Abstract
Myxococcus xanthus cells carrying the Ω4408 Tn5lac insertion at the sde locus show defects in fruiting body development and sporulation. Our analysis of sde expression patterns showed that this locus is induced early in the developmental program (0 to 2 h) and that expression increases approximately fivefold after 12 h of development. Further studies showed that expression of sde is induced as growing cells enter stationary phase, suggesting that activation of the sde locus is not limited to the developmental process. Because the peak levels of sde expression in both an sde+ and an sde mutant background were similar, we conclude that the sde locus is not autoregulated. Characterization of the sde locus by DNA sequence analysis indicated that the Ω4408 insertion occurred within the sdeK gene. Primer extension analyses localized the 5′ end of sde transcript to a guanine nucleotide 307 bp upstream of the proposed start for the SdeK coding sequence. The DNA sequence in the −12 and −24 regions upstream of the sde transcriptional start site shows similarity to the ς54 family of promoters. The results of complementation studies suggest that the defects in development and sporulation caused by the Ω4408 insertion are due to an inactivation of sdeK. The predicted amino acid sequence of SdeK was found to have similarity to the sequences of the histidine protein kinases of two-component regulatory systems. Based on our results, we propose that SdeK may be part of a signal transduction pathway required for the activation and propagation of the early developmental program.
In response to starvation, cells of the gram-negative soil bacterium Myxococcus xanthus initiate a developmental program that culminates in the formation of a dome-shaped structure called a “fruiting body.” Construction of the multicellular fruiting body requires nutrient deprivation, the coordinated effort of approximately 105 cells, and a solid surface. Within the fruiting body, individual cells differentiate into environmentally resistant and metabolically quiescent spores (for reviews, see references 17, 18, and 47).
Fruiting body development proceeds through a series of steps that require cell-cell signaling. At least five extracellular signals (designated A-signal through E-signal) are needed for construction of a spore-filled fruiting body (6, 10, 28, 29, 40). Several studies have examined the effects of extracellular signaling mutations on expression of developmentally regulated Tn5lac reporter gene fusions (22–24, 27). The results of these studies suggest that each extracellular signal coordinates the temporal expression of a unique set of developmentally regulated genes.
The genes tagged by Tn5lac fusions have been positioned on the M. xanthus developmental pathway based on their temporal regulation by extracellular signaling events (23). In the early portion of the developmental pathway (between 0 and 6 h), there are two distinct classes of development genes: those which require the extracellular A-signal for full expression, and those which do not. Thus, there appears to be a bifurcation of the early developmental pathway into an A-signal-dependent branch and an A-signal-independent branch.
Characterization of Tn5lac insertion mutants showed that the A-signal-independent fusion Ω4408 inhibits construction of normal-looking fruiting bodies (24, 25). This result suggests that the Ω4408 Tn5lac insertion defines a locus that is required for fruiting body formation and that the A-signal-independent pathway is essential for propagating the developmental program. Consistent with this proposal is the finding that the Ω4408 insertion reduces expression of a second developmentally regulated locus called “devRS” (25). Recent studies have shown that a wild-type copy of the devRS locus is needed for progression through the later stages of the developmental process (24, 25, 53).
We are interested in the regulation of gene expression in the A-signal-independent pathway and how the products of these genes propagate the developmental program. In this paper, we describe our analysis of the locus defined by Ω4408 Tn5lac insertion, designated sde (for starvation-induced, development essential). To begin to understand how the sde genes are regulated, we examined the patterns of sde expression in developing cells and in vegetatively growing cells and localized the transcriptional start site for the sde operon. DNA flanking Ω4408 was cloned and characterized by DNA sequence analysis and insertion mutagenesis to determine which gene in the sde locus is required for fruiting body development.
MATERIALS AND METHODS
Bacterial strains and plasmids.
A complete list of the strains and plasmids used in this study is shown in Table 1. Plasmids were propagated in Escherichia coli DH5α (11) or JM101 (34). DK101 is wild type for fruiting body development and sporulation, and it was chosen as the parent of all strains used in this study because it carries the sglA1 mutation (14). The sglA1 allele allows for dispersed growth in liquid cultures. Previous work by Kroos et al. (24) demonstrated that the developmental defects produced by the Ω4408 insertion are identical in both an sglA1 background and an sglA+ background, indicating that these defects are caused by the Ω4408 insertion itself and are independent of the sglA1 allele.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | supE44 ΔlacU169 φ80ΔlacZM15 hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 11 |
| JM101 | Δ(lac pro) thi supE F′ traD36 proAB lacIq ΔlacZM15 | 34 |
| M. xanthus | ||
| DK101 | sglA1 (wild-type development) | 14 |
| DK4408 | sglA1 sdeK::Tn5lac Ω4408 | 24 |
| MS1503 | sglA1 ΔsdeK | This study |
| MS1506 | sglA1 ΔsdeK attB::pJEF4 | This study |
| MS1510 | sglA1 sde::pELF1 | This study |
| MS2010 | sglA1 trcF::pBAR101 | This study |
| Plasmids | ||
| pBJ114 | KanrgalK | 16 |
| pBGS18 | Kanr | 49 |
| pBluescript SKII | Ampr | Stratagene |
| pPLH343 | Kanr | 19 |
| pBAR101 | Kanr, pBGS18 containing 1 kb of trcF on a StuI-SalI fragment | This study |
| pIMK50 | Ampr Kanr, pBluescript SKII containing 9.5 kb of Ω4408 Tn5lac and 12.5 kb of upstream DNA on a XhoI fragment | This study |
| pIMK51 | Ampr, pBluescript SKII containing 50 bp of Ω4408 Tn5lac and 6 kb of upstream DNA on a BamHI fragment | This study |
| pELF1 | Kanr, pBGS18 containing 50 bp of Ω4408 Tn5lac and 6 kb of upstream DNA on a BamHI fragment | This study |
| pJEF1 | Kanr, pBGS18 containing sdeK and 6 kb of upstream DNA on a BamHI-PstI fragment | This study |
| pJEF2 | Kanr, pBGS18 containing the internal sdeK 1.2-kb NcoI-PstI fragment | This study |
| pJEF2.5 | Kanr, pBGS18 containing a 25-kb NcoI (within sdeK)-EcoRI fragment | This study |
| pJEF3 | Kanr, pPLH343 containing sdeK and 1 kb of upstream DNA on a StuI-PstI fragment | This study |
| pJEF4 | Kanr, pPLH343 containing sdeK and 1 kb of upstream DNA on a StuI-HindIII fragment | This study |
| pJEF9 | Kanr, pBJ114 containing a 1.9-kb fragment of the sde locus (with a 700-bp ApaI internal deletion) | This study |
Strain MS1503 is a derivative of DK101 that carries a 700-bp deletion in the sde locus. The deletion removes 550 bp from the 5′ end of the predicted SdeK coding sequence and an additional 150 bp of upstream DNA. The 700-bp deletion in MS1503 was constructed by a two-step gene replacement procedure with a derivative of plasmid pBJ114 (16), called pJEF9. Plasmid pJEF9 carries the galK gene, which can be used as a counterselection for the loss of an integrated plasmid, and a 1.9-kb internal EcoRI-HindIII fragment from the sde locus. This 1.9-kb insert was generated by deleting 700 bp of DNA from the internal portion of a 2.6-kb fragment of the sde locus. Plasmid pJEF9 was introduced into DK101 cells as described below, and Kanr transformants that contain a single copy of pJEF9 integrated by homologous recombination at the sde locus were identified by Southern blot analysis (43). The single integration event produced a tandem duplication with one copy of sdeK intact and one copy of sdeK with its 5′ end deleted. After identification of Kanr transformants with a single plasmid insertion, cells were inoculated into flasks containing 5 ml of CTT broth (components given below) and incubated at 32°C overnight with vigorous agitation. Cells were harvested from overnight cultures by centrifugation, resuspended in 500 μl of TPM buffer (components given below), and aliquoted onto CTT plates supplemented with 2% galactose. In the presence of galactose, expression of the galK gene from plasmid pJEF9 is lethal to M. xanthus, and Galr colonies can arise from cells which have undergone a plasmid excision event. To identify isolates that had excised the chromosomal copy of plasmid pJEF9, the Galr colonies were replica plated onto CTT in the presence and absence of kanamycin. Colonies that were only able to grow in the absence of kanamycin (Kans) were analyzed further. Excision of plasmid pJEF9 can yield a strain with a wild-type copy of sdeK or a strain with a deletion at the 5′ end of sdeK. Chromosomal DNA was isolated from Kans colonies (43) and used for Southern blot analysis (43) to identify strains carrying the deletion.
