Abstract
The thin pili of IncI1 plasmid R64, which is required for conjugation in liquid media, belong to the type IV pilus family. They consist of a major subunit, the pilS product, and a minor component, one of the seven pilV products. The pilS product is first synthesized as a 22-kDa prepilin, processed to a 19-kDa mature pilin by the function of the pilU product, and then secreted outside the cell. The mature pilin is assembled to form a thin pilus with the pilV product. To reveal the relationship between the structure and function of the pilS product, 27 missense mutations, three N-terminal deletions, and two C-terminal deletions were constructed by PCR and site-directed mutagenesis. The characteristics of 32 mutant pilS products were analyzed. Four pilS mutant phenotype classes were identified. The products of 10 class I mutants were not processed by prepilin peptidase; the extracellular secretion of the products of two class II mutants was inhibited; from 11 class III mutants, thin pili with reduced activities in liquid mating were formed; from 9 class IV mutants, thin pili with mating activity similar to that of the wild-type pilS gene were formed. The point mutations of the class I mutants were distributed throughout the prepilin sequence, suggesting that processing of the pilS product requires the entire prepilin sequence.
Type IV pili are flexible, rod-like, polarly inserted surface appendages protruding from the cell surface of gram-negative bacteria including Pseudomonas aeruginosa, Bacteroides nodosus, Neisseria gonorrhoeae, Moraxella bovis, Vibrio cholerae, and enteropathogenic and enterotoxigenic Escherichia coli (9, 19, 20, 23, 27, 32). Type IV pili promote the attachment of bacterial pathogens to receptors of host cells during colonization, and they mediate the bacterial locomotion called twitching motility of P. aeruginosa (35) and the social gliding motility of Myxococcus xanthus (36). In addition, they act as receptors for pilus-specific bacteriophage (6).
Type IV pili are polymers of type IV pilin subunits (23, 27), which are produced from type IV prepilins by the function of prepilin peptidases (18). In many cases, the N-terminal amino acid of mature pilin is phenylalanine and is N-methylated. In P. aeruginosa, both processing of prepilin and N-methylation of mature pilin are catalyzed by a single bifunctional enzyme, the PilD protein (28). Among all type IV pilins, the N-terminal region including the cleavage site is highly conserved. Particularly, the C-terminal amino acid of the prepeptide is invariantly glycine, and the fifth amino acid of mature pilin is always glutamic acid. The C-terminal one-third of mature pilin forms a disulfide loop between two conserved cysteine residues (21, 25).
During bacterial conjugation, the donor cells harboring self-transmissible plasmids synthesize sex pili encoded by the genes on the plasmids (6). Sex pili of donor cells create a specific contact with recipient cells, leading to the formation of a mating pair. IncI1 plasmids such as R64 and ColIb-P9 form two types of sex pili, a thick rigid pilus and a thin flexible one (1, 2). Thick rigid pili are required for both surface and liquid mating, while thin flexible pili are required only for liquid mating. Cells producing R64 thin pili become sensitive to bacteriophages Iα and PR64FS, which adsorb to the shaft and tip of IncI1 thin pilus, respectively (4, 5).
DNA sequence analysis of the R64 pil region responsible for thin-pilus formation revealed that the pil region consists of 14 genes, pilI through pilV, and that several pil products contain amino acid sequence homology with proteins involved in type IV pilus biogenesis (11) (Fig. 1A). Thus, the R64 thin pilus was shown to belong to the type IV family, specifically group IVB, of pili.
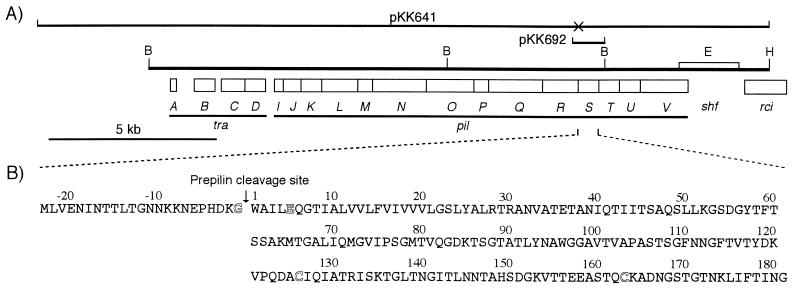
FIG. 1.
(A) Organization of the tra-pil region of plasmid R64. The horizontal bold line represents a restriction map. B, BglII; E, EcoRI; H, HindIII. The open bar above the map represents the extent of movement of the EcoRI site through DNA rearrangement of the shufflon. Below the map, the open reading frames are represented by open bars. tra, transfer; pil, formation of thin pilus; shf, shufflon; rci, recombinase for the shufflon. DNA regions of pKK641 and pKK692 are indicated above the map. The cross on pKK641 marks the location of the pilS1 mutation. (B) Amino acid sequence of the PilS protein. The downward arrow indicates the type IV prepilin cleavage site. The conserved glycine, glutamic acid, and two cysteine residues are indicated by the outline letters.
R64 and ColIb-P9 thin pili were sedimented by ultracentrifugation from the culture medium, in which E. coli cells harboring R64- and ColIb-P9-derived plasmids had grown, and purified by CsCl density gradient centrifugation (13, 37). In negatively stained thin-pilus samples, long rods with a diameter of 6 nm, characteristic of type IV pili, were observed under an electron microscope. R64 and ColIb-P9 thin pili consist of a major 19-kDa pilin protein, the product of the pilS gene, and a minor 45-kDa protein, the product of the pilV gene. The amino acid sequence of the pilS product contains residues characteristic of a type IV prepilin, although its prepeptide is unusually long (Fig. 1B). The pilS product is first synthesized as a 22-kDa prepilin and then cleaved between Gly23 and Trp24 to produce a 19-kDa protein via the function of the pilU product, prepilin peptidase. The N-terminal amino group of the processed PilS protein appears to be modified. The C-terminal segments of the pilV gene are under the control of shufflon DNA rearrangement mediated by the rci product (15, 16). The shufflon determines the recipient specificity in liquid mating by converting seven C-terminal segments of the pilV product (13, 14). The pilV product also carries a type IV prepilin cleavage site. Formation of PilV-specific cell aggregates by ColIb-P9 and R64 thin pili was shown and suggested to play an important role in liquid mating (37).
