Abstract
Human papillomavirus (HPV) is the major causative agent for cervical and many head and neck cancers (HNCs). HPVs randomly acquire single nucleotide polymorphisms (SNPs) that may become established via positive selection. Within an HPV type, viral isolates differing by <2% in the L1 region are termed “variants” and classified based on combinations of SNPs. Studies in cervical cancer demonstrate clear differences between HPV16 intratypic variants in terms of persistence of infection, tumor histology, cancer risk, and death. Much less is known about the frequency of HPV16 variants in HNC, and their effects on clinical outcomes. We combined HPV16 positive (HPV16+) HNC samples from a local Southwestern Ontario, Canada cohort with those from the Cancer Genome Atlas to create a larger North American cohort of 149 cases with clinical data and determined the distribution of intratypic variants and their impact on clinical outcomes. Most isolates were lineage A, sublineage A1, or A2, with roughly half exhibiting the T350G polymorphism in E6. Univariable analysis identified significant differences between 350T and 350G intratypic variants in clinical T, N, and O staging, as well as disease-free survival. Multivariable analysis failed to identify any clinical factor as a statistically significant covariate for disease-free survival differences between 350T and 350G. Significant differences in several measures of B-cell mediated immune response were also observed between 350T and 350G intratypic variants. We suggest that HPV genetic variation may be associated with HNC clinical characteristics and may have prognostic value.
Keywords: human papillomavirus, intratypic variants, E6, TCGA, head and neck squamous cell carcinoma
1. Introduction
Human papillomaviruses (HPVs) are non-enveloped viruses with a small double-stranded DNA (dsDNA) genome of about 8 kilobases in size [1]. There are over 400 HPV types that have been identified to date, with all identified types having exclusive tropism for either cutaneous or mucosal epithelia. The mucosa-associated HPVs are dichotomized as high-risk (HR) or low-risk (LR) based on their propensity to induce carcinogenesis following infection [2,3,4]. In 2018, HR HPVs were estimated to be responsible for 4.1% of the global burden of cancer, causing virtually all cases of cervical and anal cancers, a large proportion of vaginal, vulvar, and penile cancers, and a significant subset of head and neck cancers (HNCs) [5,6]. HR HPV type 16 (HPV16) is the most frequently detected HR HPV at the population level, and its high association with carcinogenesis, in comparison to other HPV types, makes HPV16 the most clinically relevant [7,8,9].
HNCs are a heterogeneous group of malignancies in the head and neck region that includes the oral cavity, oropharynx, hypopharynx, and larynx [6,10,11]. It is the 7th most diagnosed cancer worldwide, with approximately 890,000 new cases and 453,000 deaths in 2018 [5]. HR HPVs are responsible for approximately 25% of all HNCs, with 83% of the HPV-positive (HPV+) subtype being caused by HR HPV16 [10,12]. In comparison to the HPV-negative (HPV−) subtype—caused by excessive drinking and smoking—HPV+ HNCs are a distinct epidemiological, molecular, and clinical entity with patients exhibiting strikingly better responses to treatment and clinical outcomes [10,11,13,14,15,16,17]. Interestingly, HNCs caused by other HR HPVs other than HPV16 have been associated with different patient outcomes [18,19,20].
During infection, HPVs usurp their host cell’s DNA replication machinery to replicate their small dsDNA genome. This strategy of viral DNA replication exploits the proofreading capabilities of the host cell’s DNA polymerase, leading to a very low rate of nucleotide polymorphisms [21,22,23]. However, nucleotide polymorphisms can still arise through random mutations and can become established in a population over time or arise because of genome editing by APOBECs as part of the host’s innate immune response [24]. This genetic drift has been observed through the identification of HPV16 intratypic variants, signifying their coevolution with humans [25,26]. Notably, the E6 oncogene from HR HPV16 is a hotspot for naturally occurring polymorphisms, whereas this seems to be rare for the E7 oncogene [27,28,29]. The E6 oncoprotein interacts with and subsequently inhibits the activity of p53—a tumor suppressor [30]. This thwarts the triggering of p53-mediated cell cycle arrest or apoptosis upon the unsanctioned initiation of the cell cycle induced by E7 [31]. Together, these activities deregulate the cell cycle, creating an environment conducive to viral replication, which can ultimately lead to oncogenic transformation [3,32].
HPV16 is a member of the Papillomaviridae family of viruses, the Alphapapillomavirus genus, and Alphapapillomavirus 9 species [33]. Different genera of HPVs are defined as having sequence differences of more than 40%, whereas sequence differences between 30 and 40% define HPV species [33]. Different types of HPVs have sequence differences between 10 and 30%. HPV intratypic variants have smaller genetic differences within the viral genome within a given type. Sequence differences between 1 and 10% define the main HPV16 variant lineages (A, B, C, D) and differences between 0.5 and 1% define the sublineages (A1–A4, B1–B2, D1–D3) [30]. HPV16 intratypic variants are classified into 4 major phylogenetic lineages: A (sublineages A1–A4; A1 is the reference genome/sequence), B (sublineages B1–B2), C, and D (sublineages D1–D3) [34]. Intratypic classification is based on different combinations of single nucleotide polymorphisms (SNPs) in the non-coding regulatory long control region (LCR) and/or the E6 oncogene [34,35]. Historically, intratypic variant nomenclature was derived from the geographical origin of the populations in which they were identified [25,36]. The variants were referred to as European (A1–A3), Asian (A4), African-1 (B1–B2), African-2 (C), North American (D1), and Asian-American (D2–D3) [37,38].
In addition, there are non-lineage-specific HPV variants that have minor genetic variations that do not fit a phylogenetic tree. The prime example is HPV16 T350G which has a non-synonymous nucleotide change at position 350 of the E6 oncogene from thymine (T) to guanine (G). This SNP at position 350 changes the original amino acid residue at position 83, within the E6 oncoprotein, from leucine (L) to valine (V) [33]. Infections with the non-lineage-specific HPV16 T350G have been associated with elevated rates of persistent infection and progression to high-grade cervical lesions [39]. Furthermore, it has been suggested that HPV16 T350G carries a higher risk for the development of cervical cancer and may be more oncogenic than the A1 reference genome [40].
Naturally occurring polymorphisms within the E6 oncogene leading to amino acid changes could lead to alterations in the steady state levels or activity of its product, thereby having a major influence on carcinogenesis or prognosis. Alternatively, these amino acid changes could influence the antigenicity of viral-derived peptides, as demonstrated for the viral capsid proteins [41]. Perhaps the best-studied polymorphism is a T to G transversion at bp 350, which alters a leucine to valine at position 83 in the E6 oncoprotein [40,42,43,44]. Since different HPV types exhibit distinctive oncogenic potential, it is, therefore, reasonable to hypothesize that HPV16 intratypic variants also display a difference in their oncogenicity. In fact, there is compelling data that HPV16 intratypic variants influence viral persistence, progression to premalignant lesions, development of malignant lesions, and histological type of lesion in the context of cervical cancers [45,46,47,48]. Indeed, the same HPV16 intratypic variant-specific effects observed in HPV16+ cervical cancers could be true for HPV+ HNCs; however, an extensive literature search shows that this is a severely understudied area of research, particularly in North America. Due to the rarity of some of the intratypic variants in the general North American population, we combined HPV16+ HNC samples from a local Southwestern Ontario, Canada cohort (SWO) with those from the Cancer Genome Atlas (TCGA) HNC cohort to create a large North American cohort (CANUSA)—the increase in sample size contributes to an increase in the statistical power of this study. Then, with the large CANUSA cohort of HPV16+ HNC samples, we determined the distribution of intratypic variants, as well as their impact on clinical variables, tumor immune response, and patient survival outcomes.
2. Materials and Methods
2.1. Southwestern Ontario (SWO) Cohort
Approval for the study was obtained from Western University’s Ontario Research Ethics Board (LHSC HSREB #7182—9 September 2010). A retrospective search of the London Regional Cancer Program (LRCP) database was completed to identify patients diagnosed with oropharyngeal squamous cell carcinomas (OSCC) from 2003 to 2009. The following was required for patient eligibility: (1) histological confirmation of squamous cell carcinoma, (2) no prior history of head and neck cancer, and (3) the availability of a pre-treatment primary site biopsy specimen for analysis. Patient data were extracted from a retrospective chart review, which included age at diagnosis, use of tobacco and alcohol, AJCC TNM stage, treatment regimen, and post-treatment follow-up information.
After completion of cancer therapy, patients were followed at 3–6 months intervals by either a radiation oncologist or head and neck surgeon. Treatment response was evaluated by physical exam as well as computed tomography imaging as needed. All the patients were HIV-negative.
2.2. DNA Extraction
2.2.1. Formalin-Fixed Paraffin-Embedded (FFPE) Samples
Deparaffinization and DNA extraction were performed as previously described [49]. In brief, the FFPE blocks from each patient’s primary site were sectioned and mounted on slides. The slides were then deparaffinized with washes in xylene, followed by a 1:1 xylene:ethanol mix, then ethanol twice, followed by single washes in ethanol. Lastly, the slides were washed in water for 5 min. The deparaffinized tissue was scraped into a 1.5 mL microcentrifuge tube that contained 50–100 μL of TE and proteinase K (2 mg/mL; Qiagen, Hilden, Germany) and then incubated overnight at 65 °C. Following proteinase K treatment, the samples were heated at 95 °C for 10 min and allowed to cool to room temperature.
