Abstract
The product of the Escherichia coli modE gene, ModE, is a member of a unique class of molybdate-responsive DNA binding proteins. Here we investigated the roles of the N- and C-terminal domains of ModE in mediating DNA binding and protein dimerization, respectively. Compared to the full-length protein, the N-terminal half of ModE has a greatly diminished capacity to bind the modA promoter in vitro and to repress expression from a modA-lacZ operon fusion in vivo. Fusing a protein dimerization domain, encoded by the C terminus of λ CI repressor protein, to the truncated ModE protein generated a ModE-CI fusion protein that not only displayed a greatly increased in vivo repressor activity but could also substitute for ModE at the moaA and dmsA promoters. In the reciprocal experiment, we restored repressor activity to a truncated CI protein by addition of the C-terminal domain of ModE, which is comprised of two MopI-like subdomains. By an in vivo competition assay, we also demonstrated that the CI-ModE chimeric protein retained the ability to interact with wild-type ModE. Finally, specific deletions within the ModE portion of the CI-ModE protein chimera abolished both in vivo repression and the ability to interact with wild-type ModE. Together, these data demonstrate that the N-terminal domain of ModE is sufficient to mediate DNA binding, although efficient binding requires that ModE form a dimer, a function that is supplied by the C-terminal MopI-like subdomains.
The molybdate-responsive transcription factor, ModE, regulates the expression of a number of operons in Escherichia coli in response to changes in the intracellular levels of molybdate. ModE-regulated operons identified to date in E. coli encode either proteins involved in molybdate uptake (modABCD [3, 11, 14, 16, 18]), and molybdenum cofactor synthesis (moaABCDE [12]) or enzymes that require incorporation of a molybdenum cofactor (dmsABC [13]) and napF (unpublished data). DNase I footprinting identified a consensus binding site at the promoters of all four operons (1, 12, 13; unpublished data), and this site confirms a consensus sequence proposed from sequence comparisons of promoter regions of a number of molybdate-responsive operons in E. coli and in other organisms (8).
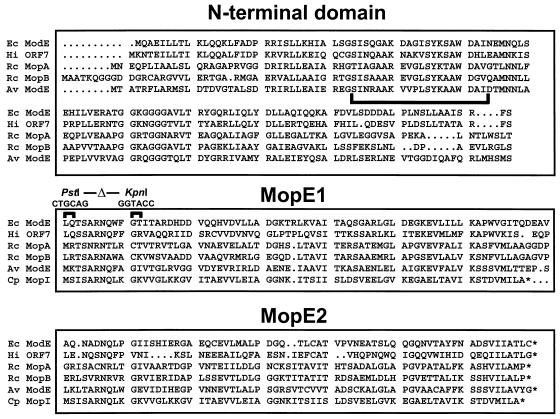
Sequence alignments of ModE homologs from a variety of organisms suggest that the proteins all have a common bipartite structure (3, 9, 11). The C-terminal domain of ModE is comprised of two conserved tandem repeats, designated MopE1 and MopE2 (Fig. 1), each of which is similar to a small molybdopterin binding protein, MopI, from Clostridium pasteurianum (6). The Mop family of proteins in C. pasteurianum, of which there are three highly homologous members (MopI, -II, and -III [7]), have been implicated in molybdate storage within the cell and are proposed to multimerize in the presence of molybdate (5, 8). The possibility that the MopI-like domains in ModE from E. coli are involved in molybdate sensing and regulation of DNA binding was suggested by the finding that either partially or completely deleting the last domain (MopE2 [Fig. 1]) abolished the requirement for molybdate in effecting repression of modA-lacZ expression in vivo (3, 11). These C-terminal deletions also reduced the ability of ModE to function as a repressor, and the removal of both MopE1 and MopE2 domains virtually abolished repressor activity (11). These findings suggest that the C-terminal domain of ModE may both negatively regulate DNA binding and be required for efficient binding. These data also suggest that the N terminus of ModE may be sufficient to mediate DNA binding, albeit weakly. Consistent with these findings, a search of the BLOCKS database indicated that the first 54 residues of ModE have a distant homology with the same region of the LysR family of transcriptional regulators (1), and inspection of this sequence revealed the presence of a weak helix-turn-helix motif (Fig. 1).