Recently, the three-dimensional structure of the N. gonorrhoeae pilin was determined by X-ray crystallography (21). The monomer structure was an α-β-roll fold with an 85-Å N-terminal α-helical spine. The gross monomer structure resembles a ladle with the N-terminal half of the α-helical spine forming the handle. From the monomer structure, a model of fiber structure with a parameter of five turns per helix, 41-Å pitch, and 60-Å diameter (34) was proposed. In the model, the N-terminal α helices gather in the center of the fiber, forming a core of coiled α helices banded by a β sheet. Slight similarities including two conserved cysteine residues are noted between the amino acid sequences of R64 and N. gonorrhoeae pilins, suggesting that the two proteins fold similarly and then assemble to form similar fibers.
In N. gonorrhoeae, P. aeruginosa, and V. cholerae, amino acid substitutions were introduced into the prepeptide and highly conserved N-terminal regions of prepilin genes (3, 22, 26). The mutant genes were analyzed with respect to processing, secretion, and function. The importance of the conserved glycine in the prepeptide and some hydrophobic amino acids in the N-terminal region has been established.
This work was performed to reveal the relationship between the structure and function of the pilS product. Thirty-two missense and deletion mutations were introduced throughout the entire sequence of the pilS product by PCR and site-directed mutagenesis. The characteristics of the mutant pilS products were analyzed in terms of processing, secretion, and assembly to active thin pili with the pilV product. The activities of the thin pili composed of the mutant pilS genes were determined as the transfer frequency in liquid mating and the sensitivity to IncI1-specific phages.
MATERIALS AND METHODS
E. coli strains, plasmids, phages, and media.
E. coli strains, plasmids, and phages used in this study are listed in Table 1.
TABLE 1.
E. coli strains, plasmids, and phages used in this study
| Strain, plasmid, or phage | Relevant genotype or phenotype | Reference or source |
|---|---|---|
| E. coli strains | ||
| JM83 | ara Δ(lac-proAB) rpsL φ80dlacZΔM15 | 33 |
| JM109 | recA1 gyrA96 thi hsdR17 relA1 supE44 Δ(lac-proAB)/F′ traD36 proAB lacIqZΔM15 | 33 |
| CJ236 | dut-1 ung-1 thi-1 relA1/pCJ105 (Cmr) | 17 |
| BMH71-18 | Δ(lac-proAB) thi supE mutS215::Tn10 (Tcr)/F′ traD36 proAB lacIqZΔM15 | 17 |
| NF83 | recA56 ara Δ(lac-proAB) rpsL φ80 dlacZΔM15 | 7 |
| TN102 | W3110 Nalr | 12 |
| BL21(DE3) | ompT hsdS gal/λ DE3 lacUV5-T7 gene 1 | 29 |
| Plasmids | ||
| pUC119 | Apr, lacZ′ | 33 |
| pET11a | Apr, T7 promoter | 29 |
| pKK641A′ | Kmr, R64drd-11 derivative carrying 18.5-kb BglII-HindIII pil segment and rep sequence | 12 |
| pKK641A′ pilS1 | pKK641A′ carrying the pilS1 mutation | This work |
| pKK661 | Cmr, pHSG576 derivative carrying 35.6-kb HindIII-BglII tra segment of R64drd-11 | 12 |
| pKK692 | 0.8-kb HincII-BglII pilS+ fragment in pUC119 | 37 |
| Phages | ||
| Iα | 4 | |
| PR64FS | 5 |
Luria-Bertani (LB) medium was prepared as previously described (24). The solid medium contained 1.5% agar. In the experiments using pET11a-derived plasmids, 10 μM isopropyl-β-d-thiogalactopyranoside was added to LB medium. Antibiotics were added to the liquid and solid media at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; chloramphenicol, 25 μg/ml; tetracycline, 12 μg/ml; and nalidixic acid, 20 μg/ml.
Designation of mutant pilS genes.
Mutant pilS genes were designated as amino acid replacements: original amino acid-position-replaced amino acid. Since the coordinate of the N-terminal tryptophan of the mature pilS product was defined as +1, the initiating methionine was numbered −23. For example, replacement of the fifth glutamic acid (E) with valine (V) is denoted pilSE5V. The N-terminal deleted pilS genes were designated as the number of amino acids remaining in the prepeptide region. For example, the deletion resulting in a 10-amino-acid prepeptide is denoted as pilSΔsp10.
Construction of pilS mutants.
The pilS1 mutant of pKK641A′ was constructed by the introduction and removal of a tetracycline resistance gene cassette (10). A 22-bp DNA sequence, AATTCCCCGGATCCGGGGAATT, remaining at the SspI site and corresponding to the sixth codon of the pilS gene gave rise to the pilS1 frameshift mutation.
Mutagenesis of the pilS gene was performed by PCR (24) using pKK692 as the template DNA. PCR amplification was performed with a PCR kit (Takara Shuzo) containing 0.1 μg of pKK692 DNA and 50 pmol each of M13 primers M4 and RV. In some experiments, 0.5 nM MnCl2 was added into the reaction mixture to increase the mutation frequency. The pilSG-1A, -G-1R, -E5V, -Y118F, -C126A, -C163A, and -K174(Am) mutations were constructed by site-directed mutagenesis with synthetic oligonucleotides (17).
To construct pilS mutants carrying prepeptides shorter than the wild-type gene (pilSΔsp10, -Δsp6, and -Δsp2), PCR amplification was performed with synthetic oligonucleotides containing an NdeI site at the desired ATG initiation codon. The NdeI-BamHI segment of the amplified DNA was inserted into the NdeI-BamHI sites of pET11a to give pKK693 to pKK695.
Conjugal transfer and phage sensitivity.
Liquid mating was performed as described previously (12). E. coli NF83 donor cells that harbored pKK641A′ pilS1, pKK661, and pKK692 with or without pilS mutations were grown to log phase and then mixed with an overnight culture of E. coli TN102 recipient cells. In the cases of pKK693 to pKK695, E. coli BL21 was used as the donor. The mixture was incubated for 90 min at 37°C and then plated at various dilutions onto selective media. Transfer frequency is presented as the ratio (expressed as a percentage) of the number of transconjugant to the number of donor cells.
Sensitivity of E. coli cells harboring pKK641A′ pilS1 and the mutant pilS plasmids to phages Iα and PR64FS was determined as described previously (12).
Preparation of thin-pilus fraction.
Crude thin-pilus fraction was prepared as described by Yoshida et al. (37), with a slight modification. E. coli cells harboring pKK641A′ pilS1 and the mutant pilS plasmids were grown to optical density at 620 nm of 0.8 with shaking at 37°C. The culture medium was centrifuged three times at 9,200 × g for 10 min to remove cells. The supernatant was centrifuged at 140,000 × g for 1 h. The pellet is referred to as the thin-pilus fraction.
Western blot analysis.