2.2.2. Fresh-Frozen (FF) Samples
DNA extraction from the FF samples was performed using the AllPrep DNA/RNA kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. In brief, the patient tumor samples (20–30 mg) were placed in Buffer RLT Plus (Qiagen, Hilden, Germany) and homogenized with a fine-motorized tissue homogenizer (VWR 200, VWR, Radnor, Pennsylvania). The disrupted tissue was centrifuged through a QiaShredder (Qiagen, Hilden, Germany) at full speed for 3 min. The filtrate was then used to extract RNA and DNA.
2.3. HPV Typing, PCR, Sequencing, and Variant Identification
The HPV status and HPV type were determined with HPV type-specific primers as described previously [19,49,50]. The DNA extracted from 57 FFPE or 49 FF HPV16+ patient samples was used as a template to amplify the full-length HPV16 E6 gene by PCR using 3 primer pairs as previously described (Table 1) [51]. Since formalin fixation is known to cause DNA fragmentation, we used primer pairs that generated overlapping amplicons of less than 250 base pairs [27,51]. A phusion high-fidelity PCR kit (New England Biolabs, Ipswich, MA, USA) was utilized with the following program: 98 °C for 30 s for the initial denaturation step, followed by 30 cycles that consisted of 98 °C for 5 s, 60 °C for 10 s, 72 °C for 15 s, and a final extension at 72 °C for 5 min.
Table 1.
Primers used for the amplification of HPV16 E6.
| Primers | Sequences (5′–3′) | Position (nt) | Product Size (bp) |
|---|---|---|---|
| E6-1 Forward | TTGAACCGAAACCGGTTAGT | 46–65 | 211 |
| E6-1 Reverse | GCATAAATCCCGAAAAGCAA | 237–256 | |
| E6-2 Forward | GCAACAGTTACTGCGACGTG | 204–224 | 235 |
| E6-2 Reverse | GGACACAGTGGCTTTTGACA | 419–438 | |
| E6-3 Forward | CAGCAATACAACAAACCGTTG | 371–391 | 220 |
| E6-3 Reverse | TCATGCAATGTAGGTGTATCTCC | 568–590 |
nt = nucleotides; bp = base pairs.
The amplicons were analyzed by DNA gel electrophoresis, purified using a commercial PCR clean-up kit (GeneJET PCR Purification Kit, Thermo Scientific, Waltham, MA, USA), and Sanger sequenced by Bio Basic’s DNA sequencing service (Bio Basic, Markham, Ontario, Canada). Sequencing was performed separately with both forward and reverse primers. Only data with no discrepancies were used for analysis. Sequences were then aligned (Snapgene, San Diego, CA, USA; MUSCLE algorithm) to the reference HPV16 sequence (A1), and differences in the E6 gene (nt: 83–559) were recorded. Samples were classified into phylogenetic branches using diagnostic SNPs in the E6 gene as previously described (Table 2) [34,35]. It is important to note that variant lineage classification restricted to the E6 ORF is highly correlated with those based on the HPV16 long-control region (LCR) [35].
Table 2.
HPV16 lineage reference sequences and diagnostic SNPs within E6.
| Lineage | Sublineage | Variant Genome ID |
GenBank Accession Numbers | E6 Nucleotide Position | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 0 9 |
1 3 1 |
1 3 2 |
1 4 3 |
1 4 5 |
1 7 8 |
2 7 6 |
2 8 6 |
2 8 9 |
3 3 5 |
3 5 0 |
4 0 3 |
4 3 3 |
5 3 2 |
||||
| A | A1 | Ref | K02718 | T | A | G | C | G | T | A | T | A | C | T | A | G | A |
| A2 | W0122 | AF536179 | — | —/G | — | — | — | — | — | — | — | — | —/G | — | — | — | |
| A3 | AS411 | HQ644236 | — | — | — | — | — | — | G | — | — | — | — | — | — | — | |
| A4 | W0724 | AF534061 | — | — | — | — | — | G/A | — | — | — | — | — | — | — | — | |
| B | B1 | W0236 | AF536180 | — | — | C | G | T | — | — | A | G | T | — | — | — | — |
| B2 | Z109 | HQ644298 | — | G | — | G | T | — | — | A | G | T | — | — | — | — | |
| C | R460 | AF472509 | C | — | T | G | T | — | — | A | G | T | — | G | — | — | |
| D | D1 | QV00512 | HQ644257 | — | — | — | — | T | — | — | A | G | T | G | — | — | — |
| D2 | QV15321 | AY686579 | — | — | — | — | T | — | — | A | G | T | G | — | — | G | |
| D3 | QV00995 | AF402678 | — | — | — | — | T | — | — | A | G | T | G | — | A | G | |
SNP = single nucleotide polymorphism.
2.4. The Cancer Genome Atlas (TCGA) Cohort
All the data from the Cancer Genome Atlas (TCGA) was downloaded via the Broad Genome Data Analysis Center’s Firehose server (https://gdac.broadinstitute.org/, accessed on 27 June 2018). These samples were treatment-naïve before surgical resection. The TCGA HNSC survival data sets were sourced from Liu et al. [52], with the data sets manually annotated for HPV variant status. All the patients were HIV-negative.
Variant calling was performed on the TCGA RNA-seq data using bcftools mpileup and call functions. bcftools mpileup was run with the max-depth parameter set to 10,000. bcftools call was run with the ploidy parameter set to 1. The HPV16 sublineage was established by comparing nucleotide identities (SNPs) at positions 109, 131, 132, 143, 145, 178, 276, 286, 289, 335, 350, 403, 433, and 532 within the E6 gene [34,35].
The immune landscape features for the TCGA HNSC data set were sourced from Thorsson et al. [53]. This data included 53 immune landscape features of various predicted measures of immune infiltration by various cell types, innate and adaptive immune inflammation scores, and antigen presentation scores. The correlation of immune landscape features and HPV16+ variant subsets was performed via R’s built-in cor.test function, with the function being run with the linear relationship and Spearman correlation coefficient arguments. q values were calculated for each comparison group with an FDR of 10%.
2.5. Statistical Analysis
The HPV16 variant lineages were correlated with patient clinical variables using either Fisher’s exact test or Freeman–Halton extension of Fisher’s exact test (Fisher–Freeman–Halton test). Five-year overall and disease-free survival outcomes were compared to the HPV16 variants as indicated. Log-rank statistical analysis was performed using GraphPad Prism v7.0 (Graphpad Software, Inc., San Diego, CA, USA). The figures were assembled using Adobe Illustrator 2023 (Adobe Systems Inc., San Jose, CA, USA). The univariate analysis was performed for the indicated HPV16 variants and the following clinical variables: age, sex, subsite, T stage, N stage, overall stage, smoking status, smoking frequency, and treatment, through RStudio (version 1.2.1335) based on a Cox proportional-hazards model with the survival package (version 2.41-3). In addition, Multivariate analyses were performed using a stepwise bidirectional method. The smoking frequency clinical variable was stratified as heavy smokers (>20 pack year history), light smokers (≤20 pack year history), or non-smokers. The statistical p values were derived from the Wald test on hazard ratios.
3. Results
3.1. Study Cohorts and Distribution of HPV16 Intratypic Variants in HNCs from North America
Utilizing our local Southwestern Ontario, Canada cohort (SWO) of HPV16+ HNC patient samples, we PCR amplified extracted DNA, subjected the amplified DNA to Sanger sequencing, and aligned the sequencing results to the reference HPV16 sequence A1. Of the 94 samples that were analyzed from the SWO cohort, 38 were classified into the A1 sublineage, 54 into the A2 sublineage, and 2 into the D2/D3 sublineage (Table 3).
Table 3.
