Abstract
The integration of the human immunodeficiency virus type 1 DNA into the host cell genome is catalysed by the viral integrase (IN). The reaction consists of a 3′-processing [dinucleotide released from each 3′ end of the viral long terminal repeat (LTR)] followed by a strand transfer (insertion of the viral genome into the human chromosome). A 17 base pair oligonucleotide d(GGAAAATCTCTAGCAGT), d(ACTGCTAGAGATTTTCC) reproducing the U5-LTR extremity of viral DNA that contains the IN attachment site was analysed by NMR using the classical NOEs and scalar coupling constants in conjunction with a small set of residual dipolar coupling constants (RDCs) measured at the 13C/15N natural abundance. The combination of these two types of parameters in calculations significantly improved the DNA structure determination. The well-known features of A-tracts were clearly identified by RDCs in the first part of the molecule. The binding/cleavage site at the viral DNA end is distinguishable by a loss of regular base stacking and a distorted minor groove that can aid its specific recognition by IN.
INTRODUCTION
The acquired immunodeficiency syndrome (AIDS) epidemic, due to rapid spreading of human immunodeficiency virus type 1 (HIV-1), has caused an estimated 20 million worldwide deaths. Vaccines are in clinical tests, but none has lived up to its early promise and we still need treatments inhibiting the retrovirus replication. Replication and chronic infection require integration of the retroviral DNA (9000 bp) into the host cell genome. The mechanism of this key reaction has been extensively studied during the last decade, essentially in order to promote the development of new antiviral compounds (1,2). Integration is a two-step reaction that is catalysed by viral integrase (IN). In the first step, called 3′-processing, a phosphodiester bond is hydrolysed within the 5′CA⇓GT3′ site at both the U3- and the U5-LTR (long terminal repeat) extremities of the viral cDNA. During the second step, termed DNA strand transfer, which is a concerted cleavage-ligation reaction, the 3′-hydroxyl group of each cDNA strand attacks a strand of the cellular DNA and becomes joined by transesterification (3–5).
IN belongs to the polynucleotidylphosphotransferase family and exerts an endonuclease-like activity. Its functional form is likely a tetramer, although it is experimentally found in a dynamic equilibrium of monomers to high order oligomers (3,6–8). The monomer (32 kDa) is folded into three distinct functional domains: N-terminal (residues 1–50), C-terminal (residues 213–288) and central [catalytic core (CC), residues 51–212]. CC that contains the catalytic trio of acidic amino acids DDX35E embedded within an RNase H fold is organized in five β-strands surrounded by six α-helices (3,9–12). The helix labelled α4 is amphipathic and protrudes at the protein surface. It is involved in several essential functions as the folding and the stabilization of the monomer and the dimer (8,11,12), and, above all, in the selective recognition of the viral cDNA at its U3- and U5-LTR ends (13–15). It should also contribute to the assemblage of dimers in functional tetramers, as recently suggested by Podtelezhnikov et al. (16).
A good understanding of the DNA–protein recognition events needs detailed information on the fine conformations of partners both unbound and bound. The specific recognition of viral DNA by IN initiates the integration reaction, which is a complex two-step reaction. The two steps of the reaction (3′-processing and strand transfer) are separated in time and space, the 3′-processing step occurring in the cytoplasm and the strand transfer in the nucleus. Several conformational changes and rearrangements of both protein and DNA are expected to happen all along the process that requires the formation of a pre-integration complex imported into the nucleus. Here, we focused on the properties of the LTR extremities which are cleaved with a high efficiency in the 3′-processing reaction (5–6 orders of magnitude for specific versus no specific sequences). We performed an NMR analysis of a 17 bp oligonucleotide termed U5-term, since it reproduces the U5-LTR extremity of the HIV-1 cDNA. According to our previous fluorescence anisotropy results, the α4 helix of IN binds this oligonucleotide with a high affinity, provided the sequence integrity of the six outermost base pairs 5′AGCAGT3′/5′ACTGCT3′ is not altered (15). The biological relevance of these in vitro results has been recently confirmed by the in vivo work of Engelman et al. and Masuda et al. (17,18).
The impact of the viral DNA on the binding event has not yet been weighed up. Elucidating a 17mer DNA structure (U5-term), however, remains a major challenge especially as very subtle sequence-dependent variations, not always detectable by conventional structural methods, may affect the double helix. Our previous simulation work has suggested that the determination of the U5-term structure can be substantially improved with residual dipolar couplings (RDCs) combined to NOEs (19), although the degree of structure improvement depends on factors, such as the number and the sort of collected RDCs, as well as the size of the oligonucleotide analysed (20–22). The current experimental study has confirmed that a sparse number of 1H–13C RDCs, accessible from spectra of unlabelled U5-term, significantly improves the structure determination when combined to conventional proton–proton distance and torsion angle restraints (19,21,23). The use of RDCs has required an assessment of both the possible interactions between the oligonucleotide and the orienting molecules and of the effect of DNA flexibility on the interpretation of results (24–26). The obtained U5-term structure is strongly heterogeneous with structural distortions that clearly demarcate the binding/cleavage site from the rest of the molecule. These could be detected by IN in the presence of Mg2+ and aid to initiate a specific complex formation.
MATERIALS AND METHODS
Sample preparation
The oligonucleotide d(GGAAAATCTCTAGCAGT), d(ACTGCTAGAGATTTTCC), termed U5-term, (Eurogentec Inc.) was synthesized at a 10 μM scale by solid-phase using the standard phosphoramidite procedure (27) and purified by high-performance liquid chromatography and precipitation in LiClO4-Acetone. Two NMR samples at 2 mM duplex concentration were prepared by dissolving U5-term in 0.4 ml sodium phosphate buffer (10 mM) containing 1 mM EDTA, pH 6.6 and ionic strength 0.1. After lyophilization, one sample was dissolved in 0.4 ml 90% H2O/10% 2H2O for acquisition of exchangeable proton spectra. The other was lyophilized twice in 2H2O before dissolution in 0.4 ml 99.9% 2H2O for acquisition of non-exchangeable proton experiments.
The Pf1 phage media (ASLA labs, http://www.asla-biotech.com/asla-phage.htm) were prepared as described previously (28). The oriented sample was obtained by exchanging Pf1 into the 2H2O-phosphate buffer, repeating pelleting in a tabletop ultracentrifuge and resuspension in the same buffer. A small amount of the so-prepared phage was added to the oligonucleotide samples in the appropriate 2H2O-buffer. Final phage concentration was ∼13 mg ml−1.
NMR spectroscopy
The NMR experiments were performed at 500 MHz on a Bruker Avance-500 spectrometer. Data were processed and analysed on an Octane Silicon Graphics Workstation using XWINNMR (Bruker) and Felix 98 (Accelrys Inc., San Diego, CA). Spectra consisted of NOESY, P-COSY and clean-TOCSY. The NOESY spectra were recorded at different mixing times (40, 80, 120 and 200 ms). Most spectra were recorded at 20°C, but several were also recorded at 5, 30 and 40°C. The imino proton resonances of the penultimate base pairs were visible at 20°C and below. The imino proton of the ultimate T17A18 base pair was observed at 5°C. One bond 13C–1H couplings were determined from 2D heteronuclear gradient sensitivity enhanced HSQC spectra (29–32). Coupling constants from aromatic and sugar anomeric regions were collected separately. Each experiment (aromatic region and anomeric region) was repeated three times in the presence and absence of orienting phage Pf1.