FIG. 1.
Alignment of the various ModE homologs. Shown are the amino acid sequences, aligned with the BLAST program, of ModE homologs from Haemophilus influenzae (Hi), Rhodobacter capsulatus (Rc), and Azotobacter vinelandii (Av). Also shown is the MopI protein (also aligned with the BLAST program) from C. pasteurianum (Cp). The various ModE domains, N-terminal, MopE1, and MopE2 domains, are boxed, and the putative helix-turn-helix motif is indicated by a bracket below the sequence. Stop codons are indicated by stars, and gaps are indicated by periods. The region of ModE that corresponds to those DNA sequences that were deleted in the cI-modE2 fusion (i.e., between the PstI and KpnI sites) is labeled. Ec, E. coli.
In this study, we employed fusions to the λ CI repressor protein to determine the individual roles of the N- and C-terminal domains of ModE. We demonstrate that a weak DNA binding activity of the N-terminal domain is enhanced 11-fold by appending the dimerization domain of CI. By constructing the reciprocal protein chimera, we show that the C-terminal domain of ModE is able to substitute for the dimerization domain of CI, implying that it plays a similar role in the native protein. Through the construction of defined deletions, we also demonstrate that ModE dimerization requires both MopE1 and MopE2 subdomains.
MATERIALS AND METHODS
Bacterial strains, phages, plasmids, and culture conditions.
Strains, phages, and plasmids used are listed in Table 1. For β-galactosidase assays, cells were grown aerobically at 37°C in glucose (20 mM) minimal medium (pH 7); sodium molybdate and sodium nitrate were added as required at 100 μM and 40 mM concentrations, respectively (2).
TABLE 1.
E. coli K-12 strains, bacteriophages, and plasmids used in this study
| Strain, phage, or plasmid | Origin | Relevant genotype or phenotype | Source or reference |
|---|---|---|---|
| Strains | |||
| MC4100 | F Δ(argF-lac)U169 | Laboratory stock | |
| RCC42 | MC4100 | modC | 16 |
| PM6 | MC4100 | modC modE::Kan | 11 |
| PM8 | MC4100 | modE::Kan | 11 |
| Phages | |||
| λ202 | Φ(Pr-lacZ) lacY+ lacA+ (operon fusion) | 17 | |
| λUD4 | λRS45 | Φ(modA-lacZ) lacY+ lacA+ (operon fusion) | 16 |
| λPM53 | λRS45 | Φ(moaA-lacZ) lacY+ lacA+ (operon fusion) | This study |
| λPM40 | λRS45 | Φ(dmsA-lacZ) lacY+ lacA+ (operon fusion) | 13 |
| Plasmids | |||
| pACYC184N | pACYC184 | tetA gene modified to place an NdeI site at start codon; Cmr | Laboratory stock |
| pPM6 | pACYC184N | modE+ Cmr | 11 |
| pPM80 | pPM6 | modE+; His tag at N terminus and engineered PstI site; Cmr | This study |
| pPM81 | pACYC184N | modE; truncated at Q-120 with a His tag at N terminus; Cmr | This study |
| pPM82 | pPM80 | modE-cI gene fusion; His tag at N terminus; Cmr | This study |
| pPM83 | pPM80 | cI-modE2 gene fusion: fusion is at KpnI site; Cmr | This study |
| pPM84 | pPM80 | cI-modE1 gene fusion: fusion is at PstI site; Cmr | This study |
| pPM85 | pACYC184N | cI; truncated at R-116; Cmr | This study |
| pPM86 | pACYC184N | Full-length cI indI ts857; Cmr | This study |
| pPM87 | pPM84 | cI-modE3 gene fusion (PstI site) with deletion of MopE2 domain; Cmr | This study |
Recombinant DNA techniques.