Clinical patient variables and cohort comparisons.
| Clinical Variables | SWO Cohort | TCGA Cohort | CANUSA Cohort |
|---|---|---|---|
| n = 94 a | n = 55 | n = 149 | |
| Age at diagnosis | |||
| Median (IQR) | 57 (52–66) | 57 (50.5–61) | 57 (50.5–66) |
| Sex | |||
| Female | 15 | 6 | 21 |
| Male | 79 | 49 | 128 |
| Subsite | |||
| Tonsil | 61 | 30 | 91 |
| BOT | 28 | 10 | 38 |
| Other | 5 | 15 | 20 |
| T Stage b | |||
| T1 | 14 | 5 | 19 |
| T2 | 36 | 28 | 64 |
| T3 | 17 | 9 | 26 |
| T4 | 19 | 12 | 31 |
| N Stage c | |||
| N0 | 10 | 15 | 25 |
| N1 | 14 | 4 | 18 |
| N2 | 54 | 33 | 87 |
| N3 | 8 | 2 | 10 |
| O Stage d | |||
| I | 0 | 1 | 1 |
| II | 6 | 9 | 15 |
| III | 11 | 6 | 17 |
| IV | 58 | 39 | 97 |
| Smoking Status | |||
| Never | 28 | 19 | 47 |
| Former | 26 | 26 | 52 |
| Current | 40 | 10 | 50 |
| Smoking Frequency e | |||
| Non-Smoker | 28 | 19 | 47 |
| Light Smoker | 22 | 10 | 32 |
| Heavy Smoker | 39 | 20 | 59 |
| HPV16 Sublineages | |||
| A1 | 38 | 20 | 58 |
| A2 | 54 | 20 | 74 |
| A3 | 0 | 0 | 0 |
| A4 | 0 | 4 | 4 |
| B1 | 0 | 3 | 3 |
| B2 | 0 | 1 | 1 |
| C | 0 | 1 | 1 |
| D1 | 0 | 0 | 0 |
| D2/D3 | 2 | 6 | 8 |
| D3 | 0 | 0 | 0 |
| HPV16 E6 | |||
| 350T | 33 | 16 | 49 |
| 350G | 40 | 9 | 49 |
SWO = Southwestern Ontario; TCGA = The Cancer Genome Atlas; CANUSA = Canada–USA; IQR = interquartile range; BOT = base-of-tongue; HPV16 = human papillomavirus type 16. a Clinical data was available for 94/106 samples with variant calls. b T stage data missing for 8 patients from the SWO cohort and undefined for 1 patient from the TCGA cohort. c N stage data missing for 8 patients from the SWO cohort and 1 patient from the TCGA cohort. d O stage data missing for 19 patients from the SWO cohort. e Smoking frequency data missing for 5 patients from the SWO cohort and 6 patients from the TCGA cohort.
Next, we utilized whole-genome sequencing (WGS) data from a secondary cohort of 55 HPV16+ HNC patient samples from the Cancer Genome Atlas (TCGA), which were collected from patients in Canada and the USA [54]. Similarly, the HPV16 genomes were classified into sublineages based on combinations of SNPs in the viral E6 oncogene. Of the 55 samples analyzed from this second cohort, 20 were classified into the A1 sublineage, 20 into A2, 4 into A4, 3 into B1, 1 into B2, 1 into C, and 6 into D2/D3 (Table 3). In addition, we also analyzed the E6 oncogene in both cohorts for a SNP at position 350. This is the most frequent SNP in HPV16 and leads to a leucine-to-valine change at residue 83 in the E6 oncoprotein. This SNP has been reported to alter the oncogenic properties of E6 and has been correlated with disease outcomes in cervical cancer [40,41,42,43]. The SWO cohort had 33 samples that were classified as 350T and 40 that were 350G. Whereas the TCGA cohort had 16 samples that were classified as 350T and 9 that were 350G (Table 3).
To increase the statistical power of our study and account for the rarity of some of the intratypic variants in the populations of Canada and the United States, we increased the sample size of our cohort by combining both the SWO and TCGA cohorts (CANUSA). This new cohort now contained 149 HPV16+ HNC samples of which 58 were classified into the A1 sublineage, 74 into A2, 4 into A4, 3 into B1, 1 into B2, 1 into C, and 8 into D2/D3 (Table 3). Furthermore, 49 samples were classified as 350T, and 49 were 350G.
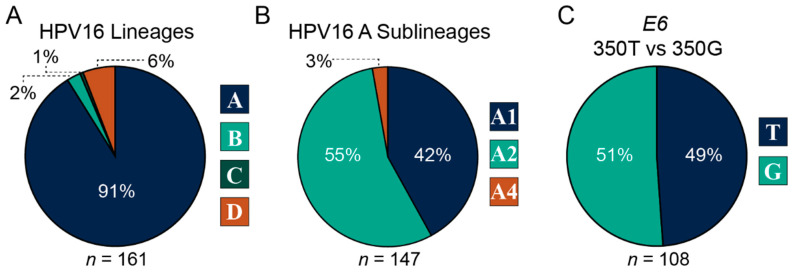
When analyzing the distribution of HPV16 intratypic variants in HNCs, we also included samples that had clinical data missing from the SWO cohort (Table 3). This increased the size of the CANUSA cohort from 149 samples to a total of 161 samples with variant calls that were classified into the 4 major phylogenetic lineages A, B, C, and D (Figure 1). The distribution of the major lineages in the CANUSA cohort was 91% A, 2% B, 1% C, and 6% D (Figure 1A). Since most of the samples were classified into lineage A, we specifically analyzed the distribution of sublineages within lineage A. The distribution of the A sublineages was 42% A1 (reference genome), 55% A2, and 3% A4 (Figure 1B). Notably, there were no samples that were classified into the A3 sublineage. Finally, we analyzed the distribution of samples that contained either thymine (T) at position 350 (reference genome) or the guanine (G) SNP at position 350. The distribution was 49% 350T and 51% 350G (Figure 1C). Taken together, the CANUSA cohort is predominantly made up of the A lineage, specifically the sublineages A1 and A2. Furthermore, roughly the same number of samples were segregated as 350T or 350G.
Figure 1.
Distribution of HPV16 intratypic variants. Pie charts depicting the distribution of HPV16 lineages (A), HPV16 A sublineages (B), and single nucleotide polymorphism at position 350 of the HPV16 oncogene E6 (C). The numbers below charts (n) indicate total numbers per analysis.
3.2. Impact of HPV16 Intratypic Variants in HNCs on Clinical Variables
Next, we wanted to determine the association of HPV16 intratypic variants with various clinical variables that include age, sex, smoking history, anatomical subsite of the cancer, T stage, N stage, and O stage. Since most of the samples in the CANUSA cohort were classified into major lineage A, we were unable to assess the association of clinical variables between all 4 major lineages. We first assessed the impact of lineage A vs. lineages B/C/D on clinical variables and found no statistically meaningful associations (Table S1A). We then assessed lineage A vs. lineage B vs. lineage D and the results indicated that there were no statistically meaningful associations with those comparisons (Table S1B). A comparison of the impact of lineage A vs. lineage D on clinical variables detected no statistically significant results (Table S1C).
Since most of the samples are predominantly classified into the A sublineage, we analyzed the association of clinical variables with sublineages A1, A2, and A4. Our results showed no significant association with any of the clinical variables analyzed (Table S1D). However, when we analyzed the impact of sublineage A1 vs. sublineage A2 on clinical variables, the association with the sublineages and O stage trended towards significance (p = 0.05544), and smoking status was significant (p = 0.029; Table 4). Finally, we focused on the SNP at position 350 of the E6 viral ORF and analyzed the association of 350T (reference genome) vs. 350G on clinical variables. The results showed a statistically significant association with T stage (p = 0.01129), N stage (p = 0.004325), O stage (p = 0.001462), and smoking status (p = 0.017; Table 5). Specifically, more patients with the 350T HPV16 genome (reference genome) were staged with T4, while more with the 350G genome were staged into T3. Similarly, there were more patients with the 350T HPV16 reference genome that were staged with N0 and N3 compared to those with 350G that were more frequently staged at N1. Notably, there were no patients with the 350G HPV16 intratypic variant that had O stage I or II. Taken together, this data suggests that specific intratypic HPV16 variants may impact clinical variables and identify significant associations between SNPs in the E6 viral oncogene and the clinical parameters analyzed.
Table 4.
Association of clinical variables with HPV16 sublineages A1 and A2.
| Clinical Variables | Sublineage A1 | Sublineage A2 | Total | p Value | |
|---|---|---|---|---|---|
| Age | ≤60 | 41 | 43 | 84 | 0.149 |
| >60 | 17 | 31 | 48 | ||
| Sex | Female | 10 | 10 | 20 | 0.628 |
| Male | 48 | 64 | 112 | ||
| Subsite | Tonsil | 34 | 46 | 80 | 0.907 |
| BOT | 16 | 18 | 34 | ||
| Other | 8 | 10 | 18 | ||
| T Stage | T1 | 8 | 9 | 17 | 0.210 |
| T2 | 27 | 28 | 55 | ||
| T3 | 6 | 17 | 23 | ||
| T4 | 15 | 13 | 28 | ||
| N Stage | N0 | 11 | 11 | 22 | 0.165 |
| N1 | 4 | 14 | 18 | ||
| N2 | 35 | 38 | 73 | ||
| N3 | 6 | 4 | 10 | ||
| O Stage | I | 1 | 0 | 1 | 0.055 |
| II | 9 | 4 | 13 | ||
| III | 4 | 12 | 16 | ||
| IV | 41 | 42 | 83 | ||
| Smoking Status | Never | 24 | 19 | 43 | 0.029 |
| Former | 20 | 21 | 41 | ||
| Current | 14 | 34 | 48 | ||
| Smoking Frequency | Non-Smoker | 24 | 19 | 43 | 0.089 |
| Light Smoker | 11 | 14 | 25 | ||
| Heavy Smoker | 18 | 36 | 54 | ||
BOT = base-of-tongue. p ≤ 0.05 are in red.
Table 5.
