Abstract
Aromatic amino and nitro compounds are potent carcinogens found in the environment that exert their toxic effects by reacting with DNA following metabolic activation. One important adduct is N-(deoxyguanosin-8-yl)-2-acetylaminofluorene (dG-AAF), which has been extensively used in studies of the mechanisms of DNA repair and mutagenesis. Despite the importance of dG-AAF adducts in DNA, an efficient method for its incorporation into DNA using solid-phase synthesis is still missing. We report the development of a modified ‘ultra-mild’ DNA synthesis protocol that allows the incorporation of dG-AAF into oligonucleotides of any length accessible by solid-phase DNA synthesis with high efficiency and independent of sequence context. Key to this endeavor was the development of improved deprotection conditions (10% diisopropylamine in methanol supplemented with 0.25 M of β-mercaptoethanol) designed to remove protecting groups of commercially available ‘ultra-mild’ phosphoramidite building blocks without compromising the integrity of the exquisitely base-labile acetyl group at N8 of dG-AAF. We demonstrate the suitability of these oligonucleotides in the nucleotide excision repair reaction. Our synthetic approach should facilitate comprehensive studies of the mechanisms of repair and mutagenesis induced by dG-AAF adducts in DNA and should be of general use for the incorporation of base-labile functionalities into DNA.
INTRODUCTION
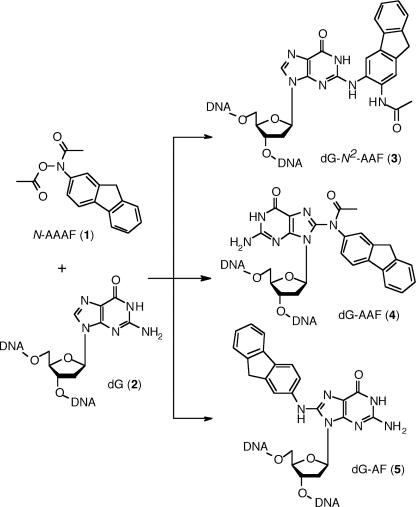
The preparation of defined single site-specific DNA damages is often the limiting step in understanding the repair characteristics or the mutagenic properties of a given lesion. The toxicological studies of potent aromatic nitro or amino carcinogens present in cooked food, tobacco smoke or diesel exhaust (1–3) illustrate this point; they are typically performed by exposing cells or oligonucleotides to the compound of interest, usually in an activated form (4–9). These approaches, however, lead invariably to inextricable mixtures of DNA lesions containing multiple and heterogeneous adducts at random sites, and are of limited use for studies concerning the molecular mechanisms underlying mutagenesis or DNA repair. For instance, N-acetoxy-2-acetylaminofluorene (N-AAAF) (1), a compound commonly used as reference in mutagenesis studies, is known to react predominantly with 2′-deoxyguanosine (dG) (2) residues in DNA to produce at least three different adducts (2,10–12) (Figure 1): N-(deoxyguanosin-N2-yl)-2-acetylaminofluorene (dG-N2-AAF) (3), N-(deoxyguanosin-8-yl)-2-acetylaminofluorene (dG-AAF) (4) and N-(deoxyguanosin-8-yl)-2-aminofluorene (dG-AF) (5). To generate defined single AAF adducts, the commonly used strategy consists in the treatment of a short oligonucleotide containing a unique reactive dG residue with N-AAAF (6,13–16). Although this approach is limited in terms of accessible sequences and yields to heterogeneous mixtures of adducts (11,12,17), it has been used in conjunction with extensive high-performance liquid chromatography (HPLC) purification to prepare defined single dG-AAF adducts in oligonucleotides containing up to three guanine residues (18–21). Biochemical studies using these well-defined damaged oligonucleotides have allowed invaluable insights into the impact of these lesions on DNA structure (22–24), replication (24–26), mutagenesis (14,15,20,27–29) and repair (6,19,30,31). Despite these advances, the generation of AAF-containing oligonucleotides by solid-phase DNA synthesis would greatly facilitate further biological investigations. Early studies along those lines quickly revealed that the ammonia deprotection step required after solid-phase synthesis compromised the integrity of the N8 acetyl group of the dG-AAF adduct (32). In 1993, Zhou and Romano reported the successful incorporation of an Fmoc-protected dG-AAF phosphoramidite in a 12mer oligonucleotide (33,34). The use of Fmoc as a protecting group for the exocyclic amines of all the bases, made it possible to perform a mildly basic deprotection step, devised to preserve the N8 acetyl group of dG-AAF (33). Unfortunately, the fact that Fmoc-protected phosphoramidites are not commercially available and that their preparation is cumbersome has prevented this method from finding widespread use. The post-synthetic modification strategy has thus so far remained the standard procedure for the preparation of AAF-modified oligonucleotides (21,29,35). In the context of our studies on damage recognition and catalysis in nucleotide excision repair (NER) (36–38), we wished to develop a general approach for the synthesis of oligonucleotides containing defined, site-specific dG-AAF adducts. Our goal required two main issues to be resolved: (i) the efficient preparation of C8-amine and acetylamine adducts of dG and (ii) the development of a general and straightforward solid-phase DNA synthesis protocol for the incorporation of dG-AAF residues that does not compromise the integrity of the base-labile N8 acetyl group. We (39) and some others (40,41) have previously described a solution to the first issue by preparing arylamine adducts at the C8 position of dG using a Buchwald–Hartwig coupling reaction between various aromatic amines and a protected 8-bromo-2′-deoxyguanosine. We have further established that these adducts can be specifically acetylated at the N8 position and can be suitably protected for DNA synthesis (39). Here, we describe a new protocol for the site-specific incorporation of exquisitely base-labile residues into DNA and its application for the synthesis of dG-AAF-containing oligonucleotides, ranging from 9mer to 120mer. Our approach allows the generation of dG-AAF and dG-AF modified oligonucleotides of any length and sequence. As an example of an application, these oligonucleotides are incorporated into plasmids and the repair efficiencies of dG-AAF and dG-AF residues were compared. The approach described here should be generally applicable for the preparation of oligonucleotides containing base-labile modifications.
Figure 1.
Commonly formed adducts of N-acetoxy-2-acetylaminofluorene with 2′-deoxyguanosine. N-AAAF can react with dG residues in DNA to form at least three different adducts: 3-(deoxyguanosin-N2-yl)-2-acetylaminofluorene (3) (dG-N2-AAF); N-(deoxyguanosin-8-yl)-2-acetylaminofluorene (4) (dG-AAF); and N-(deoxyguanosin-8-yl)-2-aminofluorene (5) (dG-AF). dG, dG-N2-AAF and dG-AF are believed to have the base mainly in the syn conformation, while the anti conformation is favored in dG-AAF.
