Abstract
The Tn10 transpososome is a DNA processing machine in which two transposon ends, a transposase dimer and the host protein integration host factor (IHF), are united in an asymmetrical complex. The transitions that occur during one transposition cycle are not limited to chemical cleavage events at the transposon ends, but also involve a reorganization of the protein and DNA components. Here, we demonstrate multiple pathways for Tn10 transposition. We show that one series of events is favored over all others and involves cyclic changes in the affinity of IHF for its binding site. During transpososome assembly, IHF is bound with high affinity. However, the affinity for IHF drops dramatically after cleavage of the first transposon end, leading to IHF ejection and unfolding of the complex. The ejection of IHF promotes cleavage of the second end, which is followed by restoration of the high affinity state which in turn regulates target interactions.
INTRODUCTION
Transposons have been found in all organisms examined with notable exceptions, such as specialized intracellular parasites with highly degenerate genomes (1). They are important agents in the spread of antibiotic resistance and the mobilization of pathogenicity islands in bacteria (2–4). Prokaryotic and eukaryotic transposons can also be regarded as ‘natural genomic engineers’ as they are the single most important source of genomic rearrangements, such as deletions, inversions, duplications and translocations (5). In free living bacteria, transposons usually represent ∼1% of the genome, but may be far more numerous in pathogenic species (2–4,6). In eukaryotes, DNA transposons are far more numerous and retro-elements frequently represent up to 50% of the genome (7–9). During the course of evolution, transposons have diversified to provide functions, such as HIV integrase (10), centromere binding protein (11), insect telomerase (12) and the V(D)J recombination machinery responsible for generating immunoglobulin diversity (13,14).
DNA transactions generally take place within higher-order nucleoprotein complexes in which the components are engaged in a wide range of different interactions (15–17). In addition to specific enzymes, these complexes frequently contain protein cofactors that function as architectural catalysts. Examples of this type include the ubiquitous integration host factor (IHF) protein in bacteria and the HMG box proteins in eukaryotes (16–19). The large number of interactions within these complexes is thought to provide a number of functions, such as regulating the start of the reaction, providing specificity, imposing directionality and delivering the activation energy for intermediate steps.
Recently, there has been significant progress in understanding the structure and composition of the higher-order complexes involved in DNA replication, recombination and repair (20–23). In eukaryotes, it is not uncommon for such complexes to contain 5 or 10 different protein components. Determination of the inventory of protein components and their pairwise interactions has provided great insight into the structure and function of such complexes. Elegant models have also been developed to illustrate the dynamics of the complexes as components enter or exit the reaction at different stages. However, very little may be known about the precise function or molecular mechanism of the various components, or the conformational changes that may take place at different stages of the reaction. Fortunately, several unique characteristics of the Tn10 transposition system have allowed us to address the nature and function of conformational changes at different stages of the reaction using traditional molecular biological techniques (24–27). Key to this progress has been the insight provided by the crystal structure of the related Tn5 transpososome (28).
Tn10 is a non-replicative composite transposon in which the flanking IS10 elements cooperate to mobilize the intervening genes encoding tetracycline resistance [reviewed in (29)]. The flanking IS10 elements are present in opposite orientations. This means that the terminal inverted repeats at either end of IS10, which are the recognition sites for transposase, can therefore be defined as ‘inside’ or ‘outside’ with respect to their position in Tn10. The major difference between the two types of ends is the presence of a strong binding site for the host protein IHF adjacent to the inverted repeat at the outside end. This means that IS10 transposition involves synapsis of an inside and an outside end, while Tn10 transposition involves the synapsis of two identical outside ends.
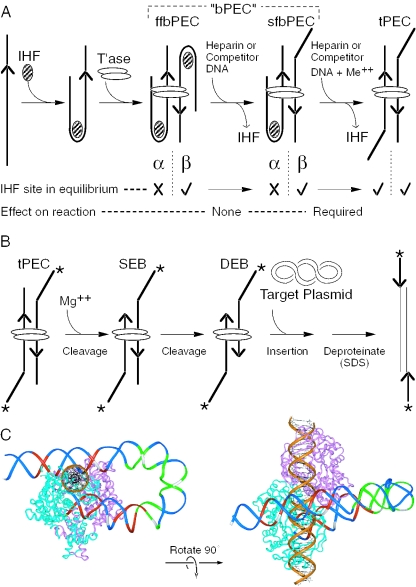
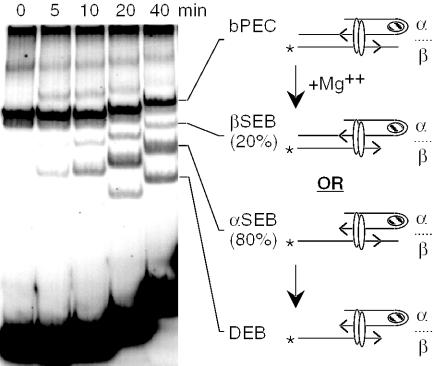
Tn10 transposition has been reconstituted in vitro using purified transposase and transposon ends encoded on short linear fragments of DNA (30,31). The synapsis of the transposon ends by transposase into a paired end complex (PEC) requires binding of the host protein IHF to the specific binding site next to the inverted repeat at the outside end of the element (Figure 1A). The 180° bend in the DNA imposed by IHF provides a set of ‘subterminal’ transposase contacts, located distal to the IHF binding site. The terminal and subterminal transposase contacts, therefore, define a loop that constrains the behavior of the bound IHF (see below).
Figure 1.
Assembly of the Tn10 transpososome and the chemical steps of the reaction. (A) Assembly and unfolding of the synaptic complex. Synapsis of two IHF-bound transposon arms by transposase produces an ffbPEC. Treatment with competitor DNA or heparin strips IHF from the β side of the complex to produce sfbPEC. IHF remains bound to the α side of the complex, presumably due to the subterminal transposase contacts with the transposon end. Divalent metal ion unlocks the IHF binding site on the α side of the complex. Dissociation of IHF produces the tPEC. Hatched oval, IHF; open ovals, transposase; and arrowhead, transposon end. In the tPEC, unoccupied IHF binding sites are indicated by a kink in the transposon end. (B) The chemical steps of the transposition reaction are illustrated using the tPEC as the starting point. In the presence of Mg++, the flanking DNA is cleaved to produce a SEB, followed by a DEB. Non-covalent interactions with a target site are followed by the strand transfer step that produces an insertion product. Asterisk, location of the 32P-label on the transposon end; other elements are as described in (A). (C) The Tn10 transpososome was modeled by superimposing the DNA from the IHF co-crystal structure onto the structure of the Tn5 transpososome (28,32,44,45). Superimposition of the IHF-folded DNA was achieved by minimizing the RMS difference in the position of the equivalent atoms in the Tn5 DNA. One transposon end, IHF and flanking DNA have been omitted for clarity. A section of B DNA (gold) has been docked in the target binding groove to illustrate its spatial relationship to the IHF-folded transposon arm. Regions of transposase and IHF mediated hydroxyl radical protection are shown in red and green, respectively. Every tenth nucleotide on the transferred strand is shown in white. The transposon end is seen embedded in the turquoise monomer of transposase. The subterminal transposase contacts are located on the top of the structure illustrated on the left.