Media for growth and development.
M. xanthus strains were grown at 32°C in CTT broth containing 1% Casitone (Difco Laboratories), 10.0 mM Tris hydrochloride (pH 8.0), 1 mM KH2PO4, and 8 mM MgSO4 or on plates containing CTT broth and 1.5% Difco Bacto-Agar. CTT broth and CTT plates were supplemented with 40 μg of kanamycin sulfate per ml (Sigma) or 2% galactose (Sigma) as needed. CTT soft agar is CTT broth containing 0.7% Difco Bacto-Agar. E. coli DH5α and JM101 were grown at 37°C in Luria-Bertani broth (LB) containing 1% tryptone (Difco), 0.5% yeast extract (Difco), and 0.5% NaCl or on plates containing LB and 1.5% Difco Bacto-Agar. LB and LB plates were supplemented with 50 μg of ampicillin per ml (Sigma) or 40 μg of kanamycin sulfate per ml (Sigma) as needed. Fruiting body development was carried out at 32°C on TPM plates containing TPM buffer (10.0 mM Tris hydrochloride [pH 8.0], 1 mM KH2PO4, and 8 mM MgSO4) and 1.5% Difco Bacto-Agar.
β-Galactosidase, developmental, and sporulation assays.
To harvest vegetatively growing cells, M. xanthus strains were inoculated into a flask containing 10 ml of CTT broth, and the culture was placed at 32°C with vigorous swirling. At various times after the culture reached a density of 2 × 108 cells per ml, 500-μl aliquots were removed and quick-frozen in liquid nitrogen. Developmental cells were harvested from TPM agar plates as described previously (24), and β-galactosidase assays were performed with quick-frozen cell extracts (20). β-Galactosidase specific activity is defined as nanomoles of o-nitrophenol (ONP) produced minute−1 milligram of protein−1.
Fruiting body development on TPM agar plates was monitored visually with a dissecting microscope (Wild-Heerbrug). Two methods were used to examine sporulation efficiency. First, the number of ovoid-shaped, phase-bright spores produced by developing cells was determined with a Petroff-Hausser counting chamber and phase-contrast microscopy. Second, the viability of these spores was determined as described previously (53). Due to day-to-day variation in the absolute spore number, spore assays are reported as percentage of that of the wild type for each experiment. One hundred percent wild-type sporulation was between 2 × 106 and 2 × 107 spores/ml, which corresponds to 1 to 10% sporulation efficiency based on the number of input cells.
Analysis of RNA.
To isolate RNA during growth, M. xanthus cells were inoculated into CTT broth, and the culture was placed at 32°C with vigorous shaking. Cells were harvested at various times after the culture reached a density of 2 × 108 cells per ml. To isolate developmental RNA, approximately 5 × 1010 cells were harvested from TPM agar plates as previously described (20). After harvesting, vegetatively growing cells and developing cells were quick-frozen in liquid nitrogen as described above for the β-galactosidase assays. RNA was isolated from quick-frozen cells by the hot phenol method (43). Total cellular RNA was used in slot blot hybridizations as described by Kaplan et al. (20). Two probes were used for the slot blot hybridization experiments: an ApaI fragment containing 300 bp of DNA immediately upstream of the Ω4408 insertion site and an NcoI-PstI fragment containing 1.2 kb of DNA downstream of the Ω4408 insertion site. Similar results were found with both probes. The specificity of these probes was confirmed by using yeast mRNA, which yielded no detectable signal. Primer extension analyses were carried out as described previously (15) with the modifications of Mirel and Chamberlin (36). The primers used for these analyses were 5′-GCCACCCTAACCCCGCAGCA-3′ and 5′-ACACTCCCTTCATACAGACGCAG-3′. Both primers were synthesized by Operon Technologies, Inc., (Alameda, Calif.).
Plasmid transfer to M. xanthus.
Plasmids containing DNA fragments from the sde locus were electroporated into DK101 or MS1503 cells by the technique of Plamann et al. (41). Southern blot analysis (43) was used to identify electroporants that contain a single copy of the appropriate plasmid integrated by homologous recombination into the sde locus or a single copy of the appropriate plasmid integrated by site-specific recombination into the chromosomal Mx8 phage attachment site (attB). Kanr electroporants carrying a single plasmid insertion were scored for development and sporulation efficiency as needed.
Cloning the sde locus.
To isolate chromosomal DNA upstream (with respect to lacZ transcription) of the Ω4408 Tn5lac fusion and insertion, genomic DNA was prepared from strain DK4408 (43), digested with XhoI, and ligated into XhoI-digested plasmid pBluescript SKII. The DNA ligation mixture was electroporated into E. coli cells as described previously (43), and Kanr colonies were identified on LB plates containing kanamycin. Plasmid DNA was isolated from overnight cultures grown in kanamycin-supplemented LB plates as described by Sambrook et al. (43). One clone, plasmid pIMK50, that contains a 22-kb insert was identified by digestion of the plasmid DNA with the appropriate restriction enzymes. Subsequently, a BamHI fragment from pIMK50 with 50 bp of the 5′ end of Ω4408 Tn5lac and 6 kb of chromosomal DNA upstream of the insertion was identified by restriction analysis, purified, and ligated into BamHI-digested pBluescript SKII to yield plasmid pIMK51. To confirm that the approximately 6 kb of DNA cloned into pIMK51 is identical to DNA located upstream of the chromosomal Ω4408 insertion, this 6-kb BamHI fragment was used as a probe for Southern blot analysis (43). Chromosomal DNA prepared from strain DK4408 was digested with SmaI, SalI, XhoI, EcoRI, or PstI, separated on an 0.8% agarose gel, and blotted onto a nylon membrane. The 6-kb BamHI probe made from pIMK51 showed a hybridization pattern identical to that obtained when the same blot was probed with the 5′ end of Tn5lac. The restriction map generated from this Southern blot analysis was similar to the one constructed previously by Kroos et al. (24) for DNA upstream of the Ω4408 insertion.
An in situ cloning technique was used to isolate chromosomal DNA downstream (with respect to lacZ transcription) of the Ω4408 Tn5lac fusion and insertion as described by Gill et al. (9) and Stephens and Kaiser (50). Kanr transformants of strain DK101 containing a single insertion of plasmid pELF1 or pJEF2 in the sde locus were isolated as described above. After identification of Kanr transformants with the single insertion event, chromosomal DNA was isolated and digested with PstI or EcoRI for pELF1 and pJEF2, respectively. Each chromosomal digest was then self-ligated and electroporated into E. coli DH5α (43). Electroporated cells were plated on LB plates containing kanamycin and incubated at 37°C overnight. Plasmid DNA was isolated and digested with the appropriate restriction enzymes to confirm the composition of the clones. One clone (plasmid pJEF1) carries a 7.7-kb insert with 6 kb of chromosomal DNA upstream of the Ω4408 insertion site and 1.7 kb of chromosomal DNA downstream of the Ω4408 insertion site. The second clone (plasmid pJEF2.5) carries a 25-kb insert, with 1.2 kb of sdeK DNA and approximately 24 kb of downstream DNA. Neither plasmid contains Tn5lac DNA.