MATERIALS AND METHODS
Reagents and equipment
Chemicals and solvents were purchased from Fluka-Sigma-Aldrich. Reagents for DNA synthesis were purchased from Applied Biosystem, except for 5-ethylthio-1H-tetrazole, which was from Sigma and was recrystallized from dry toluene before use. The ‘ultra-mild’ phosphoramidites were available from Glen Research and the 1000 Å ‘Q-columns’ from Biosearch Technologies. DNA syntheses were performed on a PerSeptive Biosystems Expedite 8909. HPLC analysis and purifications were performed on a JASCO system equipped with a BGB Analytik column: Nucleosil 100 Å, C18, 5 μm, 250 × 4.0 mm2. The C18-SepPak cartridges were from Micropore. Snake venom phosphodiesterase I (SVPD) was from Worthington Biochemical; calf intestine phosphatase, T4 PNK, T4 DNA polymerase, T4 DNA ligase from New England Biolabs. The sequenase enzyme (T7 DNA polymerase) was purchased from USB and [α-32P]dCTP from Amersham.
NMR spectra were recorded on a Bruker ARX-300 MHz, the HR-MALDI on an Ionspec FT MS Ultima. The 9mer and 24mer were analyzed by nano-ESI on a Micromass quadrupole time-of-flight (Q-TOF) mass spectrometer in negative mode; the 60mer, 90mer and 120mer were analyzed by LC-ESI-MS in negative mode on a Q-TOF-Ultima coupled to Cap-LC (see Supplementary Material). The mass deconvolution was realized with the MaxEnt1 software.
Synthesis of the iPrPac-dG-AAF building block
(5′-O-Dimethoxytrityl)-N-[(N2-isopropylphenoxyacetyl)-deoxyguanosin-8-yl]-2-acetylaminofluorene
To N-[(N2-isopropylphenoxyacetyl)-deoxyguanosin-8-yl]-2-acetylaminofluorene (6) (39) (671 mg, 1.0 mmol) dissolved in pyridine (5 ml), 4,4-dimethoxytrityl chloride (514 mg, 1.5 mmol) was added. The reaction mixture was stirred for 2 h at room temperature, concentrated and purified by chromatography on silica gel (CH2Cl2/MeOH 40:1) to provide 741 mg (0.77 mmol, 76%) of a slightly yellow powder.
Rf = 0.50 (CH2Cl2/MeOH 10:1).
1H NMR [DMSO-d6, δ (p.p.m.)]: 11.76 (s, 1H, N1-H), 11.30 (s, 1H, N2-H), 7.84–7.95 (m, 2H, AF-H2), 7.44–7.78 (m, 3H, AF-H3), 7.25–7.41 (m, 4H, AF-H2 + DMTr-H2), 7.05–7.21 (m, 9H, Pac-H2 + DMTr-H7), 6.89 (d, J = 8.7, 2H, Pac-H2), 6.56–6.73 (m, 4H, DMTr-H4), 6.32 (dd, J = 6.9, 7.2, 1H, C1′-H), 5.19 (d, J = 5.3, 1H, C3′-OH), 4.79 (m, 2H, Pac-CH2), 4.61 (m, 1H, C3′-H), 4.07 (m, 1H, C4′-H), 3.86 (m, 2H, AF-C9-H2), 3.66 (s, 3H, OMe), 3.64 (s, 3H, OMe), 3.39 (m, 1H, C5′-H), 3.16 (m, 1H, C5′-H), 2.99 (m, 1H, C2′-H), 2.85 (sept, J = 6.9, 1H, iPr-H), 2.30 (m, 1H, C2′-H), 2.11 (br s, 3H, N-Ac), 1.18 (s, 3H, iPr-CH3), 1.16 (s, 3H, iPr-CH3).
13C-NMR [DMSO-d6, δ (p.p.m.)]: 170.5, 157.8, 157.7, 155.6, 154.2, 147.3, 146.4, 144.7, 143.9, 143.2, 141.4, 140.0, 135.5, 129.6, 129.5, 127.7, 127.2, 127.1, 126.9, 126.7, 126.2, 124.9, 123.7, 120.3, 120.1, 119.1, 114.4, 112.6, 112.5, 86.8, 85.1, 83.7, 78.9, 70.1, 66.5, 64.9, 54.8, 54.7, 37.5, 36.3, 32.4, 30.5, 23.9, 22.5.
HR-MALDI (m/z): calcd for C57H54N6O9·Na+ 989.3845; found, 989.3831.
[3′-O-(2′-Cyanoethoxydiisopropylaminophosphino)]-(5′-O-dimethoxytrityl)-N-[(N2-isopropylphenoxyacetyl)-deoxyguanosin-8-yl]-2-acetylaminofluorene (7)
To (5′-O-dimethoxytrityl)-N-[(N2-isopropylphenoxyacetyl)-deoxyguanosin-8-yl]-2-acetylaminofluorene (679 mg, 0.7 mmol) in CH2Cl2 (5 ml) N-ethyldiisopropylamine (480 μl, 2.8 mmol) and 2-cyanoethoxydiisopropylchloro-phosphoramidite (313 μl, 1.4 mmol) were added. The reaction mixture was stirred for 2 h at room temperature, concentrated to dryness and redissolved in the miminal amount of CH2Cl2. 40 ml of hexane was then added under smooth stirring and the product was further precipitated for 2 h at −20°C. The clear supernatant was then removed and the residue was purified by chromatography on silica gel (CH2Cl2/MeOH 40:1) to provide 710 mg (0.61 mmol, 86%) of a slightly yellow powder.
Rf = 0.54 (CH2Cl2/MeOH 10:1).
1H NMR [DMSO-d6, δ (p.p.m.)]: 11.80 (s, 1H, N1-H), 10.97 (s, 1H, N2-H), 7.86–7.94 (m, 2H, AF-H2), 7.50–7.81 (m, 3H, AF-H3), 7.26–7.41 (m, 4H, AF-H2 + DMTr-H2), 7.09–7.19 (m, 9H, Pac-H2 + DMTr-H7), 6.88–6.93 (m, 2H, Pac-H2), 6.60–6.72 (m, 4H, DMTr-H4), 6.34 (m, 1H, C1′-H), 4.65–4.88 (m, 3H, Pac-CH2 + C3′-H), 4.17 (m, 1H, C4′-H), 3.81–3.95 (m, 2H, AF-C9-H2), 3.61–3.69 (m, 7H, 2xOMe + C5′-H2), 3.37–3.58 (m, 4H, O-CH2-CH2-CN), 3.13–3.27 (m, 2H, C5′-H2 + C2′-H), 2.84 (sept, J = 6.9, 1H, Pac-iPr-H), 2.59–2.75 (m, 2H, N-iPr2-H2), 2.39 (m, 1H, C2′-H), 2.10 (br s, 3H, N-Ac), 0.94–1.29 (m, 18H, Pac-iPr-(CH3)2 + N-iPr2-(CH3)4).
13C-NMR [DMSO-d6, δ (p.p.m.)]: 157.8, 157.7, 155.6, 144.7, 144.6, 144.5, 143.2, 141.3, 141.1, 140.0, 138.9, 135.4, 135.4, 129.6, 129.5, 127.7, 127.6, 127.6, 127.6, 127.5, 127.3, 127.1, 127.0, 127.0, 126.7, 126.3, 126.2, 125.0, 120.1, 118.8, 118.6, 118.5, 118.0, 114.4, 114.3, 112.7, 112.6, 107.6, 85.3, 85.2, 60.3, 58.1, 58.0, 57.2, 57.1, 56.4, 54.8, 54.8, 54.7, 46.1, 44.4, 44.4, 42.6, 42.5, 36.3, 32.5, 32.4, 24.2, 24.1, 24.0, 24.0, 23.9, 23.9, 23.8, 22.5, 22.5, 22.4, 20.9, 19.7, 19.6, 19.5, 19.5, 19.3, 19.2, 19.1, 19.1, 19.0, 18.7.