E. coli cells harboring pKK641A′ pilS1 and the mutant pilS plasmids were recovered by centrifugation from the overnight culture (1.5 ml), resuspended in water (1 ml), and broken by sonication. Total proteins in 1.5 μl of lysate were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and electrophoretically transferred to a nitrocellulose membrane (BA85; Schleicher & Schuell). The membrane was incubated with antipilin antiserum in Tris-buffered saline (50 mM Tris-HCl [pH 7.5], 150 mM NaCl), and the antiserum-bound proteins were then detected with a horseradish peroxidase-labeled goat anti-rabbit antiserum by using an ABC-POD kit (Wako).
To analyze the pilS and pilV products in the thin-pilus fraction, aliquots of the thin-pilus fractions were subjected to SDS-PAGE followed by immunoblot detection using antipilin and anti-PilV antisera, respectively, as described above.
RESULTS
Construction of mutations in the pilS gene.
Mutations in the pilS gene were constructed by the PCR method. Since Taq polymerase does not contain 3′-to-5′ exonuclease activity, the frequency of errors in DNA replication is high during PCR amplification using Taq polymerase. One amber and 21 missense mutations in the pilS coding sequence were obtained by PCR. In addition, one amber and six missense mutations were constructed by site-directed mutagenesis to analyze the function of specific amino acid residues in the pilS product. To determine the minimal prepeptide length required for processing of prepilin at the specific cleavage site, three mutant pilS genes (pilSΔsp10, -Δsp6, and -Δsp2) encoding prepeptides shorter than that of the wild-type pilS gene were constructed to yield plasmids pKK693, pKK694, and pKK695, respectively. In summary, 27 missense mutations, three N-terminal deletions, and two C-terminal deletions were constructed and characterized (Table 2; see also Fig. 4).
TABLE 2.
Characteristics of the products of various mutant pilS genes
| Plasmid | Whole-cell extracta
|
Thin-pilus fractionb
|
Transfer frequencyc
|
Sensitivity to phagesd
|
||||
|---|---|---|---|---|---|---|---|---|
| Prepilin | Processing | PilS | PilV | pilS activity | Transdominance | PR64FS | Iα | |
| pKK692 | + | + | + | + | 1.2 ± 0.4 | 1.4 ± 0.4 | s | s |
| None | − | <0.0001 | 1.2 | r | r | |||
| pKK693 (pilSΔsp10) | + | + | + | NT | 1.2 | NT | s | s |
| pKK694 (pilSΔsp6) | + | + | ± | NT | 0.4 | NT | s | s |
| pKK695 (pilSΔsp2) | + | ? | − | NT | <0.0001 | NT | r | r |
| pKK692 pilSE-20Q | + | + | + | + | 1.1 | 1.1 | s | s |
| pKK692 pilSN-7D | + | + | + | + | 1.4 | 1.6 | s | s |
| pKK692 pilSN-7K | + | + | + | + | 0.85 | 1.2 | s | s |
| pKK692 pilSK-2R | + | + | + | + | 0.81 | 1.25 | s | s |
| pKK692 pilSG-1A | + | − | − | − | <0.0001 | 0.43 | r | r |
| pKK692 pilSG-1R | + | − | − | − | <0.0001 | 0.66 | r | r |
| pKK692 pilSE5V | + | + | − | − | <0.0001 | 0.76 | r | r |
| pKK692 pilSG7E | + | + | + | + | 0.0046 | 0.97 | s | r |
| pKK692 pilSI9L | + | + | + | + | 0.062 | 1.4 | s | ps |
| pKK692 pilSL14F | + | + | + | + | 0.062 | 1.5 | s | ps |
| pKK692 pilSV16D | ± | − | − | − | <0.0001 | 0.05 | r | r |
| pKK692 pilSL21Q | + | + | + | + | 0.13 | 0.68 | s | r |
| pKK692 pilSA47P | + | − | − | − | <0.0001 | 0.18 | r | r |
| pKK692 pilSL51Q | + | + | + | + | 0.03 | 1.1 | s | s |
| pKK692 pilSK64R | + | + | + | + | 1.6 | 1.6 | s | s |
| pKK692 pilSM65T | + | + | + | + | 1.6 | 1.7 | s | s |
| pKK692 pilSQ71(Am) | ± | − | − | − | <0.0001 | 0.38 | r | r |
| pKK692 pilSN112S | + | + | + | + | 0.29 | 1.2 | s | s |
| pKK692 pilSY118N | + | − | − | − | <0.0001 | 0.12 | r | r |
| pKK692 pilSY118F | + | + | + | + | 1.1 | 1.1 | s | s |
| pKK692 pilSC126A | + | + | − | − | <0.0001 | 0.09 | r | r |
| pKK692 pilSI127F | + | + | + | + | 0.32 | 0.98 | s | s |
| pKK692 pilST147S | + | + | + | + | 0.15 | 1.2 | s | s |
| pKK692 pilSE157D | + | + | + | + | 0.39 | 1.25 | s | s |
| pKK692 pilSC163A | + | − | − | − | <0.0001 | 0.17 | r | r |
| pKK692 pilSS169G | + | + | + | + | 0.29 | 1.1 | s | s |
| pKK692 pilSK174(Am) | ± | − | − | − | <0.0001 | 0.08 | r | r |
| pKK692 pilSL175P | + | − | − | − | <0.0001 | 0.37 | r | r |
| pKK692 pilSI179T | + | + | + | + | 1.6 | 1.7 | s | s |
Presence of 22-kDa (prepilin) and 19-kDa (processing) proteins reacted with antipilin antiserum in whole-cell extract; summarized from data in Fig. 2.
Presence of 19-kDa (PilS) and 45-kDa (PilV) proteins reacted with antipilin and anti-PilVA′ antisera, respectively, in thin-pilus fraction; summarized from data in Fig. 3. +, detected; −, not detected; ±, small amount detected; NT, not tested.
Ratio (expressed as percentage) of number of transconjugants to number of donor cells. E. coli NF83 donor cells carried pKK661, pKK641A′ pilS1, and the indicated plasmid (pilS activity) or pKK661, pKK641A′ and the indicated plasmid (transdominance).
s, sensitive; r, resistant; ps, partially sensitive.
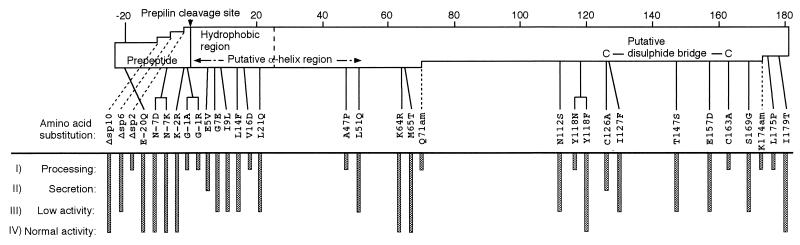
FIG. 4.