During Tn10 transposition, synapsis of two identical IHF-folded transposon arms produces a fully folded bottom PEC (ffbPEC). However, the IHF-folded transposon arms behave differently and this defines the α and the β sides of the complex (25) (Figure 1A). In the absence of the divalent metal ion cofactor, competitor DNA or heparin treatment strips IHF from the β side of the complex to produce the semi-folded bottom PEC (sfbPEC). On the α side of the complex, IHF remains locked in position until released by the addition of divalent metal ion. Since IHF freely associates and dissociates from DNA in the presence or absence of metal ion, locking of the α IHF binding site is probably mediated by the subterminal transposase contacts. In Figure 1A, this is illustrated by the slightly asymmetric position of the transposase dimer that provides contacts with the α transposon arm. Complete dissociation of IHF produces the top-PEC (tPEC), so called because the extended structure of the unfolded transposon arms causes reduced mobility in the electrophoretic mobility shift assay (EMSA).
In Tn10 transposition, Ca++ can serve as a non-catalytic analog of the physiological cofactor Mg++. The tPEC species can, therefore, be prepared artificially by treating the bPEC with Ca++ to unlock the α transposon arm and using competitor DNA or heparin to strip IHF from the complex (Figure 1A and B). Addition of Mg++ initiates the reaction producing a single end break (SEB), followed by a double end break (DEB) complex. If a closed circular plasmid is provided as a target site, insertion of the transposon ends on opposite strands of the DNA produces a linear insertion product.
The protein–DNA contacts of several intermediates in the Tn10 transposition reaction were previously determined by hydroxyl radical footprinting (32). To display the protection pattern, a molecular model of IHF-folded arm of Tn10 was developed by combining the structure of the related Tn5 transposase dimer with that of the IHF co-crystal (Figure 1C). The contacts in the model fit well with the hydroxyl radical protection pattern and together with several other lines of evidence support the model as an accurate representation of the general shape of the Tn10 transpososome (25–27,32). The molecular model also illustrates the spatial relationship between the terminal and subterminal transposase contacts that define the IHF-loop. This highlights a fascinating structural congruence between the role of IHF in Tn10 excision and phage λ recombination. In phage λ, IHF provides an identical DNA-loop spanned by a single subunit of the integrase protein. In both systems, the site at which recombination is initiated is exactly 30 bases 3′ to the IHF consensus sequence. Since Tn10 and phage λ are unrelated systems, this suggests that the involvement of IHF may impose powerful structural and functional constraints on recombination reactions in general. Indeed, there are further conceptual similarities between the IHF-dependent conformational changes in phage λ and Tn10 (26,33,34).
Tn10 transposition in vitro depends on the presence of negative supercoiling in the DNA. As alluded to above, this requirement is relieved by the presence of IHF and the specific IHF binding site in the outside end of the transposon. Indeed, IHF also functions as a ‘supercoiling relief factor’ in transposition of phage Mu during the lytic cycle of infection (35). From the molecular model in Figure 1C, it is easy to visualize why assembly of the Tn10 transpososome would require a contribution from the energy of IHF binding to bend the DNA and establish the subterminal contacts. However, in addition to its role during transpososome assembly, IHF dissociation is specifically required during two or more steps later in the reaction. The first reason for this is purely structural and is again clearly illustrated by the molecular model in Figure 1C: IHF dissociation is required to unfold the transposon arm that would otherwise block access to the target binding groove and inhibit subsequent target interactions (24,32,36).
The second reason why IHF dissociation from the complex is important is because it promotes conformational changes required during the cleavage steps of the reaction. Cleavage at each transposon end is achieved in three steps: nicking of the transposon end to generate a 3′-OH is followed by a nucleophilic attack on the opposite strand to generate a DNA hairpin, followed by resolution of the hairpin intermediate (37). After unfolding of the α transposon arm as IHF dissociates, two adjacent nucleotides on the transposon end become hypersensitive to hydroxyl radicals (26). Although the precise significance of this is unknown, it almost certainly reflects a conformational change in preparation for the first chemical step of the reaction, a nick located between the two hypersensitive nucleotides. Further evidence that unfolding of the IHF-loop promotes conformational changes is provided by the point mutations RA182 and RA184, which both have altered unfolding properties and block the formation of the hairpin intermediate (27). However, the most direct evidence is that truncation of the α transposon arm, to deprive the complex of the most distal set of subterminal contacts, blocks hairpin resolution and stalls the reaction at the SEB stage (26).
In this work, we have extended our investigation of the role of the IHF-loop in driving the cleavage steps of the reaction and controlling the selection of target sites. We find that the SEB stage of the reaction is most favorable for unfolding of the IHF-loop. This is because the affinity of the IHF binding site appears to change at different intermediate stages of the reaction. The affinity is high during the initial PEC-assembly phase, followed by a drastic reduction during the SEB stage of the reaction. After completion of cleavage, the change is reversed and the DEB intermediate attains a very high affinity for IHF. If IHF is reacquired at this stage of the reaction, the IHF-loop is essentially resistant to unfolding and inhibits target interactions. This provides an additional level of regulation that probably serves to couple the rate of transposition to the physiology of the host cell.
MATERIALS AND METHODS
Materials were generally of the best quality available. Chemicals were from Sigma or VWR. Restriction enzymes and the exo− Klenow fragment of DNA polymerase used for 32P-labeling were from New England Biolabs. The 32P-radiolabeled nucleotides used for end filling of restriction fragments were from Amersham Biosciences.
DNA, proteins and assembly of complexes
Transposase and IHF were expressed and purified as described previously (30,38). The expression plasmids were as follows: wild-type transposase was from pRC60, which contains the transposase gene on an NdeI–BamHI fragment cloned into pET11a; IHF was expressed from pRC188, which is identical to pPR204 obtained from Phoebe Rice. This plasmid contains the IHF operon cloned downstream of the bacteriophage T7 promoter in pET27b (Novagen). Details of the plasmids and restriction digests used to generate the transposon end DNA fragments are given in the respective Figure legends. With the exception of the even-end, all of the linear transposon ends had the natural sequence of the outside end of IS10. The even-end was created by replacing the DNA between bp +19 and +47 of the natural outside end with a tandem repeats of the bases 5′-CTGA (32). This was used as an analog of the inside end of IS10 for the reasons given in the text. Following restriction enzyme digestion as indicated in the respective Figure legends, the DNA was labeled by end filling using a single 32P-labeled deoxynucloetide and the exo− Klenow fragment of DNA polymerase. The transposon end DNA fragment was then purified by electrophoresis in TAE-buffered 5% polyacrylamide gels and recovered by the crush and soak method as described previously (31,39).