31P-NMR {1H} NMR [DMSO-d6, δ (p.p.m.)]: 148.4 (s), 148.0 (s).
HR-MALDI (m/z): calcd for C66H71N8O10P·Na+ 1189.4923; found, 1189.4907.
Modified ‘ultra-mild’ solid-phase DNA synthesis and deprotection conditions
The sequences prepared, with the exception of the 9mer, were designed to be complementary to the (+) strand of pBluescript II SK and are as follows: 9mer, d(CGATG*CAGT); 24mer, d(GTATCGATAAG*CTTGATATCGAAT); 60mer, d(CCCCCCTCGAGGTCGACGGTATCGATAAG*CTTGATATCGAATTCCTGCAGCCCGGGGGAT); 90mer, d(CAAAAGCTGGGTACCGGGCCCCCCCTCGAGGTCGACGGTATCGATAAG*CTTGATATCGAA TTCCTGCAGCCCGGGGGATCCACTAGTTCT); and 120mer, d(CGTAATACGACTCACTATAGGGCGAATTGGGTACCGGGCCCCCCCTCGAGGTCGACGGTAT CGATAAG*CTTGATATCGAATTCCTGCAGCCCGGGGGATCCACTAGTTCTAGAGCGGCCG); where G* denotes dG (unmodified), dG-AAF or dG-AF. All DNA syntheses were performed on 1 μM scale, 1000 Å ‘Q-columns’. The ‘ultra-mild’ phosphoramidites (T, Ac-dC, Pac-dA and iPrPac-dG) were dissolved to 0.1 M in CH3CN. The iPrPac-dG-AAF phosphoramidite (7) was dissolved to 0.1 M in CH2Cl2 and its coupling time during the DNA synthesis extended to 12 min. A 0.25 M solution of 5-(ethylthio)-1H-tetrazole in CH3CN was used as an ‘activator’. A 0.5 M solution of phenoxyacetyl anhydride [prepared according to a published procedure (42)] in THF was used as ‘capping A’ solution with an extended capping time of 6 s and double amount of reagent delivered on the solid support in comparison with the standard 1 μM scale DNA synthesis protocol. A 3% (v/v) dichloroacetic acid solution in CH2Cl2 was used in the deblocking step. The final 5′-DMTr protective group was retained for all the syntheses (‘DMTr-ON’ synthesis). After completion of the synthesis, the solid support was dried and treated overnight at 55°C with a solution containing 10% (v/v) of diisopropylamine (iPr2NH) and 0.25 M of β-mercaptoethanol in MeOH. The supernatant containing the released oligonucleotide was decanted and concentrated, taken up in 1 ml of 1 M triethylamoniumacetate (TEAA) buffered at pH 7, passed through a 0.45 μm filter and purified by HPLC.
HPLC purification
All HPLC elutions were performed at a flow rate of 1 ml/min, with the following gradient: linear 5–20% B over 15 min, linear 20–75% B until 30 min, isocratic 75% B until 35 min, linear 75–5% B until 36 min, isocratic 5% B until 40 min; buffer A, 0.1 M TEAA (pH 7); and buffer B, CH3CN. The peak of the ‘DMTr-ON’ oligonucleotide, eluting between 20 and 22 min, was collected (see Supplementary Material), concentrated and treated with an 80% acetic acid solution for 40 min at room temperature to remove the 5′-DMTr group. After concentration, the oligonucleotide was redissolved in 1 ml of 1 M TEAA buffered at pH 7 and repurified on HPLC. The major peak—eluting between 15 and 17 min—was collected, concentrated, redissolved in 0.1 M TEAA (pH 7), desalted on a C18-SepPak cartridge and lyophilized. The lyophilizate was redissolved in 300–400 μl of milli-Q water to typically yield concentrations of 100–600 pmol/μl (μM). With protocols optimized for the generation of highly pure oligonucleotides, yields were as follows: 9-AAF, 57–103 nmol (6–10%); 24-AAF, 24–62 nmol (2.6%); 60-AAF, 28–57 nmol (3–6%); 90-AAF, 15 nmol (1.5%); and 120-AAF, 0.2 nmol (0.02%). For the preparation of the corresponding dG-AF adduct, the oligonucleotide containing the dG-AAF modification was incubated for 3 h at 37°C in a 1 M solution of NaOH containing 0.25 M of β-mercaptoethanol, according to a published procedure (12). The solution was then neutralized with 1 M HCl and directly desalted on a C18-SepPak cartridge and lyophilized.
Enzymatic digestion analysis
The modified DNA (25 μl; between 2 and 10 nmol) was incubated for 7 h at 37°C with 20 μl of snake venom phosphodiesterase (2 U) in a buffered solution containing 20 mM Tris–HCl, 10 mM MgCl2, 100 mM NaCl at pH 9 (5 μl of a pre-made 10× buffer). The reaction mixture was then incubated for 15 min at 37°C with 0.5 μl (5 U) of calf intestine phosphatase and directly injected into the HPLC using the gradient described above. For the digestion of oligonucleotide containing the dG-AF adduct, the addition of 1 mM of DTT to the digestion buffer was necessary to avoid complete oxidation and degradation of the dG-AF nucleoside (5) (32,43,44). The resulting peaks were identified by co-injection with the corresponding standards and eluted at the following time periods: dC (3.4 min), dG (5.5 min), T (6.3 min), dA (8.2 min), dG-AAF (24.2 min), dG-AF (24.8 min) and iPrPac-dG-AAF (30.8 min).
Primer extension and in vitro NER assay
An aliquot of 120 pmol of 24-AAF (or 24-AF) oligonucleotide was 5′-phosphorylated by incubation with 20 U of T4 PNK enzyme and 2 mM of ATP for 2 h. After annealing with 30 pmol of single-stranded pBluescript II SK+, further incubation with dNTPs, T4 DNA polymerase and T4 DNA ligase (45) yielded covalently closed circular DNA containing a single AAF (or AF) adduct. The closed circular DNA was purified by caesium chloride/ethidium bromide density gradient centrifugation, by consecutive butanol extractions to remove the ethidium bromide and finally concentrated on a Centricon YM-30 (Millipore). In addition to the published purification procedure (45), the plasmid was repurified by 1% agarose gel electrophoresis to remove traces of the 24-AAF (or 24-AF) oligonucleotides that remained associated with the plasmid during CsCl purification. The band of the closed circular plasmid was excised from the agarose gel and purified with the QIAquick Gel Extraction Kit from Qiagen. The dG-AAF and dG-AF containing plasmids were aliquoted and stored at −80°C. HeLa whole-cell extracts were prepared as described (46), aliquoted and stored at −80°C. The in vitro NER assay was performed as described (45) and the excised NER products detected after annealing with the following complementary 3′-phosphorylated oligonucleotide: d(GGGGGATATCAAGCTTATCGATACCGTCGACCTCG). Subsequent fill-in reaction of [α-32P]dCTP was performed in the presence of sequenase enzyme as described previously (45). The reactions were analyzed on a 14% denaturing polyacrylamide sequencing gel (migrating for 2 h at 50 W) and exposed overnight to a BioMax MS (Kodak) film at −80°C.