Phenotypes and locations of the 32 pilS mutations of plasmid R64. The horizontal open bar represents the entire PilS sequence, with deletion mutations indicated. The prepeptide region, the hydrophobic region, the putative α-helix region, and the putative disulfide bridge are indicated in the horizontal open bar. The triangle indicates the prepilin cleavage site. The classification of the phenotype of each mutation is indicated by vertical bars.
Production and processing of prepilin molecules.
To examine the activity of mutant pilS genes, we first constructed pilS1 mutation by inserting a 22-bp sequence into the sixth codon of the pilS gene on pKK641A′ (Fig. 1A). The pilS product was shown to be synthesized as a 22-kDa protein, prepilin, and then processed to a 19-kDa protein by the function of the pilU product (37). Formation of the nonprocessed and processed products from the various pilS mutants was studied by immunoblot analysis using antipilin antiserum (Fig. 2; Table 2). Complementation of pilS1 mutation by pilS+ plasmid pKK692 was demonstrated, since the 22- and 19-kDa proteins were detected as the wild-type pilS gene products in E. coli cells harboring pKK641A′ pilS1 and pKK692, while neither protein was detected in cells harboring pKK641A′ pilS1 (Fig. 2, lanes 1 and 2). Only the 22-kDa protein was detected in cells harboring pKK692 (lane 3). Less than half of the pilS product from the multicopy pilS gene was processed by prepilin peptidase in E. coli cells harboring pKK641A′ pilS1 and pKK692, while most of the prepilin was processed in cells harboring pKK641A′ (data not shown). In both cases, approximately the same amount of mature pilin was detected from the same amount of cells. These results indicate that the surplus prepilin produced from the multicopy pilS gene was not processed, suggesting that the pilU product is saturated.
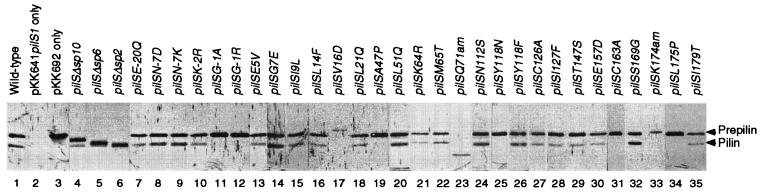
FIG. 2.
Expression and processing of the products of various pilS mutants. Whole extracts of E. coli cells harboring pKK641A′ pilS1 and the mutant pilS plasmids were separated by SDS-PAGE and subjected to Western blot analysis using antipilin antiserum. Lanes: 1, pKK692 (wild-type pilS); 2, without pilS plasmid; 3, without pKK641A′ pilS1; 4 to 6, pKK693 to pKK695; 7 to 35, pKK692 with the indicated pilS mutations. The positions of prepilin (22 kDa) and pilin (19 kDa) are indicated on the right.
The products of the mutant pilS genes in E. coli cells harboring pKK641A′ pilS1 and the mutant pilS plasmids were detected by immunoblot analysis (Fig. 2). Note that extracts from similar numbers of E. coli cells were loaded in each lane of Fig. 2. Similar amounts of the PilS-related proteins (prepilin and mature pilin) were observed from all pilS mutants except pilSV16D and two amber mutants, while the amounts of the products from pilSV16D and two amber mutants were less than those from the wild-type pilS gene or the other pilS mutants. The electrophoretic mobilities of prepilins in SDS-PAGE from all pilS mutants were similar to that of the wild-type prepilin, with the exception of three N-terminal deletions, pilSV16D, and two amber mutants. The mobilities of the products from the three N-terminal deletion mutants and pilSQ71(Am) mutant were faster than that of wild-type prepilin, as expected, while the mobilities of the products from pilSV16D and K174(Am) mutants were slower than that of the wild-type prepilin.
Among six missense mutants in the prepeptide region, both the 22- and 19-kDa proteins were produced from the four mutants (pilSE-20Q, -N-7D, -N-7K, and -K-2R), indicating that their products were processed normally (Fig. 2). In contrast, the products of the pilSG-1A and -G-1R mutants could not be processed at all. Among three N-terminal deletion mutants, the 10-amino-acid-residue prepeptide was cleaved off normally from the pilSΔsp10 product. The six-amino-acid-residue prepeptide was also cleaved off from the pilSΔsp6 product but at a reduced rate. The processing of the pilSΔsp2 product was obscure, since the sizes of the precursor and processed forms from this mutant were too close to be separated by SDS-PAGE. Among 21 missense mutations in the mature pilin region, the products of five mutants (pilSV16D, -A47P, -Y118N, -C163A, and -L175P) could not be processed, while those of the other mutants were processed normally (Fig. 2). Neither of the products of two amber mutants was processed. It is noteworthy that in the three mutants [pilSV16D, -Q71(Am), and -K174(Am)] producing nonprocessed products, the levels of expression of the products were also suppressed as described above. Formation of nonprocessed products from the mutants located in the mature pilin region is of particular interest, since it indicates the importance of mature pilin region amino acids for processing of prepilin.
Formation of thin pilus from the mutant pilS genes.
The pilS product may be processed to lose the prepeptide, be translocated across the cell membrane, and polymerize to form thin pili. R64 thin pili detached from E. coli cells harboring pKK641A′ could be recovered by ultracentrifugation from the culture medium in which the cells had grown (thin-pilus fraction) (37). The 19-kDa protein, mature pilin, was detected in the thin-pilus fraction from the culture media of E. coli cells harboring pKK641A′ pilS1 and pKK692 by immunoblot analysis using antipilin antiserum, while the 22-kDa prepilin was not (Fig. 3A, lane 1). The pilS product was not detected in the thin-pilus fraction from cells harboring pKK641A′ pilS1 or those with pKK692 (lanes 2 and 3).
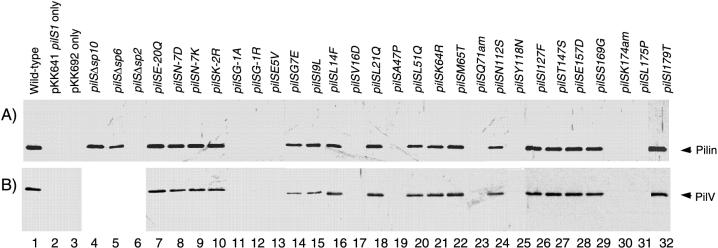
FIG. 3.