Transposase–DNA complexes were assembled and visualized using the EMSA described previously (31,32). The standard reaction was 20 μl and contained 50 fmol of radioactively labeled transposon end, 20 fmol transposase and 300 fmol IHF. When mixed complexes were assembled, 50 fmol of the labeled transposon end was mixed with 200 fmol of the unlabeled end. In experiments with mixed complexes, IHF was the standard 300 fmol, but transposase was increased to 100 fmol. The reactions were assembled at room temperature and the components were mixed before the addition of transposase. Assembly of the complexes is very rapid and they can be visualized in the EMSA after 1 min or after overnight incubation. Where indicated the following additions were made to reactions: 4 mM MgCl2, 4 mM CaCl2, 25 mM EDTA and EGTA, and 500 ng/ml heparin. Quantification was performed using a Fuji phosphorImager.
Transposition reactions with supercoiled plasmid substrates were performed as described previously (24). The plasmid substrates encoded mini-Tn10 transposons, in which two identical outside ends were present in the inverted repeat configuration on either side of a kanamycin resistance marker. A description of the mutations is given in the Figure legend.
Hydroxyl radical footprinting
DNA footprints were generated by hydroxyl radical treatment of protein–DNA complexes as described previously (32). The resultant ladders were compared with Maxam–Gilbert G + A sequence ladders and plotted using NIH image software available from the US National Institutes of Health web server (http://rsb.info.nih.gov/nih-image/).
RESULTS
Slow unfolding of PEC
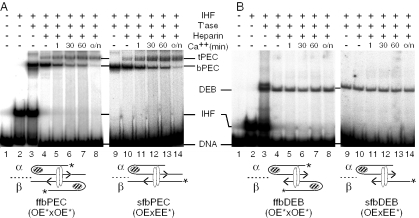
Assembly of the Tn10 transpososome in the presence of IHF produces the bPEC that can be converted to a tPEC by the ejection of the IHF from the complex (25,32,36). The efficiency of bPEC unfolding was addressed by performing a kinetic analysis (Figure 2A). bPEC was assembled by mixing radioactively labeled transposon ends with IHF and transposase. Since bPEC assembly is not 100% efficient, the free and IHF-shifted DNA fragments are also detected (lanes 1–3). When heparin is added, IHF is immediately stripped from the free transposon ends, but most of the bPEC remains in position and is not converted to tPEC (lane 4). As illustrated in Figure 1A, heparin treatment will strip IHF from the β side of the complex, converting the ffbPEC to an sfbPEC. However, since these species comigrate in the gel, this change is not visible (25). When the reaction is supplemented with Ca++, the bPEC unfolds slowly producing the tPEC, reaching 50% completion after half an hour (lanes 5–8). Even after overnight incubation, some folded bPEC still remains.
Figure 2.
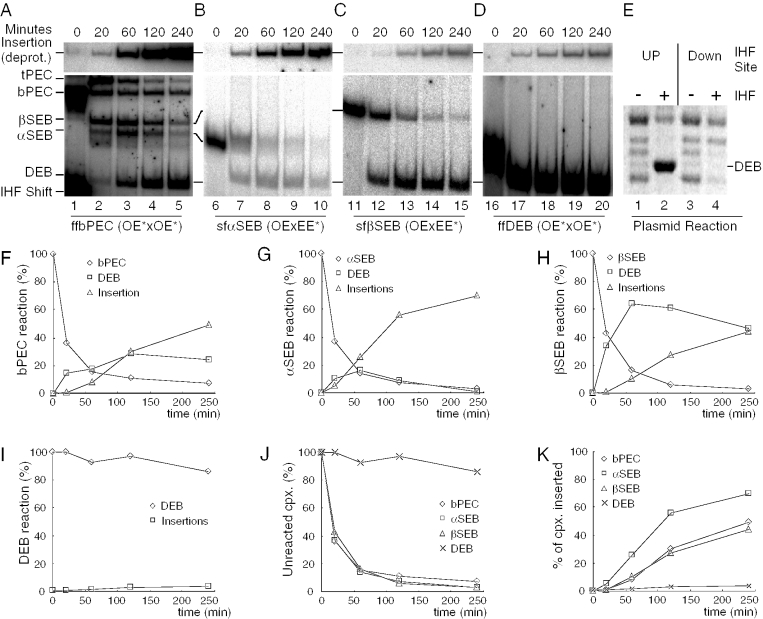
Slow unfolding of the bPEC and DEB complexes. (A) bPEC was assembled and treated with heparin and Ca++ to initiate unfolding. The kinetics of unfolding was monitored using the EMSA. Autoradiograms are shown. Left panel: the transpososome contains two identical outside ends of Tn10. In this and most other experiments, the IHF concentration was limited so that just over 50% of the transposon end DNA fragments are shifted (lanes 2 and 3). The bPEC therefore represents a mixture of ffbPEC and sfbPEC which comigrate in the gel (see Figure 1A). Right panel: the sfbPEC was prepared directly by mixing a radioactively labeled even-end with an unlabeled outside end. The even-end is analogous to the inside end of IS10 that lacks an IHF binding site. The even-end will not form complex without an outside end partner. Since the label is present on the even-end, only mixed complexes are detected. Also, since the even-end lacks an IHF binding site, no IHF-shifted DNA is observed. In the left panel, the outside end was prepared by digesting pRC98 with XbaI+HincII (85 bp transposon arm/62 bp flanking DNA). In the right panel, the outside end was prepared by digesting pRC98 with AccI+SacII (84 bp transposon arm/75 bp flanking DNA). The even-end DNA fragment was prepared by digesting pRC100 with AccI+BamHI (73 bp transposon arm/39 bp flanking DNA). Details of the additions made to the reactions, indicated above the gel, are given in Materials and Methods. OE, outside end; EE, even-end; other details are as given in Figure 1. (B) The DEB complexes were prepared as described in (A) using precleaved transposon ends. The outside end was prepared by digesting pRC35 with BstEII+PvuII (85 bp transposon arm). The even-end DNA fragment was prepared by digesting pRC99 with AccI+PvuII (73 bp transposon arm). Other details are as given in (A).
To demonstrate that the unfolding behavior of the bPEC is owing to the dissociation of the αIHF protomer, mixed complexes were assembled in which only one of the two transposon arms contains an IHF binding site (Figure 2A, right panel). For this purpose, we used an artificial transposon end in which the DNA between bp +19 and +47 was replaced by tandem repeats of the bases 5′-CTGA (32). This is referred to as an ‘even-end’ because the hydroxyl radical cleavage profile in the presence of a saturating concentration of IHF is completely even, indicating that IHF binding is negligible. The even-end fails to form PEC on its own because it lacks an IHF binding site. However, it is efficiently recruited into a mixed PEC when the assembly reaction is supplemented with an outside end fragment. In these experiments, the even-end is the equivalent of the inside end of IS10 which also lacks an IHF binding site. In such mixed complexes, the outside end DNA fragment with its IHF binding site is always located on the α side of the PEC. When these complexes are challenged with heparin and Ca++, unfolding has the same slow kinetics and some bPEC still remains after overnight incubation (Figure 2A, right panel).