RESULTS AND DISCUSSION
Development of an improved ‘ultra-mild’ deprotection procedure
The exquisite base sensitivity of the acetyl group in dG-AAF provides a challenge for the conditions used in the deprotection step following the solid-phase DNA synthesis. Previous studies have revealed that this N8 acetyl group is readily cleaved upon treatment with ammonia (32) or K2CO3/MeOH under the standard deprotection conditions described for ‘ultra-mild’ DNA synthesis (33,47). To date, the only solution to this problem was provided by Zhou and Romano (33), who used Fmoc as protecting group for the exocylic amines of all the phosphoramidites and 1:9 piperidine/MeOH or 1:1 diisopropylamine/MeOH as a mildly basic deprotection solution. Since the Fmoc-protected building blocks are not commerically available and are not trivial to synthesize, we sought to develop conditions that would cleave the protecting groups of the commercial ultra-mild phosphoramidites (iPrPac-dG, Pac-dA and Ac-dC) but would retain the acetyl group on dG-AAF. As a first step toward this goal, we had previously reported that treatment of iPrPac protected dG-AAF (6) with a 5% diisopropylamine (iPr2NH) solution in methanol (12 h, room temperature), led to deprotection of the N2-iPrPac group without detectable loss of the N8 acetyl group (39). This suggested that deprotection of oligonucleotides prepared with the commercial ultra-mild phosphoramidites could be achieved under conditions compatible with the maintenance of the acetyl group of AAF. We, therefore, assessed further the stability of the iPrPac protected dG-AAF (6) under prolonged incubation with the various reagents used in DNA synthesis, to determine whether additional modifications were necessary. Preliminary tests showed that iPrPac-dG-AAF (6) was stable toward the capping reagent [phenoxyacetic anhydride was used rather than acetic anhydride to avoid transamidation reactions on the Pac/iPrPac protected exocyclic amines (48)], the activator [5-(ethylthio)-tetrazole was used rather than tetrazole to avoid precipitation and for improved coupling yields] and the oxidizing solutions were used during the DNA synthesis (data not shown). However, the modified nucleoside (6) underwent complete depurination almost instantly upon incubation with the deblocking solutions: 3–5% (v/v) trichloroacetic acid or dichloroacetic acid (DCA) in CH2Cl2. Propensity similar to depurination had also been described for Fmoc-dG-AAF on the mononucleoside level (33). Further tests demonstrated, however, that the use of 3% (v/v) DCA/CH2Cl2 did not lead to any depurination of iPrPac-dG-AAF during solid-phase DNA synthesis and this mixture was thus used in the deblocking step.
The last issue to be addressed was the choice of a solid-support. Initial experiments revealed that, whereas a solution of 0.05 M K2CO3/MeOH readily released an oligonucleotide from a solid support with a succinimid linker, a 5% iPr2NH solution in methanol led only to a marginal recovery of the expected oligonucleotide. Resorting to the more labile hydroquinone-based ‘Q-support’ (49), however, adequately addressed this issue. We then carried out a solid-phase synthesis of a model 9mer oligonucleotide using all of these modifications (summarized in Table 1). Following deprotection and purification of the oligonucleotide by using HPLC, nano-ESI MS revealed that all the protective groups of the bases and of the phosphate backbone had been released (Table 2 and Supplementary Material), thus validating our synthesis and deprotection conditions.
Table 1.
Modified ‘ultra-mild’ DNA synthesis
| Modified ‘ultra-mild’ protocol | |
|---|---|
| Solid support | Hydroquinone based (‘Q-column’) |
| Phosphoramidites | dT, Ac-dC, Pac-dA, iPrPac-dG (0.1 M in CH3CN) |
| Modification | iPrPac-dG-AAF (0.1 M in CH2Cl2) |
| • Coupling time extended to 12 min | |
| Activator | 5-(Ethylthio)-1H-tetrazole (0.25 M in CH3CN) |
| Oxidizer | I2 (0.1 M in THF/pyridine/H2O) |
| CappingA | iPrPac anhydride (0.5 M in THF) |
| • Capping time extended to 6 s | |
| • Delivered double volume of solution | |
| Capping B | 1-Methylimidazole (2 M in THF) |
| Deblock | DCA (3%, v/v) in CH2Cl2 |
| Deprotection | iPr2NH (10%, v/v), 0.25 M β-mercaptoethanol in MeOH |
| • Overnight treatment at 55°C |
Table 2.
MS analyses of the synthesized oligonucleotides
| Oligonucleotide | Calcd mass | Measured |
|---|---|---|
| 9-Unmod | 2738.9 | 2739 |
| 9-AAF | 2960.2 | 2960 |
| 9-AF | 2918.1 | 2918 |
| 24-AAF | 7612.2 | 7611 |
| 24-AF | 7570.1 | 7569 |
| 60-AAF | 18 674.4 | 18 672 |
| 90-AAF | 27 928.4 | 27 927 |
| 120-AAF | 37 262.4 | 37 262 |
Preparation and analysis of dG-AAF and dG-AF modified 9mer oligonucleotides
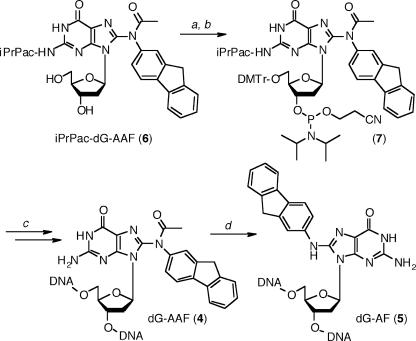
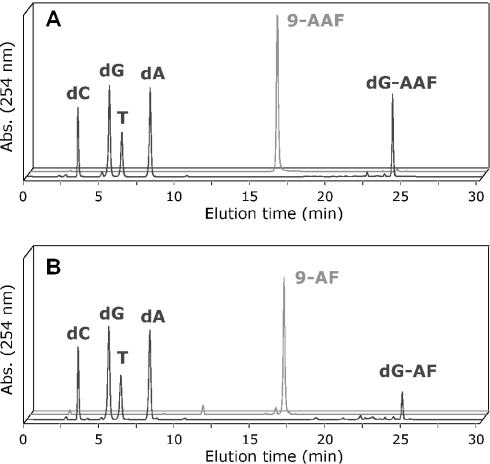
With the improved ultra-mild DNA synthesis and deprotection protocol in hand, we assessed its usefulness to incorporate dG-AAF residues into oligonucleotides. To be consistent with the ‘ultra-mild’ iPrPac protected dG, we had previously reported the preparation of N2-iPrPac protected dG-AAF (6) (39). 5′-Dimethoxytrityl protection of iPrPac-dG-AAF (6) and subsequent 3′-phosphitylation to (7) were performed according to the standard procedures (Figure 2). The iPrPac-dG-AAF phosphoramidite (7) was dissolved to 0.1 M in CH2Cl2, which provided improved stability and coupling efficiency over acetonitrile. We prepared then a 9mer (9-AAF) containing a single dG-AAF adduct using the conditions summarized in Table 1, and purified the oligonucleotide by HPLC. The HPLC trace revealed the homogeneity and the purity of the dG-AAF modified oligonucleotide (Figure 3A). Treatment of 9-AAF for 3 h at 37°C in a 1 M NaOH solution containing 0.25 M of β-mercaptoethanol (12) led to selective cleavage of the N8 acetyl and to the formation of corresponding dG-AF modified 9mer (9-AF). The 9-AAF and 9-AF oligomers eluted at two distinct retention times on HPLC (Figure 3), confirming the effective removal of the acetyl group from 9-AAF to yield the less polar 9-AF (12), and proving that the purified 9-AAF was really exempt of decomposition into the deacetylated 9-AF. Nano-ESI analyses confirmed the identity and purity of the 9-AAF and 9-AF (Table 2 and Supplementary Material).