Transformation of E. coli and plasmid isolation and manipulations were performed as described previously (10). DNA sequencing, with the Sequitherm Excel kit (Epicentre Technologies), and PCR amplification, with a GeneAmp PCR system (Perkin-Elmer Cetus), were performed according to the manufacturers’ instructions. One strand of all PCR products was sequenced entirely to verify accurate amplification.
Construction of modE-cI chimera.
The modE gene was amplified with primers 683 (5′-GGGCATATG[CAT]6CAGGCCGAAATCCTTCTCA-3′) and 723 (5′-ATGGTACCGAACCACTGGTTACGGGCGCTGGTCTGCAGT-3′) to introduce six histidine residues at the N terminus of ModE and a unique PstI site midway (the PstI site corresponded to amino acid residues 123 and 124 and required one conservative mutation [Fig. 1]) between the sequences encoding the N- and C-terminal domains. This fragment was restricted with NdeI and KpnI and used to replace the corresponding fragment in pPM6, generating pPM80. In a similar manner, modE was amplified with primers 683 and 726 (5′-GGGGATCCTTATTGCAGTGAAAAACGTGAGAT-3′) to change the sequence encoding residue 124 (glutamine) to a nonsense codon; the resultant fragment was cloned into pACYC184N, generating pPM81. The region of cI encoding amino acid residues 117 to 237 was amplified from λ cI indI ts857 (Promega) with primers 724 (5′-GGAACTGCAGACCTTTACCAAAGGTGATGCGA-3′) and 725 (5′-GGGGATCCTTAGCCAAACGTCTCTTCAGGCCACT-3′) to introduce flanking PstI and BamHI sites and was used to replace the corresponding fragment of modE in pPM80, generating pPM82.
Construction of cI-modE chimeras.
The region of cI encoding amino acid residues 1 to 115 was amplified with primers 727 (5′-GGGGCATATGAGCACAAAAAAGAAACCATTA-3′) and 729 (5′-GGAACTGCAGAAGCTTAGGTGAGAACATCCCT-3′) to introduce flanking NdeI and PstI sites. This fragment was used to replace the corresponding fragment of modE in pPM80, generating pPM84 (encoding CI-ModE1). The same region of cI was amplified with primers 727 and 728 (5′-GGGGGTACCAAGCTTAGGTGAGCCCATCCCT-3′) to introduce flanking NdeI and KpnI sites and cloned in pPM80, generating pPM83 (encoding CI-ModE2). Arginine 116 of CI was changed to a nonsense codon with primers 727 and 730 (5′-GGGGATCCTTAAAGCTTAGGTGAGAACATCCCT-3′) and cloned into pACYC184N, generating pPM85. Full-length cI was amplified with primers 725 and 727 and cloned into the NdeI-BamHI sites in pACYC184N, generating pPM86. The cI-modE chimera in pPM84 was modified as follows: modE was amplified with primers 683 and 740 (5′-CCCGGATCCTTACTGAGTAATACCTACCCACGGCGCTTT-3′) to change codon 192 to a nonsense codon and place a BamHI site immediately downstream of the change. This fragment was restricted with PstI and BamHI and used to replace the corresponding fragment in pPM84, generating pPM87 (encoding CI-ModE3).
Construction of moaA-lacZ operon fusion.
The moaA promoter region was amplified with primers 639 (5′-GGGATCCGCAATATATTGAATT-3′) and 631 (5′-CGAATTCGACAGGCGCAAGTAGTAA-3′), and the resultant fragment was cloned into the BamHI-EcoRI sites in pRS415, generating pPM53. The fusion was transferred to λRS45, generating the prophage λPM53, and integrated into the chromosome of strains MC4100 and PM8 in single copy.
β-Galactosidase assays.
β-Galactosidase levels were determined by hydrolysis of 2-nitrophenyl-β-d-galactopyranoside (ONPG), and units of activity are expressed as nanomoles of ONPG hydrolyzed per minute per milligram of protein (2). The values presented are the averages of at least three independent experiments, and the standard deviation between experiments was less than 10%.
RESULTS
Efficient DNA binding requires that ModE form a dimer.