Unfolding is blocked in DEB complexes
Tn10 DEB complexes can be prepared directly from DNA fragments in which the flanking DNA has been removed by restriction digestion (40). When supplemented with Mg++, the catalytic metal ion, these complexes can interact with a target site and perform integration. Furthermore, hydroxyl radical footprinting has demonstrated that the transposase and IHF contacts in the DEB complex are identical to those in the bPEC (32). To further investigate the elements required for unfolding, we performed a kinetic analysis of the DEB complex (Figure 2B). The DEB complex was assembled using precleaved transposon ends, in which all the DNA flanking the transposon sequence has been removed. The left panel shows complexes containing two precleaved outside end DNA fragments, while the right panel shows mixed complexes containing one precleaved outside end and one precleaved even-end. Surprisingly, and in complete contrast to the PEC, the DEB complex fails to unfold even after overnight incubation with Ca++ and heparin. This finding suggests that flanking DNA has a key role in the unfolding of the PEC.
Two isomers of the SEB complex have different unfolding characteristics
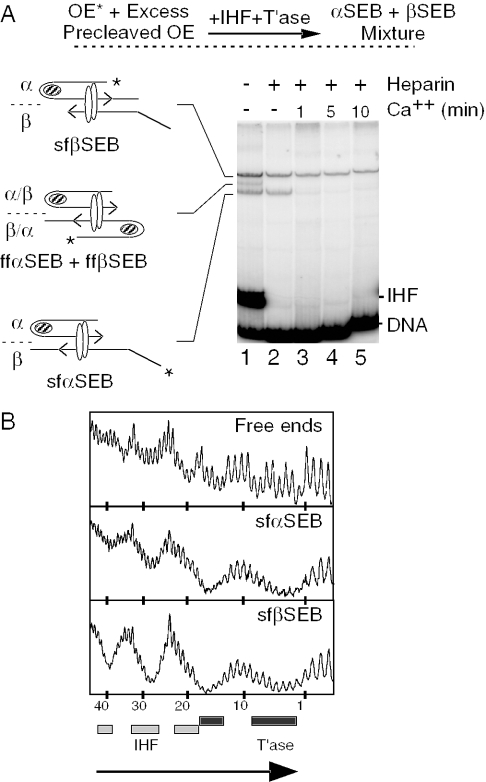
Since the PEC unfolds when challenged with Ca++ plus heparin, while the DEB complex lacking flanking DNA is resistant, we were particularly interested to determine the unfolding behavior of the SEB complex. SEB complexes were assembled by mixing a radiolabeled outside end with an excess of unlabeled precleaved outside end (Figure 3A). In such a mixture, we would expect to assemble two structural isomers of the SEB complex: the αSEB when the cleaved transposon end is on the α side of the complex and the βSEB when it is on the β side of the complex. In fact, three complexes were obtained (lane 1). As will be demonstrated below, this is due to the fact that the SEB complex can exist in the fully folded and semi-folded form, depending on whether one or both IHF sites are occupied. In contrast to the α IHF site, occupation of the β IHF site is irrelevant to the reaction as it is always in equilibrium with IHF in solution (Figure 1A). Furthermore, in contrast to the bPEC, the fully folded and semi-folded SEB complexes are resolved in the EMSA.
Figure 3.
Identification of two isomers of the SEB complex. (A) SEB complex was assembled by mixing a radioactively labeled uncleaved outside end with an excess of unlabeled precleaved outside end. This biases the reaction strongly in favor of the assembly of mixed complexes. The uncleaved outside end was prepared by digesting pRC167 with PstI+XhoI (96 bp transposon arm/77 bp flanking DNA). The precleaved outside end was prepared by digesting pRC35 with PvuII+BstEII (87 bp transposon arm). sfαSEB, semi-folded α-single-end-break; sfβSEB, semi-folded β-single-end-break. Other details are as given in Figures 1 and 2. (B) The sfαSEB and sfβSEB were footprinted with hydroxyl radicals. Briefly, complexes were assembled, treated in solution with hydroxyl radicals and separated using the EMSA. The complexes were recovered from the gel and the footprints were displayed on a DNA sequencing gel. Dark and light shaded boxes represent the transposase and IHF footprints as described previously (32). The large arrowhead indicates the location of the transposon end. The number of base pair inside the transposon is indicated.
The lower and upper bands of the triplet represent the semi-folded αSEB isomer (sfαSEB) and the semi-folded βSEB isomer (sfβSEB), respectively. This was demonstrated by hydroxyl radical footprinting of the complexes (Figure 3B). In the sfαSEB, the radioactively labeled uncleaved-transposon end is on the unfolded β side of the complex. As expected, it shows little or no evidence of an IHF footprint. In the sfβSEB, the radioactively labeled uncleaved-transposon end is on the folded α side of the complex. As expected, this complex has the strong footprint of IHF.
The central band of the triplet represents a mixture of the fully folded (ff) αSEB and βSEB which comigrate in the gel. This was demonstrated by two observations: first, the upper and lower band of the triplet are converted to the central band at progressively higher IHF concentrations (data not shown); and second, the central band disappears at low IHF concentration (data not shown) and after heparin treatment to strip IHF from the β side of the complex (Figure 3A, lane 2).
At this point, it should be noted that unfolding of the SEB complex does not produce a top-SEB in the EMSA in the same way that unfolding of the bPEC produces a tPEC. Instead, the SEB complex disappears from the gel when it is completely unfolded. However, the unfolded SEB are still present in the reaction mixture because they reappear in the EMSA if the IHF lost during unfolding is added back (36). This behavior can be explained if the unfolded SEB complexes are simply unstable during electrophoresis.
To determine the unfolding behavior of the αSEB and βSEB isomers, the complexes were treated with heparin and Ca++, and the kinetics of unfolding were followed for 10 min (Figure 3A, lanes 3–5). The αSEB disappeared within 1 min, indicating that it unfolds very rapidly. In contrast, the βSEB was almost completely resistant to unfolding. This result suggests that the presence of flanking DNA on the β side of the complex is somehow required to drive or license unfolding of the transpososome.
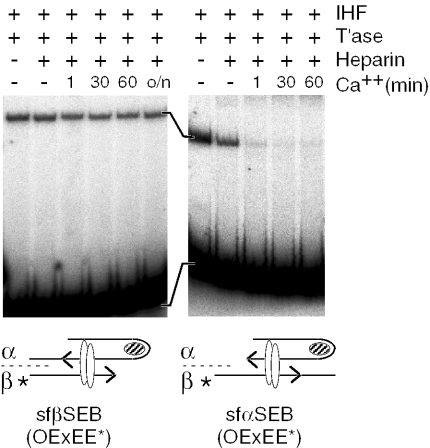
To further confirm the identity of the SEB complexes and to validate their surprising unfolding behavior, the two isomers of the SEB complex were prepared directly by mixing outside end and even-end DNA fragments (Figure 4). Recall that the absence of an IHF binding site on the even-end ensures that it is always incorporated on the β side of the complex. As before, the radioactive label was present only on the even-end, so that only mixed complexes are detected. The sfβSEB was prepared by mixing an uncleaved outside end with a precleaved even-end (left panel). Counter wise, the sfαSEB was prepared by mixing an uncleaved even-end and a precleaved outside end (right panel). When the complexes are challenged with Ca++ to unlock the α side of the complex and with heparin to strip out the IHF, only the αSEB complex unfolds. The βSEB complex was highly resistant to unfolding and 80% remained after overnight incubation (Figure 4, left panel). These results confirm the unfolding behavior shown in Figure 3 where the αSEB and βSEB complexes were prepared in a single mixture.