Formation of thin pili with PilVA′ protein from various pilS mutants. Thin-pilus fractions were prepared from the culture media of E. coli cells harboring pKK641A′ pilS1 and the mutant pilS plasmids. Proteins were separated by SDS-PAGE and subjected to Western blot analysis using antipilin antiserum (A) and anti-PilVA′ antiserum (B). Lanes: 1, pKK692 (wild-type pilS); 2, without pilS plasmid; 3, without pKK641A′ pilS1; 4 to 6, pKK693 to pKK695; 7 to 32, pKK692 with the indicated pilS mutations. The positions of pilin (19 kDa) and PilVA′ protein (45 kDa) are indicated on the right.
The thin-pilus fractions from E. coli cells harboring pKK641A′ pilS1 and the mutant pilS plasmids were subjected to immunoblot analysis. Among the pilS mutants whose products were processed normally, all except pilSΔsp6, pilSE5V, and pilSC126A produced thin pili outside the cells at levels similar to that of the wild-type gene (Fig. 3A; Table 2). The pilSΔsp6 mutant produced thin pili outside the cells at a lower level. Thin pili were not detected from the culture media of the pilSE5V and C126A mutants. The results of whole-cell enzyme-linked immunosorbent assay analysis paralleled that of immunoblot analysis (data not shown). Electron microscopic observation indicated that thin-pilus fractions from cells with mutant pilS contained fibrous structures similar to those from cells with wild-type pilS (data not shown). In the pilSΔsp2 and pilSG-1A, -G-1R, -V16D, -A47P, -Q71(Am), -Y118N, -C163A, -K174(Am), and -L175P mutants, for which mutant prepilins were not processed, no thin pili were detectable in the culture media (Fig. 3A).
R64 thin pili were shown to contain a minor subunit, the pilVA′ product (37). The pilVA′ product was detected in the thin-pilus fraction prepared from the culture media of E. coli cells harboring pKK641A′ pilS1 and pKK692 as a 45-kDa protein by immunoblot analysis using anti-PilVA′ antiserum, while the pilVA′ product was not detected in the same fraction from cells harboring pKK641A′ pilS1 or those harboring pKK692 (Fig. 3B). The thin-pilus fractions from the culture media of E. coli cells harboring pKK641A′ pilS1 and the mutant pilS plasmids were subjected to immunoblot analysis. The thin pili from all of the piliated mutants (see above) contained the pilVA′ product; the pilSΔsp10 and pilSΔsp6 mutants were not analyzed (Fig. 3B; Table 2).
Activity of the mutant pilS genes in R64 transfer.
To determine the activity of the mutant pilS genes during conjugation, the pKK641-pKK661 system was used (12). In this system, since a lack of the rci gene prevents DNA rearrangement of the shufflon, the transfer frequency of R64 in liquid mating can be accurately estimated. E. coli donor cells harboring pKK641 and pKK661 transmitted pKK661 carrying the oriT sequence into the recipient cells by conjugation. The activity of the mutant pilS genes was determined by measuring the effects of the addition of the mutant pilS plasmids on the transfer frequency in liquid mating from donor cells harboring pKK641A′ pilS1 and pKK661. The transfer frequency from donor cells harboring pKK641A′ pilS1 and pKK661 was less than 0.0001% (Table 2). When pKK692 carrying the wild-type pilS gene was introduced into donor cells, the transfer frequency was increased to 1.2%.
Different levels of recovery in the transfer frequency were observed by introducing various mutant pilS genes into donor cells harboring pKK641A′ pilS1 and pKK661 (Table 2). Among three N-terminal deletion mutants, donor cells harboring the pilSΔsp10 mutant exhibited a transfer frequency similar to that of the wild-type gene during liquid mating. For the pilSΔsp6 mutant, which formed thin pili at a lower level, the transfer frequency was 10-fold lower than that of the wild-type gene. The pilSΔsp2 mutant did not exhibit pilS activity.
For nine missense mutants (pilSG-1A, -G-1R, -E5V, -V16D, -A47P, -Y118N, -C126A, -C163A, and -L175P) as well as two nonsense mutants [pilSQ71(Am) and K174(Am)], the products of which were not processed or secreted outside the cells, no transfer was observed, as expected (Table 2). For eight piliated mutants (pilSE-20Q, -N-7D, -N-7K, -K-2R, -K64R, -M65T, -Y118F, and -I179T), transfer frequencies similar to that of the wild-type gene were observed. For 10 piliated mutants (pilSG7E, -I9L, -L14F, -L21Q, -L51Q, -N112S, -I127F, -T147S, -E157D, and -S169G), transfer frequencies were lower than that of the wild-type pilS gene. The transfer frequencies of these mutants varied from allele to allele. In particular, the transfer frequency of the pilSG7E mutant was 100-fold lower than that of the wild-type gene.
Transdominance of the mutant pilS genes over the wild-type pilS gene.
Addition of pKK692 (multicopy wild-type pilS) to donor cells harboring pKK641A′ and pKK661 had no effect on the transfer frequency in liquid media (Table 2). The effects of introducing multiple copies of mutant pilS genes to donor cells harboring pKK641A′ and pKK661 on the transfer frequency were studied to test whether the pilS mutants were dominant negative over the wild-type pilS gene.
The introduction of 12 mutants [pilSG-1A, -G-1R, -E5V, -V16D, -L21Q, -A47P, -Q71(Am), -Y118N, -C126A, -C163A, -K174am(Am), and -L175P] into donor cells harboring pKK641A′ and pKK661 decreased the transfer frequencies, indicating the transdominant character of these mutants. All of these mutants except pilSL21Q lacked pilS activity during liquid mating.
Sensitivity to phages Iα and PR64FS.
Upon infection of bacterial cells harboring IncI1 plasmids, phages Iα and PR64FS specifically adsorb to the shaft and tip of the thin pili formed by IncI1 plasmids, respectively, and subsequently the infected cells produce progeny phage (4, 5). Therefore, the sensitivity of cells to phages Iα and PR64FS can be used as the indication of thin-pilus formation. Although E. coli cells harboring pKK641A′ pilS1 were resistant to phages Iα and PR64FS, cells harboring pKK641A′ pilS1 and pKK692 were sensitive to them (Table 2).