Previously, we observed that expression of the N-terminal domain of ModE resulted in a twofold decrease in modA-lacZ expression in vivo (11). One explanation for the impaired in vivo repression is that ModE binds the modA operator as a dimer and that removal of the C-terminal domain abolished dimerization. To test this hypothesis, we fused the C terminus of λ CI (residues 117 to 237), which encodes a well-characterized protein dimerization domain, to the N-terminal domain of ModE (residues 1 to 124 [Fig. 1 and 2]) and expressed the resultant protein in vivo. All the genes and gene fusions used in this study were fused precisely at the start codon of the tetA gene on pACYC184 so that the transcription and translation signals were precisely the same for each construct. The ModE-CI fusion protein, encoded by pPM82, displayed an 11-fold increase in repressor activity in vivo compared to that of the truncated ModE protein (pPM81 [Table 2]). The DNA binding activity of ModE, both in vitro and in vivo, is increased in the presence of molybdate (1, 12). To determine if the repressor activity of the fusion protein was similarly dependent on molybdate, we repeated these assays with a modC strain; the modC mutation abolishes molybdate uptake, and unless the medium is supplemented with large amounts of molybdate (ca. 100 μM), the levels of molybdate in the cell are negligible and wild-type ModE is unable to repress modA-lacZ expression. The absence of molybdate had no effect on the repressor activity of the fusion protein (data not shown).
FIG. 2.
Schematic representation of the various wild-type, truncated, and chimeric proteins. Hatched bars represent ModE sequences, and solid bars represent sequences from CI. Approximate locations of the DNA binding domains in both ModE and CI are shown. Also shown are the locations of the tandem MopI-like repeats (designated MopE1 and MopE2) in ModE and the dimerization domain in CI.
TABLE 2.
Dimerization of ModE improves repressor activity
| Strain (plasmid) | Relevant genotypea | Protein under test | β-Galactosidase activityb | Repression ratioc |
|---|---|---|---|---|
| PM8(pACYC184N) | modE λUD4 | 8,050 | 1 | |
| PM8(pPM6) | modE λUD4 (modE+) | ModE | 30 | 268 |
| PM8(pPM80) | modE λUD4 (modE+) | ModE | 15 | 537 |
| PM8(pPM81) | modE λUD4 [modE(Am)] (truncation at Q-120 of gene product) | ModEN | 1,950 | 4 |
| PM8(pPM82) | modE λUD4 (modE-cI) | ModE-CI | 185 | 44 |
| PM8(pPM86) | modE λUD4 (cI) | CI | 8,100 | 1 |
λUD4 is a modA-lacZ-carrying prophage, inserted in the chromosome of strain PM8 in single copy. Genes present on multicopy plasmids are shown in parentheses.
Units are given in nanomoles of ONPG hydrolyzed per minute per milligram of protein. Cells were grown in minimal glucose medium under aerobic conditions as described in the text.
Repression ratio is defined as the units of β-galactosidase exhibited by the control strain, PM8(pACYC184N), divided by the units of activity in the strains carrying the various test plasmids.
The C terminus of ModE mediates dimerization.
The above data demonstrate that, although the N terminus of ModE encodes all the determinants necessary for DNA recognition and binding, efficient repression is achieved only when ModE is able to dimerize. To determine if the C-terminal domain of ModE is able to mediate dimerization, we fused this domain to the DNA binding domain of CI (residues 1 to 115). It is important to note that the fusion was made at the PstI site in modE (Fig. 1) and therefore preserves both MopE1 and MopE2 domains intact. As noted previously (17), the DNA binding domain of CI alone (encoded by pPM85) functioned poorly as a repressor of Pr-lacZ expression in vivo (Table 3). In contrast, the CI-ModE1 fusion protein (encoded by pPM84) repressed expression from a Pr-lacZ operon fusion to the same degree as did full-length CI (encoded by pPM86) and exhibited a 14-fold increase over the level with the truncated CI protein (Table 3). Previous data suggested that the two MopI-like domains in full-length ModE negatively regulate ModE’s DNA binding activity in response to low molybdate availability (11). To determine if they functioned in the same manner in the CI-ModE1 fusion protein, we repeated the assay in a modC background. The absence of molybdate had no effect on the ability of the CI-ModE1 fusion protein to repress Pr-lacZ expression (data not shown). We conclude that ligand is not essential for dimerization by this assay.