DNA sequence analysis.
DNA was sequenced by the dideoxynucleotide chain-termination method (44) with the Sequi-Therm cycle sequencing kit (Epicentre Technologies, Madison, Wis.) and custom-designed oligonucleotide primers synthesized by Operon Technologies, Inc. The nucleotide sequence of both strands of a 6.3-kb region in the sde locus was determined in this manner. The DNA sequence was analyzed with ABI prism software and assembled with Deneba Sequencher software. Protein sequences were analyzed with the TMpred program to search for hydrophobicity and potential membrane-spanning domains.
RESULTS
Phenotypes of the Ω4408 mutant.
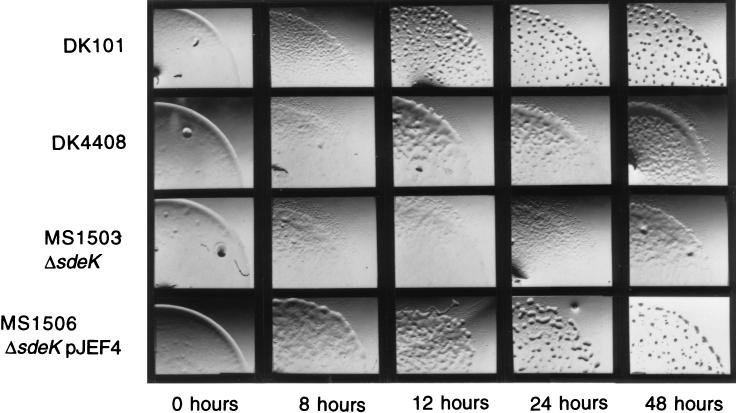
Fruiting body development in M. xanthus is accompanied by a series of morphological and behavioral changes that can be easily observed (Fig. 1). These changes include cellular aggregation, mound formation, and spore maturation. Kroos et al. (24, 25) reported that the Ω4408 Tn5lac insertion in the sde locus blocks fruiting body development and sporulation. To further examine these behavioral and morphological defects caused by Ω4408, cells from isogenic strains DK101 and DK4408 (DK101::Tn5lacΩ4408) were spotted on TPM agar for developmental assays. A comparison of the developmental phenotypes for each strain is shown in Fig. 1. Aggregation of DK4408 cells began at about the same time (8 h) as aggregation of wild-type DK101 cells. However, the aggregates of DK101 cells formed mound-shaped structures after 24 h of development, while the aggregates of DK4408 cells failed to compact into mounds throughout the 48 h that development was observed. Between 12 and 24 h of development, the DK4408 aggregates dissociated into featureless, translucent mats of cells, whereas the mounds of DK101 cells developed into darkened, spore-filled fruiting bodies.
FIG. 1.
Behavior of M. xanthus cells during development on TPM agar plates. The developmental progress of wild-type DK101 cells, DK4408 cells, MS1503 cells, and MS1506 cells is shown. Cells were spotted on TPM starvation agar and monitored visually as described in Materials and Methods. Photographs were taken at the times indicated.
Two methods were used to assay the sporulation efficiency of DK4408 and DK101 cells developing on TPM agar plates for 5 days. To determine the number of heat-resistant and sonication-resistant ovoid spores produced by developing cells, we used a Petroff-Hausser counting chamber and phase-contrast microscopy. To examine the viability of these spores, we placed them on CTT agar and counted the number of colonies that arose. The results from the sporulation assays are summarized in Table 2. The two techniques yielded similar results; the number of spores produced by DK4408 cells was about 0.8% of that produced by DK101 cells and the number of viable spores from DK4408 cells was about 0.1% of that produced by DK101 cells.
TABLE 2.
Developmental phenotypes of wild-type and mutant strainsa
| Strain | Presence of fruiting body developmentb | % of wild-type sporesc | % of wild-type viable sporesd |
|---|---|---|---|
| DK101 | + | 100.0 ± 7.7 | 100.0 ± 19.5 |
| DK4408 | − | 0.8 ± 0.3 | 0.1 ± 0.1 |
| MS1503 | − | 0.1 ± 0.1 | 0.2 ± 0.1 |
| MS1506 | + | 117.1 ± 9.7 | 90.0 ± 39.2 |
| MS2010 | + | NDe | 100f |
Cells were placed on TPM agar and allowed to develop for 5 days. Development was monitored visually as described in Materials and Methods. Direct spore counts and spore viability assays were used to determine the sporulation efficiency of a strain. Spore assays were performed three times for each strain except where indicated. The mean values (± standard deviations) for the spore assays are shown as a percentage of that of DK101 (wild type). The number of spores produced by wild-type cells ranged from 2 × 106 to 2 × 107.
+, fruiting bodies detected visually; −, fruiting bodies not detected visually.
Values were determined by counting the number of heat- and sonication-resistant spores on a Petroff-Hausser chamber by phase-contrast microscopy.
Values were determined by transferring heat- and sonication-resistant spores to CTT agar plates, incubating the plates for 5 days, and counting the number of colonies that arose from the spores.
ND, not determined.
This value was derived from one spore assay.
Expression of sde during growth and development.
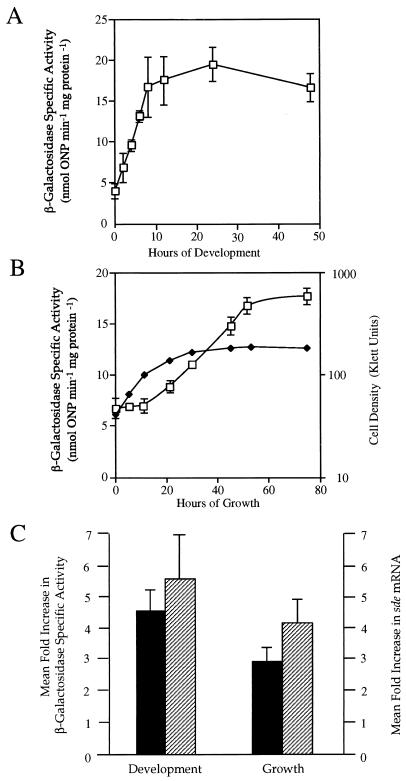
The Ω4408 Tn5lac insertion in DK4408 places lacZ under transcriptional control of a promoter in the sde locus. Using this lacZ reporter-gene fusion, we examined sde expression in DK4408 cells developing on TPM agar (Fig. 2A). β-Galactosidase specific activity began to increase within 2 h after starvation-initiated fruiting body development. Peak β-galactosidase specific activity was observed around 12 h of development, and these levels remained relatively unchanged over the next 36 h. The level of β-galactosidase specific activity after 12 h of development is approximately fivefold higher than that at the onset of the developmental program. To examine expression of sde during vegetative growth, we assayed β-galactosidase specific activity in DK4408 cells placed in CTT liquid media. Figure 2B shows that β-galactosidase specific activity started to increase during the transition from exponential growth to stationary phase and that β-galactosidase specific activity continued to increase well into stationary phase. The peak β-galactosidase specific activity observed in stationary phase is approximately threefold higher than the peak observed in exponential phase. These findings show that expression of sde is induced in both developing cells and in vegetative cells entering stationary phase.
FIG. 2.
Patterns of sde expression. The mean β-galactosidase specific activities found in DK4408 cells and the mean sde mRNA levels found in DK101 cells were determined from three independent experiments. Error bars are the standard deviations of the mean. The open squares represent β-galactosidase specific activity, and solid diamonds represent cell density. The mean β-galactosidase specific activities found at the indicated times during development on TPM agar (A) and during vegetative growth in CTT broth (B) are shown. The mean fold increase in β-galactosidase specific activity (■) and the mean fold increase in sde mRNA (▨) are compared for 0 to 12 h of development on TPM agar plates and 0 to 53 h of growth in CTT liquid (C).