Association of clinical variables with HPV16 E6 350T and 350G single nucleotide polymorphism.
| Clinical Variables | E6 350T | E6 350G | Total | p Value | |
|---|---|---|---|---|---|
| Age | ≤60 | 34 | 27 | 61 | 0.211 |
| >60 | 15 | 22 | 37 | ||
| Sex | Female | 9 | 7 | 16 | 0.785 |
| Male | 40 | 42 | 82 | ||
| Subsite | Tonsil | 28 | 31 | 59 | 0.840 |
| BOT | 14 | 12 | 26 | ||
| Other | 7 | 6 | 13 | ||
| T Stage | T1 | 7 | 7 | 14 | 0.011 |
| T2 | 22 | 15 | 37 | ||
| T3 | 4 | 16 | 20 | ||
| T4 | 14 | 7 | 21 | ||
| N Stage | N0 | 8 | 3 | 11 | 0.004 |
| N1 | 2 | 13 | 15 | ||
| N2 | 31 | 27 | 58 | ||
| N3 | 6 | 2 | 8 | ||
| O Stage | I | 1 | 0 | 1 | 0.001 |
| II | 6 | 0 | 6 | ||
| III | 2 | 10 | 12 | ||
| IV | 37 | 28 | 65 | ||
| Smoking Status | Never | 20 | 15 | 35 | 0.017 |
| Former | 20 | 12 | 32 | ||
| Current | 9 | 22 | 31 | ||
| Smoking Frequency | Non-Smoker | 20 | 15 | 35 | 0.346 |
| Light Smoker | 10 | 9 | 19 | ||
| Heavy Smoker | 15 | 22 | 37 | ||
BOT = base-of-tongue. p ≤ 0.05 are in red.
3.3. Impact of HPV16 Intratypic Variants on Clinical Outcomes
There is evidence, in the context of cervical cancer (CC), that HPV16 intratypic variants can influence virus persistence, infection recurrence, disease risk, and cancer survival [43,55,56,57]. To determine if those intratypic-specific influences hold true for HPV16+ HNCs, we grouped the CANUSA cohort by lineages, sublineages, and SNP present at position 350 of the viral E6 oncogene and correlated those groups with overall and disease-free survival (Figures S1 and S2).
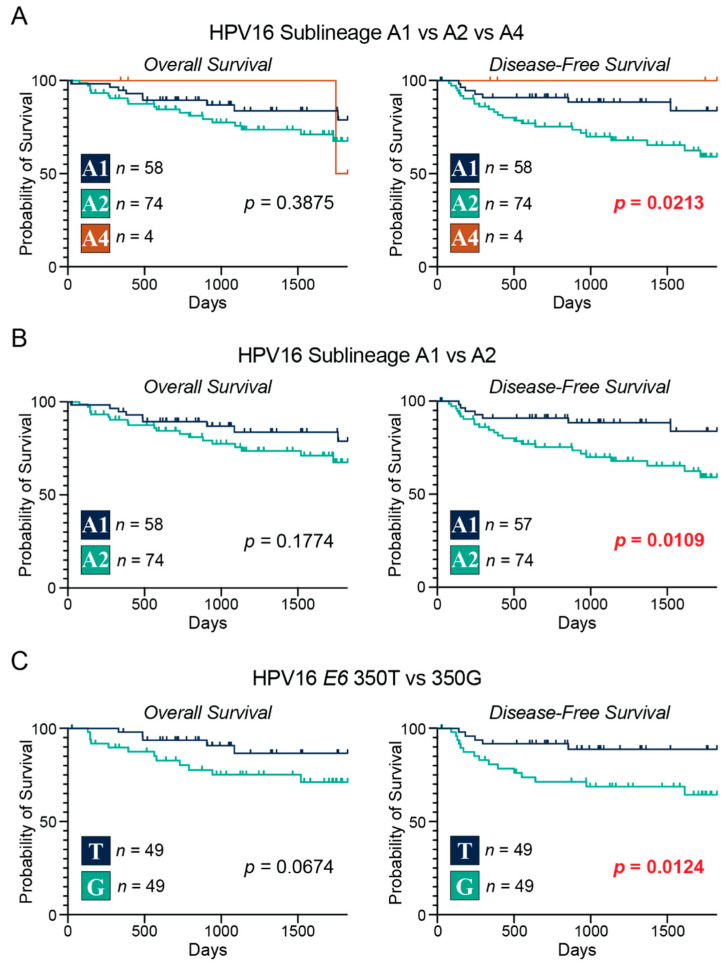
We began our analysis by correlating the major lineages with overall and disease-free survival. The results from lineage A vs. lineages B/C/D yielded no significant survival differences between the 2 groups (Figure S1A). Likewise, the correlations with survival between lineage A vs. lineage B vs. lineage D (Figure S1B) or lineage A vs. lineage D (Figure S1C) were also non-significant. We then focused on the sublineages of lineage A and correlated them to our survival metrics. When comparing sublineage A1 to A4 (Figure S1D) or sublineage A2 to A4 (Figure S1E) there were no statistically significant correlations with survival. However, when we compared sublineages A1 vs. A2 vs. A4 (Figure 2A) there was a significant correlation with disease-free survival (p = 0.0213). We then did a pairwise comparison between sublineage A1 and A2 and found that those patients with the A1 sublineage of HPV16 had a significant correlation with disease-free survival (p = 0.0109; Figure 2B). Finally, we compared patients with the HPV16 genomes that had the SNP at position 350 of the E6 viral oncogene and correlated the association of 350T vs. 350G with survival (Figure 2C). The results indicated that those patients with the 350T genome (reference genome) had statistically better disease-free survival compared to their 350G counterparts (p = 0.0124).
Figure 2.
Correlation of HPV16 intratypic variants with overall and disease-free survival in patients with head and neck cancers. Overall and disease-free survival of patients grouped by HPV16 sublineages A1, A2, and A4 (A); sublineages A1 and A2 (B); HPV16 E6 350T and 350G single nucleotide polymorphism (C). Comparison between groups was calculated with the 2-sided log-rank test (p-value). The number of samples included in each group is indicated with n. p ≤ 0.05 are in red.
Results from univariable Cox proportional hazards regression analysis similarly identified 350G as a significant risk factor for disease-free survival (HR = 3.55, p = 0.030), but not overall survival (Table 6 and Table 7). The multivariable analysis did not identify any clinical feature or 350G as statistically significant independent predictors of worse disease-free survival. Although not significant, 350G (HR = 2.78, p = 0.090), along with age (HR = 1.03, p = 0.144), and T-stage (HR = 2.86, p = 0.082) were the top three covariates in the predicted model of survival (Table 7). Univariable and multivariable analysis did not identify any significant correlations between sublineage A1 vs. A2 for either disease-free or overall survival (Tables S2A,B), although A2 trended towards worse disease-free survival (HR = 2.52, p = 0.054) and was identified as a covariate in that model. Taken together, these results show that there are differences in disease-free survival between patients that harbor different A sublineages of the HPV16 genome, as well as those with HPV16 genomes that had a SNP at position 350 of the E6 viral oncogene.
Table 6.
Univariable and multivariable Cox proportional hazards regression for overall survival with HPV16+ 350T and 350G in the CANUSA cohort.
| Variable | Univariable Analysis | Multivariable Analysis | ||
|---|---|---|---|---|
| HR (95% CI) | p Value | HR (95% CI) | p Value | |
| Age at diagnosis | 1.02 (0.97–1.07) | 0.423 | ||
| Sex | ||||
| Male vs. Female | 0.41 (0.12–1.32) | 0.134 | ||
| Subsite | ||||
| Tonsil vs. base-of-tongue | 0.74 (0.22–2.52) | 0.629 | ||
| Other vs. base-of-tongue | 1.06 (0.19–5.77) | 0.950 | ||
| T stage | ||||
| T3–T4 vs. T1–T2 | 4.62 (1.27–16.8) | 0.020 | ||
| N stage | ||||
| N2–N3 vs. N0–N1 | 0.43 (0.14–1.27) | 0.126 | ||
| Overall Stage | ||||
| III–IV vs. I–II | 7.59 × 107 (0–Inf) | 0.998 | 4.62 (1.27–16.8) | 0.020 |
| Smoking status | ||||
| Former vs. Never | 0.47 (0.09–2.40) | 0.360 | ||
| Current vs. Never | 1.35 (0.41–4.45) | 0.620 | ||
| Smoking Frequency | ||||
| Light vs. Never | 0.66 (0.13–3.40) | 0.619 | ||
| Heavy vs. Never | 1.05 (0.32–3.45) | 0.938 | ||
| HPV16 T350G | ||||
| 350G vs. 350T | 2.91 (0.89–9.46) | 0.076 | ||
| Treatment | ||||
| Chemotherapy + Radiation | 8.98 × 10−1 (0.27–2.95) | 0.859 | ||
| Surgery + Chemotherapy + Radiation | 1.29 × 10−8 (0–Inf) | 0.999 | ||
| Surgery + Radiation | 1.30 × 10−8 (0–Inf) | 0.999 | ||
| Surgery Alone | 1.11 (0.34–3.66) | 0.859 | ||
| Radiation Alone | 1.29 × 10−8 (0–Inf) | 0.999 | ||
p ≤ 0.05 are in red.