TABLE 3.
The C terminus of ModE promotes dimerization of lambda repressor CI
| Strain (plasmid) | Relevant genotypea | Protein under test | β-Galactosidase activityb | Repression ratioc |
|---|---|---|---|---|
| PM8(pACYC184N) | modE λ202 | 20,500 | 1 | |
| PM8(pPM86) | modE λ202 (cI+) | CI | 980 | 21 |
| PM8(pPM85) | modE λ202 [cI(Am)] (truncation at R-116 of gene product) | CIN | 12,500 | 1.6 |
| PM8(pPM84) | modE λ202 (cI-modE1) | CI-ModE1 | 950 | 22 |
| PM8(pPM83) | modE λ202 (cI-modE2) | CI-ModE2 | 15,900 | 1.3 |
| PM8(pPM87) | modE λ202 (cI-modE3) | CI-ModE3 | 20,000 | 1 |
| PM8(pPM80) | modE λ202 (modE+) | ModE | 21,000 | 1 |
λ202 is a prophage carrying a λPr-lacZ operon fusion that contains a wild-type set of λ CI binding sites. It is inserted in the chromosome of strain PM8 in single copy. Genes present on multicopy plasmids are shown in parentheses.
Units are given in nanomoles of ONPG hydrolyzed per minute per milligram of protein. Cells were grown in minimal glucose medium under aerobic conditions as described in the text.
Repression ratio is defined as the units of β-galactosidase exhibited by the control strain, PM8(pACYC184N), divided by the units of activity in the strains carrying the various test plasmids.
To determine if dimerization required the presence of both MopE1 and MopE2 domains, we constructed a second protein fusion, CI-ModE2 (encoded by pPM83), in which we utilized the same region of cI but moved the fusion point further downstream in modE (from the PstI site to the KpnI site [Fig. 1]). This deletion was specifically engineered to remove the first 10 amino acids of MopE1, which were proposed to encode the molybdate binding domain (3). The deletion almost completely abolished the ability of the chimeric protein to function as a repressor (pPM83 [Table 3]). Previously, we demonstrated that deletion of the last MopI-like domain, MopE2 (Fig. 1 and 2), from full-length ModE abolished the requirement for molybdate in effecting repression (11). When we deleted the MopE2 domain from the CI-ModE1 protein fusion, generating CI-ModE3 (Fig. 2), it completely abolished repressor activity (pPM87 [Table 3]). Taken together, these data argue that the C-terminal domain of ModE alone is sufficient to promote dimerization and that dimerization requires that both MopI-like domains be intact.
The CI-ModE1 chimeric protein is negatively dominant over wild-type ModE.
The above data imply that the ModE C-terminal domain is sufficient to mediate dimerization. However, as we are working with chimeric, rather than native, proteins, we cannot rule out the possibility that dimerization is somehow an artifactual result of the recombinant constructions. We therefore sought to determine if the CI-ModE1 chimeric protein could interact with wild-type ModE. We reasoned that, if the two proteins interacted and formed stable heterodimers, then the resultant dimer, having two different DNA binding specificities, would be unable to bind and repress modA-lacZ expression in vivo. This was found to be the case (Table 4); the presence of pPM84, which expresses the CI-ModE1 fusion protein, in a wild-type strain resulted in a 33-fold increase in modA-lacZ expression compared to that of either the vector or a plasmid expressing wild-type CI. We repeated this assay with plasmids which express either CI-ModE2 (pPM83) or CI-ModE3 (pPM87); these fusion proteins have deletions at the proximal and distal end of the ModE portion of the chimera, respectively. Neither plasmid had any effect on modA-lacZ expression (Table 4). These data, taken together with the previous observations, strongly suggest that efficient dimerization of ModE requires two intact MopI-like domains (Fig. 1).
TABLE 4.