To determine whether the increases in β-galactosidase specific activity observed during growth and development of DK4408 cells occur at the level of sde mRNA accumulation, RNA slot blot hybridization analysis was used. Total RNA was purified from DK101 cells during development on TPM agar and growth in CTT broth. The RNA was transferred to nylon membranes and probed with a 300-bp ApaI fragment containing DNA immediately upstream of the Ω4408 insertion site (Fig. 3), and the relative levels of sde mRNA were quantified. The results shown in Fig. 2C indicate that the relative increases in sde mRNA during growth and development are similar to relative increases in β-galactosidase specific activity. Furthermore, these findings show that sde expression does not change in an sde+ or sde mutant background, suggesting that this locus is not autoregulated.
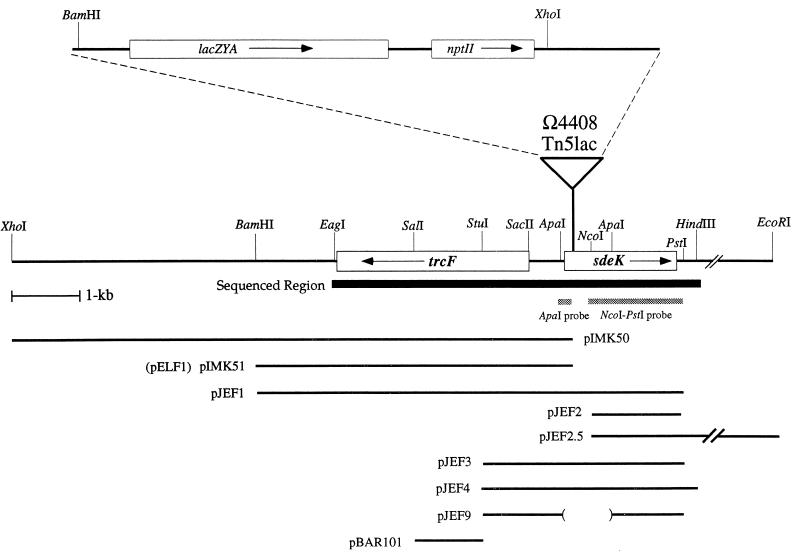
FIG. 3.
Physical map of the Ω4408 insertion region. The broadened black line indicates the region that has been characterized by DNA sequence analysis. Boxes show the locations of the indicated ORF. The arrow inside each box shows the predicted orientation of transcription of the ORF. The location of the Ω4408 insertion (triangle) was determined by DNA sequencing and Southern blot analysis as described in Materials and Methods. An enlargement of Tn5lac is shown above the triangle. Plasmids containing the indicated DNA fragments of the sde locus are shown below the map of the Ω4408 insertion region. The parentheses indicate the extent of the deleted region in plasmid pJEF9, and the parallel slashes indicate contiguous DNA sequence that extends to the EcoRI site, approximately 24 kb downstream from the end of the sdeK ORF. The 300-bp ApaI fragment and the 1.1-kb NcoI-PstI used for slot blot hybridization analysis of sde mRNA are represented by stippled boxes.
Cloning the sde locus.
In a previous study, Kroos et al. (24) mapped several restriction sites in DNA upstream (with respect to lacZ transcription) of the Ω4408 insertion. The restriction enzyme XhoI cuts once within Ω4408 Tn5lac and once a short distance upstream of the insertion, generating a 22-kb chromosomal fragment with 9.5 kb of Ω4408 Tn5lac and approximately 12.5 kb of upstream DNA (Fig. 3). The fact that the 9.5 kb of Ω4408 Tn5lac on this XhoI fragment contains the gene that confers kanamycin resistance (nptII) allowed us to identify a plasmid (pIMK50) carrying the 22-kb XhoI insert. A 6-kb BamHI fragment from plasmid pIMK50 was subcloned into pBluescript SKII and into pBGS18 to yield plasmids pIMK51 and pELF1, respectively. This 6-kb BamHI fragment was subsequently used as a probe for Southern blot analysis (data not shown), and DNA sequence analysis was used to confirm that it contained 50 bp from the 5′ end of Ω4408 Tn5lac and 6 kb of chromosomal DNA upstream of this insertion.
An in situ cloning technique was used to isolate chromosomal DNA downstream of Ω4408 Tn5lac. Plasmid pELF1 was introduced into strain DK101 by electroporation, and Kanr colonies were isolated for further analysis. One Kanr isolate (MS1510) was shown by Southern blot analysis to carry a single copy of plasmid pELF1 integrated in the chromosome by homologous recombination (data not shown). The findings from the Southern blot analysis indicated that the restriction enzyme PstI cuts once within the multicloning site of pELF1 and once 1.7 kb downstream of the plasmid insertion site. Thus, by digesting chromosomal DNA prepared from MS1510 with PstI and ligating the pool of PstI fragments, we generated a plasmid (pJEF1) that carries 6 kb of chromosomal DNA upstream of the Ω4408 insertion site, 1.7 kb of chromosomal DNA downstream of the Ω4408 insertion site, and no Ω4408 Tn5lac DNA (Fig. 3). The composition of pJEF1 was confirmed by restriction analysis and by DNA sequencing. To obtain an additional 24 kb of DNA downstream of the PstI site, a similar procedure was used (see Materials and Methods). The resulting clone containing this additional DNA was designated pJEF2.5.
DNA sequence analysis of the sde locus.
A 6.3-kb region of the native sde locus from the upstream EagI site to the downstream HindIII site was characterized by DNA sequence analysis (Fig. 3 [GenBank accession no. AF031084]). This region of DNA covers 4.1 kb upstream and 2.2 kb downstream of the Ω4408 insertion site. Two open reading frames (ORFs), initially designated ORF1 and ORF2, were identified. Both ORFs have a strong bias toward G + C nucleotides in the third position of the codons (92.7% for ORF1 and 89.2% for ORF2), which is typical for genes in M. xanthus and other high-G + C organisms (3, 47).
The 3.6-kb ORF1 is located upstream of the Ω4408 insertion site, and it is predicted to be transcribed in the opposite orientation relative to lacZ from the Ω4408 Tn5lac fusion. This suggests that ORF1 is not part of the operon defined by Ω4408. The deduced amino acid sequence of the ORF1 product shows 44% identity and 64% similarity to the E. coli transcription repair coupling factor, TrcF (45, 46). In E. coli, TrcF is involved in strand-specific repair of DNA lesions that block transcription by RNA polymerase. We have renamed ORF1 “trcF” based on this strong sequence similarity.