Table 7.
Univariable and multivariable Cox proportional hazards regression for disease-free survival with HPV16+ 350T and 350G in the CANUSA cohort.
| Variable | Univariable Analysis | Multivariable Analysis | ||
|---|---|---|---|---|
| HR (95% CI) | p Value | HR (95% CI) | p Value | |
| Age at diagnosis | 1.04 (0.99–1.08) | 0.124 | 1.03 (0.99–1.08) | 0.144 |
| Sex | ||||
| Male vs. Female | 0.47 (0.15–1.48) | 0.198 | ||
| Subsite | ||||
| Tonsil vs. base-of-tongue | 0.70 (0.23–2.14) | 0.530 | ||
| Other vs. base-of-tongue | 0.89 (0.17–4.58) | 0.888 | ||
| T stage | ||||
| T3–T4 vs. T1–T2 | 3.86 (1.23–12.14) | 0.021 | 2.86 (0.88–9.35) | 0.082 |
| N stage | ||||
| N2–N3 vs. N0–N1 | 0.74 (0.25–2.17) | 0.586 | ||
| Overall Stage | ||||
| III–IV vs. I–II | 7.63 × 107 (0–Inf) | 0.998 | ||
| Smoking status | ||||
| Former vs. Never | 0.79 (0.22–2.80) | 0.712 | ||
| Current vs. Never | 0.98 (0.30–3.21) | 0.968 | ||
| Smoking Frequency | ||||
| Light vs. Never | 0.84 (0.21–3.35) | 0.799 | ||
| Heavy vs. Never | 0.91 (0.29–2.82) | 0.867 | ||
| HPV16 T350G | ||||
| 350G vs. 350T | 3.55 (1.13–11.17) | 0.030 | 2.78 (0.85–9.05) | 0.090 |
| Treatment | ||||
| Chemotherapy + Radiation | 6.17 × 10−1 (0.22–1.77) | 0.369 | ||
| Surgery + Chemotherapy + Radiation | 1.38 × 10−8 (0–Inf) | 0.998 | ||
| Surgery + Radiation | 1.42 × 10−8 (0–Inf) | 0.999 | ||
| Surgery Alone | 1.62 (0.57–4.64) | 0.369 | ||
| Radiation Alone | 1.41 × 10−8 (0–Inf) | 0.999 | ||
p ≤ 0.05 are in red.
3.4. Impact of HPV16 Intratypic Variants on Immune Characteristics
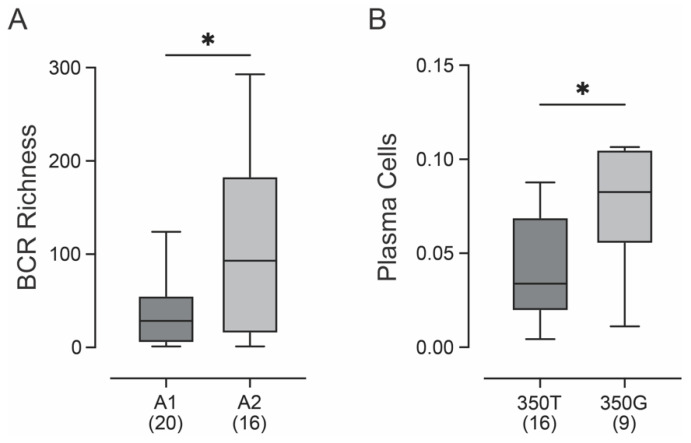
Recent breakthroughs in cancer immunotherapy have clearly demonstrated the critical role of the immune system in controlling malignancy. Thorsson et al. [53] recently calculated 53 distinct immune signatures related to the tumor immune microenvironment for all the TCGA samples with RNA-seq data. These include various predicted measures of immune infiltration by various cell types, innate and adaptive immune inflammation scores, and antigen presentation scores. We used this data to look for potential differences in the tumor immune microenvironment between the A1 and A2 sublineage samples from the TCGA head and neck cancer cohort, as no RNA-seq data is available for the SWO cohort. Of the wide variety of immune signatures, only B-cell receptor (BCR) richness was significantly different between these two sublineages after false-discovery correction (Figure 3A). This measurement reflects the relative abundance of unique clones of BCRs, which was significantly higher in A2 samples compared to A1 samples. The increased diversification of the BCR suggests that there may be minor differences in the immune response in HNCs caused by these two sublineages. We similarly compared the samples stratified as 350T or 350G and identified an increase in plasma cells, which are terminally differentiated cells that arise from antigenically stimulated B lymphocytes, in those samples with the 350G polymorphism (Figure 3B).
Figure 3.
Correlation of HPV16 intratypic variants with immune characteristics of patients with head and neck cancer. Predicted B-cell receptor (BCR) richness was significantly increased in HPV16+ HNC associated with A2 vs. A1 sublineage (A). Predicted plasma cell presence in HPV16+ HNC expressing the T350G polymorphism was significantly increased (B). Comparison between groups was performed with a two-tailed non-parametric Mann–Whitney U test. The number of samples included in each group is indicated in brackets. * p ≤ 0.05.
4. Discussion
HPV16 is responsible for an estimated 83% of all HPV+ head and neck cancers, and 50–60% of CCs [10,12,58]. The most predominant HPV16 variant lineage in CC is the reference/prototype A1. Notably, studies have revealed that the non-A variant lineages of HPV16 are associated with a higher risk of precancerous lesions and development of CC [34,59], or associated with higher rates of persistent infection and progression to CC when compared to the other lineage variants [59,60,61]. In addition, the well-characterized HPV16 T350G, which leads to the L83V E6 substitution, has been associated with higher rates of persistent infection and progression to cervical cancers [39,44,45,62,63]. In the context of HNCs, the same HPV16 variant lineage-specific effects observed in HPV16+ cervical cancers could be true; however, few studies reporting on the distribution of HPV16 variant lineages in HNC or the non-lineage-specific HPV16 T350G were available [37].
In this study, we combined our local cohort of HPV16+ HNC samples from Southwestern Ontario, Canada (SWO) with those from the Cancer Genome Atlas (TCGA) HPV16+ HNC cohort to create a larger North American cohort (CANUSA). This cohort, representative of Canada and the United States of America, increases the statistical power of the study and increases the diversity of the samples analyzed (Table 3). Utilizing our CANUSA cohort of 161 HPV16+ HNC samples—only 149 samples had clinical data available—we determined the distribution of variant lineages, their association with patient clinical variables, and their impact on patient survival outcomes.
The distribution of HPV16 intratypic variant lineages in HNCs was 91% A, 2% B, 1% C, and 6% D (Figure 1A). Furthermore, within the A lineage, the distribution was 42% A1 (reference/prototype sequence), 55% A2, and 3% A4 (Figure 1B). When analyzing the distribution of non-lineage-specific HPV16 T350G, we found that 49% of the samples were classified as 350T (reference/prototype sequence), whereas 51% were 350G (Figure 1C). These results show that the CANUSA cohort is predominately made up of variant lineage A, specifically sublineages A1 and A2, and the non-lineage-specific variant T350G is present in roughly the same number of samples compared to the reference/prototype genome. These results are very similar to recent studies from various sites in the USA (Table 8) [64,65,66], although the study from the University of North Carolina with 107 HNC samples differed in that it reported a much larger fraction of the A lineage were A1 (85%), with only 10% A2 [64].
Table 8.
The distribution of HPV16 intratypic variants from recent studies in the USA.
| Study | CANUSA | Vanderbilt [66] | North Carolina [64] | Vanderbilt/Pittsburgh [65] | |||||
|---|---|---|---|---|---|---|---|---|---|
| HPV16 Variant | A | 136 (91.3%) | A1: 58 (42.6%) | 191 (90.1%) | A1: 112 (58.33%) | 91 (85%) | A1: 77 (84.6%) | 347 (90.4%) | A1: 215 (62%) |
| A2: 74 (54.4) | A2: 63 (32.8%) | A2: 9 (9.89%) | A2: 107 (30.8%) | ||||||
| A3: 0 (0%) | A3: 3 (1.56%) | A3: 3 (3.3%) | A3: 3 (0.865%) | ||||||
| A4: 4 (2.94%) | A4: 14 (7.29%) | A4: 2 (2.2%) | A4: 22 (6.34%) | ||||||
| B | 4 (2.68%) | 1 (0.472%) | 1 (0.934%) | 1 (0.26%) | |||||
| C | 1 (0.671%) | 4 (1.89%) | 2 (1.87%) | 6 (1.56%) | |||||
| D | 8 (5.37%) | 16 (7.55%) | 13 (12.1%) | 30 (7.81%) | |||||
| Total | 149 (100%) | 136 (100%) | 212 (100%) | 192 (100%) | 107 (100%) | 91 (100%) | 384 (100%) | 347 (100%) | |
When we analyzed the association of sublineages A1 or A2 with patient clinical variables, the association with the sublineages and O stage was trending towards significance (p = 0.05544), and smoking status was significant (p = 0.029; Table 4). When we focused on the non-lineage-specific variant T350G compared to the reference/prototype genome, the results showed a statistically significant association with T stage (p = 0.01129), N stage (p = 0.004325), O stage (p = 0.001462), and smoking status (p = 0.017; Table 5). Specifically, there were more patients with the reference/prototype genome that were staged with T4, N0, or N3, whereas more patients with the T350G variant were staged into T3 or N1. These results identified a significant association between SNPs in the E6 viral oncogene and the clinical parameters analyzed, illustrating the association of HPV16 intratypic variants on HNC patient clinical factors.