Heterodimer formation between wild-type ModE and a CI-ModE fusion protein is abolished by disruption of either MopE1 or MopE2
| Strain (plasmid) | Relevant genotypea | Protein under test | β-Galactosidase activityb | Induction ratioc |
|---|---|---|---|---|
| MC4100(pACYC184N) | λUD4 | 230 | 1 | |
| PM8(pACYC184N) | λUD4 modE | 9,500 | 41 | |
| MC4100(pPM80) | λUD4 (modE+) | ModE | 25 | 0.1 |
| MC4100(pPM84) | λUD4 (cI-modE1) | CI-ModE1 | 7,500 | 33 |
| MC4100(pPM83) | λUD4 (cI-modE2) | CI-ModE2 | 250 | 1 |
| MC4100(pPM87) | λUD4 (cI-modE3) | CI-ModE3 | 230 | 1 |
| MC4100(pPM86) | λUD4 (cI) | CI | 220 | 1 |
λUD4 is a modA-lacZ-carrying prophage, inserted in the chromosome of strains MC4100 and PM8 in single copy. Genes present on multicopy plasmids are shown in parentheses.
Units are given in nanomoles of ONPG hydrolyzed per minute per milligram of protein. Cells were grown in minimal glucose medium under aerobic conditions as described in the text.
Induction ratio is defined as the units of β-galactosidase exhibited by the strains carrying the various test plasmids divided by the units of activity in the control strain, MC4100(pACYC184N).
The ModE-CI chimeric protein can substitute for ModE at the moaA and dmsA promoters.
We previously demonstrated that ModE is required for optimal expression from the moaA and dmsA promoters (12, 13). To determine if the ModE-CI chimera can substitute for ModE at these promoters, we introduced pPM82 into strains carrying the relevant reporter fusions. The absence of modE resulted in a sixfold decrease in expression (Table 5) from the moaA-lacZ fusion under both aerobic and anaerobic growth conditions (Note that the fusion carried on λPM53 differed from the fusion described previously [12] in that it contained an additional 1.2 kb of upstream DNA [see Materials and Methods for details]). Provision of either modE+ or modE-cI in trans restored expression to the level observed in a wild-type strain. Similarly, utilizing a dmsA-lacZ fusion we observed that provision of modE-cI in trans restored both optimal anaerobic expression and nitrate-dependent repression (Table 5).
TABLE 5.
The ModE-CI fusion protein can substitute for ModE at the moaA and dmsA promoters
| Strain | Relevant genotypea | β-Galactosidase activityb
|
||
|---|---|---|---|---|
| + O2 (none) | − O2
|
|||
| None | + NO3 | |||
| PM8(pACYC184) | modE λPM53 | 1,850 | 1,700 | ND |
| PM8(pPM6) | modE λPM53 (modE+) | 7,200 | 8,900 | ND |
| PM8(pPM82) | modE λPM53 (modE-cI) | 7,100 | 7,450 | ND |
| PM8(pACYC184) | modE λPM40 | 25 | 50 | 55 |
| PM8(pPM6) | modE λPM40 (modE+) | 15 | 145 | 20 |
| PM8(pPM82) | modE λPM40 (modE-cI) | 20 | 140 | 25 |
λPM53 and λPM40 are prophages, inserted in the chromosome of PM8 in single copy, carrying moaA-lacZ and dmsA-lacZ operon fusions, respectively. Genes present on multicopy plasmids are shown in parentheses.
Units are given in nanomoles of ONPG hydrolyzed per minute per milligram of protein. Cells were grown in minimal glucose medium under aerobic and anaerobic conditions as described in the text. Sodium nitrate (NO3) was added as indicated at 40 mM. ND, not determined.
DISCUSSION
The ModE homologs identified to date all appear to have a similar bipartite structure in which the N and C termini are predicted to mediate DNA and molybdate binding, respectively. In this study, we further analyzed these binding properties by using ModE from E. coli as a model.