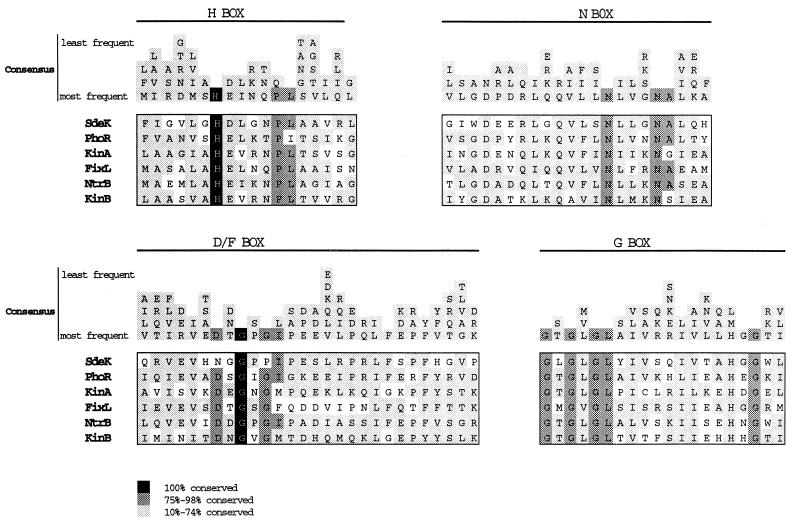
The 1.6-kb ORF2 is predicted to be transcribed in the same orientation relative to lacZ from the Ω4408 Tn5lac fusion. In addition, the DNA sequence analysis revealed that the Ω4408 insertion site is in the 5′ end of ORF2, indicating that ORF2 is part of the operon defined by Ω4408. The C-terminal 228 residues of the putative ORF2 product show strong similarity and identity to the transmitter domain in histidine kinase proteins. Histidine kinase proteins function as sensors in two-component signal transduction systems, and a conserved histidine residue, which is often found in the transmitter domain of these proteins, is phosphorylated in response to a particular intracellular or extracellular signal. Four highly conserved regions have been identified in the transmitter domain of these sensor proteins (38, 51). Region H contains the histidine residue that serves as the site of autophosphorylation, and region G is thought to function as a nucleotide binding site (38, 51). A specific function has not yet been assigned to region N or region D/F. The sequence alignments shown in Fig. 4 indicate that ORF 2 (designated SdeK for starvation-inducible, development-essential kinase) contains all four of these conserved regions.
FIG. 4.
Alignment of the SdeK C terminus with conserved regions in the transmitter domain of known histidine kinase proteins. Conserved sequences of FixL from Rhizobium meliloti (1, 5), KinA from Bacillus subtilis (2, 39), KinB from B. subtilis (54), NtrB from E. coli (35), PhoR from E. coli (30), and SdeK from M. xanthus are included. The four highly conserved regions (H-Box, N-Box, D/F-Box, and G-Box) in histidine kinase proteins are shown (38, 51). Amino acid residues found in 10 to 100% of the histidine kinase proteins are presented above the sequence alignment as presented by Stock et al. (51). Black boxes indicate that a residue is absolutely conserved among all histidine kinases. The dark gray boxes indicate that a residue is found in 75 to 98% of histidine kinases. The light gray boxes indicate that a residue is found in 10 to 74% of histidine kinases.
The deduced amino acid sequence in the N-terminal portion of SdeK yielded no significant similarities to protein sequences in the GenBank database. However, previous analyses have shown that the amino acid sequences of the N-terminal region of histidine kinases are not well conserved (38). This lack of conservation is consistent with the proposed function of the N-terminal region as input domains that change the activity of each protein in response to a specific signal. Further analysis of the N-terminal region of SdeK revealed no hydrophobic membrane-spanning regions, suggesting that SdeK may localize to the cytoplasmic compartment of M. xanthus cells. Thus, SdeK may belong to the family of cytoplasmic histidine kinase proteins with an N-terminal input domain followed by a C-terminal transmitter domain. Some of the members of this family of proteins are DegS from Bacillus subtilis (13, 26), KinA from Bacillus subtilis (2, 39), NifL from Azotobacter vinelandii (4, 42), and NtrB from E. coli (35).
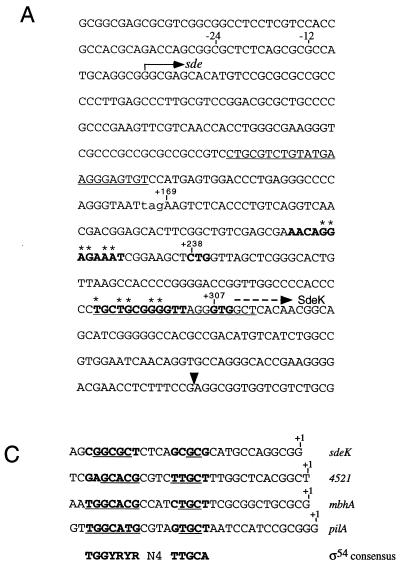
The 5′ end of the sdeK ORF is defined by a TAG stop codon (+169, Fig. 5A). Because the N-terminal domain is not conserved in histidine kinases, the sequence analysis does not aid in assigning the start of SdeK translation. The 5′ end of the sdeK ORF contains two potential ribosome binding sites for translation initiation of the SdeK protein. The first putative ribosome binding site is separated from a downstream GTG codon (+307, Fig. 5A) by 3 nucleotides, whereas the second putative site is separated by 8 nucleotides from a CTG codon (+238, Fig. 5A). Because GTG is more commonly used as a translation initiation codon than CTG (7), we propose that the downstream site at +307 is the probable start of the SdeK protein coding sequence.
FIG. 5.
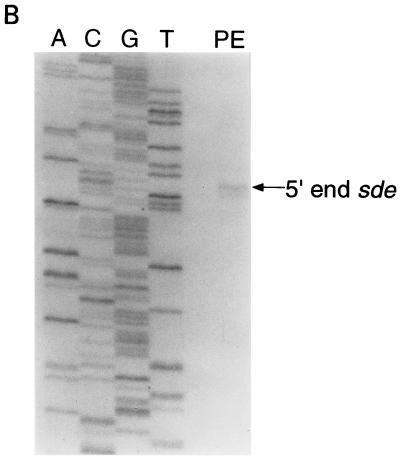
(A) DNA sequence of the region upstream of the Ω4408 Tn5lac insertion. The black triangle shows the location of the Ω4408 insertion. The bent arrow above the +1 shows the 5′ end of the predicted sde transcript, and the −12 and −24 regions are indicated. The stop codon (TAG) that defines the 5′ end of the sdeK ORF (+169) is shown in lowercase. Potential ribosome binding sites and translational start codons for the SdeK protein are in boldface. Nucleotides marked with an asterisk are complementary to the 3′ end of M. xanthus 16S rRNA (37). The dashed arrow above the GTG at position +307 is the proposed start of the SdeK coding sequence. The underlined regions show the sequences that are complementary to the two primers (ORF12 and ORF12-2) used to identify the 5′ end of the sde transcript. (B) Mapping the 5′ end of the sde developmental transcript by primer extension analysis. A, C, G, and T show the DNA sequencing ladders. PE, primer extension with total RNA prepared from DK101 cells developing on TPM agar for 12 h and primer ORF12 (Materials and Methods). (C) Comparison of the sde promoter region to the ς54 consensus sequence and three M. xanthus ς54 promoters: mbhA (21), 4521 (21), and pilA (55). The proposed start of transcription is designated by the +1.
Localizing the 5′ end of the sde developmental transcript.
The DNA sequence of the sde locus strongly suggests that the sdeK gene is the first gene in the sde transcriptional unit. To identify the 5′ end of the sde developmental transcript, primer extension analysis was performed with 12-h (the time at which peak levels of sde mRNA were detected) developmental RNA and a primer (ORF12) that is complementary to the region around the putative start codon for SdeK (Fig. 5A). Figure 5B shows that the 5′ end of the sde mRNA mapped to a guanine nucleotide 307 bp upstream of the putative start of the SdeK coding region. In a similar primer extension experiment with a second primer (ORF12-2), the same 5′ end was detected for sde mRNA (data not shown). Analysis of this 307-bp region upstream of sdeK revealed no obvious ORF, suggesting that sdeK is the first gene in the sde transcript.
Examination of the 5′ region preceding the putative transcriptional start site for the sde operon revealed a sequence with similarities to the ς54 family of promoters (52) (Fig. 5C). The similarity is strongest in the −24 region, with 5 out of 7 nucleotides identical to the ς54 consensus sequence. The similarity is less conserved around the −12 region, with only 2 out of 5 matches to the consensus. However, the highly conserved dinucleotides GG in the −24 region and GC in the −12 region are found. No similarities to other families of bacterial promoters were observed.
Identifying the gene in the sde locus that is required for development and sporulation.