Determination of distance and angular restraints
Sequence-specific resonance assignments were carried out using COSY and NOESY spectra (33). Distance constraints for non-exchangeable protons were determined using the Felix program from the NOESY spectra, with the cytosine H5–H6 distance (2.5 Å) taken as reference. They were given as described in the text, either with uncertainties of ±20%, or with the limits of 2–2.8 Å (strong), 2–3.8 Å (medium), 2.8–4.4 Å (weak) and 3–6 Å (very weak). Distance constraints involving exchangeable protons were introduced using generous boundaries. Hydrogen bond restraints corresponded to distances in standard Watson–Crick base pairs, i.e. between the two heavy atoms of the hydrogen bond and between the heavy atom and the hydrogen atom, using the procedure described previously by Mauffret et al. (34). During the torsion angle and Cartesian dynamics periods weak planarity constraints (weight of 25 kcal mol−1 Å −2) were added (35). Sugar conformations were estimated from COSY spectra. Rotamer domains for backbone torsion angles α, β, γ, δ, ε and ζ were determined using 1H- and 31P-NMR data. These were restricted to 300, 175, 60, 135, −110 and 275°, respectively, with relatively loose margins of ±50° except for δ and χ (±40 and 55°). Three-dimensional structures, back-calculations of NOESY spectra were determined using the rate matrix approach provided by MORASS 2.1 (36).
Structure refinement with residual dipolar couplings
The structures were calculated with CNS 0.9 (37) that includes the RDC refinement. Fifty starting structures were thus generated by randomizing the backbone torsion angles. Torsion-angle dynamics were used at the initial stages of calculations and were followed by Cartesian molecular dynamics at high temperature according to the classical protocols (19). Calculations with RDCs were made at low temperature (200K) using torsion-angle molecular dynamics. In a first step, we used a simplified repulsive van der Waals term with the dipolar coupling force constant set to a low value of 0.01 kcal mol−1 Hz−1. In a second step of 20 ps, the dipolar coupling force constant was ramped to 10 kcal mol−1 Hz−1. An additional torsion-angle dynamics period of 7 ps was then run at 100K with a complete force field, followed by 2000 steps of Powell minimization. During these calculations, the remaining constraints were maintained with the force constants set to their final values.
Helical parameters
The U5-term structures obtained with RDCs were checked by INSIGHT II (Accelrys Inc.) and MOLMOL (38). CURVES (39) was used for the determination of the helical parameters. CURVES fits a curved or a straight global helix axis that follows the double helix. It allows both global and local geometrical analysis, providing the values of helical parameters and of backbone dihedral angles. All the helical parameters used in this study are local except the inclination. For each system of two consecutive base pairs, CURVES can calculate the parameters for a base pair (inter base pair parameters) or for a single base, i.e. within one strand (inter base parameters). Zp values and minor groove width were calculated with 3DNA (40–42). Zp values are the average of the z-component of the phosphorus atoms of the two strands from an xy-plan defined as the middle frame for each base pair step (40,42).
RESULTS
Structure resolution with classical NOE and scalar coupling restraints
Classical 2D NMR methods, consisting of NOESY, COSY, TOCSY and gradient sensitivity enhanced HSQC (PFG-PEP-HSQC) (30–32) experiments, were used to assign the proton and carbon resonances of U5-term: 5′G1G2A3A4A5A6T7C8T9C10T11A12G13C14A15G16T173′, 5′A18C19T20G21C22T23A24G25A26G27A28T29T30T31T32C33C343′. Despite the large number of signals owing to both the size of U5-term and the absence of autocomplementary strands in the duplex, the good scattering of the resonances in the homonuclear spectra allowed the assignments of all the non-exchangeable protons, except those H5′ and H5″. The NOE spectra recorded at different mixing times (from 40 up to 200 ms) provided the distances useful for structure determination. Assignments of imino and amino exchangeable protons in H2O solutions allowed the determination of the hydrogen bond network involved in base pair stabilization. Assignments of 13C resonances needed for measurements of 13C–1H RDCs were achieved for all the aromatic positions and sugar 1′ positions, and also for some sugar 3′ and 4′ positions. According to the values of scalar couplings and of proton chemical shifts of sugar, and also to the NOE H6/8-H1′ pattern (Supplementary Material Figure 1), the oligonucleotide U5-term globally adopts a B-DNA type structure.
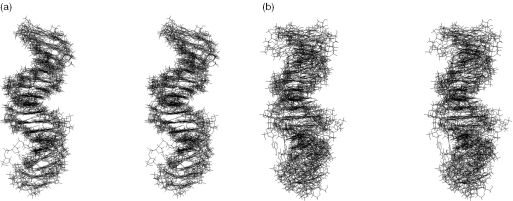
We obtained an average of 24.2 restraints (13.3 distances, 6.8 torsion angles, 0.5 planarity, 1.2 RDCs and 2.4 hydrogen bonds) per residue (Table 1). Distance restraints involving non-exchangeable protons were optimized by relaxation matrix refinement with MORASS using either uncertainties of ±20% or very weak, weak, medium and strong intensities. A set of the nine lowest energy structures with low violation values was selected from the base set of 50 calculated structures, applying a torsion angle dynamics (TAD) protocol (43). The so-obtained set corresponded to an average overall energy of −956 kcal mol−1. Although resulting in a moderately precise structure [heavy-atom root-mean-square deviation (r.m.s.d.) of 1.75 ± 0.58 Å relatively to the average structure], the approach with distance uncertainties restraints of ±20% (Figure 1a) provided better results than the one using distance restraints classified in very weak, weak, medium and strong categories (heavy-atom r.m.s.d. of 3.07 ± 0.92 Å) (Figure 1b). The gain on precision is visible all along the double helix but is particularly marked in the last third part (base pairs T11–A24 to G16–C19).
Table 1.
Structure statistics of the U5-term oligonucleotide
| Number of NMR restraints | |
| Intra-residue distancesa | 171 |
| Inter-residue distances | 282 |
| Sequential distances | 236 |
| Cross-strand distances | 46 |
| Residual dipolar coupling restraints | 41 |
| Hydrogen bond restraints | 82 |
| Dihedral angle restraints | 230 |
| Planarity restraints | 17 |
| Total restraints | 823 |
| Restraints/residue | 24.2 |
| Structure analysis of the 36 best conformersb | |
| Average deviation from ideal covalent geometry | |
| Bond length (×10−3 Å) | 2.347 ± 0.039 |
| Bond angles (°) | 0.425 ± 0.007 |
| Improper angles (°) | 0.383 ± 0.012 |
| NOE violationsc (>0.2 Å) | 4.39 |
| Dihedral angles violationsd (>10°) | 0 |
| r.m.s.d.e of the structures (Å) | 0.900 ± 0.155 |
| r.m.s.d pairwise (Å) | 1.295 ± 0.226 |
aOnly meaningful distance constraints were used (intrasugar distances independent of the conformation were discarded).
bThirty-six best structures obtained with classical and RDC restraints (four tensor values).
cForce constant K = 50 kcal mol−1 Å−2.
dForce constant K = 100 kcal mol−1 rad−2.
eThe r.m.s.d. is calculated by superposing each structure to the average structure.
Figure 1.
Stereo views of the nine lowest-energy structures of U5-term generated from classical scalar coupling and NOE restraints. These were optimized by relaxation matrix refinement using: (a) uncertainties of ±20%; (b) very weak, weak, medium and strong distance restraint categories.
Residual dipolar couplings for DNA structure determination
The last third portion of U5-term is of great biological relevance as it contains both the IN attachment site and the 3′-processing site, which actually coincide at the end of viral DNA. Thus, a good assessment of sequence-dependent variations in this portion could help us to understand the binding requirements and the correlated hydrolysis of DNA by IN, especially if the double helix is pre-organized for a specific recognition (44,45).