When we correlated the HPV16 sublineages of lineage A with overall survival and disease-free survival, we observed that patients with the A1 reference/prototype genome of HPV16 had significantly improved disease-free survival compared to their A2 counterparts (Figure 2). Although not directly comparable, a smaller study reported that sublineage A1 exhibited worse relapse-free survival compared to the collective aggregation of all other sublineages [64]. No direct comparison between sublineage A1 and A2 was performed in that cohort, as there were relatively few A2 samples.
In addition to survival differences based on sublineage, when correlating the non-lineage-specific variant T350G and the reference/prototype genome (350T) with survival, we found that those patients with the reference/prototype genome (350T) had significantly greater disease-free survival (p = 0.0124; Figure 2C, Table 7). No changes in overall survival were noted, likely related to the low level of mortality in this cohort. These results suggest that even minor nucleic acid changes in the coding region of the E6 oncogene can impact patient survival outcomes. A larger cohort study using 384 HPV16+ HNC samples from several centers in the USA assessed genetic variation across the entire viral genome for correlation with overall survival. While a few SNPs were associated with overall survival, T350G was not, and this study did not report on disease-free survival or compare sublineages directly for patient outcomes [65].
Given the critical role of the immune system in controlling malignancy, we looked for differences in immune signatures that reflect differences in the tumor immune microenvironment between the A1 and A2 sublineage samples or 350T and 350G samples from the TCGA. After false-discovery correction, only B-cell receptor (BCR) richness was significantly different between the A1 and A2 sublineages (Figure 3A) and plasma cell abundance between 350T and 350G (Figure 3B). The increased diversification of the BCR is consistent with differences in the immune response in HNCs caused by these two sublineages. We similarly compared the samples stratified as 350T or 350G and identified an increase in plasma cells in those samples with the 350G polymorphism (Figure 3B). B-cells are important players in immune responses to cancer and differences in B-cell infiltration in HPV+ and HPV− HNC have been reported [67]. Additionally, HPV antigen-specific activated and germinal center B cells, as well as plasma cells can be found in the HPV16+ HNC tumor microenvironment [68], the differences we identified in B-cell richness and plasma cell frequency could contribute mechanistically to altered patient outcomes associated with HPV variants.
In conclusion, our findings suggest that minor sequence variations within HPV16 appear to be associated with HPV+ HNC patient characteristics and prognosis. Even straightforward targeted sequencing of small portions of the HPV genome may be sufficient to obtain clinically relevant information that can help stratify patient risk. However, this study was limited by the number of available sequences, and powering future investigations with much larger cohort sizes will be necessary to unequivocally establish if intratypic variant typing is of prognostic value for HNC.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/v15122411/s1, Table S1. Association of clinical variables with (A): HPV16 lineages A and B/C/D, (B): HPV16 lineages A, B, and D, (C): HPV16 lineages A and D, (D): HPV16 sublineages A1, A2, and A4; Table S2. Univariable and multivariable Cox proportional hazards regression for (A) overall survival with HPV16+ A1 and A2 in the CANUSA cohort. (B) disease-free survival with HPV16+ A1 and A2 in the CANUSA cohort.
Author Contributions
Conceptualization, S.F.G., A.C.N. and J.S.M.; formal analysis, S.F.G., M.Y.S. and P.Y.F.Z.; data curation, S.F.G., M.Y.S., P.Y.F.Z. and J.W.B.; writing—original draft preparation, S.F.G. and J.S.M.; writing—review and editing, M.Y.S., P.Y.F.Z., J.W.B. and A.C.N.; visualization, S.F.G. and M.Y.S.; supervision, A.C.N. and J.S.M.; funding acquisition, A.C.N. and J.S.M. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Research Ethics Board of the University of Western Ontario (LHSC HSREB #7182—9 September 2010).
Informed Consent Statement
Informed consent was obtained from all the subjects involved in the study.
Data Availability Statement
Data are contained within the article and Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This work was supported by a grant from the Canadian Institutes of Health Research (PJT-173496) to J.S.M. and A.C.N. M.Y.S. was supported by a Canadian Institutes of Health Research Canada Graduate Scholarship. A.C.N. was supported by the Wolfe Surgical Research Professorship in the Biology of Head and Neck Cancers Fund. P.Y.F.Z. was supported by a CIHR Vanier Canada Graduate Scholarship and a PSI Foundation Fellowship.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Doorbar J., Egawa N., Griffin H., Kranjec C., Murakami I. Human papillomavirus molecular biology and disease association. Rev. Med. Virol. 2015;25((Suppl. S1)):2–23. doi: 10.1002/rmv.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burd E.M. Human papillomavirus and cervical cancer. Clin. Microbiol. Rev. 2003;16:1–17. doi: 10.1128/CMR.16.1.1-17.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moody C.A., Laimins L.A. Human papillomavirus oncoproteins: Pathways to transformation. Nat. Rev. Cancer. 2010;10:550–560. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- 4.zur Hausen H. Papillomaviruses and cancer: From basic studies to clinical application. Nat. Rev. Cancer. 2002;2:342–350. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]
- 5.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 6.de Martel C., Georges D., Bray F., Ferlay J., Clifford G.M. Global burden of cancer attributable to infections in 2018: A worldwide incidence analysis. Lancet Glob. Health. 2020;8:e180–e190. doi: 10.1016/S2214-109X(19)30488-7. [DOI] [PubMed] [Google Scholar]
- 7.Schiffman M., Doorbar J., Wentzensen N., de Sanjose S., Fakhry C., Monk B.J., Stanley M.A., Franceschi S. Carcinogenic human papillomavirus infection. Nat. Rev. Dis. Primers. 2016;2:16086. doi: 10.1038/nrdp.2016.86. [DOI] [PubMed] [Google Scholar]
- 8.Pesut E., Dukic A., Lulic L., Skelin J., Simic I., Milutin Gasperov N., Tomaic V., Sabol I., Grce M. Human Papillomaviruses-Associated Cancers: An Update of Current Knowledge. Viruses. 2021;13:2234. doi: 10.3390/v13112234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Martel C., Plummer M., Vignat J., Franceschi S. Worldwide burden of cancer attributable to HPV by site, country and HPV type. Int. J. Cancer. 2017;141:664–670. doi: 10.1002/ijc.30716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leemans C.R., Snijders P.J.F., Brakenhoff R.H. The molecular landscape of head and neck cancer. Nat. Rev. Cancer. 2018;18:269–282. doi: 10.1038/nrc.2018.11. [DOI] [PubMed] [Google Scholar]
- 11.Gameiro S.F., Evans A.M., Mymryk J.S. The tumor immune microenvironments of HPV+ and HPV− head and neck cancers. WIREs Mech. Dis. 2022;14:e1539. doi: 10.1002/wsbm.1539. [DOI] [PubMed] [Google Scholar]
- 12.Kreimer A.R., Clifford G.M., Boyle P., Franceschi S. Human papillomavirus types in head and neck squamous cell carcinomas worldwide: A systematic review. Cancer Epidemiol. Biomark. Prev. 2005;14:467–475. doi: 10.1158/1055-9965.EPI-04-0551. [DOI] [PubMed] [Google Scholar]
- 13.Kang H., Kiess A., Chung C.H. Emerging biomarkers in head and neck cancer in the era of genomics. Nat. Rev. Clin. Oncol. 2015;12:11–26. doi: 10.1038/nrclinonc.2014.192. [DOI] [PubMed] [Google Scholar]