Figure 4.
Kinetics and requirements for unfolding of the SEB isomers. Mixed complexes were assembled using the transposon ends illustrated below each panel. There was a 4-fold molar excess of the outside end present. The outside end was prepared by digesting pRC98 with AccI+ScaI (84 bp transposon arm/75 bp flanking DNA). The precleaved outside end was prepared by digesting pRC35 with BstEII+PvuII (85 bp transposon arm). The even-end DNA fragment was prepared by digesting pRC100 with AccI+BamHI (73 bp transposon arm/39 bp flanking DNA). The precleaved even-end was prepared by digesting pRC99 with AccI+PvuII (73 bp transposon arm). Other details are as given in Figure 2.
The α side of the complex is cleaved preferentially
The unfolding of the αSEB in less than a minute is extremely fast compared with the bPEC that takes several hours or to the βSEB and DEB complexes which are essentially resistant to unfolding. Since unfolding is required for target interactions later in the reaction, the cleavage reaction might be expected to have a bias toward the αSEB intermediate. This would occur if the α flanking DNA is cleaved in preference to the β flanking DNA.
To address this question, a bPEC was assembled from a mixture of outside and even-end substrates that have different lengths of flanking DNA (Figure 5). The different lengths of the flanking DNA ensures that the fully folded forms of the α and βSEB are resolved in the EMSA at a saturating concentration of IHF which would otherwise cause them to comigrate (see Figure 3A). Since no target DNA was provided, the DEB complex is the end product in these reactions. Also, since no heparin was added, IHF is able to reassociate with the outside end fragment even if it has been ejected during a previous step in the reaction.
Figure 5.
The cleavage step is biased toward the αSEB isomer. The standard complex assembly reaction was scaled up and initiated at time zero by the addition of Mg++. Aliquots were removed at the indicated times and loaded directly onto the gel that was under electrical tension. The outside end and even-end DNA fragments were prepared by digesting pRC167 and pRC100 with PstI+XhoI and HindIII+SpeI, respectively. To preclude any potential influence on the rate of cleavage by the sequence of bases flanking the transposon, the flanking DNA on each fragment is isogenic. The outside and even-ends are also isogenic out to bp 19 of the transposon end. Other details are as given in Figure 2.
As before, the radioactive label was present only on the even-end DNA fragment so that only mixed complexes are detected in the EMSA (Figure 5). This also ensures that the outside end, with the long flanking DNA, is always incorporated on the α side of the complex. The bPEC was assembled in the absence of divalent metal ion and the reaction was initiated by the addition of Mg++. At the indicated time points, aliquots were removed from the reaction and loaded onto the gel that was under electrical tension. The fact that electrophoresis was underway throughout the time course accounts for the displacement of the bands upwards at later time points. The αSEB is the first cleavage intermediate to appear after only 5 min. A faint signal representing the βSEB appears after 10 min and the DEB appears at 20 min. At 40 min, 80% of the SEB intermediate is the αSEB isomer. This is the bias expected if the order of cleavage is to favor unfolding of the transpososome at the SEB stage of the reaction.
The efficiency of unfolding dictates the efficiency of insertions
All of the unfolding experiments described above were performed under non-catalytic conditions. We therefore sought to determine whether the unfolding kinetics of the various complexes is correlated with the efficiency of the insertion reaction (Figure 6). A single large reaction was assembled for each of the complexes and initiated at time zero by addition of the catalytic metal ion in a mixture with a closed circular target plasmid. The target plasmid also served as a competitor that will sequester IHF after a single cycle of dissociation from the transpososome. At each time point, an aliquot was removed from the reaction and stopped by the addition of EDTA to chelate the Mg++. Half of each aliquot was used to visualize the complexes, while the other half was deproteinated by SDS treatment to allow detection of the large insertion product. For each type of complex, the native and denatured sets of samples were electrophoresed on the same gel so that the quantification of the native complexes and deproteinated insertion products would be directly comparable.
Figure 6.
Insertion kinetics of the different complexes. (A–D) and (F–K) The standard 20 μl reaction was scaled up to 200 μl and initiated at time zero by the addition of Mg++ in a mixture with 10 μg of pBluescript as target DNA. Aliquots (40 μl) were withdrawn at the indicated time and stopped by the addition of EDTA. Half of the reaction was deproteinated by SDS treatment to allow detection of the large insertion product. For each type of complex, the native and deproteinated aliquots (20 μl) were electrophoresed on the same gel so that the quantification of the various complexes and products was exactly comparable and did not require normalization. The sequence of bases flanking the transposon ends on each fragment was isogenic to preclude any potential effect on the rate of cleavage. bPEC panel; the outside end was prepared by digesting pRC98 with AccI+SacII (84 bp transposon arm/75 bp flanking DNA). αSEB panel: the precleaved outside end was prepared by digesting pRC35 with BstEII+PvuII (85 bp transposon arm); the uncleaved even-end DNA fragment was prepared by digesting pRC100 with AccI+BamHI (73 bp transposon arm/39 bp flanking DNA). βSEB panel: the uncleaved outside end was prepared by digesting pRC98 with AccI+SacII (84 bp transposon arm/75 bp flanking DNA); the precleaved even-end was prepared by digesting pRC99 with AccI+PvuII (73 bp transposon arm). DEB panel: the precleaved outside end was prepared by digesting pRC35 with BstEII+PvuII (85 bp transposon arm). (E) Transposition reactions (200 μl) contained 1 nM plasmid substrate encoding a mini-Tn10 transposon, 6 nM transposase and 35 nM IHF. The reactions were incubated at 30°C for 3 h, concentrated by ethanol precipitation and analyzed on a TBE-buffered 1.1% agarose gel (30,39). Excision of the transposon segment from the supercoiled plasmid substrate produces the DEB product (referred to in previous publications as the ‘excised transposon fragment’ or ETF). Auto-integration of the DEB product produces a topologically complex set of knot and catenane products (39). Apart from the DEB product, these are the only products present in the section of the gel shown. The IHF-up and IHF-down mutations were encoded on plasmids pNK2588 and pNK2590, respectively. IHF-up (single point mutation) and IHF-down (triple point mutation) move the IHF binding site closer to or further away from the IHF consensus binding site as described in Figure 1 of ref. (46). A reverse contrast photograph of an ethidium bromide stained agarose gel is shown.