The sensitivities to these phages of E. coli cells harboring pKK641A′ pilS1 and the mutant pilS plasmids were determined. Cells harboring 16 mutant pilS genes (pilSΔsp10, -Δsp6, -E-20Q, -N-7D, -N-7K, -K-2R, -L51Q, -K64R, -M65T, -N112S, -Y118F, -I127F, -T147S, -E157D, -S169G, and -I179T) in addition to pKK641A′ pilS1 exhibited Iα and PR64FS phage sensitivity similar to that of cells with the wild-type pilS gene (Table 2). Cells with 12 mutant genes [pilSΔsp2, -G-1A, -G-1R, -E5V, -V16D, -A47P, -Q71(Am), -Y118N, -C126A, -C163A, -K174(Am), and -L175P], which exhibited no pilS activity in liquid mating, were resistant to both phages. Cells with two mutations (pilSI9L and -L14F) were sensitive to PR64FS but partially sensitive to Iα. Cells with two mutations (pilSG7E and -L21Q) were sensitive to PR64FS but resistant to Iα. These four mutations were located within the N-terminal hydrophobic region of mature pilin.
DISCUSSION
Genetic analysis was performed to reveal the relationship between the structure and function of the R64 pilS product. Thirty-two pilS mutants were characterized with respect to the processing, extracellular secretion, and assembly to active thin pilus (Table 2). The activities of thin pili produced from these pilS mutants were analyzed as the transfer frequency in liquid media and the sensitivity to phages Iα and PR64FS. From these results, the pilS mutants can be classified into four classes (Fig. 4). The products of 10 mutants [pilSΔsp2, -G-1A, -G-1R, -V16D, -A47P, -Q71(Am), -Y118N, -C163A, -K174(Am), and -L175P] were not processed by prepilin peptidase (class I). The extracellular secretion of the products of two mutants (pilSE5V and -C126A) was inhibited, although these products were processed at the normal rate (class II). From 11 mutants (pilSΔsp6, -G7E, -I9L, -L14F, -L21Q, -L51Q, -N112S, -I127F, -T147S, -E157D, and -S169G), thin pili with reduced activities during liquid mating were formed (class III). From nine mutants (pilSΔsp10, -E-20Q, -N-7D, -N-7K, -K-2R, -K64R, -M65T, -Y118F, and -I179T), thin pili with activities similar to that of the wild-type pilS gene were formed (class IV).
Prepeptide region.
The products of four mutants in the prepeptide region (pilSE-20Q, -N-7D, -N-7K, and -K-2R) were processed as efficiently as that of the wild-type strain and used in the formation of normal thin pili. Since these mutants are expected to produce normal pilin after processing, it is reasonable that thin pili from these mutants exhibited normal transfer frequency. A series of mutations were introduced into the prepeptide sequence of the pilA gene in P. aeruginosa (26). Normal pili were produced from all single or double pilA mutants except Gly−1, in fair agreement with the results of the R64 pilS mutants.
The products of the R64 pilSG-1A and pilSG-1R mutants could not be processed. All type IV prepilins have a glycine residue at the C terminus of the prepeptide. In P. aeruginosa pilA and V. cholerae tcpA, the products of all Gly−1 mutants except pilAG-1A could not be processed, while the pilAG-1A product was partially processed (3, 26). In contrast, in the R64 system, even the pilSG-1A product could not be processed.
The prepeptides of typical type IV prepilins consist of 6 to 7 amino acids (23, 27), while R64 pilS product has a longer prepeptide consisting of 23 amino acids. The product of the pilSΔsp10 mutant carrying a 10-amino-acid prepeptide was processed normally, indicating that the entire sequence of prepeptide is not required for the processing of the R64 pilS product. The product of the pilSΔsp6 mutant carrying a six-amino-acid prepeptide was processed partially and produced thin pili at a reduced level. This mutant is the only class III mutant which exhibits a quantitative reduction in thin-pilus production. The product of the pilSΔsp2 mutant carrying only glycine most likely is not processed, as was found for P. aeruginosa pilA (26).
Putative α-helix region.
By analogy with N. gonorrhoeae pilin (21), approximately 53 N-terminal amino acids of R64 pilin are predicted to form an α-helical spine with the N-terminal half protruding from a C-terminal globular head. Among eight mutations located within this region, the pilSV16D and -A47P were defective in processing. The pilSE5V product was susceptible to processing but could not be secreted extracellularly. Similar phenotypes of the E5K and E5V mutations were reported for N. gonorrhoeae pilE and P. aeruginosa pilA genes (22, 26). The remaining five mutant genes (pilSG7E, -I9L, -L14F, -L21Q, and -L51Q) formed extracellular thin pili with the PilVA′ protein. However, thin pili from these mutants exhibited a transfer frequency lower than that of the wild-type pilS gene in liquid mating (Table 2). These results suggest an important role of the putative α-helix region in the function of R64 thin pili. In fiber formation, the N-terminal hydrophobic regions of pilin are predicted to assemble with one another to form coiled α helices (21). It is noteworthy that even an isoleucine-to-leucine replacement was not tolerated in this region. No class IV mutants were obtained within this region.
C-terminal region.
As was found for N. gonorrhoeae pilin (21), the C-terminal two-thirds of R64 pilin is predicted to fold into a globular structure. The products of two amber mutant genes [pilSQ71(Am) and pilSK174(Am)] could not be processed. The pilSK174(Am) mutant lacks only seven C-terminal amino acids. The inability to be processed in pilSK174(Am) and pilSL175P indicates the importance of the C-terminal segment of the pilS product in processing. In addition, two class I mutants (pilSY118N and pilSC163A) are present in this region. Although the pilSY118N mutant was a class I mutant, the product of the pilSY118F mutant produced active thin pili, suggesting that Tyr118 is important for the formation of a hydrophobic core in the pilin molecule as in N. gonorrhoeae pilin (21).
Various type IV pilins are thought to be stabilized by a disulfide bridge between two conserved cysteine residues (21, 25). In R64 pilin, Cys126 and Cys163 are predicted to form a disulfide bridge (Fig. 1B). The pilSC126A and pilSC163A mutations disrupting the disulfide bridge between the two cysteine residues were classified as class II and I mutants, respectively, signifying the important role of the disulfide bridge in protein folding of the R64 pilS product. In contrast to our results, in bundle-forming pili of enteropathogenic E. coli, two cysteine-to-serine mutations as well as introduction of dsbA mutation resulted in reduced levels of pilin (38). The reason for the apparent discrepancy is not known.
Characteristics of the four mutant classes.
The class I and II mutants exhibited nonpiliated phenotype. To our surprise, the mutation points of the class I mutants were distributed throughout the prepilin sequence. The presence of class I mutants in the pilS C-terminal region indicates that the entire prepilin must be made prior to cleavage of the prepeptide. These results exhibit a striking contrast to the case of the general secretory pathway, for which the importance of the signal peptide has been established (23). By using the signal peptide of the E. coli ompA gene, a plasmid vector for the secretion of foreign protein was constructed (8). In the case of type IV prepilin, replacements of many amino acids in the prepeptide sequence, but not of Gly−1, were permissible (reference 26 and this study). Many class I and II mutations appear to disrupt PilS protein folding by the introduction of deletions, the introduction of proline, or the removal of the disulfide bridge. It is possible that the folded PilS protein structure is required for cleavage of its prepeptide.