The DNA sequence analysis showed that the Ω4408 Tn5lac insertion is located within the sdeK coding sequence. Because sdeK and trcF appear to be in diverging operons, this finding implies that the developmental block caused by Ω4408 does not result from inactivation of the trcF gene. The most plausible explanation for the Ω4408 phenotype is an inactivation of sdeK function or a polar effect on a gene or genes further downstream of sdeK.
To confirm that inactivation of trcF is not responsible for the Ω4408 phenotype, we constructed strain MS2010 and examined its ability to form fruiting bodies and to sporulate (Table 2). MS2010 is a derivative of strain DK101 containing an insertion of plasmid pBAR101 in the chromosome. The insertion in MS2010 produces two incomplete copies of the trcF gene. When placed on TPM agar plates, cells of MS2010 formed normal-looking fruiting bodies after 24 h of development, and the number of viable spores produced by these fruiting bodies was similar to the number of viable spores produced by wild-type DK101 fruiting bodies. These findings indicate that inactivation of the trcF gene is not responsible for the developmental arrest observed for the Ω4408 insertion mutant.
To determine whether the developmental defects caused by the Ω4408 insertion are due to an inactivation of the sdeK gene, we constructed a strain, designated MS1503, that carries a deletion in sdeK (ΔsdeK). This deletion removes 550 bp from the 5′ end of the putative SdeK coding region, including the putative translation initiation sites, as well as an additional 150 bp of upstream DNA. When a fragment of DNA located downstream of the 700-bp deletion was used as a probe for slot blot hybridization analysis, no sde mRNA was detected in the MS1503 mutant (data not shown), indicating that the deletion has a polar effect on downstream transcription. To examine the developmental defects caused by ΔsdeK, MS1503 cells were assayed for development and compared to DK101 and DK4408 cells (Fig. 1). Cells from all three strains began to aggregate at about the same time, but DK4408 cells and MS1503 cells failed to compact into mounds after 48 h of development. In addition, the number of spores and the number of viable spores produced by strain MS1503 were about 1,000-fold lower than those in wild-type strain DK101 and similar to those in strain DK4408 (Table 2). Thus, the defects in development and sporulation observed by the ΔsdeK strain are similar to the defects caused by the Ω4408 insertion.
To demonstrate that the loss of sdeK function was responsible for the developmental phenotypes observed for strain MS1503, we attempted to complement the mutation with a plasmid, pJEF4, that contains a wild-type copy of sdeK, as well as the upstream DNA sequences required to drive expression of this gene. In addition, plasmid pJEF4 carries the Mx8attP site for integration at the chromosomal Mx8 phage attachment site, attB. This plasmid was introduced into strain MS1503, giving rise to strain MS1506. MS1506 cells were assayed for development and compared to DK101 and MS1503 cells to determine whether a wild-type copy of the sdeK gene can rescue the defects caused by the ΔsdeK mutation (Fig. 1). MS1506 cells aggregated at approximately the same time as wild-type DK101 cells. However, these aggregates took 48 h to fully compact into darkened fruiting bodies, while DK101 cells took only 24 h. When we examined the sporulation efficiency of strain MS1506 after 5 days, we found that it was similar to that of strain DK101 (Table 2). These findings show that the defects in development and sporulation caused by the ΔsdeK mutation can be complemented by providing a wild-type copy of the sdeK gene at an ectopic site, although development appears to be slightly delayed. Our results suggest that the developmental block produced by the Ω4408 insertion is due to an inactivation of the sdeK gene and not to a polar effect on a gene downstream of sdeK.
DISCUSSION
Tn5lac transcriptional fusions were used by Kroos and Kaiser (22) to identify approximately 30 developmentally regulated loci in the M. xanthus chromosome. Expression of eight of the Tn5lac fusions is induced within a few hours after starvation initiates the developmental program (24). Among these eight early fusions, Ω4408 Tn5lac is unique because the insertion itself produces a developmental block (24, 25). Thus, the Ω4408 insertion defines a genetic locus that is absolutely required for progression through the developmental cycle. We have designated this locus sde, and in this report we have examined its regulation and its potential function in development.
Expression of sde is induced when cells initiate development and when cells enter stationary phase.
Expression from the Ω4408 Tn5lac fusion is induced early in the M. xanthus development program (0 to 2 h), well in advance of the first visible signs of cellular aggregation. Peak expression of sdeK was not observed until approximately 12 h into development, when the aggregates of cells are compacting into mounds. After 12 h of development, the level of sde expression was found to be about fivefold higher than at the onset of development. Further analysis of Ω4408 expression showed that induction of sde occurs as growing cells begin to enter stationary phase. Expression of sde continued to increase well into stationary phase, giving a peak induction of about threefold. These results show that induction of sde is not limited to the M. xanthus developmental program. Our finding that levels of expression of sde are similar in both an sdeK+ and an sdeK mutant background suggests that sdeK is not autoregulated.
What is the event that leads to activation of the sde locus? The results of our expression studies suggest that sde may be induced when nutrient levels are insufficient to maintain sustained growth. Previous findings are consistent with this hypothesis. For example, Singer and Kaiser (48) and Harris et al. (12) showed that expression of Ω4408 Tn5lac is dependent on high levels of the intracellular starvation signal (p)ppGpp. Furthermore, the conditions that activated expression of sde in our studies correlate with the conditions previously shown to generate relatively high levels of (p)ppGpp (31–33). Thus, the sde locus may be expressed either directly or indirectly in response to the starvation signal (p)ppGpp.
The 5′ end of the sde transcript is located 307 bp upstream of the putative start codon for SdeK.
The results of primer extension analyses with developmental RNA showed that the 5′ end of the sde transcript is located 307 bp upstream of the putative SdeK coding region. Because this 307-bp region does not appear to contain a bona fide ORF, we believe that sdeK is the first gene in the sde operon. This finding suggests that sde mRNA has a relatively long leader sequence. A similar result was observed by Fisseha et al. (8), who proposed that the C-signal-dependent Ω4403 mRNA has a 142-bp leader sequence. The sde leader could potentially be involved in regulation of sde expression at either the transcriptional or translational level. Alternatively, it could be involved in regulating the stability of sde mRNA.
The DNA sequence upstream of the transcriptional start for sde has similarity to the ς54 family of promoters. This class of promoters has sequence identity in the −12 and −24 regions, with the GC dinucleotide in the −12 region and the GG dinucleotide in the −24 region being highly conserved (52). The putative promoter for sde contains these two conserved dinucleotides, and it has strong identity to the consensus sequence in the −24 region (5 out of 7 nucleotides). Recent work by Keseler and Kaiser (21) suggests that two developmentally regulated genes, mbhA and 4521, have ς54-like promoters. Based on their findings, they proposed that development in M. xanthus could be regulated by a cascade or series of positive activators all utilizing the same ς54 RNA polymerase holoenzyme (21). Although the temporal regulation of these three genes during development is similar, each has unique requirements for full expression. For example, mbhA and 4521 require the extracellular A-signal for full expression, while sde does not (27, 29). Furthermore, mbhA requires a solid surface for expression, while 4521 and sde do not (21). Thus, the contrast between levels of expression of these genes may reflect the differences in their utilization of upstream activator proteins.
The product of the sdeK gene is required for fruiting body development and sporulation.
The results of the DNA sequence analysis with cloned fragments of the sde locus localized the site of the Ω4408 insertion to the sdeK ORF, 96 bp downstream of the putative translation start codon of the SdeK protein. A second ORF, designated trcF, was identified upstream of the sdeK gene in what appears to be a diverging operon. An insertion within the trcF ORF has no detectable effect on development or sporulation, indicating that the product of this gene is not required for fruiting body formation.