When the reaction with the bPEC is initiated by adding the mixture of Mg++ and target plasmid, the IHF-retarded DNA located at the bottom left hand corner disappears (Figure 6A). This indicates that the IHF is effectively competed away from the free transposon ends by the target plasmid. At the first time point, 37% of the bPEC has been converted to tPEC due to the release of IHF. During the course of the reaction, both isomers of the SEB intermediate accumulate and then disappear as they are converted to DEB. The rapid appearance of the βSEB isomer contradicts the results in Figure 5, where 80% of the SEB complexes are contributed by the αSEB isomer. This discrepancy is due to the presence of competitor DNA in this experiment and the fact that αSEB unfolds much more rapidly than the βSEB (Figures 3 and 4). At the end of the time course, 50% of the bPEC has been converted to insertion products, 25% remains at the DEB stage and the remainder is accounted for by the tPEC, bPEC and SEB species (Figure 6A and F).
The kinetics of the two SEB isomers is significantly different from each other. In the reactions with the αSEB complex, the DEB behaves like a reaction intermediate; it accumulates at early time points and then diminishes as it is converted to insertion product (Figure 6B and G). However, in reactions with the βSEB complex, the DEB accumulates more rapidly but it appears to be trapped and is converted only very slowly to the insertion product (Figure 6C and H). Finally, the kinetics of the reaction with the DEB complex is completely different from the others and only 4% is converted to insertion product during the 4 h time course (Figure 6D and I). Overall, the kinetics of the insertion reactions fits very well with the unfolding kinetics under non-catalytic conditions: αSEB > bPEC > βSEB ≫ DEB (Figures 2–4).
The most important aspect of these results is that IHF is locked into the DEB complex and inhibits the integration step of the reaction. To confirm that this behavior is not an artifact due to the short linear DNA substrates, another experiment was performed with supercoiled plasmid substrates in the presence or absence of IHF (Figure 6E). With supercoiled substrates, initiation of the reaction is independent of IHF (39) and a set of topologically complex integration products are detected (lanes 1 and 3). When the transposon end has a strong IHF binding site, the DEB product of the reaction accumulates (lane 2) (24). However, inhibition of the integration step is relieved when the IHF binding site is abolished by the introduction of a triple point mutation that destroys the IHF consensus binding site (lane 4).
When the amount of unreacted complex is plotted, the bPEC, αSEB and βSEB disappear with identical kinetics (Figure 6J). However, this observation is difficult to interpret because these complexes may disappear due to unfolding and/or cleavage. The kinetics of insertion accumulation is more informative (Figure 6K). The bPEC and βSEB have almost identical kinetics. However, since the βSEB starts at a later point on the reaction pathway, this suggests that the kinetics of the bPEC reaction is probably slightly faster. The reaction with the αSEB complex goes closer to completion than the others with 70% of the starting material converted to insertion products. Furthermore, this difference is even greater at early time points, indicating that the kinetics of the reaction is also faster. This suggests that the most favorable pathway for cleavage is via the αSEB intermediate. The difference between the kinetics of the αSEB and βSEB reactions is largely owing to the amount of material that remains trapped at the DEB stage. Overall, it therefore appears that the transposition reaction stalls at the DEB stage unless unfolding is achieved before cleavage.
DISCUSSION
Multiple pathways to unfold the IHF-loop
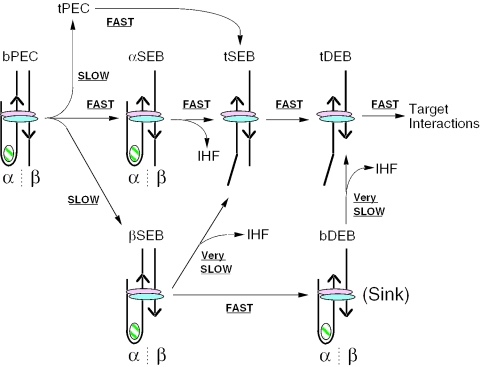
The IHF-loop has an important structural role during assembly of the Tn10 transpososome. However, it is also required to promote conformational changes during the cleavage steps of the reaction. This was demonstrated previously because disruption of the loop blocks resolution of the DNA hairpin intermediate on the first transposon end to be cleaved and stalls the reaction at the SEB stage (26). The results presented here provide further insight into this mechanism by showing that there are multiple pathways for unfolding of the IHF-loop and cleavage of the transposon ends. The pathways are summarized in Figure 7 where they are illustrated for a mixed complex that represents transposition of IS10 in which only one transposon end has an IHF binding site.
Figure 7.
Multiple pathways for the unfolding and cleavage of the Tn10 transpososome. The model summarizes the potential of the transpososome to unfold at different stages of the cleavage reaction. At the start of the reaction, the bPEC can follow one of the three pathways. The least productive is via the βSEB intermediate. Transpososomes that arrive at the DEB stage of the reaction with IHF still associated, or reacquire IHF at this stage of the reaction, unfold and perform the strand transfer step very slowly.
IHF must be ejected from the complex at some point during the reaction to allow unfolding and subsequent target interactions. In principle, this could take place before cleavage (bPEC), after cleavage (DEB) or during cleavage (SEB). We found that under non-catalytic conditions, the most favorable point in the reaction was during the SEB stage. The capacity of the various complexes to unfold under catalytic conditions was also determined (Figure 6). However, the interpretation of these experiments is complicated because the unfolded SEB intermediates are invisible in the EMSA. Thus, the disappearance of an SEB intermediate could be either due to cleavage of the flanking DNA or due to unfolding. This problem was circumvented by measuring the efficiency of strand transfer into a target plasmid that requires both unfolding of the transposon arms and cleavage of the flanking DNA (36,40). The efficiency with which the different complexes produced insertions was identical to the efficiency of unfolding under non-catalytic conditions: αSEB > bPEC > βSEB ≫ DEB.
Cyclic changes in IHF-binding affinity
Previously, the molecular spring model for unfolding of the Tn10 transpososome proposed that IHF was ejected by micro-mechanical movements that produce a torsional force in the IHF-loop (24). The cyclic changes in the affinity of the IHF binding site demonstrated in the present work supports this view and for the first time suggests that ejection occurs specifically at the SEB stage of the reaction. Since Tn10 transposition involves no high energy cofactors, the energy to drive the conformational changes is presumably derived from the binding energy of the protein components. This is supported by the fact that transposition is irreversible and that the transpososome becomes progressively more stable as the reaction progresses. This raises the question of where in the complex the energy is stored? In the original molecular spring model, energy was stored in the distortion of the DNA in the IHF-loop. However, the present work raises the possibility that the driving energy is stored in the distortion of the DNA flanking the transposon sequences.