When cautiously used with other known NMR restraints, RDCs can provide a fine description of the double helix morphology (46,47). Many studies show that local and long-range information can be obtained (29,46–49), provided the alignment tensor of the molecule is known. For a given alignment tensor, the dipolar coupling between nuclei i and j, Dij, can be calculated using:
where θ and φ describe the internuclear vector ij orientation in the principal axis system of the alignment tensor, Da is a function of the degree of the molecule alignment and of gyromagnetic ratios, the internuclear distance, and the order parameter for i and j, while R is the rhombic component of the alignment tensor. The Da and R values, the axial and rhombic components of the alignment tensor, are determined by the orientation that molecules tend to assume in the orienting medium.
Here, the 13C–1H RDC values were extracted from the HSQC spectra of unlabelled U5-term recorded in the presence and absence of orienting material. Addition of Pseudomonas aeruginosa bacteriophage Pf1 quantities (13 mg ml−1) to the U5-term sample in 2H2O provoked an RDC related splitting of nearby 9 Hz of the HOD deuterium signal, together with a global line broadening in the spectra (28). U5-term, which is a non self-complementary heptadecamer, displays rather complicated heteronuclear spectra, and despite obvious ambiguities on signal assignments, 41 1H–13C RDCs varying from −1.1 to 18.2 Hz were determined by subtracting the 1H–13C splittings in the absence of phages from those in the presence of phages. Errors from three independent measurements were estimated to be ±2.0 Hz. A total of 24 RDCs were measured in sugars (59%) and 17 in bases (41%), leading to an average of 1.2 restraints per residue (Table 1). They were homogeneously distributed along the oligonucleotide sequence and therefore allowed a good probing of the overall double helix structure. The significance of uncertainties on helical parameters has been carefully analysed, similar to the analysis carried out by other authors (23,50). We applied the SSIA program (51,52) that allows to predict the sterically induced alignment of molecules with known structures, whatever is the type of orienting medium (here, the Pf1 phage). The best conformers issued from calculations with NOE parameters were used as input structures. The mean values for the 9 best conformers, Da = −20 Hz and R = 0.13, were incrementally varied in a grid search procedure (53), from −24 to −16 for Da and from 0 to 0.3 for R. The set of the 50 NOE-derived structures was submitted to a calculation with each (Da, R) combination. Results, presented in Table 2, establish that the pair Da = −22 Hz and R = 0.1 is the most appropriate in terms of both energy (−875 kcal mol−1) and structural convergence. Three other combinations (Da = −18 Hz, R = 0.0; Da = −18 Hz, R = 0.1; Da = −22 Hz, R = 0.0) showed close overall energy terms (−868, −864 and −860 kcal mol−1, respectively), while the remaining combinations displayed a noticeable increase of energy. The fact that the magnitude and rhombicity values of the alignment tensor obtained with the SSIA program are close to values providing a minimal energy gives us confidence in the accuracy of our measurements. We considered 36 structures composed of the four families (9 members of each) selected in varying the tensor values, as similarly considered by Tjandra et al. (23). Our small set of RDCs did not permit the obtention of U5-term structures as precisely as the ones determined by Tjandra et al. (23), who have used an oligonucleotide both 13C, 15N labelled and shorter than U5-term (dodecamer versus heptadecamer). Despite these differences, the U5-term structures have been greatly improved by the addition of RDCs, as shown by the various statistics on the DNA parameters (Table 1). Considering the 36 structures as an ensemble, the pairwise r.m.s.d. value for the 12 last base pairs of U5-term was 0.97 ± 0.24 Å, which when compared with the r.m.s.d. value (0.58 ± 0.07 Å) of Tjandra's molecules (pdb 1DUF) can be accepted as a good value. The mean pairwise r.m.s.d. calculated by taking the four families treated separately was found equal to 0.84 ± 0.15 Å, while the structures obtained by Tjandra et al. (23) were nearly indistinguishable inside each family (r.m.s.d. < 0.1 Å).
Table 2.
Characteristics of the nine best structures obtained with various NMR constraints
| Constraints | Energy (kcal mol−1) | r.m.s.d.a (Å) |
|---|---|---|
| NOEs (strong, medium, weak and very weak) + 3J | −901.08 ± 6.13 | 3.073 ± 0.92 |
| NOEs (±20%) + 3J | −956.26 ± 3.95 | 1.754 ± 0.58 |
| NOEs (±20%) + 3J + RDCs (Da = −22 Hz; R = 0.1) | −875.91 ± 7.43 | 0.809 ± 0.11 |
| NOEs (±20%) + 3J + RDCs (Da = −18 Hz; R = 0.0) | −868.62 ± 8.93 | 0.841 ± 0.13 |
| NOEs (±20%) + 3J + RDCs (Da = −18 Hz; R = 0.1) | −864.72 ± 5.14 | 0.863 ± 0.18 |
| NOEs (±20%) + 3J + RDCs (Da = −22 Hz; R = 0.0) | −860.33 ± 6.67 | 0.836 ± 0.18 |
aThe r.m.s.d. is calculated by superposing each structure to the average structure.
U5-term structures refined with RDCs
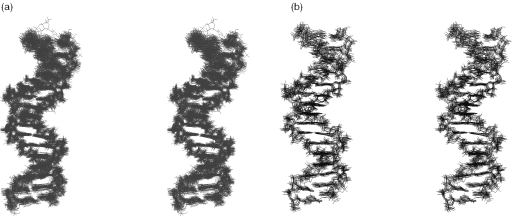
The U5-term structures were calculated using the TAD protocols. The efficiency of the TAD protocols for obtaining a strong structural convergence has been demonstrated by Kuszewski et al. (35). TAD protocols are also well suited to structure refinements using RDC parameters. According to Vermeulen et al. (21), introduction of RDCs in refinements can induce noticeable changes within the global structures through the modification of only a small number of torsion angles. The present study showed that the TAD protocols provide better results than the usually employed Cartesian dynamics protocols. Also of importance, different assays with the NOE-derived structures at low temperature indicated that the structure definition cannot be improved by the TAD protocols without the addition of RDCs. The best 36 RDC-derived structures (i.e. four families of nine structures as described above) (Figure 2a) obtained with the TAD protocols displayed satisfying statistics (Table 1). Among these, nine structures (Da = −22 Hz; R = 0.1) (Figure 2b) demonstrated a highly enhanced overall precision when compared with the NOE-structures (Figure 1a and b). Further calculations carried out with the electrostatic term in CNS turned off led to similar results.
Figure 2.
Stereo views of the U5-term structures corresponding to: (a) 36 RDC-structures, i.e. four families of nine best structures each family being generated from a different value of alignment tensor (see Table 2); (b) nine lowest-energy structures generated from classical NOE and scalar coupling restraints combined to RDC restraints (Da = −22 Hz; R = 0.1).