Cells carrying the Ω4408 insertion or the 700-bp deletion in sdeK form loose aggregates, but these aggregates fail to compact into visible mound-shaped structures. Furthermore, these mutations in sdeK reduce the number of spores and the number of viable spores from about 100- to 1,000-fold. Thus, both mutations cause similar defects in development and in sporulation. When a wild-type copy of the sdeK gene is provided at an ectopic site, the developmental defects caused by the ΔsdeK mutation could be complemented, although fruiting body formation was slightly delayed. Because other studies have found that gene expression from the attB site can be substantially reduced compared to expression from native sites (8, 21), we propose that this delay may be due to lower levels of sdeK expression from attB. These findings suggest that the defects in development and sporulation observed for the Ω4408 insertion mutant are due to an inactivation of the sdeK gene, implying that the product of sdeK is required for normal fruiting body formation.
The C terminus of SdeK has similarity to the transmitter domain of histidine kinase proteins.
When we searched the GenBank database for similarities to the putative product of the sdeK gene, we found that the C-terminal 228 amino acids of SdeK have similarity to the transmitter domain of histidine kinase proteins of two-component regulatory systems. In the two-component paradigm, the histidine kinase component senses a specific intracellular or extracellular signal. In response to this change, it autophosphorylates on a conserved histidine residue within the transmitter domain. The phosphoryl group can then be transferred to an aspartate residue within the receiver domain of the cognate response regulator component, modulating its activity. Typically, the response regulator is a DNA-binding protein that alters gene expression. Thus, two-component regulatory systems allow cells to monitor environmental and physiological changes and then respond accordingly.
The finding that SdeK may be a cytoplasmic histidine kinase has several implications. First, it suggests that SdeK may be part of a signal transduction pathway. Disruption of the sdeK gene blocks development prior to mound formation, indicating that this signal transduction pathway is needed relatively early in the developmental program. Second, the activity of SdeK may be modulated by an intracellular signal or by association with a membrane-bound component which senses some extracellular change. Third, SdeK may propagate the input signal by altering the activity of a response regulator, allowing this protein to change developmental gene expression or some other cellular process. Because a wild-type copy of the sde locus is needed for developmental expression of the devRS genes (25), it is tempting to speculate that one potential target of the SdeK signal transduction pathway is the devRS operon. Further understanding of the function of the proposed SdeK signaling pathway requires that we identify the input signal for SdeK, its response regulator partner, and the target or targets of the response regulator.
ACKNOWLEDGMENTS
We thank Peggy Baer and Jodi Nunnari for helpful discussions and for critical reading of the manuscript. In addition, we thank Stacia Hoover and Dean Lavell for technical assistance with sequencing and for loaning computer software.
This work was supported (in part) by a National Institutes of Health postdoctoral fellowship (GM19080) to A.G.G. and a National Institutes of Health grant (GM54592) to M.S.
REFERENCES
- 1.Anthamatten D, Hennecke H. The regulatory status of the fixL- and fixJ-like genes in Bradyrhizobium japonicum may be different from Rhizobium meliloti. Mol Gen Genet. 1991;225:38–48. doi: 10.1007/BF00282640. [DOI] [PubMed] [Google Scholar]
- 2.Antoniewski C, Savelli B, Stragier P. The spoIIJ gene, which regulates early developmental steps in Bacillus subtilis, belongs to a class of environmentally responsive genes. J Bacteriol. 1990;172:86–93. doi: 10.1128/jb.172.1.86-93.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bibb M J, Findlay P R, Johnson M W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein coding sequences. Gene. 1984;30:157–166. doi: 10.1016/0378-1119(84)90116-1. [DOI] [PubMed] [Google Scholar]
- 4.Blanco G, Drummond M, Woodley P, Kennedy C. Sequence and molecular analysis of the nifL gene of Azotobacter vinelandii. Mol Microbiol. 1993;9:869–879. doi: 10.1111/j.1365-2958.1993.tb01745.x. [DOI] [PubMed] [Google Scholar]
- 5.David M, Daveran M L, Batut J, Dedieu A, Domergue O, Ghai J, Hertig C. Cascade regulation of nif gene expression in Rhizobium meliloti. Cell. 1988;54:671–683. doi: 10.1016/s0092-8674(88)80012-6. [DOI] [PubMed] [Google Scholar]
- 6.Downard J, Ramaswamy S V, Kil K-S. Identification of esg, a genetic locus involved in cell-cell signaling during Myxococcus xanthus development. J Bacteriol. 1993;175:7762–7770. doi: 10.1128/jb.175.24.7762-7770.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Draper D E. Translation initiation. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: American Society for Microbiology; 1996. pp. 902–908. [Google Scholar]
- 8.Fisseha M, Gloudemans M, Gill R E, Kroos L. Characterization of the regulatory region of a cell interaction-dependent gene in Myxococcus xanthus. J Bacteriol. 1996;178:2539–2550. doi: 10.1128/jb.178.9.2539-2550.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gill R E, Cull M G, Fly S. Genetic identification and cloning of a gene required for developmental cell interactions in Myxococcus xanthus. J Bacteriol. 1988;170:5279–5288. doi: 10.1128/jb.170.11.5279-5288.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hagen D C, Bretscher A P, Kaiser D. Synergism between morphogenetic mutants of Myxococcus xanthus. Dev Biol. 1978;64:284–296. doi: 10.1016/0012-1606(78)90079-9. [DOI] [PubMed] [Google Scholar]
- 11.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 12.Harris, B. Z., D. Kaiser, and M. Singer. 1998. The guanosine nucleotide (p)ppGpp initiates development and A-factor production in Myxococcus xanthus. 12:1022–1035. [DOI] [PMC free article] [PubMed]
- 13.Henner D J, Yang M, Ferrari E. Localization of Bacillus subtilis sacU(Hy) mutations to two linked genes with similarities to the conserved procaryotic family of two-component signaling systems. J Bacteriol. 1988;170:5102–5109. doi: 10.1128/jb.170.11.5102-5109.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hodgkin J, Kaiser D. Cell-to-cell stimulation of movement in nonmotile mutants of Myxococcus. Proc Natl Acad Sci USA. 1977;74:2938–2942. doi: 10.1073/pnas.74.7.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones K A, Yamamoto K R, Tjian R. Two distinct transcription factors bind the HSV thymidine kinase promoter in vitro. Cell. 1985;42:559–572. doi: 10.1016/0092-8674(85)90113-8. [DOI] [PubMed] [Google Scholar]
- 16.Julien, B., and D. Kaiser. Personal communication.