Figure 2.
Site-specific incorporation of dG-AAF and dG-AF into oligonucleotides. Reaction conditions: (a) DMTr-Cl, pyridine (76%); (b) N-ethyldiisopropylamine, 2-cyanoethoxydiisopropylchloro-phosphoramidite, CH2Cl2 (86%); (c) modified ‘ultra-mild’ DNA synthesis as shown in Table 1; and (d) 1 M NaOH, 0.25 M β-mercaptoethanol.
Figure 3.
Nucleoside composition analyses of 9-AAF (A) and 9-AF (B). The oligonucleotides 9-AAF and 9-AF were digested with snake venom phosphodiesterase and calf intestine phosphatase and the resulting nucleosides were separated using the HPLC gradient described in Materials and Methods. The elution times were as follows: dC = 3.4 min, dG = 5.5 min, T = 6.3 min, dA = 8.2 min, 9-AAF = 16.3 min, 9-AF = 16.7 min, dG-AAF = 24.2 min and dG-AF = 24.8 min.
We performed an enzymatic digestion analysis of the 9-AAF and 9-AF oligonucleotides to further prove the presence of dG-AAF and dG-AF, respectively, and to rule out the possibility that the additional acetyl group in 9-AAF was located on a dC residue because of incomplete deprotection of an acetyl-dC. The modified oligonucleotides were incubated with a large excess of snake venom phosphodiesterase I at 37°C for 7 h, since it was described that this enzyme is partially blocked at the site of the dG-AAF lesion (50). Following incubation with calf intestine phosphatase, the resulting nucleosides were separated by HPLC and the different peaks assigned by co-elution with the corresponding reference compounds (Figure 3). These studies confirmed the unique presence of dG-AAF in 9-AAF and of dG-AF in 9-AF, in addition to the four normal nucleosides. This thus demonstrated that the additional acetyl group measured by nano-ESI in 9-AAF, was indeed located on the expected N8 position of the modified dG adduct. Taken together, the analytical data fully validate the modified ‘ultra-mild’ deprotection protocol for the preparation of dG-AAF and dG-AF modified 9mer.
Optimization of the deprotection conditions and preparation of 24-AAF, 24-AF, 60-AAF, 90-AAF and 120-AAF
When we applied the same protocol for the preparation of longer oligonucleotides (24mer to 120mer) containing a single dG-AAF modification, we obtained, to our surprise, HPLC elution profiles presenting broader peaks, suggesting that a heterogeneous mixture of oligonucleotides had been formed. Indeed, MS analysis confirmed that we had purified a combination of oligonucleotides containing up to four additional acetyl groups. Since the Ac-dC residues are the only plausible source of acetyl groups, it appeared that our deprotection conditions were not strong enough to remove all the protecting groups from these residues.
We decided thus to use harsher conditions and to treat the oligonucleotides with a methanolic solution containing 10% (v/v) iPr2NH and 0.25 M β-mercaptoethanol for 20 h at 55°C. β-Mercaptoethanol was added to avoid aerial oxidation of dG-AAF arising during long incubation at higher temperatures (43,44). Although preliminary stability tests at the nucleoside level with iPrPac-dG-AAF (6) revealed some cleavage of the N8 acetyl group of dG-AAF under these conditions, no decomposition was observed at the oligonucleotide level. Using these conditions, we could indeed prepare a 24mer, a 60mer, a 90mer and even a 120mer containing each a single dG-AAF modification, with yields ranging from 10% (103 nmol, 9-AAF) to 0.05% (0.5 nmol, 120-AAF) from a 1 μM scale DNA synthesis under conditions optimized to obtain highly pure oligonucleotides. The identity of each of these oligonucleotides was confirmed by MS analysis (Table 2 and Supplementary Material), thus validating this modified ‘ultra-mild’ DNA synthesis and deprotection protocol (summarized in Table 1) as the most straightforward method to achieve the single site-specific incorporation of dG-AAF into oligonucleotides of any desired length or sequence.
Test of the dG-AAF and dG-AF substrates during an in vitro NER assay
To demonstrate the usefulness of the dG-AAF- and dG-AF-modified oligonucleotides, we used them as substrates for in vitro NER assays. This assay consists in mixing a plasmid or an oligonucleotide containing a single lesion, with a NER proficient cell extract. The excision reaction (36–38) results in the release in solution of short oligonucleotides (24mer to 32mer) containing the lesion, which can be indirectly detected with an appropriate ‘fill-in’ labeling (see Figure 4 for details). We wished to use, directly from the DNA synthesizer, the 120mer containing the dG-AAF modification as substrate for the NER reaction. Although 120-AAF was devised to meet the size and damage-position requirements to allow the assembly of the NER machinery (51), we were unable to detect any significant signal of repair activity after incubation with HeLa cell extract (data not shown). We do not presently know whether the 120mer was too short or whether an unrelated activity in the cell extract such as a non-specific nuclease or a protein that binds to DNA ends (52–54) prevented the NER reaction from occurring.
Figure 4.
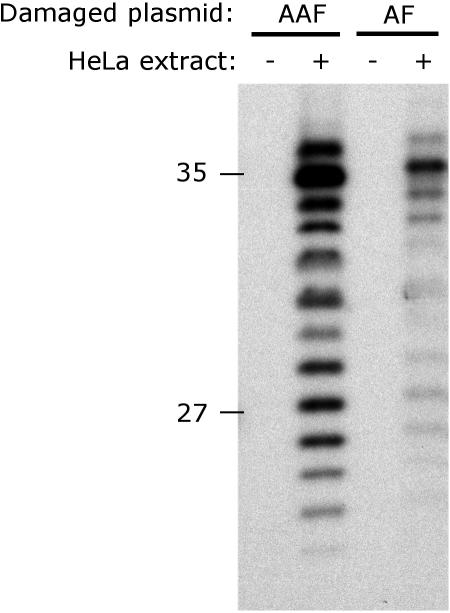
NER dual incision activity on dG-AAF and dG-AF-containing plasmids. Plasmids containing site-specific dG-AAF or dG-AF residues were incubated with cell extracts prepared from HeLa cells. The 24mer to 32mer excision products containing dG-AAF or dG-AF were detected by annealing to a complementary oligonucleotide with a 5′-GpGpGpG overhang, which served as a template for end-labeling with [α-32P]dCTP with sequenase. The reaction products were resolved on a 14% denaturing polyacrylamide gel. An MspI digest of pBR322 was used as a size marker and the position of the 27 and 35 nt bands are indicated.