Remarkable structural features
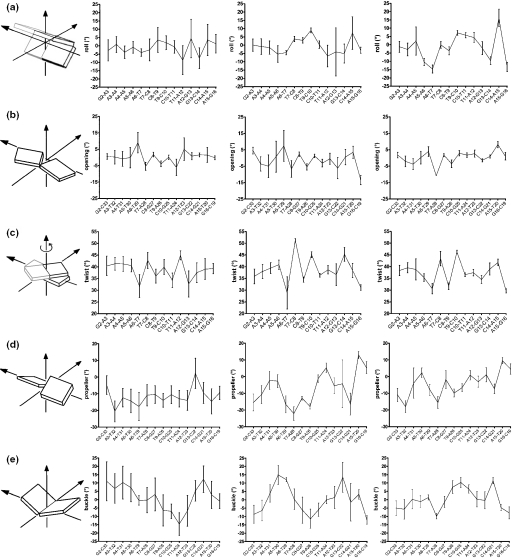
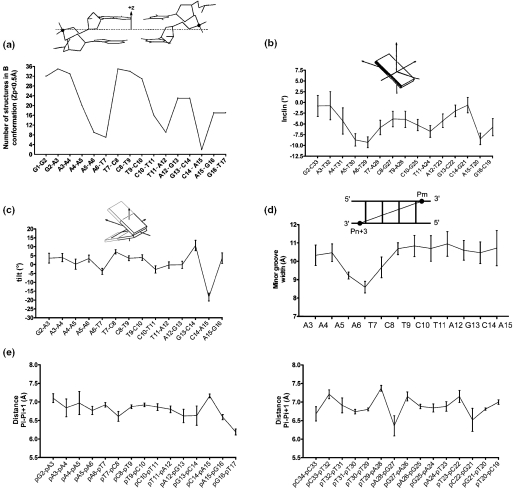
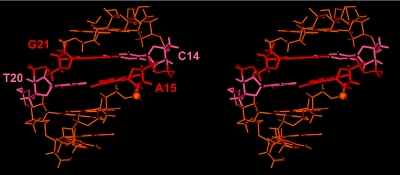
The profiles of parameters presented in Figures 3 and 4 highlighted the existence of important variations in U5-term generated by its particular sequence. The beneficial effect of RDCs on the structure determination became evident from the comparison of the parameter profiles (roll, opening, twist, propeller twist and buckle) presented in Figure 3. These correspond, from left to right, to NOEs classified according to their intensities (from very weak to strong), to NOEs using ± 20% uncertainties and to NOEs plus RDCs. Generally, introduction of RDCs modifies the profiles, but the effects are more or less important according to the parameter or the oligonucleotide region examined. The first portion of the upper strand, containing four successive adenines flanked by G2 and T7, showed that the introduction of RDCs produces a progressive 5′ to 3′ decrease of its intrastrand roll and twist values, typical of A-tract motifs (Figure 3a and c) (22,54,55). The roll of the A6-T7 step ending the A-tract adopts a negative value as large as −15°. At the same time, the minor groove shows the expected 5′ to 3′ narrowing (Figure 4d). The Zp (actually, the number of B-DNA structures) and inclination parameters (Figure 4a and b) follow the same decrease. Altogether these events translate the emergence of a B′-DNA structure in the A-tract portion, which is known for its implication in recognition of viral DNA by the C-terminal domain of IN (14). The central portion of U5-term, TC rich, displays important twist variations already discerned in the NOE (±20% uncertainties) profiles. The third portion, corresponding to the six terminal base pairs exhibits, according to almost all the parameters, more heterogeneous features, in conformity with its pyrimidine–purine step content. Important deviations from regular B-DNA occur at the level of the dinucleotide pair 5′C14-A153′/5′T20-G213′ that carries the conserved C14–A15 step, a pyrimidine–purine step known for its particular malleability (56). In the context of viral DNA, the C14–A15 step exhibits a low B-DNA content, as illustrated by the Zp value of 0.8 Å (Zp is negative in B-DNA and equals ∼2 Å in A-DNA) (Figure 4a), although its sugar pucker and glycosyl angle still present characteristics of B-DNA. It also displays a strongly negative tilt (Figure 4c) and a very large positive intrastrand roll, while its facing T20–G21 step raises a normal intrastrand roll (data not shown). This particular arrangement results in a large positive propeller twist and opening at the T20–A15 base pair, associated to an important buckle at the preceding C14–G21 base pair (Figure 3d and e and Figure 5). Consecutive to these variations, a strong cross-strand stacking takes place between the A15 base and the G21 base. This atypical cross-chain stacking of purines is made possible by the shift of the G21 base into the minor groove as observed in A-DNA. The anomalous lowfield chemical shifts experienced by the aromatic H8 and sugar H1′ protons of G21, and also by the sugar H1′ proton of T20, are certainly caused by this event (Supplementary Figure 1 and Supplementary Table 1) (33). The whole translates the particular malleability of this DNA part (41,57,58).
Figure 3.
Structural parameters of U5-term. Profiles of: (a) inter base (intrastrand) rolls in the upper strand; (b) global base–base openings; (c) inter base twists in the upper strand; (d) global base–base propellers; (e) global base–base buckles. Values were extracted from outputs of CURVES (39) for: in left, a set of nine structures generated from NOE restraints classified into very weak, weak, medium and strong distance categories; in middle, a set of nine structures generated from NOE restraints with uncertainties of ±20%; and in right, the 36 RDC structures (see legend in Figure 2).
Figure 4.
(a) Profiles of the local number of B-DNA structures (Zp value inferior to 0.5 Å) among the 36 RDC structures [calculated with 3DNA (42)]. A schematic definition of Zp is given in insert (41); (b) Profile of the global base pair axis inclinations of the 36 RDC structures (see legends in Figure 3 and Table 2), values being calculated with CURVES (39); (c) Profiles of local inter base (intrastrand) tilts in the upper strand of the 36 RDC structures (see legend in Figure 3), values being calculated with CURVES (39); (d) Profile of the average interstrand phosphorus–phosphorus (Pm–Pn+3) distances (illustrating the minor groove width) from the 36 RDC structures (see legend in Figure 3), calculated with 3DNA (42); (e) Profiles of intrastrand phosphate–phosphate Pi–Pi+1 distances along the upper strand (left) and the lower strand (right).
Figure 5.
A stereo view of the average structure of the five outermost base pairs obtained from the 36 RDC structures of U5-term (see legend in Figure 2) featuring the particular interstrand stacking of purines A15 and G21 in red thick lines and the phosphorus of the scissile bond in CPK.
The backbone geometry and motions also appear singular in the 5′G13C143′/5′G21C223′ dinucleotide base pairs. The low-fielded chemical shifts of the sugar H1′ proton of G13, besides that already mentioned for G21, can be caused by distortions affecting the backbone of the G13–C14 step as reflected by the large positive tilt value adopted at this position (Figure 4c). Actually, the epsilon and zeta angle values of the paired 5′G13C143′/5′G21C223′steps are clearly distinguishable from the other epsilon and zeta angle values of the molecule (data not shown). Moreover, the H8 proton of G21 displays a broadened signal at >30°C (data not shown), testifying to slow local motions (μs–ms) at this nucleotide position.
The other point of interest regarding the backbone geometry concerns the distance between two consecutive phosphates in the IN binding and cleavage site (Figure 4e). For instance, the PG16–PT17 distance (6.2 Å) appears considerably shorter compared with the distance between two consecutive phosphates in a standard B-DNA (6.8 Å) and also that of the PC19–PT20 step (7.0 Å) on the facing strand as well as the other P–P distances in the U5-term. The shortening of PG16-PT17 step cannot be assigned to fraying effects from the oligonucleotide ends as the facing step is not affected. It is also noteworthy that the A-like DNA character gained by the 5′C14A153′/5′T20G213′ dinucleotide base pair makes the ordinary buried A15 3′-oxygen atom submitted to the cleavage now more exposed.