We first addressed the role of the N terminus in mediating DNA binding. Expression of the N terminus of ModE in vivo resulted in weak repression of a modA-lacZ fusion, and in vitro studies confirmed that a similar truncated ModE polypeptide bound the modA operator with low affinity (unpublished results). The latter finding argues that the failure to repress in vivo is not due solely to differences in the in vivo stability of the wild-type and truncated proteins. An alternative explanation, suggested by the dyad symmetry of the ModE binding site (1, 8, 12, 13), is that ModE must dimerize in order to efficiently bind DNA. This appears to be the case since fusing the dimerization domain from CI to the N terminus of ModE resulted in an 11-fold increase in modA-lacZ repression in vivo. As was predicted from the absence of the putative molybdate binding domains, repressor activity was molybdate independent. Interestingly, the ModE-CI chimera could also substitute for ModE at the moaA and dmsA promoters. Since the chimeric protein lacks the entire C terminus of ModE, these data imply that regulation is not dependent on interactions between the missing domain and other proteins bound at either the dmsA or the moaA promoter.
We next addressed the role that the C terminus of ModE plays in mediating dimerization. Fusing this domain to the DNA binding domain of CI generated a chimera, CI-ModE1, that both functioned as an efficient repressor in vivo and retained the ability to interact with wild-type ModE. This latter finding allows us to speculate on when the dimerization occurs in the cell. By one model, ModE monomers interact and dimerize in the cytoplasm; by another, the monomers bind the ModE operator independently of one another and then interact to form stable dimers. The finding that CI-ModE1 and ModE are able to form stable heterodimers, despite the fact that they recognize and interact with totally different DNA binding sites, strongly suggests that dimerization takes in the cytoplasm.
We also addressed the role of the SARNQ motif in mediating ModE dimerization. This motif, located at the start of MopE1 (MopE2 carries a much poorer match), is strongly conserved among the various ModE homologs (Fig. 1) and has been identified in several molybdoenzymes (3). This led to the proposition that this sequence mediates molybdate binding (3, 4). A second fusion protein, CI-ModE2, which had the SARNQ motif deleted, was unable to mediate repression in vivo or to interact with wild-type ModE. Thus, the SARNQ motif does appear to be required to mediate dimerization. However, since the original CI-ModE1 fusion protein mediated repression in the absence of molybdate, these studies did not allow us to determine if the motif influenced molybdate binding. Dimerization, as measured by both in vivo repressor activity and negative dominance over wild-type ModE, was also abolished by completely deleting the MopE2 domain. We therefore conclude that both MopE1 and MopE2 are required to mediate dimerization. One future question is the role that molybdate plays in regulating DNA binding activity of ModE. The above-mentioned studies imply that molybdate does not influence dimerization directly. Another possibility, suggested by the finding that molybdate binding to ModE induces a conformational change (1), is that the molybdate-induced conformational change unmasks a previously occluded DNA binding domain; experiments are in progress to test this model.
ACKNOWLEDGMENTS
This work was supported in part by a grant from the National Institutes of Health, AI21678.
We thank K. Hantke for supplying λ202.