Coupling the rate of transposition to the physiological state of the cell
In vivo, IHF regulates Tn10 transposition positively and negatively depending on the length of the transposon and whether it is located on the chromosome or a multi-copy plasmid (41). These observations can be rationalized based on the effects of IHF and supercoiling on reactions performed in vitro (24). In the absence of IHF, supercoiling promotes assembly of the transpososome and the cleavage steps of the reaction. However, after excision, the presence of supercoiling in the excised transposon reduces the rate of transposition by favoring non-productive intramolecular target interactions (39). Likewise, low concentrations of IHF stimulate transpososome assembly and the cleavage steps, while high concentrations favor non-productive intramolecular target sites. The effects of IHF and supercoiling, therefore, both act homeostatically by stimulating the early steps of transposition but inhibiting the late steps. This homeostatic mechanism is further reinforced because IHF and supercoiling may respond in opposite directions during environmental changes. When cells enter stationary phase, for example, supercoiling declines up to 50% while the level of IHF increases 10-fold (42,43).
The present work supports this view of the mechanism of IHF regulation and provides an additional insight. Even if transposome assembly is stimulated by supercoiling without the involvement of IHF, there is a window of opportunity at the DEB stage during which the reaction will be sensitive to the concentration of IHF in the host cell. Indeed, most of the difference between the insertion efficiency of the various complexes can be accounted for by the amount of material that remains trapped as folded-DEB complex at the end of the time course (Figure 6): DEB (86%) > βSEB (46%) > PEC (25%) > αSEB (1%).
Conformational coupling between opposite sides of the transpososome
We have shown previously that the bPEC is asymmetric even when it contains two identical outside ends (Figure 1A). The asymmetry is revealed by the different unfolding behavior of the transposon ends that defines the α and β sides of the complex. The biological rationale for the asymmetry is that the molecular mechanism of transposition is tailored to accommodate the structure of IS10 which has an IHF binding site at only one end. The unfolding behavior of the two SEB isomers represents an additional aspect of the structural asymmetry. Specifically, the presence of flanking DNA on the β side of the complex is communicated to the opposite side of the complex, promoting or licensing the unfolding of the α transposon arm.
Acknowledgments
This work was funded by The Wellcome Trust grant 065119 to R.C. Funding to pay the Open Access publication charges for this article was provided by The Wellcome Trust and a grant from JISC to Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Katinka M.D., Duprat S., Cornillot E., Metenier G., Thomarat F., Prensier G., Barbe V., Peyretaillade E., Brottier P., Wincker P., et al. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature. 2001;414:450–453. doi: 10.1038/35106579. [DOI] [PubMed] [Google Scholar]
- 2.Mahillon J., Chandler M. Insertion sequences. Microbiol. Mol. Biol. Rev. 1998;62:725–774. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis B.M., Waldor M.K. Mobile genetic elements and bacterial pathogenesis. In: Craig N.L., Craigie R., Gellert M., Lambowitz A.M., editors. Mobile DNA II. Washington, DC: American Society for Microbiology; 2002. pp. 1040–1059. [Google Scholar]
- 4.Campbell A. Eubacterial genomes. In: Craig N.L., Craigie R., Gellert M., Lambowitz A.M., editors. Mobile DNA II. Washington, DC: American Society for Microbiology; 2002. pp. 1024–1039. [Google Scholar]
- 5.Chalmers R., Blot M. Insertion sequences and transposons. In: Charlebois R.L., editor. Organization of the Prokaryotic Genome. Washington, DC: American Society for Microbiology; 1999. pp. 151–169. [Google Scholar]
- 6.Buisine N., Tang C.M., Chalmers R. Transposon-like Correia elements: structure, distribution and genetic exchange between pathogenic Neisseria sp. FEBS Lett. 2002;522:52–58. doi: 10.1016/s0014-5793(02)02882-x. [DOI] [PubMed] [Google Scholar]
- 7.Jiang N., Bao Z., Zhang X., Hirochika H., Eddy S.R., McCouch S.R., Wessler S.R. An active DNA transposon family in rice. Nature. 2003;421:163–167. doi: 10.1038/nature01214. [DOI] [PubMed] [Google Scholar]
- 8.Jiang N., Bao Z., Zhang X., Eddy S.R., Wessler S.R. Pack-MULE transposable elements mediate gene evolution in plants. Nature. 2004;431:569–573. doi: 10.1038/nature02953. [DOI] [PubMed] [Google Scholar]
- 9.Lipkow K., Buisine N., Lampe D.J., Chalmers R. Early intermediates of mariner transposition: catalysis without synapsis of the transposon ends suggests a novel architecture of the synaptic complex. Mol. Cell. Biol. 2004;24:8301–8311. doi: 10.1128/MCB.24.18.8301-8311.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Plasterk R.H. The HIV integrase catalytic core. Nature Struct. Biol. 1995;2:87–90. doi: 10.1038/nsb0295-87. [DOI] [PubMed] [Google Scholar]
- 11.Kipling D., Warburton P.E. Centromeres, CENP-B and Tigger too. Trends Genet. 1997;13:141–145. doi: 10.1016/s0168-9525(97)01098-6. [DOI] [PubMed] [Google Scholar]
- 12.Pardue M.-L., DeBaryshe P.G. Telomeres and transposable elements. In: Craig N.L., Craigie R., Gellert M., Lambowitz A.M., editors. Mobile DNA II. Washington, DC: American Society for Microbiology; 2002. pp. 870–887. [Google Scholar]
- 13.Gellert M. V(D)J recombination: rag proteins, repair factors, and regulation. Annu. Rev. Biochem. 2002;71:101–132. doi: 10.1146/annurev.biochem.71.090501.150203. [DOI] [PubMed] [Google Scholar]
- 14.van-Gent D.C., Mizuuchi K., Gellert M. Similarities between initiation of V(D)J recombination and retroviral integration [see comments] Science. 1996;271:1592–1594. doi: 10.1126/science.271.5255.1592. [DOI] [PubMed] [Google Scholar]
- 15.Rice P.A., Baker T.A. Comparative architecture of transposase and integrase complexes. Nature Struct. Biol. 2001;8:302–307. doi: 10.1038/86166. [DOI] [PubMed] [Google Scholar]
- 16.Segall A.M., Goodman S.D., Nash H.A. Architectural elements in nucleoprotein complexes: interchangeability of specific and non-specific DNA binding proteins. EMBO J. 1994;13:4536–4548. doi: 10.1002/j.1460-2075.1994.tb06775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Travers A., Muskhelishvili G. DNA microloops and microdomains: a general mechanism for transcription activation by torsional transmission. J. Mol. Biol. 1998;279:1027–1043. doi: 10.1006/jmbi.1998.1834. [DOI] [PubMed] [Google Scholar]