DISCUSSION
Use of the RDCs to assess the U5-term structure
We have used a small and easily detectable number of 13C–1H–RDCs (one bond base and sugar C1′–H1′) in conjunction with scalar and NOE couplings to determine the structure of a 17mer oligonucleotide that reproduces the sequence of the U5-LTR extremity. One difficulty in calculations that incorporate RDCs is the correct estimation of the axial and rhombic components of the alignment tensor (51–53,59,60). In the case where the number of RDCs is small and anisotropically distributed, such as in nucleic acids, the components are determined by selecting the lowest energy structures derived from refinement rounds using different input values of Da and R. Although the procedure is highly time-consuming and presents some pitfalls (53,59), values are generally found in close agreement with those determined by the SSIA program using NOE-derived structures. Obviously, the number of RDCs introduced in calculations may have a large impact on the structure determination, this number depending on whether the oligonucleotide has or not been labelled with 13C and 15N. Many papers have addressed this issue and results have always revealed the beneficial effect of RDCs, including small sets of RDCs (19,49). The degree of structure improvement also depends on the size of the oligonucleotide. In many cases, the structure of small oligonucleotides has been found less improved than that of long oligonucleotides by the addition of RDCs (20–22). In the present work, the gain of precision produced by the small set of 13C–1H–RDCs combined to NOEs with ±20% uncertainties has significantly improved the U5-term structure determination. For instance, the mean uncertainty on the roll was lowered from ±4.5° to ±3°, while Tjandra et al. (23) have obtained a value of ±0.8° with a larger set of RDC restraints applied to a shorter oligonucleotide. We are aware of the fact that such uncertainties represent the r.m.s. distribution over an ensemble of structures and underestimate the true uncertainty, as MD simulations clearly indicate that due to the innate flexibility of DNA, variations on the roll could attain ±10° (61). In fact to be fully assessed, the influence of the conformational averaging on RDC values in systems like double-stranded DNA needs additional data, but such a work is beyond the scope of this study. Yet, the analysis seems feasible in systems presenting clear evidence of strong dynamics (25,26). Just to vindicate the relevance of our small set of 13C–1H RDCs as a powerful tool to probe the conformation of oligonucleotides, note that the well-known 5′ to 3′ narrowing of the minor groove exhibited by A-rich regions (22,54) is perfectly reproduced in the current work (Figure 4d). However, the impact of RDCs is not equal on all the helical parameters as already mentioned in our simulation study on the same oligonucleotide (19). For instance, parameters, such as the roll and the twist, generally present a very good resolution, but the roll seems better improved by RDCs than the twist comparatively with the starting structure. According to our simulations, the number of collected NOEs has an important impact on the structure determination. For instance, the number of NOEs involving the base protons of AT base pairs is smaller compared with the GC base pairs (the adenine H2 protons being poor supplier of internucleotide NOEs), so that the impact of RDCs on the AT base pair parameters (roll, twist, Zp and minor groove width) appears larger than on the GC base pair parameters. Thus, the degree of improvement produced by RDCs on helical parameters depends, certainly among other factors, on the number and the quality of NOEs (uncertainties) that have been used to generate the starting structure.
Structural variations along the U5-term helix
Together with the distortions affecting the third portion of U5-term, the 5′ to 3′ narrowing of the minor groove displayed by the A-tract could also be of biological relevance, as demonstrated by Esposito and Craigie (14). The A-tract would be recognized by the C-terminal domain of IN and contribute to the tight binding of the enzyme to viral DNA (16), while the six outermost residues, i.e. the third portion, could be recognized specifically by the catalytic core domain (13–15).
The distortions occurring in the third portion of U5-term reflect a tendency of the conserved 5′C14A153′/5′T20G213′ dinucleotide pair, perceptible in some structural parameters [cross-strand stacking of purines, roll (∼15°)-slide (−1.6 Å) (data not shown) correlation, Zp value, tilt value, etc.] to adopt a structure intermediate between B-DNA and A-DNA (Figure 4a and c and Figure 5). The highly conserved C14pA15 step in the upper strand is known for its involvement in the viral DNA integration (62,63). The distortions that affect this step could be the foundation upon which the LTR-end of viral DNA is specifically recognized and cleaved by IN. They could be in large part due to the innate ‘bistability’ of stackings (two preferential stackings either intrastrand or cross-strand) which generally characterizes the pyrimidine–purine steps and, in particular, those CpAs (56,64). The tendency of the CpA/TpG dinucleotide pair to display an A-like conformation could be necessary for the good adjustment of IN to viral DNA as suggested by inspection of a great number of protein-bound DNAs (41). The decrease in base pair overlap (Figure 5) is similar to that observed in the so-called fragile DNA regions, including the MARs (matrix attachment regions) (65), which are generally highly sensitive to DNase I (66). Moreover, A-DNA selectively exposes its sugar–phosphate atoms ordinarily buried within the backbone chain. Such events have been precisely observed for complexes with enzymes performing cutting or sealing operations at the O3′-P phosphodiester linkage as polymerases, endonucleases and transposases (67). Also note that benzopyrene that forms specific adducts with the exocyclic amino group of guanines in the minor groove of DNA covers in the 5′C14A153′/5′T20G213′ base pairs. As benzopyrene hinders the access of IN to the cleavable site 5′CA⇓GT3′, the result is deleterious for the 3′-processing, and hence, the viral DNA integration (68).
Variations in the backbone
Distance shortening between the scissile phosphate and the succeeding phosphate as shown in Figure 4e has been already observed in the complexes of endonucleases EcoRI and EcoRV (69) and PvuII (70). In these three examples, the two phosphates next to the scissile bond are bridged by the ε amino group of a catalytic lysine residue. Contacts implying basic amino acid side chains and two consecutive phosphate groups have also been encountered in the damaged DNA–glycosylase complexes (71). In that case, the bringing closer of phosphates pinches the DNA backbone and forces the sugar moiety of the lesion into the catalytic site of the enzyme. In U5-term, the shortening of the PG16–PT17 distance could aid the anchoring of the IN lysine K159 side chain, which is known to participate to the specific recognition and cleavage of viral DNA (15–18,72,73). The importance of the phosphate–phosphate distance shortening cannot be neglected since, as pointed by Rauch et al. (70) for the PvuII endonucleases, this creates a cleft within the minor groove. A catalytic water molecule serving as the incoming nucleophile for the phosphodiester hydrolysis could be held in the cleft, while other water molecules in its surrounding could participate to a network of hydrogen bonds with DNA and possibly one or two Mg2+ liganded to the protein catalytic centre. Moreover, the gain of A-like character by the 5′C14A153′/5′T20G213′ dinucleotide base pair makes the 3′-oxygen of the CA⇓GT phosphodiester linkage more exposed to the IN attack from the minor groove. In fact, the insertion of a (Rp)-phosphorothioate at the ApG step of CA⇓GT impairs the cleavage reaction, while the (Sp)-phosphorothioate does not exert any significant perturbation (74). In the (Rp)-phosphorothioate configuration, the unchanged phosphorus oxygen (O1P) is more buried in the minor groove than in the (Sp)-phosphorothioate configuration (75) and is, therefore, less accessible to proteins attacking the DNA from this groove.
It can also be noted also that the current models based on experimental data (15,16) or resting on docking studies (72,73) stipulate that several phosphates of the six outermost residues of the viral DNA are contacted in the minor groove by the α4 helix that protrudes at the IN surface (11). The proposed model resembles to that of Tn5 transposase bound to its cognate DNA. In the reported crystal structure, a lysine residue similar to the IN lysine K159 interacts with a distorted phosphate backbone that accompanies displacements occurring in base pairs (67).
Pre-organized DNA-binding site
In most cases of sequence-specific DNA binding, it is not clear whether the double helix perturbations are induced by proteins or whether are intrinsic to the base sequence. In U5-term, the particular structure adopted by the binding/cleavage site results from two effects acting in conjunction: one corresponds to the particular sequence and the other to the position of the site at the DNA end. Actually, inspection of crystallographic data strongly suggests that most of the sequence-dependent distortions occurring in unbound DNAs are also found, but more accentuated in bound DNAs. One of the best examples of pre-organized structures in nucleic acids helping the formation of sequence-specific interactions is provided by the DNA-binding site of catabolite activator protein (CAP) (44). The CAP is known to bend DNA to an exceptional extent, but molecular dynamics simulations of bound and free DNA reveal that 40% of the curvature of bound DNA is due to an intrinsic strained base sequence structure. A second example concerns the reverse transcriptase RNase H, which is often considered as the paradigm for the control of protein function by nucleic acids structures. The protein degrades the RNA moiety of a DNA/RNA duplex but not the hybrid duplex formed by the polypurine tract (PPT) conserved in the genome of HIV. Crystallographic studies (76) reveal that the PPT structure is strongly distorted in the protein complex with several base pairs out of register, while a recent study using chemical footprinting suggests that pre-existing distortions intrinsic to the specific base sequence are present in the PPT nucleic acids substrate (45). A third example concerns the integrase itself. Mismatches and modified bases have been inserted in its viral DNA site. These have shown that the local destabilization of the third base pair (A–T) before the DNA end significantly increases the 3′-processing activity. Weak stacking properties at the cleavage site have been suspected by authors (62,63). Their existence has been demonstrated in our NMR study.