We, therefore, turned our focus on incorporating the dG-AAF and dG-AF adducts into a plasmid to study NER according to a well-established protocol (45). The 24-AAF and 24-AF oligonucleotides were incorporated into pBluescriptII SK+ plasmid using primer extension by T4 DNA polymerase, ligation with T4 DNA ligase and purification over a caesium chloride/ethidium bromide density gradient as described by Shivji et al. (45). We included an additional agarose gel purification step to remove the excess of 24-AAF (or 24-AF) primer, which was found to remain associated with the plasmid during CsCl gradient purification. Although this extra purification step was not required for plasmids containing a cisplatin lesion [(45) and data not shown], it is likely that the aromatic nature of the dG-AAF (or dG-AF) adducts prevented the primer from being completely eliminated during the course of the protocol established by Shivji et al. (45). The plasmid containing the lesion was then incubated with HeLa whole-cell extracts and the excised products of the NER reaction were detected with a ‘fill-in’ labeling to yield the NER-specific pattern of 28mer to 36mer oligonucleotides (Figure 4). We found that, while a non-damaged mock treated plasmid did not yield any excised product [(45,55,56) and data not shown], the plasmid containing dG-AAF was incised at least 20-fold more efficiently than the one containing dG-AF (Figure 4, compare lanes 2 and 4). Although it had been demonstrated previously that dG-AAF adducts are repaired more efficiently then dG-AF by the human NER machinery (9,57), this surprisingly represents the first direct comparison of the incision rates of these two lesions. Our approach allows thus to generate any desired site-specifically modified dG-AAF or dG-AF plasmids, making it possible to address the influence of the sequence context on the structure, mutagenesis and repair activities of AAF and AF adducts.
SUMMARY AND CONCLUSIONS
We have reported the development of a modified ‘ultra-mild’ DNA synthesis and deprotection protocol and its application to the synthesis of oligonucleotides containing dG-AAF residues. Central to this protocol is the use of a 10% diispropylamine/0.25 M β-mercaptoethanol methanolic solution that is effective in removing the protecting groups from oligonucleotides prepared with the commercially available ‘ultra-mild’ phosphoramidites and in releasing them from Q-solid support, without cleaving the base-labile acetyl group at N8 of dG-AAF. We have fine-tuned this protocol such that it can be used for the synthesis of oligonucleotides up to 120 residues in length without any decrease in coupling yields or deprotection efficiency compared with the standard DNA synthesis conditions. The ability to generate oligonucleotides containing base-labile mutagenic adducts by solid-phase synthesis is a significant advance over the synthesis of these adducts by post-synthetic modification of DNA with carcinogens. It provides straightforward access to the well-defined substrates for mutagenesis and DNA repair studies. This protocol is now routinely used in our laboratory for the preparation of oligonucleotides containing modifications that require special care during the deprotection step.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
We are grateful to Rolf Häfliger and Walter Amrein (MS service, Department of Chemistry of the ETH Zürich) for the HR-MALDI analyses, to Serge Chesnov and Peter Hunziker (Protein Analysis Unit, University of Zürich) for the analysis of the 9mer and 24mer, and especially to Philipp Wenter, Luc Reymond and Stefan Pitsch (Department of Chemistry, EPF Lausanne) for their precious help with the analysis of the 60mer, 90mer and 120mer. This work was supported by the Swiss National Science Foundation, the Human Frontier Science Program Organization (HFSPO) and the EMBO Young Investigator Program. Funding to pay the Open Access publication charges for this article was provided by the HFSPO.
Conflict of interest statement. None declared.
REFERENCES
- 1.Beije B., Moller L. 2-Nitrofluorene and related compounds: prevalence and biological effects. Mutat. Res. 1988;196:177–209. doi: 10.1016/0165-1110(88)90019-x. [DOI] [PubMed] [Google Scholar]
- 2.Heflich R.H., Neft R.E. Genetic toxicity of 2-acetylaminofluorene, 2-aminofluorene and some of their metabolites and model metabolites. Mutat. Res. 1994;318:73–174. doi: 10.1016/0165-1110(94)90025-6. [DOI] [PubMed] [Google Scholar]
- 3.Schut H.A.J., Snyderwine E.G. DNA adducts of heterocyclic amine food mutagens: implications for mutagenesis and carcinogenesis. Carcinogenesis. 1999;20:353–368. doi: 10.1093/carcin/20.3.353. [DOI] [PubMed] [Google Scholar]
- 4.Kriek E., Miller J.A., Juhl U., Miller E.C. 8-(N-2-Fluorenylacetamido)guanosine, an arylamidation reaction product of guanosine and the carcinogen N-acetoxy-N-2-fluorenylacetamide in neutral solution. Biochemistry. 1967;6:177–182. doi: 10.1021/bi00853a029. [DOI] [PubMed] [Google Scholar]
- 5.Pierce J.R., Case R., Tang M.S. Recognition and repair of 2-aminofluorene- and 2-(acetylamino)fluorene-DNA adducts by UVRABC nuclease. Biochemistry. 1989;28:5821–5826. doi: 10.1021/bi00440a018. [DOI] [PubMed] [Google Scholar]
- 6.Hansson J., Munn M., Rupp W.D., Kahn R., Wood R.D. Localization of DNA repair synthesis by human cell extracts to a short region at the site of a lesion. J. Biol. Chem. 1989;264:21788–21792. [PubMed] [Google Scholar]
- 7.Gupta P.K., Pandrangi R.G., Lee M.-S., King C.M. Induction of mutations by N-acetoxy-N-acetyl-2-aminofluorene modified M13 viral DNA. Carcinogenesis. 1991;12:819–824. doi: 10.1093/carcin/12.5.819. [DOI] [PubMed] [Google Scholar]
- 8.van Oosterwijk M.F., Filon R., Kalle W.H., Mullenders L.H., van Zeeland A.A. The sensitivity of human fibroblasts to N-acetoxy-2-acetylaminofluorene is determined by the extent of transcription-coupled repair, and/or their capability to counteract RNA synthesis inhibition. Nucleic Acids Res. 1996;24:4653–4659. doi: 10.1093/nar/24.23.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Oosterwijk M.F., Filon R., de Groot A.J., van Zeeland A.A., Mullenders L.H. Lack of transcription-coupled repair of acetylaminofluorene DNA adducts in human fibroblasts contrasts their efficient inhibition of transcription. J. Biol. Chem. 1998;273:13599–13604. doi: 10.1074/jbc.273.22.13599. [DOI] [PubMed] [Google Scholar]