In N. gonorrhoeae pilus-positive (P+)-to-P− phase transitions, a P− rp+ variant (P−, but can be reverted to P+) of pilE was characterized (30, 31). In the P− rp+ cells, type IV pilus was not produced, while a mutant protein which reacted with antipilin antibody was produced and processed. Hence, the P− rp+ phenotype is very similar to that of the R64 class II pilS mutants. The difference in the amino acid sequence between the P− rp+ strain and its P+ revertant was 9 amino acid replacements within a 12-amino-acid segment of the C-terminal hypervariable region of the pilE gene. These results suggest that the formation of class II (and most likely class I) mutations by antigenic variation in the N. gonorrhoeae pilE gene results in P+-to-P− phase transitions.
The class I and II mutants exhibited a transdominant phenotype over the wild-type allele (Table 2), suggesting that overproduced mutant proteins may competitively inhibit some step(s) in thin-pilus formation from the wild-type PilS protein.
The products of the class III mutants formed extracellular thin pili with a low transfer frequency during liquid mating. Among 11 class III mutants, all except pilSΔsp6 produced the altered pilin with various degrees of pilS activity, while the pilSΔsp6 mutant exhibited a quantitative mutant phenotype. E. coli cells harboring the class III mutants were sensitive to phage PR64FS, but their sensitivity to Iα was allele dependent, exhibiting a sensitive, partially sensitive, or resistant phenotype. Most mutants exhibiting resistance or partial sensitivity to Iα clustered within the N-terminal hydrophobic region.
Why do the class III mutants exhibit the low-frequency phenotype during liquid mating? At the beginning of liquid mating, the R64 thin pili in donor cells are predicted to attach to the surface of the recipient cells, resulting in the formation of donor-recipient cell aggregates (13, 14, 37). At that time, one of seven PilV proteins selected by the shufflon DNA rearrangement recognizes the surface receptor of the recipient cells and determines the recipient specificity. Subsequent steps promoted by thin and thick pili and other transfer genes eventually transfer R64 DNA into the recipient cells.
One possible reason for the low-frequency phenotype is that the mutant pilin forms a weak thin pilus, which is broken at a rate higher than that of the wild-type pilin. The weak thin pilus is expected to cause a defect in donor-recipient complex formation. Another possible reason for the low-frequency phenotype is related to the retraction model of pilus (6). Various pili are thought to outgrow and retract. Retraction may be important in establishing the donor-recipient complex in conjugation and phage infection. It is possible that the R64 thin pilus consisting of mutant pilin is defective in the retraction process.
It was reported that in the V. cholerae Tcp system, type IV pili were formed by two tcpA mutants (V9M and V20T) but were inactive in autoagglutination and colonization (3). The R64 thin-pilus system seems to be one of the best for such analyses, since it allows for quantitative estimation of the function of type IV pilus.
The isolation of four classes of pilS mutations leads us to propose the following model for the process of R64 thin-pilus formation: (i) the R64 pilS product is synthesized, (ii) the R64 pilS product is processed by prepilin peptidase to yield R64 pilin, and (iii) R64 pilin is secreted across the cell membrane and assembled with the PilV protein.
ACKNOWLEDGMENTS
We are grateful to N. Furuya for useful discussions and to K. Takayama for critical reading of the manuscript.
This work was supported in part by a grant from the Ministry of Education, Science, Sports and Culture of Japan.
REFERENCES
- 1.Bradley D E. Derepressed plasmids of incompatibility group I1 determine two different morphological forms of pilus. Plasmid. 1983;9:331–334. doi: 10.1016/0147-619x(83)90011-2. [DOI] [PubMed] [Google Scholar]
- 2.Bradley D E. Characteristics and function of thick and thin conjugative pili determined by transfer-derepressed plasmids of incompatibility groups I1, I2, I5, B, K, and Z. J Gen Microbiol. 1984;130:1489–1502. doi: 10.1099/00221287-130-6-1489. [DOI] [PubMed] [Google Scholar]
- 3.Chiang S L, Taylor R K, Koomey M, Mekalanos J J. Single amino acid substitutions in the N-terminus of Vibrio cholerae TcpA affect colonization, autoagglutination, and serum resistance. Mol Microbiol. 1995;17:1133–1142. doi: 10.1111/j.1365-2958.1995.mmi_17061133.x. [DOI] [PubMed] [Google Scholar]
- 4.Coetzee J N, Bradley D E, Hedges R W. Phages Iα and I2-2: IncI plasmid-dependent bacteriophages. J Gen Microbiol. 1982;128:2797–2804. doi: 10.1099/00221287-128-11-2797. [DOI] [PubMed] [Google Scholar]
- 5.Coetzee J N, Sirgel F A, Lacatsas G. Properties of a filamentous phage which absorbs to pili coded by plasmids of the IncI complex. J Gen Microbiol. 1980;117:547–551. doi: 10.1099/00221287-117-2-547. [DOI] [PubMed] [Google Scholar]
- 6.Frost L S. Conjugative pili and pilus-specific phages. In: Clewell D B, editor. Bacterial conjugation. New York, N.Y: Plenum Publishing; 1993. pp. 189–212. [Google Scholar]
- 7.Furuya N, Komano T. Nucleotide sequence and characterization of the trbABC region of the IncI1 plasmid R64: existence of the pnd gene for plasmid maintenance within the transfer region. J Bacteriol. 1996;178:1491–1497. doi: 10.1128/jb.178.6.1491-1497.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghrayeb J, Kimura H, Takahara M, Hsiung H, Masui Y, Inouye M. Secretion cloning vectors in Escherichia coli. EMBO J. 1984;3:2437–2442. doi: 10.1002/j.1460-2075.1984.tb02151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giron J A, Ho A S, Schoolnik G K. An inducible bundle-forming pilus of enteropathogenic Escherichia coli. Science. 1991;254:710–713. doi: 10.1126/science.1683004. [DOI] [PubMed] [Google Scholar]