- 17.Kaiser D. Control of multicellular development: Dictyostelium and Myxococcus. Annu Rev Genet. 1986;20:539–566. doi: 10.1146/annurev.ge.20.120186.002543. [DOI] [PubMed] [Google Scholar]
- 18.Kaiser D, Losick R. How and why bacteria talk to each other. Cell. 1993;73:873–885. doi: 10.1016/0092-8674(93)90268-u. [DOI] [PubMed] [Google Scholar]
- 19.Kalman L V, Cheng Y L, Kaiser D. The Myxococcus xanthus dsg gene product performs functions of translation initiation factor IF3 in vivo. J Bacteriol. 1994;176:1434–1442. doi: 10.1128/jb.176.5.1434-1442.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaplan H B, Kuspa A, Kaiser D. Suppressors that permit A-signal-independent developmental gene expression in Myxococcus xanthus. J Bacteriol. 1991;173:1460–1470. doi: 10.1128/jb.173.4.1460-1470.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keseler I M, Kaiser D. An early A-signal-dependent gene in Myxococcus xanthus has a ς54-like promoter. J Bacteriol. 1995;177:4638–4644. doi: 10.1128/jb.177.16.4638-4644.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kroos L, Kaiser D. Construction of Tn5lac, a transposon that fuses lacZ expression to exogenous promoters, and its introduction into Myxococcus xanthus. Proc Natl Acad Sci USA. 1984;81:5816–5820. doi: 10.1073/pnas.81.18.5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kroos L, Kaiser D. Expression of many developmentally regulated genes in Myxococcus depends on a sequence of cell interactions. Genes Dev. 1987;1:840–854. doi: 10.1101/gad.1.8.840. [DOI] [PubMed] [Google Scholar]
- 24.Kroos L, Kuspa A, Kaiser D. A global analysis of developmentally regulated genes in Myxococcus xanthus. Dev Biol. 1986;117:252–266. doi: 10.1016/0012-1606(86)90368-4. [DOI] [PubMed] [Google Scholar]
- 25.Kroos L, Kuspa A, Kaiser D. Defects in fruiting body development caused by Tn5 lac insertions in Myxococcus xanthus. J Bacteriol. 1990;172:484–487. doi: 10.1128/jb.172.1.484-487.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kunst F, Debarbouille M, Msadek T, Young M, Mauel C, Karamata D, Klier A, Rapoport G, Dedonder R. Deduced polypeptides encoded by the Bacillus subtilis sacU locus share homology with two-component sensor-regulator systems. J Bacteriol. 1988;170:5093–5101. doi: 10.1128/jb.170.11.5093-5101.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuspa A, Kroos L, Kaiser D. Intercellular signaling is required for developmental gene expression in Myxococcus xanthus. Dev Biol. 1986;117:267–276. doi: 10.1016/0012-1606(86)90369-6. [DOI] [PubMed] [Google Scholar]
- 28.Kuspa A, Plamann L, Kaiser D. Identification of heat-stable A-factor from Myxococcus xanthus. J Bacteriol. 1992;174:3319–3326. doi: 10.1128/jb.174.10.3319-3326.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.LaRossa R, Kuner J, Hagen D, Manoil C, Kaiser D. Developmental cell interactions of Myxococcus xanthus: analysis of mutants. J Bacteriol. 1983;153:1394–1404. doi: 10.1128/jb.153.3.1394-1404.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Makino K, Shinagawa H, Amemura M, Nakata A. Nucleotide sequence of the phoR gene, a regulatory gene for the phosphate regulon of Escherichia coli. J Mol Biol. 1986;192:549–546. doi: 10.1016/0022-2836(86)90275-5. [DOI] [PubMed] [Google Scholar]
- 31.Manoil C, Kaiser D. Accumulation of guanosine tetraphosphate and guanosine pentaphosphate in Myxococcus xanthus during starvation and myxospore formation. J Bacteriol. 1980;141:297–304. doi: 10.1128/jb.141.1.297-304.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manoil C, Kaiser D. Guanosine pentaphosphate and guanosine tetraphosphate accumulation and induction of Myxococcus xanthus fruiting body development. J Bacteriol. 1980;141:305–315. doi: 10.1128/jb.141.1.305-315.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manoil C C. Ph.D. thesis. Stanford, Calif: Stanford University; 1982. [Google Scholar]
- 34.Messing J, Gronenborn B, Muller-Hill B, Hopschneider P. Filamentous coliphage M13 as a cloning vehicle: insertion of a HindII fragment of the lac regulatory region in M13 replicative form in vitro. Proc Natl Acad Sci USA. 1977;74:3642–3646. doi: 10.1073/pnas.74.9.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miranda-Rios J, Sanchez-Pescador R, Urdea M, Covarrubias A A. The complete nucleotide sequence of the glnALG operon of Escherichia coli K12. Nucleic Acids Res. 1987;15:2757–2770. doi: 10.1093/nar/15.6.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mirel D B, Chamberlin M J. The Bacillus subtilis flagellin gene (hag) is transcribed by the ς28 form of RNA polymerase. J Bacteriol. 1989;171:3095–3101. doi: 10.1128/jb.171.6.3095-3101.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oyaizu H, Woese C. Phylogenetic relationships among the sulfate respiring bacteria, myxobacteria and purple bacteria. Syst Appl Microbiol. 1985;6:257–263. [Google Scholar]
- 38.Parkinson J S, Kofoid E C. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 39.Perego M, Cole S P, Burbulys D, Trach K, Hoch J A. Characterization of the gene for a protein kinase which phosphorylates the sporulation-regulatory proteins Spo0A and Spo0F of Bacillus subtilis. J Bacteriol. 1989;171:6187–6196. doi: 10.1128/jb.171.11.6187-6196.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Plamann L, Kuspa A, Kaiser D. Proteins that rescue A-signal-defective mutants of Myxococcus xanthus. J Bacteriol. 1992;174:3311–3318. doi: 10.1128/jb.174.10.3311-3318.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Plamann L, Davis J M, Cantwell B, Mayor J. Evidence that asgB encodes a DNA-binding protein essential for growth and development of Myxococcus xanthus. J Bacteriol. 1994;176:2013–2020. doi: 10.1128/jb.176.7.2013-2020.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raina R, Bageshwar U K, Das H K. The Azotobacter vinelandii nifL-like gene: nucleotide sequence analysis and regulation of expression. Mol Gen Genet. 1993;237:400–406. doi: 10.1007/BF00279444. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 44.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Selby C P, Sancar A. Molecular mechanism of transcription-repair coupling. Science. 1993;260:53–58. doi: 10.1126/science.8465200. [DOI] [PubMed] [Google Scholar]
- 46.Selby C P, Sancar A. Structure and function of transcription-repair coupling factor. J Biol Chem. 1995;270:4882–4889. doi: 10.1074/jbc.270.9.4882. [DOI] [PubMed] [Google Scholar]
- 47.Shimkets L J. Social and developmental biology of the Myxobacteria. Microbiol Rev. 1990;54:473–501. doi: 10.1128/mr.54.4.473-501.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singer M, Kaiser D. Ectopic production of guanosine penta- and tetraphosphate can initiate early developmental gene expression in Myxococcus xanthus. Genes Dev. 1995;9:1633–1644. doi: 10.1101/gad.9.13.1633. [DOI] [PubMed] [Google Scholar]
- 49.Spratt B G, Hedge P J, Heesen S T, Edelman A, Broome-Smith J K. Kanamycin-resistant vectors that are analogs of plasmids pUC8, pUC9, pEMBL8 and pEMBL9. Gene. 1986;41:337–342. doi: 10.1016/0378-1119(86)90117-4. [DOI] [PubMed] [Google Scholar]
- 50.Stephens K, Kaiser D. Genetics of gliding motility in Myxococcus xanthus: molecular cloning of the mgl locus. Mol Gen Genet. 1987;207:256–266. [Google Scholar]
- 51.Stock J B, Surette M G, Levit M, Park P. Two-component signal transduction systems: structure-function relationships and mechanisms of catalysis. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: ASM Press; 1995. pp. 25–52. [Google Scholar]
- 52.Thöny B, Hennecke H. The −24/−12 promoter comes of age. FEMS Microbiol Rev. 1989;63:341–358. doi: 10.1016/0168-6445(89)90028-4. [DOI] [PubMed] [Google Scholar]
- 53.Thöny-Meyer L, Kaiser D. devRS, an autoregulated and essential genetic locus for fruiting body development in Myxococcus xanthus. J Bacteriol. 1993;175:7450–7462. doi: 10.1128/jb.175.22.7450-7462.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trach K A, Hoch J A. Multisensory activation of the phosphorelay initiating sporulation in Bacillus subtilis: identification and sequence of the protein kinase of the alternate pathway. Mol Microbiol. 1993;8:69–79. doi: 10.1111/j.1365-2958.1993.tb01204.x. [DOI] [PubMed] [Google Scholar]
- 55.Wu S S, Kaiser D. Regulation of expression of the pilA gene in Myxococcus xanthus. J Bacteriol. 1997;179:7748–7758. doi: 10.1128/jb.179.24.7748-7758.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]