CONCLUSION
The present experimental work has totally confirmed our previous simulation studies, suggesting that a small number of RDCs, easily measurable at the 13C/15N natural abundance, could substantially improve the structure determination of an oligonucleotide as long as a heptadecamer, provided they are used in conjunction with precise NOE distances (15–20% uncertainties) (19).
Results provide a binding site structure displaying mixed B- and A-like features, especially at the highly conserved CpA step. Of course, the binding of IN (and certainly Mg2+) to its cognate DNA will certainly accentuate some of the structural features pre-existing at the binding site.
An obvious next step will be an NMR study combined to an MD simulation to determine the structure of U5-term complexed to the IN α4-helix.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
J.-G.R. was funded by ARC (Association pour la Recherche contre le Cancer) and S.C. by the French Ministry of Research. This work was supported in part by the French national research agency against AIDS (ANRS) and Sidaction. Funding to pay the Open Access publication charges for this article was provided by CNRS and Institut Gustave Roussy.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hazuda D.J., Felock P., Witmer M., Wolfe A., Stillmock K., Grobler J.A., Espeseth A., Gabryelski L., Schleif W., Blau C., Miller M.D. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science. 2000;287:646–650. doi: 10.1126/science.287.5453.646. [DOI] [PubMed] [Google Scholar]
- 2.Pommier Y., Neamati N. Inhibitors of human immunodeficiency virus integrase. Adv. Virus Res. 1999;52:427–458. doi: 10.1016/s0065-3527(08)60310-3. [DOI] [PubMed] [Google Scholar]
- 3.Esposito D., Craigie R. HIV integrase structure and function. Adv. Virus Res. 1999;52:319–333. doi: 10.1016/s0065-3527(08)60304-8. [DOI] [PubMed] [Google Scholar]
- 4.Katzman M., Katz R.A. Substrate recognition by retroviral integrases. Adv. Virus Res. 1999;52:371–395. doi: 10.1016/s0065-3527(08)60307-3. [DOI] [PubMed] [Google Scholar]
- 5.Gerton J.L., Brown P.O. The core domain of HIV-1 integrase recognizes key features of its DNA substrates. J. Biol. Chem. 1997;272:25809–25815. doi: 10.1074/jbc.272.41.25809. [DOI] [PubMed] [Google Scholar]
- 6.Jenkins T.M., Engelman A., Ghirlando R., Craigie R. A soluble active mutant of HIV-1 integrase: involvement of both the core and carboxyl-terminal domains in multimerization. J. Biol. Chem. 1996;271:7712–7718. doi: 10.1074/jbc.271.13.7712. [DOI] [PubMed] [Google Scholar]
- 7.Leh H., Brodin P., Bischerour J., Deprez E., Tauc P., Brochon J-C., LeCam E., Coulaud D., Auclair C., Mouscadet J.-F. Determinants of Mg2+-dependent activities of recombinant human immunodeficiency virus type 1 integrase. Biochemistry. 2000;39:9285–9294. doi: 10.1021/bi000398b. [DOI] [PubMed] [Google Scholar]
- 8.Maroun R.G., Krebs D., El Antri S., Deroussent A., Lescot E., Troalen F., Porumb H., Goldberg M.E., Fermandjian S. Self-association and domains of interactions of an amphipathic helix peptide inhibitor of HIV-1 integrase assessed by analytical ultracentrifugation and NMR experiments in trifluoroethanol/H(2)O mixtures. J. Biol. Chem. 1999;274:34174–34185. doi: 10.1074/jbc.274.48.34174. [DOI] [PubMed] [Google Scholar]
- 9.Chen J.C., Krucinski J., Miercke L.J., Finer-Moore J.S., Tang A.H., Leavitt A.D., Stroud R.M. Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding. Proc. Natl Acad. Sci. USA. 2000;97:8233–8238. doi: 10.1073/pnas.150220297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J.Y., Ling H., Yang W., Craigie R. Structure of a two-domain fragment of HIV-1 integrase: implications for domain organization in the intact protein. EMBO J. 2001;20:7333–7343. doi: 10.1093/emboj/20.24.7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dyda F., Hickman A.B., Jenkins T.M., Engelman A., Craigie R., Davies D.R. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science. 1994;266:1981–1986. doi: 10.1126/science.7801124. [DOI] [PubMed] [Google Scholar]
- 12.Goldgur Y., Dyda F., Hickman A.B., Jenkins T.M., Craigie R., Davies D.R. Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium. Proc. Natl Acad. Sci. USA. 1998;95:9150–9154. doi: 10.1073/pnas.95.16.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jenkins T.M., Esposito D., Engelman A., Craigie R. Critical contacts between HIV-1 integrase and viral DNA identified by structure-based analysis and photo-crosslinking. EMBO J. 1997;16:6849–6859. doi: 10.1093/emboj/16.22.6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esposito D., Craigie R. Sequence specificity of viral end DNA binding by HIV-1 integrase reveals critical regions for protein–DNA interaction. EMBO J. 1998;17:5832–5843. doi: 10.1093/emboj/17.19.5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zargarian L., Benleumi M.S., Renisio J.-G., Merad H., Maroun R.G., Wieber F., Mauffret O., Porumb H., Troalen F., Fermandjian S. Strategy to discriminate between high and low affinity bindings of human immunodeficiency virus, type 1 integrase to viral DNA. J. Biol. Chem. 2003;278:19966–19973. doi: 10.1074/jbc.M211711200. [DOI] [PubMed] [Google Scholar]
- 16.Podtelezhnikov A.A., Gao K., Bushman F.D., McCammon J.A. Modeling HIV-1 integrase complexes based on their hydrodynamic properties. Biopolymers. 2003;68:110–120. doi: 10.1002/bip.10217. [DOI] [PubMed] [Google Scholar]
- 17.Lu R., Limon A., Ghory H.Z., Engelman A. Genetic analyses of DNA-binding mutants in the catalytic core domain of human immunodeficiency virus type 1 integrase. J. Virol. 2005;79:2493–2505. doi: 10.1128/JVI.79.4.2493-2505.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ikeda T., Nishitsuji H., Zhou X., Nara N., Ohashi T., Kannagi M., Masuda T. Evaluation of the functional involvement of human immunodeficiency virus type 1 integrase in nuclear import of viral cDNA during acute infection. J. Virol. 2004;78:11563–11573. doi: 10.1128/JVI.78.21.11563-11573.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mauffret O., Tevanian G., Fermandjian S. Residual dipolar coupling constants and structure determination of large DNA duplexes. J. Biomol. NMR. 2002;24:317–328. doi: 10.1023/a:1021645131882. [DOI] [PubMed] [Google Scholar]
- 20.Padrta P., Stefl R., Kralik L., Zidek L., Sklenar V. Refinement of d(GCGAAGC) hairpin structure using one- and two-bond residual dipolar couplings. J. Biomol. NMR. 2002;24:1–14. doi: 10.1023/a:1020632900961. [DOI] [PubMed] [Google Scholar]