- 10.Kriek E. Persistent binding of a new reaction product of the carcinogen N-hydroxy-N-2-acetylaminofluorene with guanine in rat liver DNA in vivo. Cancer Res. 1972;32:2042–2048. [PubMed] [Google Scholar]
- 11.Fuchs R.P. Arylamidation and arylation by the carcinogen N-2-fluorenylacetamide: a sensitive and rapid radiochemical assay. Anal. Biochem. 1978;91:663–673. doi: 10.1016/0003-2697(78)90553-5. [DOI] [PubMed] [Google Scholar]
- 12.Shibutani S., Gentles R., Johnson F., Grollman A.P. Isolation and characterization of oligodeoxynucleotides containing dG-N2-AAF and oxidation products of dG-C8-AF. Carcinogenesis. 1991;12:813–818. doi: 10.1093/carcin/12.5.813. [DOI] [PubMed] [Google Scholar]
- 13.Johnson D.L., Reid T.M., Lee M.S., King C.M., Romano L.J. Preparation and characterization of a viral DNA molecule containing a site-specific 2-aminofluorene adduct: a new probe for mutagenesis by carcinogens. Biochemistry. 1986;25:449–456. doi: 10.1021/bi00350a026. [DOI] [PubMed] [Google Scholar]
- 14.Moriya M., Takeshita M., Johnson F., Peden K., Will S., Grollman A.P. Targeted mutations induced by a single acetylaminofluorene DNA adduct in mammalian cells and bacteria. Proc. Natl Acad. Sci. USA. 1988;85:1586–1589. doi: 10.1073/pnas.85.5.1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta P.K., Johnson D.L., Reid T.M., Lee M.S., Romano L.J., King C.M. Mutagenesis by single site-specific arylamine-DNA adducts. J. Biol. Chem. 1989;264:20120–20130. [PubMed] [Google Scholar]
- 16.O'Handley S.F., Sanford D.G., Xu R., Lester C.C., Hingerty B.E., Broyde S., Krugh T.R. Strutural characterization of an N-acetyl-2-aminofluorene (AAF) modified DNA oligomer by NMR, energy minimization and molecular dynamics. Biochemistry. 1993;32:2481–2497. doi: 10.1021/bi00061a005. [DOI] [PubMed] [Google Scholar]
- 17.Tang M.S., Lieberman M.W. Quantification of adducts formed in DNA treated with N-acetoxy-2-acetylaminofluorene or N-hydroxy-2-aminofluorene: comparison of trifluoroacetic acid and enzymatic degradation. Carcinogenesis. 1983;4:1001–1006. doi: 10.1093/carcin/4.8.1001. [DOI] [PubMed] [Google Scholar]
- 18.Koehl P., Burnouf D., Fuchs R.P. Construction of plasmids containing a unique acetylaminofluorene adduct located within a mutation hot spot. A new probe for frameshift mutagenesis. J. Mol. Biol. 1989;207:355–364. doi: 10.1016/0022-2836(89)90259-3. [DOI] [PubMed] [Google Scholar]
- 19.Seeberg E., Fuchs R.P. Acetylaminofluorene bound to different guanines of the sequence -GGCGCC- is excised with different efficiencies by the UvrABC excision nuclease in a pattern not correlated to the potency of mutation induction. Proc. Natl Acad. Sci. USA. 1990;87:191–194. doi: 10.1073/pnas.87.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shibutani S., Grollman A.P. On the mechanism of frameshift (deletion) mutagenesis in vitro. J. Biol. Chem. 1993;268:11703–11710. [PubMed] [Google Scholar]
- 21.Tan X., Suzuki N., Grollman A.P., Shibutani S. Mutagenic events in Escherichia coli and mammalian cells generated in response to acetylaminofluorene-derived DNA adducts positioned in the Nar I restriction enzyme site. Biochemistry. 2002;41:14255–14262. doi: 10.1021/bi0202878. [DOI] [PubMed] [Google Scholar]
- 22.Belguise-Valladier P., Fuchs R.P. Strong sequence-dependent polymorphism in adduct-induced DNA structure: analysis of single N-2-acetylaminofluorene residues bound within the NarI mutation hot spot. Biochemistry. 1991;30:10091–10100. doi: 10.1021/bi00106a005. [DOI] [PubMed] [Google Scholar]
- 23.Veaute X., Fuchs R.P. Polymorphism in N-2-acetylaminofluorene induced DNA structure as revealed by DNase I footprinting. Nucleic Acids Res. 1991;19:5603–5606. doi: 10.1093/nar/19.20.5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belguise-Valladier P., Fuchs R.P. N-2-Aminofluorene and N-2 acetylaminofluorene adducts: the local sequence context of an adduct and its chemical structure determine its replication properties. J. Mol. Biol. 1995;249:903–913. doi: 10.1006/jmbi.1995.0347. [DOI] [PubMed] [Google Scholar]
- 25.Thomas D.C., Veaute X., Kunkel T.A., Fuchs R.P. Mutagenic replication in human cell extracts of DNA containing site-specific N-2-acetylaminofluorene adducts. Proc. Natl Acad. Sci. USA. 1994;91:7752–7756. doi: 10.1073/pnas.91.16.7752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shibutani S., Suzuki N., Grollman A.P. Mutagenic specificity of (acetylamino)fluorene-derived DNA adducts in mammalian cells. Biochemistry. 1998;37:12034–12041. doi: 10.1021/bi981059+. [DOI] [PubMed] [Google Scholar]
- 27.Burnouf D., Koehl P., Fuchs R.P. Single adduct mutagenesis: strong effect of the position of a single acetylaminofluorene adduct within a mutation hot spot. Proc. Natl Acad. Sci. USA. 1989;86:4147–4151. doi: 10.1073/pnas.86.11.4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lambert I.B., Napolitano R.L., Fuchs R.P. Carcinogen-induced frameshift mutagenesis in repetitive sequences. Proc. Natl Acad. Sci. USA. 1992;89:1310–1314. doi: 10.1073/pnas.89.4.1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shibutani S., Suzuki N., Tan X., Johnson F., Grollman A.P. Influence of flanking sequence context on the mutagenicity of acetylaminofluorene-derived DNA adducts in mammalian cells. Biochemistry. 2001;40:3717–3722. doi: 10.1021/bi0027581. [DOI] [PubMed] [Google Scholar]
- 30.Mu D., Bertrand-Burggraf E., Huang J.C., Fuchs R.P., Sancar A., Fuchs B.P. Human and E.coli excinucleases are affected differently by the sequence context of acetylaminofluorene-guanine adduct. Nucleic Acids Res. 1994;22:4869–4871. doi: 10.1093/nar/22.23.4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hess M.T., Schwitter U., Petretta M., Giese B., Naegeli H. Site-specific DNA substrates for human excision repair: comparison between deoxyribose and base adducts. Chem. Biol. 1996;3:121–128. doi: 10.1016/s1074-5521(96)90288-4. [DOI] [PubMed] [Google Scholar]