- 18.Travers A., Muskhelishvili G. DNA supercoiling—a global transcriptional regulator for enterobacterial growth? Nature Rev. Microbiol. 2005;3:157–169. doi: 10.1038/nrmicro1088. [DOI] [PubMed] [Google Scholar]
- 19.Lilley D.M. DNA–protein interactions. HMG has DNA wrapped up. Nature. 1992;357:282–283. doi: 10.1038/357282a0. [DOI] [PubMed] [Google Scholar]
- 20.Bell S.P., Dutta A. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- 21.Nyberg K.A., Michelson R.J., Putnam C.W., Weinert T.A. Toward maintaining the genome: DNA damage and replication checkpoints. Annu. Rev. Genet. 2002;36:617–656. doi: 10.1146/annurev.genet.36.060402.113540. [DOI] [PubMed] [Google Scholar]
- 22.Sancar A., Lindsey-Boltz L.A., Unsal-Kacmaz K., Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 23.Smogorzewska A., de Lange T. Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. 2004;73:177–208. doi: 10.1146/annurev.biochem.73.071403.160049. [DOI] [PubMed] [Google Scholar]
- 24.Chalmers R., Guhathakurta A., Benjamin H., Kleckner N. IHF modulation of Tn10 transposition: sensory transduction of supercoiling status via a proposed protein/DNA molecular spring. Cell. 1998;93:897–908. doi: 10.1016/s0092-8674(00)81449-x. [DOI] [PubMed] [Google Scholar]
- 25.Sewitz S., Crellin P., Chalmers R. The positive and negative regulation of Tn10 transposition by IHF is mediated by structurally asymmetric transposon arms. Nucleic Acids Res. 2003;31:5868–5876. doi: 10.1093/nar/gkg797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crellin P., Sewitz S., Chalmers R. DNA looping and catalysis; the IHF-folded arm of Tn10 promotes conformational changes and hairpin resolution. Mol. Cell. 2004;13:537–547. doi: 10.1016/s1097-2765(04)00052-8. [DOI] [PubMed] [Google Scholar]
- 27.Humayun S., Wardle S.J., Shilton B.H., Pribil P.A., Liburd J., Haniford D.B. Tn10 transposase mutants with altered transpososome unfolding properties are defective in hairpin formation. J. Mol. Biol. 2005;346:703–716. doi: 10.1016/j.jmb.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 28.Davies D.R., Goryshin I.Y., Reznikoff W.S., Rayment I. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science. 2000;289:77–85. doi: 10.1126/science.289.5476.77. [DOI] [PubMed] [Google Scholar]
- 29.Haniford D.B. Transposon Tn10. In: Craig N.L., Craigie R., Gellert M., Lambowitz A.M., editors. Mobile DNA II. Washington, DC: American Society for Microbiology; 2002. pp. 457–483. [Google Scholar]
- 30.Chalmers R.M., Kleckner N. Tn10/IS10 transposase purification, activation, and in vitro reaction. J. Biol. Chem. 1994;269:8029–8035. [PubMed] [Google Scholar]
- 31.Sakai J., Chalmers R.M., Kleckner N. Identification and characterization of a pre-cleavage synaptic complex that is an early intermediate in Tn10 transposition. EMBO J. 1995;14:4374–4383. doi: 10.1002/j.1460-2075.1995.tb00112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crellin P., Chalmers R. Protein–DNA contacts and conformational changes in the Tn10 transpososome during assembly and activation for cleavage. EMBO J. 2001;20:3882–3891. doi: 10.1093/emboj/20.14.3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Radman-Livaja M., Shaw C., Azaro M., Biswas T., Ellenberger T., Landy A. Arm sequences contribute to the architecture and catalytic function of a lambda integrase–Holliday junction complex. Mol. Cell. 2003;11:783–794. doi: 10.1016/s1097-2765(03)00111-4. [DOI] [PubMed] [Google Scholar]
- 34.Aihara H., Kwon H.J., Nunes-Duby S.E., Landy A., Ellenberger T., Radman-Livaja M., Shaw C., Azaro M., Biswas T. A conformational switch controls the DNA cleavage activity of lambda integrase Arm sequences contribute to the architecture and catalytic function of a lambda integrase–Holliday junction complex. Mol. Cell. 2003;12:187–198. [Google Scholar]
- 35.Surette M.G., Lavoie B.D., Chaconas G. Action at a distance in Mu DNA transposition: an enhancer-like element is the site of action of supercoiling relief activity by integration host factor (IHF) EMBO J. 1989;8:3483–3489. doi: 10.1002/j.1460-2075.1989.tb08513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sakai J.S., Kleckner N., Yang X., Guhathakurta A. Tn10 transpososome assembly involves a folded intermediate that must be unfolded for target capture and strand transfer. EMBO J. 2000;19:776–785. doi: 10.1093/emboj/19.4.776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kennedy A.K., Guhathakurta A., Kleckner N., Haniford D.B. Tn10 transposition via a DNA hairpin intermediate. Cell. 1998;95:125–134. doi: 10.1016/s0092-8674(00)81788-2. [DOI] [PubMed] [Google Scholar]
- 38.Lynch T.W., Mattis A.N., Gardner J.F., Rice P.A. Integration host factor: putting a twist on protein–DNA recognition. J. Mol. Biol. 2003;330:493–502. doi: 10.1016/s0022-2836(03)00529-1. [DOI] [PubMed] [Google Scholar]
- 39.Chalmers R.M., Kleckner N. IS10/Tn10 transposition efficiently accommodates diverse transposon end configurations. EMBO J. 1996;15:5112–5122. [PMC free article] [PubMed] [Google Scholar]
- 40.Sakai J., Kleckner N. The Tn10 synaptic complex can capture a target DNA only after transposon excision. Cell. 1997;89:205–214. doi: 10.1016/s0092-8674(00)80200-7. [DOI] [PubMed] [Google Scholar]
- 41.Signon L., Kleckner N. Negative and positive regulation of Tn10/IS10-promoted recombination by IHF: two distinguishable processes inhibit transposition off of multicopy plasmid replicons and activate chromosomal events that favor evolution of new transposons. Genes Dev. 1995;9:1123–1136. doi: 10.1101/gad.9.9.1123. [DOI] [PubMed] [Google Scholar]
- 42.Ditto M.D., Roberts D., Weisberg R.A. Growth phase variation of integration host factor level in Escherichia coli. J. Bacteriol. 1994;176:3738–3748. doi: 10.1128/jb.176.12.3738-3748.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murtin C., Engelhorn M., Geiselmann J., Boccard F. A quantitative UV laser footprinting analysis of the interaction of IHF with specific binding sites: re-evaluation of the effective concentration of IHF in the cell. J. Mol. Biol. 1998;284:949–961. doi: 10.1006/jmbi.1998.2256. [DOI] [PubMed] [Google Scholar]
- 44.Lovell S., Goryshin I.Y., Reznikoff W.R., Rayment I. Two-metal active site binding of a Tn5 transposase synaptic complex. Nature Struct. Biol. 2002;9:278–281. doi: 10.1038/nsb778. [DOI] [PubMed] [Google Scholar]
- 45.Rice P.A., Yang S., Mizuuchi K., Nash H.A. Crystal structure of an IHF–DNA complex: a protein-induced DNA U-turn. Cell. 1996;87:1295–1306. doi: 10.1016/s0092-8674(00)81824-3. [DOI] [PubMed] [Google Scholar]
- 46.Huisman O., Errada P.R., Signon L., Kleckner N. Mutational analysis of IS10's outside end. EMBO J. 1989;8:2101–2109. doi: 10.1002/j.1460-2075.1989.tb03619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]