REFERENCES
- 1.Anderson L A, Palmer T, Price N C, Borneman S, Boxer D H, Pau R N. Characterization of the molybdenum-responsive ModE regulatory protein and its binding to the promoter region of the modABCD (molybdenum transport) operon of Escherichia coli. Eur J Biochem. 1997;246:119–126. doi: 10.1111/j.1432-1033.1997.00119.x. [DOI] [PubMed] [Google Scholar]
- 2.Cotter P A, Gunsalus R P. Oxygen, nitrate, and molybdenum regulation of dmsABC gene expression in Escherichia coli. J Bacteriol. 1989;171:3817–3823. doi: 10.1128/jb.171.7.3817-3823.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grunden A M, Ray R M, Rosentel J K, Healy F G, Shanmugam K T. Repression of the Escherichia coli modABCD (molybdate transport) operon by ModE. J Bacteriol. 1996;178:735–744. doi: 10.1128/jb.178.3.735-744.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grunden A M, Shanmugam K T. Molybdate transport and regulation in bacteria. Arch Microbiol. 1997;168:345–354. doi: 10.1007/s002030050508. [DOI] [PubMed] [Google Scholar]
- 5.Hinton S M, Mortenson L E. Regulation and order of involvement of molybdoproteins during synthesis of molybdoenzymes in Clostridium pasteurianum. J Bacteriol. 1985;162:485–493. doi: 10.1128/jb.162.2.485-493.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hinton S M, Merrit B. Purification and characterization of a molybdenum-pterin-binding protein (Mop) in Clostridium pasteurianum W5. J Bacteriol. 1986;168:688–693. doi: 10.1128/jb.168.2.688-693.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hinton S M, Slaughter C, Eisner W, Fisher T. The molybdenum-pterin binding protein is encoded by a multigene family in Clostridium pasteurianum. Gene. 1987;54:211–219. doi: 10.1016/0378-1119(87)90489-6. [DOI] [PubMed] [Google Scholar]
- 8.Kutsche M, Leimkühler S, Angermüller S, Klipp W. Promoters controlling expression of the alternative nitrogenase and the molybdenum uptake system in Rhodobacter capsulatus are activated by NtrC, independent of ς54, and repressed by molybdenum. J Bacteriol. 1996;178:2010–2017. doi: 10.1128/jb.178.7.2010-2017.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luque F, Mitchenall L A, Chapman M, Christine R, Pau R N. Characterization of genes involved in molybdenum transport in Azotobacter vinelandii. Mol Microbiol. 1993;7:447–459. doi: 10.1111/j.1365-2958.1993.tb01136.x. [DOI] [PubMed] [Google Scholar]
- 10.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 11.McNicholas P M, Chiang R C, Gunsalus R P. The Escherichia coli modE gene: effect of modE mutations on molybdate dependent modA expression. FEMS Microbiol Lett. 1996;145:117–123. doi: 10.1016/0378-1097(96)00398-9. [DOI] [PubMed] [Google Scholar]
- 12.McNicholas P M, Rech S A, Gunsalus R P. Characterization of the ModE DNA binding sites in the control regions of modABCD and moaABCDE of Escherichia coli. Mol Microbiol. 1997;23:515–524. doi: 10.1046/j.1365-2958.1997.d01-1864.x. [DOI] [PubMed] [Google Scholar]
- 13.McNicholas P M, Chiang R C, Gunsalus R P. Anaerobic regulation of the Escherichia coli dmsABC operon requires the molybdate responsive regulator, ModE. Mol Microbiol. 1998;27:197–208. doi: 10.1046/j.1365-2958.1998.00675.x. [DOI] [PubMed] [Google Scholar]
- 14.Mouncey N J, Mitchenall L A, Pau R N. Mutational analysis of genes of the mod locus involved in molybdenum transport, homeostasis, and processing in Azotobacter vinelandii. J Bacteriol. 1995;177:5294–5302. doi: 10.1128/jb.177.18.5294-5302.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mouncey N J, Mitchenall L A, Pau R N. The modE gene product mediates molybdenum-dependent expression of genes for the high-affinity molybdate transporter and modG in Azotobacter vinelandii. Microbiology. 1996;142:1997–2004. doi: 10.1099/13500872-142-8-1997. [DOI] [PubMed] [Google Scholar]
- 16.Rech S, Deppenmeier U, Gunsalus R P. Regulation of the molybdate transport operon, modABCD, of Escherichia coli in response to molybdate availability. J Bacteriol. 1995;177:1023–1029. doi: 10.1128/jb.177.4.1023-1029.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stojilkovic I, Hantke K. Functional domains of the Escherichia coli ferric uptake regulator protein (Fur) Mol Gen Genet. 1995;247:199–205. doi: 10.1007/BF00705650. [DOI] [PubMed] [Google Scholar]
- 18.Walkenhorst H M, Hemschemier S K, Eichenlaub R. Molecular analysis of the molybdate uptake operon, modABCD, of Escherichia coli and modR, a regulatory gene. Microbiol Res. 1995;150:347–361. doi: 10.1016/S0944-5013(11)80016-9. [DOI] [PubMed] [Google Scholar]