- 21.Vermeulen A., Zhou H., Pardi A. Determining DNA global structure and DNA bending by application of NMR residual dipolar couplings. J. Am. Chem. Soc. 2000;122:9638–9647. [Google Scholar]
- 22.Stefl R., Wu H., Ravindranathan S., Sklenar V., Feigon J. DNA A-tract bending in three dimensions: solving the dA4T4 versus dT4A4 conundrum. Proc. Natl Acad. Sci. USA. 2004;101:1177–1182. doi: 10.1073/pnas.0308143100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tjandra N., Shin-ichi T., Ono A., Kainosho M., Bax A. The NMR structure of a DNA dodecamer in an aqueous dilute liquid crystalline phase. J. Am. Chem. Soc. 2000;122:6190–6200. [Google Scholar]
- 24.Zweckstetter M., Bax A. Characterization of molecular alignment in aqueous suspensions of Pf1 bacteriophage. J. Biomol. NMR. 2001;20:365–377. doi: 10.1023/a:1011263920003. [DOI] [PubMed] [Google Scholar]
- 25.Tolman J.R. Dipolar couplings as a probe of molecular dynamics and structure in solution. Curr. Opin. Struct. Biol. 2001;11:532–539. doi: 10.1016/s0959-440x(00)00245-1. [DOI] [PubMed] [Google Scholar]
- 26.Al-Hashimi H.M., Gorin A., Majumdar A., Gosser Y., Patel D.J. Towards structural genomics of RNA: rapid NMR resonance assignment and simultaneous RNA tertiary structure determination using residual dipolar couplings. J. Mol. Biol. 2002;318:637–649. doi: 10.1016/S0022-2836(02)00160-2. [DOI] [PubMed] [Google Scholar]
- 27.Matteuci M.D., Caruthers M.H. Synthesis of deoxyoligonucleotides on a polymer support. J. Am. Chem. Soc. 1981;103:3185–3191. [Google Scholar]
- 28.Hansen M.R., Mueller L., Pardi A. Tunable alignment of macromolecules by filamentous phage yields dipolar coupling interactions. Nature Struct. Biol. 1998;5:1065–1074. doi: 10.1038/4176. [DOI] [PubMed] [Google Scholar]
- 29.Tjandra N., Bax A. Direct measurement of distances and angles in biomolecules by NMR in a dilute liquid crystalline medium [Erratum (1997) Science, 278, 1697] Science. 1997;278:1111–1114. doi: 10.1126/science.278.5340.1111. [DOI] [PubMed] [Google Scholar]
- 30.Palmer A.G., Cavanagh J., Wright P.E., Rance M. Sensitivity improvement in proton-detected two-dimensional heteronuclear correlation spectroscopy. J. Magn. Reson. 1991;93:151–170. [Google Scholar]
- 31.Kay L.E., Keifer P., Saarinen T. Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J. Am. Chem. Soc. 1992;114:10663–10665. [Google Scholar]
- 32.Schleucher J., Schwendinger M., Sattler M., Schmidt P., Schedletzky O., Glaser S.J., Sorensen O.W., Griesinger C. A general enhancement scheme in heteronuclear multidimensional NMR employing pulsed field gradients. J. Biomol. NMR. 1994;4:301–306. doi: 10.1007/BF00175254. [DOI] [PubMed] [Google Scholar]
- 33.Wijmenga S.S., Mooren M.M.W., Hilbers C.W. NMR of nucleic acid: from spectrum to structure. In: Roberts G.C., editor. NMR of Macromolecules: A Practical Approach. New York: Oxford University Press; 1993. pp. 217–283. [Google Scholar]
- 34.Mauffret O., Amir-Aslani A., Maroun R.G., Monnot M., Lescot E., Fermandjian S. Comparative structural analysis by [1H,31P]-NMR and restrained molecular dynamics of two DNA hairpins from a strong DNA topoisomerase II cleavage site. J. Mol. Biol. 1998;283:643–655. doi: 10.1006/jmbi.1998.2095. [DOI] [PubMed] [Google Scholar]
- 35.Kuszewski J., Schwieters C., Clore G.M. Improving the accuracy of NMR structures of DNA by means of a database potential of mean force describing base–base positional interactions. J. Am. Chem. Soc. 2001;123:3903–3918. doi: 10.1021/ja010033u. [DOI] [PubMed] [Google Scholar]
- 36.Post C.B., Meadows R.P., Gorenstein D.G. On the evaluation of interproton distances for three-dimensional structure determination by NMR using a relaxation rate matrix analysis. J. Am. Chem. Soc. 1990;112:6796–6803. [Google Scholar]
- 37.Brunger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.S., Kuszewski J., Nilges M., Pannu N.S., et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta. Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 38.Koradi R., Billeter M., Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. 29–32. [DOI] [PubMed] [Google Scholar]
- 39.Lavery R., Sklenar H. The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. J. Biomol. Struct. Dyn. 1988;6:63–91. doi: 10.1080/07391102.1988.10506483. [DOI] [PubMed] [Google Scholar]
- 40.El Hassan M.A., Calladine C.R. Two distinct modes of protein-induced bending in DNA. J. Mol. Biol. 1998;282:331–343. doi: 10.1006/jmbi.1998.1994. [DOI] [PubMed] [Google Scholar]
- 41.Lu X.-J., Shakked Z., Olson W.K. A-form conformational motifs in ligand-bound DNA structures. J. Mol. Biol. 2000;300:819–840. doi: 10.1006/jmbi.2000.3690. [DOI] [PubMed] [Google Scholar]
- 42.Lu X.-J., Olson W.K. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stein E.G., Rice L.M., Brunger A.T. Torsion-angle molecular dynamics as a new efficient tool for NMR structure calculation. J. Magn. Reson. 1997;124:154–164. doi: 10.1006/jmre.1996.1027. [DOI] [PubMed] [Google Scholar]
- 44.Dixit S.B., Beveridge D.L. Axis curvature and ligand induced bending in the CAP–DNA oligomers. Biophys. J. 2005;88:L04–L06. doi: 10.1529/biophysj.104.053058. Epub 2004 Nov 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kvaratskhelia M., Budihas S.R., Le Grice S.F. Pre-existing distortions in nucleic acid structure aid polypurine tract selection by HIV-1 reverse transcriptase. J. Biol. Chem. 2002;277:16689–16696. doi: 10.1074/jbc.M109914200. [DOI] [PubMed] [Google Scholar]
- 46.Bax A., Kontaxis G., Tjandra N. Dipolar couplings in macromolecular structure determination. Methods Enzymol. 2001;339:127–74. doi: 10.1016/s0076-6879(01)39313-8. [DOI] [PubMed] [Google Scholar]
- 47.Prestegard J.H. New techniques in structural NMR—anisotropic interactions. Nature Struct. Biol. 1998;5:517–522. doi: 10.1038/756. [DOI] [PubMed] [Google Scholar]
- 48.Bayer P., Varani L., Varani G. Refinement of the structure of protein–RNA complexes by residual dipolar coupling analysis. J. Biomol. NMR. 1999;14:149–155. doi: 10.1023/a:1008360331296. [DOI] [PubMed] [Google Scholar]
- 49.Lawrence D.C., Stover C.C., Noznitsky J., Wu Z., Summers M.F. Structure of the intact stem and bulge of HIV-1 Psi-RNA stem–loop SL1. J. Mol. Biol. 2003;326:529–542. doi: 10.1016/s0022-2836(02)01305-0. [DOI] [PubMed] [Google Scholar]
- 50.Wu Z., Delaglio F., Tjandra N., Zhurkin V.B., Bax A. Overall structure and sugar dynamics of a DNA dodecamer from homo- and heteronuclear dipolar couplings and 31P chemical shift anisotropy. J. Biomol. NMR. 2003;26:297–315. doi: 10.1023/a:1024047103398. [DOI] [PubMed] [Google Scholar]