- 14.Gameiro S.F., Ghasemi F., Barrett J.W., Koropatnick J., Nichols A.C., Mymryk J.S., Maleki Vareki S. Treatment-naive HPV+ head and neck cancers display a T-cell-inflamed phenotype distinct from their HPV- counterparts that has implications for immunotherapy. Oncoimmunology. 2018;7:e1498439. doi: 10.1080/2162402X.2018.1498439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gameiro S.F., Ghasemi F., Zeng P.Y.F., Mundi N., Howlett C.J., Plantinga P., Barrett J.W., Nichols A.C., Mymryk J.S. Low expression of NSD1, NSD2, and NSD3 define a subset of human papillomavirus-positive oral squamous carcinomas with unfavorable prognosis. Infect. Agents Cancer. 2021;16:13. doi: 10.1186/s13027-021-00347-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gameiro S.F., Ghasemi F., Barrett J.W., Nichols A.C., Mymryk J.S. High Level Expression of MHC-II in HPV+ Head and Neck Cancers Suggests that Tumor Epithelial Cells Serve an Important Role as Accessory Antigen Presenting Cells. Cancers. 2019;11:1129. doi: 10.3390/cancers11081129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gameiro S.F., Zhang A., Ghasemi F., Barrett J.W., Nichols A.C., Mymryk J.S. Analysis of Class I Major Histocompatibility Complex Gene Transcription in Human Tumors Caused by Human Papillomavirus Infection. Viruses. 2017;9:252. doi: 10.3390/v9090252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garset-Zamani M., Carlander A.F., Jakobsen K.K., Friborg J., Kiss K., Marvig R.L., Olsen C., Nielsen F.C., Andersen E., Gronhoj C., et al. Impact of specific high-risk human papillomavirus genotypes on survival in oropharyngeal cancer. Int. J. Cancer. 2022;150:1174–1183. doi: 10.1002/ijc.33893. [DOI] [PubMed] [Google Scholar]
- 19.Ziai H., Warner A., Mundi N., Patel K., Chung E.J., Howlett C.J., Plantinga P., Yoo J., MacNeil S.D., Fung K., et al. Does HPV Subtype Predict Outcomes in Head and Neck Cancers? Int. J. Otolaryngol. 2021;2021:6672373. doi: 10.1155/2021/6672373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bratman S.V., Bruce J.P., O’Sullivan B., Pugh T.J., Xu W., Yip K.W., Liu F.F. Human Papillomavirus Genotype Association With Survival in Head and Neck Squamous Cell Carcinoma. JAMA Oncol. 2016;2:823–826. doi: 10.1001/jamaoncol.2015.6587. [DOI] [PubMed] [Google Scholar]
- 21.Doorbar J., Quint W., Banks L., Bravo I.G., Stoler M., Broker T.R., Stanley M.A. The biology and life-cycle of human papillomaviruses. Vaccine. 2012;30((Suppl. S5)):F55–F70. doi: 10.1016/j.vaccine.2012.06.083. [DOI] [PubMed] [Google Scholar]
- 22.Bodily J., Laimins L.A. Persistence of human papillomavirus infection: Keys to malignant progression. Trends Microbiol. 2011;19:33–39. doi: 10.1016/j.tim.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Longworth M.S., Laimins L.A. Pathogenesis of human papillomaviruses in differentiating epithelia. Microbiol. Mol. Biol. Rev. 2004;68:362–372. doi: 10.1128/MMBR.68.2.362-372.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riva G., Albano C., Gugliesi F., Pasquero S., Pacheco S.F.C., Pecorari G., Landolfo S., Biolatti M., Dell’Oste V. HPV Meets APOBEC: New Players in Head and Neck Cancer. Int. J. Mol. Sci. 2021;22:1402. doi: 10.3390/ijms22031402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ho L., Chan S.Y., Burk R.D., Das B.C., Fujinaga K., Icenogle J.P., Kahn T., Kiviat N., Lancaster W., Mavromara-Nazos P., et al. The genetic drift of human papillomavirus type 16 is a means of reconstructing prehistoric viral spread and the movement of ancient human populations. J. Virol. 1993;67:6413–6423. doi: 10.1128/jvi.67.11.6413-6423.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan S.Y., Ho L., Ong C.K., Chow V., Drescher B., Durst M., ter Meulen J., Villa L., Luande J., Mgaya H.N., et al. Molecular variants of human papillomavirus type 16 from four continents suggest ancient pandemic spread of the virus and its coevolution with humankind. J. Virol. 1992;66:2057–2066. doi: 10.1128/jvi.66.4.2057-2066.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pientong C., Wongwarissara P., Ekalaksananan T., Swangphon P., Kleebkaow P., Kongyingyoes B., Siriaunkgul S., Tungsinmunkong K., Suthipintawong C. Association of human papillomavirus type 16 long control region mutation and cervical cancer. Virol. J. 2013;10:30. doi: 10.1186/1743-422X-10-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamada T., Manos M.M., Peto J., Greer C.E., Munoz N., Bosch F.X., Wheeler C.M. Human papillomavirus type 16 sequence variation in cervical cancers: A worldwide perspective. J. Virol. 1997;71:2463–2472. doi: 10.1128/jvi.71.3.2463-2472.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mirabello L., Yeager M., Yu K., Clifford G.M., Xiao Y., Zhu B., Cullen M., Boland J.F., Wentzensen N., Nelson C.W., et al. HPV16 E7 Genetic Conservation Is Critical to Carcinogenesis. Cell. 2017;170:1164–1174.e6. doi: 10.1016/j.cell.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kessis T.D., Slebos R.J., Nelson W.G., Kastan M.B., Plunkett B.S., Han S.M., Lorincz A.T., Hedrick L., Cho K.R. Human papillomavirus 16 E6 expression disrupts the p53-mediated cellular response to DNA damage. Proc. Natl. Acad. Sci. USA. 1993;90:3988–3992. doi: 10.1073/pnas.90.9.3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mantovani F., Banks L. The human papillomavirus E6 protein and its contribution to malignant progression. Oncogene. 2001;20:7874–7887. doi: 10.1038/sj.onc.1204869. [DOI] [PubMed] [Google Scholar]
- 32.McLaughlin-Drubin M.E., Meyers J., Munger K. Cancer associated human papillomaviruses. Curr. Opin. Virol. 2012;2:459–466. doi: 10.1016/j.coviro.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bernard H.U., Burk R.D., Chen Z., van Doorslaer K., zur Hausen H., de Villiers E.M. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology. 2010;401:70–79. doi: 10.1016/j.virol.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burk R.D., Harari A., Chen Z. Human papillomavirus genome variants. Virology. 2013;445:232–243. doi: 10.1016/j.virol.2013.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cornet I., Gheit T., Franceschi S., Vignat J., Burk R.D., Sylla B.S., Tommasino M., Clifford G.M., Group I.H.V.S. Human papillomavirus type 16 genetic variants: Phylogeny and classification based on E6 and LCR. J. Virol. 2012;86:6855–6861. doi: 10.1128/JVI.00483-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ho L., Chan S.Y., Chow V., Chong T., Tay S.K., Villa L.L., Bernard H.U. Sequence variants of human papillomavirus type 16 in clinical samples permit verification and extension of epidemiological studies and construction of a phylogenetic tree. J. Clin. Microbiol. 1991;29:1765–1772. doi: 10.1128/jcm.29.9.1765-1772.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Combes J.D., Franceschi S. Human papillomavirus genome variants and head and neck cancers: A perspective. Infect. Agents Cancer. 2018;13:13. doi: 10.1186/s13027-018-0185-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamada T., Wheeler C.M., Halpern A.L., Stewart A.C., Hildesheim A., Jenison S.A. Human papillomavirus type 16 variant lineages in United States populations characterized by nucleotide sequence analysis of the E6, L2, and L1 coding segments. J. Virol. 1995;69:7743–7753. doi: 10.1128/jvi.69.12.7743-7753.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grodzki M., Besson G., Clavel C., Arslan A., Franceschi S., Birembaut P., Tommasino M., Zehbe I. Increased risk for cervical disease progression of French women infected with the human papillomavirus type 16 E6-350G variant. Cancer Epidemiol. Biomark. Prev. 2006;15:820–822. doi: 10.1158/1055-9965.EPI-05-0864. [DOI] [PubMed] [Google Scholar]
- 40.Zehbe I., Wilander E., Delius H., Tommasino M. Human papillomavirus 16 E6 variants are more prevalent in invasive cervical carcinoma than the prototype. Cancer Res. 1998;58:829–833. [PubMed] [Google Scholar]
- 41.Godi A., Boampong D., Elegunde B., Panwar K., Fleury M., Li S., Zhao Q., Xia N., Christensen N.D., Beddows S. Comprehensive Assessment of the Antigenic Impact of Human Papillomavirus Lineage Variation on Recognition by Neutralizing Monoclonal Antibodies Raised against Lineage A Major Capsid Proteins of Vaccine-Related Genotypes. J. Virol. 2020;94:e01236-20. doi: 10.1128/JVI.01236-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Togtema M., Jackson R., Richard C., Niccoli S., Zehbe I. The human papillomavirus 16 European-T350G E6 variant can immortalize but not transform keratinocytes in the absence of E7. Virology. 2015;485:274–282. doi: 10.1016/j.virol.2015.07.025. [DOI] [PubMed] [Google Scholar]
