Scholar]
- Taleon V, Dykes L, Rooney WL, Rooney LW. Effect of genotype and environment on flavonoid concentration and profile of black sorghum grains. J Cereal Sci. 2012;56:470–475. doi: 10.1016/j.jcs.2012.05.001. [DOI] [Google Scholar]
- Thalmann M, Pazmino D, Seung D, et al. Regulation of leaf starch degradation by abscisic acid is important for osmotic stress tolerance in plants. Plant Cell. 2016;28:1860–1878. doi: 10.1105/tpc.16.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tugizimana F, Mhlongo M, Piater L, Dubery I. Metabolomics in plant priming research: the way forward? Int J Mol Sci. 2018;19:1759. doi: 10.3390/ijms19061759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ul SH, Khan A, Ali M, et al. Heat shock proteins: dynamic biomolecules to counter plant biotic and abiotic stresses. Int J Mol Sci. 2019;20:5321. doi: 10.3390/ijms20215321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzilday B, Ozgur R, Sekmen AH, Turkan I. Endoplasmic reticulum stress regulates glutathione metabolism and activities of glutathione related enzymes in Arabidopsis. Funct Plant Biol. 2018;45:284–296. doi: 10.1071/FP17151. [DOI] [PubMed] [Google Scholar]
- Velmurugan B, Narra M, Rudakiya DM, Madamwar D (2020) Sweet sorghum: a potential resource for bioenergy production. In: Refining biomass residues for sustainable energy and bioproducts. Elsevier, pp 215–242. 10.1016/B978-0-12-818996-2.00010-7
- Vieira MR, De LCF, Cândido MJD, et al. Produtividade e qualidade da forragem de sorgo irrigado com águas salinas. Rev Bras Eng Agrícola e Ambient. 2005;9:42–46. doi: 10.1590/1807-1929/agriambi.v9nsupp42-46. [DOI] [Google Scholar]
- Wang W, Li X, Zhu M, et al. Arabidopsis GAAP1 to GAAP3 play redundant role in cell death inhibition by suppressing the upregulation of salicylic acid pathway under endoplasmic reticulum stress. Front Plant Sci. 2019;10:1–13. doi: 10.3389/fpls.2019.01032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wind J, Smeekens S, Hanson J. Sucrose: Metabolite and signaling molecule. Phytochemistry. 2010;71:1610–1614. doi: 10.1016/j.phytochem.2010.07.007. [DOI] [PubMed] [Google Scholar]
- Wu D, Cai S, Chen M et al (2013) Tissue metabolic responses to salt stress in wild and cultivated barley. PLoS One 8. 10.1371/journal.pone.0055431 [DOI] [PMC free article] [PubMed]
- Xiang Y, Sun X, Gao S, et al. Deletion of an endoplasmic reticulum stress response element in a ZmPP2C-A gene facilitates drought tolerance of maize seedlings. Mol Plant. 2017;10:456–469. doi: 10.1016/j.molp.2016.10.003. [DOI] [PubMed] [Google Scholar]
- Yang ZT, Wang MJ, Sun L et al (2014) The membrane-associated transcription factor NAC089 controls ER-stress-induced programmed Cell Death in Plants. PLoS Genet 10. 10.1371/journal.pgen.1004243 [DOI] [PMC free article] [PubMed]
- Yu X, Wang T, Zhu M, et al. Transcriptome and physiological analyses for revealing genes involved in wheat response to endoplasmic reticulum stress. BMC Plant Biol. 2019;19:1–22. doi: 10.1186/s12870-019-1798-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeshan HMA, Lee GH, Kim HR, Chae HJ. Endoplasmic reticulum stress and associated ROS. Int J Mol Sci. 2016;17:1–20. doi: 10.3390/ijms17030327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemanová V, Pavlík M, Pavlíková D, Tlustoš P. The significance of methionine, histidine and tryptophan in plant responses and adaptation to cadmium stress. Plant, Soil Environ. 2014;60:426–432. doi: 10.17221/544/2014-pse. [DOI] [Google Scholar]
- Zhang L, Xin Z, Yu X, et al. Osmotic stress induced cell death in wheat is alleviated by tauroursodeoxycholic acid and involves endoplasmic reticulum stress-related gene expression. Front Plant Sci. 2017;8:1–14. doi: 10.3389/fpls.2017.00667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong M, Yuan Y, Shu S, et al. Effects of exogenous putrescine on glycolysis and Krebs cycle metabolism in cucumber leaves subjected to salt stress. Plant Growth Regul. 2016;79:319–330. doi: 10.1007/s10725-015-0136-9. [DOI] [Google Scholar]
- Zhu JK. Abiotic stress signaling and responses in plants. Cell. 2016;167:313–324. doi: 10.1016/j.cell.2016.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. S1 Sorghum plants CSF 18 e CSF20 grown in the absence (A) e (C) or presence of ER stress inducer TM (B) e (D) in the TM 2,5 µg mL-1 concentration. (TIF 4973 KB)
Supplementary Fig. S2 Relative gene expression of SbbZIP17. Sorghum bicolor seedlings were submitted to different concentrations of TM: 0 µg mL-1 (Control), 0.5 µg mL-1, 1.5 µg mL-1 and 2.5 µg mL-1. The expression profiles were determined in the shoot (A) and in the roots (B). Gene expression was normalized using SbUBC18 as a reference gene (Queiroz et al. 2020; Lima et al. 2022). Lowercase letters compare the two varieties of sorghum. Uppercase letters compare the treatments within each variety of sorghum. All four treatments were compared by Tukey test (p<0.05) in each TM concentration. The columns indicate the means of three biological replicates and the bars indicate the standard deviation (±). (TIF 912 KB)
Supplementary Fig. S3 The loading plot for shoot (A, B) and root (C, D) indicate the positive and negative contributions of each metabolite in two varieties of sorghum: CSF 18 on the left and CSF20 on the right. The top five most positive metabolites for each component are highlighted. (TIF 373 KB)
Supplementary Fig. S4 Orthogonal Projections to Latent Structures Discriminant Analysis (OPLS-DA) in shoots (a, b) and roots (c, d) of two sorghum varieties grown under 0.5 µg mL-1 TM. (TIF 2354 KB)
Supplementary Fig. S5 Orthogonal Projections to Latent Structures Discriminant Analysis (OPLS-DA) in shoots (a, b) and roots (c, d) of two sorghum varieties grown under 1.5 µg mL-1 TM. (TIF 2452 KB)
Supplementary Table S2 List of detected metabolites in shoots and roots of sorghum with their classification in compound types, their retention time, respectively, and with their kyoto encyclopedia of genes and genomes identifier number (KEGG ID). (XLS 12 KB)
Supplementary Table S3 Loading score of primary metabolites found 0 (control), 0.5 µg mL-1, 1.5 µg mL-1, and 2.5 µg mL-1 tunicamycin (TM) treated seedlings ranked by principal component analyses values. (CSV 10 KB)
Data Availability Statement
Data will be made available on request.