- 51.Zweckstetter M., Bax A. Prediction of sterically induced alignment in a dilute liquid crystalline phase: aid to protein structure determination by NMR. J. Am. Chem. Soc. 2000;122:3791–3792. [Google Scholar]
- 52.Zweckstetter M., Bax A. Evaluation of uncertainty in alignment tensors obtained from dipolar couplings. J. Biomol. NMR. 2002;23:127–137. doi: 10.1023/a:1016316415261. [DOI] [PubMed] [Google Scholar]
- 53.Warren J.J., Moore P.B. Application of dipolar coupling data to the refinement of the solution structure of the sarcin–ricin loop RNA. J. Biomol. NMR. 2001;20:311–323. doi: 10.1023/a:1011214214552. [DOI] [PubMed] [Google Scholar]
- 54.MacDonald D., Herbert K., Zhang X., Pologruto T., Lu P., Polgruto T. Solution structure of an A-tract DNA bend [Erratum (2001) J. Mol. Biol., 307, 1519] J. Mol. Biol. 2001;306:1081–1098. doi: 10.1006/jmbi.2001.4447. [DOI] [PubMed] [Google Scholar]
- 55.Barbic A., Zimmer D.P., Crothers D.M. Structural origins of adenine-tract bending. Proc. Natl Acad. Sci. USA. 2003;100:2369–2373. doi: 10.1073/pnas.0437877100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bertrand H., Ha-Duong T., Fermandjian S., Hartmann B. Flexibility of the B-DNA backbone: effects of local and neighbouring sequences on pyrimidine–purine steps. Nucleic Acids Res. 1998;26:1261–1267. doi: 10.1093/nar/26.5.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim J.L., Nikolov D.B., Burley S.K. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature. 1993;365:520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- 58.El Hassan M.A., Calladine C.R. Propeller-twisting of base-pairs and the conformational mobility of dinucleotide steps in DNA. J. Mol. Biol. 1996;259:95–103. doi: 10.1006/jmbi.1996.0304. [DOI] [PubMed] [Google Scholar]
- 59.McCallum S.A., Pardi A. Refined solution structure of the iron-responsive element RNA using residual dipolar couplings. J. Mol. Biol. 2003;326:1037–1050. doi: 10.1016/s0022-2836(02)01431-6. [DOI] [PubMed] [Google Scholar]
- 60.Bryce D.L., Bax A. Application of correlated residual dipolar couplings to the determination of the molecular alignment tensor magnitude of oriented proteins and nucleic acids. J. Biomol. NMR. 2004;28:273–287. doi: 10.1023/B:JNMR.0000013701.16162.0c. [DOI] [PubMed] [Google Scholar]
- 61.Derreumaux S., Fermandjian S. Bending and adaptability to proteins of the cAMP DNA-responsive element: molecular dynamics contrasted with NMR. Biophys J. 2000;79:656–669. doi: 10.1016/S0006-3495(00)76324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Agapkina J., Smolov M., Zubin E., Mouscadet J.-F., Gottikh M. HIV-1 integrase can process a 3′-end crosslinked substrate. Eur. J. Biochem. 2004;271:205–211. doi: 10.1046/j.1432-1033.2003.03921.x. [DOI] [PubMed] [Google Scholar]
- 63.Scottoline B.P., Chow S., Ellison V., Brown P.O. Disruption of the terminal base pairs of retroviral DNA during integration. Genes Dev. 1997;11:371–382. doi: 10.1101/gad.11.3.371. [DOI] [PubMed] [Google Scholar]
- 64.Calladine C.R., Drew H.R. Understanding DNA: The Molecule and How it Works. London, UK: Academic Press; 1992. [Google Scholar]
- 65.Leporc S., Mauffret O., Tevanian G., Lescot E., Monnot M., Fermandjian S. An NMR and molecular modelling analysis of d(CTACTGCTTTAG). d(CTAAAGCAGTAG) reveals that the particular behaviour of TpA steps is related to edge-to-edge contacts of their base-pairs in the major groove. Nucleic Acids Res. 1999;27:4759–4767. doi: 10.1093/nar/27.24.4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Boulikas T. Nature of DNA sequences at the attachment regions of genes to the nuclear matrix. J. Cell. Biochem. 1993;52:14–22. doi: 10.1002/jcb.240520104. [DOI] [PubMed] [Google Scholar]
- 67.Davies D.R., Goryshin I.Y., Reznikoff W.S., Rayment I. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science. 2000;289:77–85. doi: 10.1126/science.289.5476.77. [DOI] [PubMed] [Google Scholar]
- 68.Johnson A.A., Sayer J.M., Yagi H., Kalena G.P., Amin R., Jerina D.M., Pommier Y. Position-specific suppression and enhancement of HIV-1 integrase reactions by minor groove benzo[a]pyrene diol epoxide deoxyguanine adducts: implications for molecular interactions between integrase and substrates. J. Biol. Chem. 2004;279:7947–7955. doi: 10.1074/jbc.M311263200. [DOI] [PubMed] [Google Scholar]
- 69.Jeltsch A., Alves J., Wolfes H., Maass G., Pingoud A. Substrate-assisted catalysis in the cleavage of DNA by the EcoRI and EcoRV restriction enzymes. Proc. Natl Acad. Sci. USA. 1993;90:8499–8503. doi: 10.1073/pnas.90.18.8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rauch C., Trieb M., Flader W., Wellenzohn B., Winger R.H., Mayer E., Hallbrucker A., Liedl K.R. PvuII-endonuclease induces structural alterations at the scissile phosphate group of its cognate DNA. J. Mol. Biol. 2002;324:491–500. doi: 10.1016/s0022-2836(02)01089-6. [DOI] [PubMed] [Google Scholar]
- 71.Fromme J.C., Verdine G.L. Structural insights into lesion recognition and repair by the bacterial 8-oxoguanine DNA glycosylase MutM. Nature Struct. Biol. 2002;9:544–552. doi: 10.1038/nsb809. [DOI] [PubMed] [Google Scholar]
- 72.De Luca L., Pedretti A., Vistoli G., Barreca M.L., Villa L., Monforte P., Chimirri A. Analysis of the full-length integrase–DNA complex by a modified approach for DNA docking. Biochem. Biophys. Res. Commun. 2003;310:1083–1088. doi: 10.1016/j.bbrc.2003.09.120. [DOI] [PubMed] [Google Scholar]
- 73.Adesokan A.A., Roberts V.A., Lee K.W., Lins R.D., Briggs J.M. Prediction of HIV-1 integrase/viral DNA interactions in the catalytic domain by fast molecular docking. J. Med. Chem. 2004;47:821–828. doi: 10.1021/jm0301890. [DOI] [PubMed] [Google Scholar]
- 74.Gerton J.L., Herschlag D., Brown P.O. Stereospecificity of reactions catalyzed by HIV-1 integrase. J. Biol. Chem. 1999;274:33480–33487. doi: 10.1074/jbc.274.47.33480. [DOI] [PubMed] [Google Scholar]
- 75.Hartmann B., Bertrand H., Fermandjian S. Sequence effects on energetic and structural properties of phosphorothioate DNA: a molecular modelling study. Nucleic Acids Res. 1999;27:3342–3347. doi: 10.1093/nar/27.16.3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sarafianos S.G., Das K., Tantillo C., Clark A.D., Jr, Ding J., Whitcomb J.M., Boyer P.L., Hughes S.H., Arnold E. Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA. EMBO J. 2001;20:1449–1461. doi: 10.1093/emboj/20.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.