- 43.Cornet I., Gheit T., Iannacone M.R., Vignat J., Sylla B.S., Del Mistro A., Franceschi S., Tommasino M., Clifford G.M. HPV16 genetic variation and the development of cervical cancer worldwide. Br. J. Cancer. 2013;108:240–244. doi: 10.1038/bjc.2012.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Londesborough P., Ho L., Terry G., Cuzick J., Wheeler C., Singer A. Human papillomavirus genotype as a predictor of persistence and development of high-grade lesions in women with minor cervical abnormalities. Int. J. Cancer. 1996;69:364–368. doi: 10.1002/(SICI)1097-0215(19961021)69:5<364::AID-IJC2>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 45.Gheit T., Cornet I., Clifford G.M., Iftner T., Munk C., Tommasino M., Kjaer S.K. Risks for persistence and progression by human papillomavirus type 16 variant lineages among a population-based sample of Danish women. Cancer Epidemiol. Biomark. Prev. Publ. Am. Assoc. Cancer Res. Cosponsored Am. Soc. Prev. Oncol. 2011;20:1315–1321. doi: 10.1158/1055-9965.EPI-10-1187. [DOI] [PubMed] [Google Scholar]
- 46.Schiffman M., Rodriguez A.C., Chen Z., Wacholder S., Herrero R., Hildesheim A., Desalle R., Befano B., Yu K., Safaeian M., et al. A population-based prospective study of carcinogenic human papillomavirus variant lineages, viral persistence, and cervical neoplasia. Cancer Res. 2010;70:3159–3169. doi: 10.1158/0008-5472.CAN-09-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zuna R.E., Tuller E., Wentzensen N., Mathews C., Allen R.A., Shanesmith R., Dunn S.T., Gold M.A., Wang S.S., Walker J., et al. HPV16 variant lineage, clinical stage, and survival in women with invasive cervical cancer. Infect. Agents Cancer. 2011;6:19. doi: 10.1186/1750-9378-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burk R.D., Terai M., Gravitt P.E., Brinton L.A., Kurman R.J., Barnes W.A., Greenberg M.D., Hadjimichael O.C., Fu L., McGowan L., et al. Distribution of human papillomavirus types 16 and 18 variants in squamous cell carcinomas and adenocarcinomas of the cervix. Cancer Res. 2003;63:7215–7220. [PubMed] [Google Scholar]
- 49.Nichols A.C., Dhaliwal S.S., Palma D.A., Basmaji J., Chapeskie C., Dowthwaite S., Franklin J.H., Fung K., Kwan K., Wehrli B., et al. Does HPV type affect outcome in oropharyngeal cancer? J. Otolaryngol. Head Neck Surg. 2013;42:9. doi: 10.1186/1916-0216-42-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nichols A.C., Palma D.A., Dhaliwal S.S., Tan S., Theuer J., Chow W., Rajakumar C., Um S., Mundi N., Berk S., et al. The epidemic of human papillomavirus and oropharyngeal cancer in a Canadian population. Curr. Oncol. 2013;20:212–219. doi: 10.3747/co.20.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Boer M.A., Peters L.A., Aziz M.F., Siregar B., Cornain S., Vrede M.A., Jordanova E.S., Kolkman-Uljee S., Fleuren G.J. Human papillomavirus type 16 E6, E7, and L1 variants in cervical cancer in Indonesia, Suriname, and The Netherlands. Gynecol. Oncol. 2004;94:488–494. doi: 10.1016/j.ygyno.2004.05.037. [DOI] [PubMed] [Google Scholar]
- 52.Liu J., Lichtenberg T., Hoadley K.A., Poisson L.M., Lazar A.J., Cherniack A.D., Kovatich A.J., Benz C.C., Levine D.A., Lee A.V., et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell. 2018;173:400–416.e11. doi: 10.1016/j.cell.2018.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thorsson V., Gibbs D.L., Brown S.D., Wolf D., Bortone D.S., Ou Yang T.H., Porta-Pardo E., Gao G.F., Plaisier C.L., Eddy J.A., et al. The Immune Landscape of Cancer. Immunity. 2018;48:812–830.e14. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.The Cancer Genome Atlas Network Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang B., Zhang L., Zhang A., Zhou A., Yuan J., Wang Y., Sun L., Cao H., Zheng W. Variant sublineages of human papillomavirus type 16 predispose women to persistent infection characterized by a sequence analysis of the E6, L1, and LCR regions. Int. J. Clin. Exp. Pathol. 2019;12:337–343. [PMC free article] [PubMed] [Google Scholar]
- 56.Rader J.S., Tsaih S.W., Fullin D., Murray M.W., Iden M., Zimmermann M.T., Flister M.J. Genetic variations in human papillomavirus and cervical cancer outcomes. Int. J. Cancer. 2019;144:2206–2214. doi: 10.1002/ijc.32038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang L., Yang B., Zhang A., Zhou A., Yuan J., Wang Y., Sun L., Cao H., Wang J., Zheng W. Association between human papillomavirus type 16 E6 and E7 variants with subsequent persistent infection and recurrence of cervical high-grade squamous intraepithelial lesion after conization. J. Med. Virol. 2016;88:1982–1988. doi: 10.1002/jmv.24541. [DOI] [PubMed] [Google Scholar]
- 58.Mirabello L., Clarke M.A., Nelson C.W., Dean M., Wentzensen N., Yeager M., Cullen M., Boland J.F., Workshop N.H., Schiffman M., et al. The Intersection of HPV Epidemiology, Genomics and Mechanistic Studies of HPV-Mediated Carcinogenesis. Viruses. 2018;10:80. doi: 10.3390/v10020080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mirabello L., Yeager M., Cullen M., Boland J.F., Chen Z., Wentzensen N., Zhang X., Yu K., Yang Q., Mitchell J., et al. HPV16 Sublineage Associations with Histology-Specific Cancer Risk Using HPV Whole-Genome Sequences in 3200 Women. J. Natl. Cancer Inst. 2016;108:djw100. doi: 10.1093/jnci/djw100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lou H., Boland J.F., Torres-Gonzalez E., Albanez A., Zhou W., Steinberg M.K., Diaw L., Mitchell J., Roberson D., Cullen M., et al. The D2 and D3 Sublineages of Human Papilloma Virus 16-Positive Cervical Cancer in Guatemala Differ in Integration Rate and Age of Diagnosis. Cancer Res. 2020;80:3803–3809. doi: 10.1158/0008-5472.CAN-20-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Clifford G.M., Tenet V., Georges D., Alemany L., Pavon M.A., Chen Z., Yeager M., Cullen M., Boland J.F., Bass S., et al. Human papillomavirus 16 sub-lineage dispersal and cervical cancer risk worldwide: Whole viral genome sequences from 7116 HPV16-positive women. Papillomavirus Res. 2019;7:67–74. doi: 10.1016/j.pvr.2019.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zacapala-Gomez A.E., Del Moral-Hernandez O., Villegas-Sepulveda N., Hidalgo-Miranda A., Romero-Cordoba S.L., Beltran-Anaya F.O., Leyva-Vazquez M.A., Alarcon-Romero Ldel C., Illades-Aguiar B. Changes in global gene expression profiles induced by HPV 16 E6 oncoprotein variants in cervical carcinoma C33-A cells. Virology. 2016;488:187–195. doi: 10.1016/j.virol.2015.11.017. [DOI] [PubMed] [Google Scholar]
- 63.Zehbe I., Tachezy R., Mytilineos J., Voglino G., Mikyskova I., Delius H., Marongiu A., Gissmann L., Wilander E., Tommasino M. Human papillomavirus 16 E6 polymorphisms in cervical lesions from different European populations and their correlation with human leukocyte antigen class II haplotypes. Int. J. Cancer. 2001;94:711–716. doi: 10.1002/ijc.1520. [DOI] [PubMed] [Google Scholar]
- 64.Schrank T.P., Landess L., Stepp W.H., Rehmani H., Weir W.H., Lenze N., Lal A., Wu D., Kothari A., Hackman T.G., et al. Comprehensive Viral Genotyping Reveals Prognostic Viral Phylogenetic Groups in HPV16-Associated Squamous Cell Carcinoma of the Oropharynx. Mol. Cancer Res. 2022;20:1489–1501. doi: 10.1158/1541-7786.MCR-21-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lang Kuhs K.A., Faden D.L., Chen L., Smith D.K., Pinheiro M., Wood C.B., Davis S., Yeager M., Boland J.F., Cullen M., et al. Genetic variation within the human papillomavirus type 16 genome is associated with oropharyngeal cancer prognosis. Ann. Oncol. 2022;33:638–648. doi: 10.1016/j.annonc.2022.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lewis J.S., Jr., Mirabello L., Liu P., Wang X., Dupont W.D., Plummer W.D., Pinheiro M., Yeager M., Boland J.F., Cullen M., et al. Oropharyngeal Squamous Cell Carcinoma Morphology and Subtypes by Human Papillomavirus Type and by 16 Lineages and Sublineages. Head Neck Pathol. 2021;15:1089–1098. doi: 10.1007/s12105-021-01318-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lechner A., Schlosser H.A., Thelen M., Wennhold K., Rothschild S.I., Gilles R., Quaas A., Siefer O.G., Huebbers C.U., Cukuroglu E., et al. Tumor-associated B cells and humoral immune response in head and neck squamous cell carcinoma. Oncoimmunology. 2019;8:1535293. doi: 10.1080/2162402X.2018.1535293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wieland A., Patel M.R., Cardenas M.A., Eberhardt C.S., Hudson W.H., Obeng R.C., Griffith C.C., Wang X., Chen Z.G., Kissick H.T., et al. Defining HPV-specific B cell responses in patients with head and neck cancer. Nature. 2021;597:274–278. doi: 10.1038/s41586-020-2931-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are contained within the article and Supplementary Materials.