- 10.Kim S-R, Funayama N, Komano T. Nucleotide sequence and characterization of the traABCD region of IncI1 plasmid R64. J Bacteriol. 1993;175:5035–5042. doi: 10.1128/jb.175.16.5035-5042.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim S-R, Komano T. The plasmid R64 thin pilus identified as a type IV pilus. J Bacteriol. 1997;179:3594–3603. doi: 10.1128/jb.179.11.3594-3603.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Komano T, Funayama N, Kim S-R, Nisioka T. Transfer region of IncI1 plasmid R64 and role of shufflon in R64 transfer. J Bacteriol. 1990;172:2230–2235. doi: 10.1128/jb.172.5.2230-2235.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Komano T, Kim S-R, Yoshida T. Mating variation by DNA inversions of shufflon in plasmid R64. Adv Biophys. 1995;31:181–193. doi: 10.1016/0065-227x(95)99391-2. [DOI] [PubMed] [Google Scholar]
- 14.Komano T, Kim S-R, Yoshida T, Nisioka T. DNA rearrangement of the shufflon determines recipient specificity in liquid mating of IncI1 plasmid R64. J Mol Biol. 1994;243:6–9. doi: 10.1006/jmbi.1994.1625. [DOI] [PubMed] [Google Scholar]
- 15.Komano T, Kubo A, Nisioka T. Shufflon: multi-inversion of four contiguous DNA segments of plasmid R64 creates seven different open reading frames. Nucleic Acids Res. 1987;15:1165–1172. doi: 10.1093/nar/15.3.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kubo A, Kusukawa A, Komano T. Nucleotide sequence of the rci gene encoding shufflon-specific DNA recombinase in the IncI1 plasmid R64: homology to the site-specific recombinases of integrase family. Mol Gen Genet. 1988;213:30–35. doi: 10.1007/BF00333394. [DOI] [PubMed] [Google Scholar]
- 17.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 18.Nunn D N, Lory S. Product of the Pseudomonas aeruginosa gene pilD is a prepilin leader peptidase. Proc Natl Acad Sci USA. 1991;88:3281–3285. doi: 10.1073/pnas.88.8.3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ogierman M A, Zabihi S, Mourtzios L, Manning P A. Genetic organization and sequence of the promoter-distal region of the tcp gene cluster of Vibrio cholerae. Gene. 1993;126:51–60. doi: 10.1016/0378-1119(93)90589-u. [DOI] [PubMed] [Google Scholar]
- 20.Ottow J C G. Ecology, physiology, and genetics of fimbriae and pili. Annu Rev Microbiol. 1975;29:79–108. doi: 10.1146/annurev.mi.29.100175.000455. [DOI] [PubMed] [Google Scholar]
- 21.Parge H E, Forest K T, Hickey M J, Christensen D A, Getzoff E D, Tainer J A. Structure of the fiber-forming protein pilin at 2.6Å resolution. Nature. 1995;378:32–38. doi: 10.1038/378032a0. [DOI] [PubMed] [Google Scholar]
- 22.Pasloske B L, Paranchych W. The expression of mutant pilins in Pseudomonas aeruginosa: fifth position glutamate affects pilin methylation. Mol Microbiol. 1988;2:289–295. doi: 10.1111/j.1365-2958.1988.tb00055.x. [DOI] [PubMed] [Google Scholar]
- 23.Pugsley A P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 25.Schoolnik G K, Fernandez R, Tai J Y, Rothbard J, Gotschlich E C. Gonococcal pili. Primary structure and receptor binding domain. J Exp Med. 1984;159:1351–1370. doi: 10.1084/jem.159.5.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strom M S, Lory S. Amino acid substitutions in pilin of Pseudomonas aeruginosa. J Biol Chem. 1991;25:1656–1664. [PubMed] [Google Scholar]
- 27.Strom M S, Lory S. Structure-function and biogenesis of the type IV pili. Annu Rev Microbiol. 1993;47:565–596. doi: 10.1146/annurev.mi.47.100193.003025. [DOI] [PubMed] [Google Scholar]
- 28.Strom M S, Nunn D N, Lory S. A single bifunctional enzyme, PilD, catalyzes cleavage and N-methylation of proteins belonging to the type IV pilin family. Proc Natl Acad Sci USA. 1993;90:2404–2408. doi: 10.1073/pnas.90.6.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Studier F W, Rosenberg A H, Dunn J J, Dubendorff W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 30.Swanson J, Bergström S, Robbins K, Corwin D, Koomey J M. Gene conversion involving the pilin structural gene correlates with pilus+→pilus− changes in Neisseria gonorrhoeae. Cell. 1986;47:267–276. doi: 10.1016/0092-8674(86)90449-6. [DOI] [PubMed] [Google Scholar]
- 31.Swanson J, Robbins K, Barrera O, Koomey J M. Gene conversion variations generate structurally distinct polypeptides in Neisseria gonorrhoeae. J Exp Med. 1987;165:267–276. doi: 10.1084/jem.165.4.1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taniguchi T, Fujino Y, Yamamoto K, Miwatani T, Honda T. Sequencing of the gene encoding the major pilin of pilus colonization factor antigen III (CFA/III) of human enterotoxigenic Escherichia coli and evidence that CFA/III is related to type IV pili. Infect Immun. 1995;63:724–728. doi: 10.1128/iai.63.2.724-728.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vieira J, Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- 34.Watts T H, Kay C M, Paranchych W. Spectral properties of three quaternary arrangements of Pseudomonas pilin. Biochemistry. 1983;22:3640–3646. doi: 10.1021/bi00284a016. [DOI] [PubMed] [Google Scholar]
- 35.Whichurch C B, Hobbs M, Livingston S P, Krishnapillai V, Mattick J S. Characterization of a Pseudomonas aeruginosa twitching motility gene and evidence for a specialized protein export system widespread in eubacteria. Gene. 1991;101:33–44. doi: 10.1016/0378-1119(91)90221-v. [DOI] [PubMed] [Google Scholar]
- 36.Wu S S, Kaiser D. Genetic and functional evidence that type IV pili are required for social gliding motility in Myxococcus xanthus. Mol Microbiol. 1995;18:547–558. doi: 10.1111/j.1365-2958.1995.mmi_18030547.x. [DOI] [PubMed] [Google Scholar]
- 37.Yoshida T, Furuya N, Ishikura M, Isobe T, Haino-Fukushima K, Ogawa T, Komano T. Purification and characterization of thin pili of IncI1 plasmids ColIb-P9 and R64: formation of PilV-specific cell aggregates by type IV pili. J Bacteriol. 1998;180:2842–2848. doi: 10.1128/jb.180.11.2842-2848.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang H-Z, Donnenberg M S. DsbA is required for stability of the type IV pilin of enteropathogenic Escherichia coli. Mol Microbiol. 1996;21:787–797. doi: 10.1046/j.1365-2958.1996.431403.x. [DOI] [PubMed] [Google Scholar]