- 32.Stöhrer G., Osband J.A., Alvarado-Urbina G. Site-specific modification of the lactose operator with acetylaminofluorene. Nucleic Acids Res. 1983;11:5093–5101. doi: 10.1093/nar/11.15.5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou Y., Romano L.J. Solid-phase synthesis of oligonucleotides containing site-specific N-(2′-deoxyguanosin-8-yl)-2-(acetylamino)fluorene adducts using 9-fluorenylmethoxycarbonyl as the base-protecting group. Biochemistry. 1993;32:14043–14052. doi: 10.1021/bi00213a038. [DOI] [PubMed] [Google Scholar]
- 34.Zhou Y., Chladek S., Romano L.J. Synthesis of oligonucleotides containing site-specific carcinogen adducts. Preparation of the 2-cyanoethyl N,N-diisopropylphosphoramidite of N-(2′-deoxyguanosin-8-yl)-2-(acetylamino)fluorene with Fmoc as the base-protecting group. J. Org. Chem. 1994;59:556–563. [Google Scholar]
- 35.Guo D., Xie Z., Shen H., Zhao B., Wang Z. Translesion synthesis of acetylaminofluorene-dG adducts by DNA polymerase zeta is stimulated by yeast Rev1 protein. Nucleic Acids Res. 2004;32:1122–1130. doi: 10.1093/nar/gkh279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Laat W.L., Jaspers N.G., Hoeijmakers J.H.J. Molecular mechanism of nucleotide excision repair. Genes Dev. 1999;13:768–785. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- 37.Petit C., Sancar A. Nucleotide excision repair: from E. coli to man. Biochimie. 1999;81:15–25. doi: 10.1016/s0300-9084(99)80034-0. [DOI] [PubMed] [Google Scholar]
- 38.Batty D.P., Wood R.D. Damage recognition in nucleotide excision repair of DNA. Gene. 2000;241:193–204. doi: 10.1016/s0378-1119(99)00489-8. [DOI] [PubMed] [Google Scholar]
- 39.Gillet L.C.J., Schärer O.D. Preparation of C8-amine and acetylamine adducts of 2′-deoxyguanosine suitably protected for DNA synthesis. Org. Lett. 2002;4:4205–4208. doi: 10.1021/ol026474f. [DOI] [PubMed] [Google Scholar]
- 40.Wang Z., Rizzo C.J. Synthesis of the C8-deoxyguanosine adduct of the food mutagen IQ. Org. Lett. 2001;3:565–568. doi: 10.1021/ol006968h. [DOI] [PubMed] [Google Scholar]
- 41.Elmquist C.E., Stover J.S., Wang Z., Rizzo C.J. Site-specific synthesis and properties of oligonucleotides containing C8-deoxyguanosine adducts of the dietary mutagen IQ. J. Am. Chem. Soc. 2004;126:11189–11201. doi: 10.1021/ja0487022. [DOI] [PubMed] [Google Scholar]
- 42.Reese C.B., Stewart J.C.M., van Boom J.H., de Leeuw H.P.M., Nagel J., de Rooy J.F.M. The synthesis of oligoribonucleotides. Part XI. Preparation of ribonucleoside 2-acetal 3-esters by selective deacylation. J. Chem. Soc. Perkin Trans. 1975;1:934–942. doi: 10.1039/p19750000934. [DOI] [PubMed] [Google Scholar]
- 43.Shibutani S., Gentles R., Johnson F. Aerial oxidation of acetylaminofluorene-derived DNA adducts. In: Howard P.C., Hecht S.S., Beland F.A., editors. Nitroarenes. New York: Plenum Press; 1990. pp. 135–147. [Google Scholar]
- 44.Johnson F., Huang C.Y., Yu P.L. Synthetic and oxidative studies on 8-(arylamino)-2′-deoxyguanosine and -guanosine derivatives. Environ. Health Perspect. 1994;102(Suppl. 6):143–149. doi: 10.1289/ehp.94102s6143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shivji M.K., Moggs J.G., Kuraoka I., Wood R.D. Dual-incision assays for nucleotide excision repair using DNA with a lesion at a specific site. Methods Mol. Biol. 1999;113:373–392. doi: 10.1385/1-59259-675-4:373. [DOI] [PubMed] [Google Scholar]
- 46.Biggerstaff M., Wood R.D. Assay for nucleotide excision repair protein activity using fractionated cell extracts and UV-damaged plasmid DNA. Methods Mol. Biol. 1999;113:357–372. doi: 10.1385/1-59259-675-4:357. [DOI] [PubMed] [Google Scholar]
- 47.Schulhof J.C., Molko D., Teoule R. The final deprotection step in oligonucleotide synthesis is reduced to a mild and rapid ammonia treatment by using labile base-protecting group. Nucleic Acids Res. 1987;15:397–416. doi: 10.1093/nar/15.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu Q., Delaney M.O., Greenberg M.M. Observation and elimination of N-acetylation of oligonucleotides prepared using fast-deprotecting phosphoramidites and ultra-mild deprotection. Bioorg. Med. Chem. Lett. 2001;11:1105–1107. doi: 10.1016/s0960-894x(01)00161-5. [DOI] [PubMed] [Google Scholar]
- 49.Pon R.T., Yu S. Hydroquinone-O,O′-diacetic acid as a more labile replacement for succinic acid linkers in solid-phase oligonucleotide synthesis. Tetrahedron Lett. 1997;38:3327–3330. doi: 10.1093/nar/25.18.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson F., Habus I., Gentles R., Shibutani S., Lee H., Iden C., Rieger R. Site-specific adduct formation in oligomeric DNA using a new protecting group. J. Am. Chem. Soc. 1992;114:4923–4924. [Google Scholar]
- 51.Huang J.C., Sancar A. Determination of minimum substrate size for human excinuclease. J. Biol. Chem. 1994;269:19034–19040. [PubMed] [Google Scholar]
- 52.Huang J.C., Svoboda D.L., Reardon J.T., Sancar A. Human nucleotide excision nuclease removes thymine dimers from DNA by incising the 22nd phosphodiester bond 5′ and the 6th phosphodiester bond 3′ to the photodimer. Proc. Natl Acad. Sci. USA. 1992;89:3664–3668. doi: 10.1073/pnas.89.8.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang J.C., Zamble D.B., Reardon J.T., Lippard S.J., Sancar A. HMG-domain proteins specifically inhibit the repair of the major DNA adduct of the anticancer drug cisplatin by human excision nuclease. Proc. Natl Acad. Sci. USA. 1994;91:10394–10398. doi: 10.1073/pnas.91.22.10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Calsou P., Frit P., Salles B. Double strand breaks in DNA inhibit nucleotide excision repair in vitro. J. Biol. Chem. 1996;271:27601–27607. doi: 10.1074/jbc.271.44.27601. [DOI] [PubMed] [Google Scholar]
- 55.Evans E., Moggs J.G., Hwang J.R., Egly J.M., Wood R.D. Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. EMBO Rep. 1997;16:6559–6573. doi: 10.1093/emboj/16.21.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hess M.T., Schwitter U., Petretta M., Giese B., Naegeli H. Bipartite substrate discrimination by human nucleotide excision repair. Proc. Natl Acad. Sci. USA. 1997;94:6664–6669. doi: 10.1073/pnas.94.13.6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gunz D., Hess M.T., Naegeli H. Recognition of DNA adducts by human nucleotide excision repair. Evidence for a thermodynamic probing mechanism. J. Biol. Chem. 1996;271:25089–25098. doi: 10.1074/jbc.271.41.25089. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.