Abstract
The novel fatty acid trans-9-methyl-10-octadecenoic acid was isolated from the coryneform bacterial strain LMG 3820 (previously misidentified as Arthrobacter globiformis) and identified by spectroscopic methods and chemical derivatization. This fatty acid is attached to the unusual lipid acyl phosphatidylglycerol. Five different species of this lipid type were identified; their structures were elucidated by tandem mass spectrometry and are reported here for the first time. Additionally, we identified three different cardiolipins, two bearing the novel fatty acid. The characteristic 10-methyl-octadecanoic acid was present only in phosphatidylinositol. Because of the unusual fatty acid pattern of strain LMG 3820, the 16S rDNA sequence was determined and showed regions of identity to sequences of Corynebacterium variabilis DSM 20132T and DSM 20536. All three strains possessed the novel fatty acid, identifying trans-9-methyl-10-octadecenoic acid as a potential biomarker characteristic for this taxon. Surprisingly, the fatty acid and relative abundances of phospholipids of Corynebacterium sp. strain LMG 3820 were similar to those of the type strain but different from those of Corynebacterium variabilis DSM 20536, although all three strains possessed identical 16S rDNA sequences and strains DSM 20132T and DSM 20536 have 90.5% DNA-DNA homology. This is one of the rare cases wherein different organisms with identical 16S rDNA sequences have been observed to present recognizably different fatty acid and lipid compositions. Since methylation of a fatty acid considerably lowers the transition temperature of the corresponding lipid resulting in a more flexible cell membrane, the intraspecific variation in the lipid composition, coinciding with the morphological and Gram stain reaction variability of this species, probably offers an advantage for this species to inhabit different environmental niches.
Coryneform bacteria are gram-positive microorganisms which are widely distributed in the environment. Among the explanations advanced for the numerical predominance of some of these bacteria in soil are their extreme resistance to drying and to starvation and the nutritional versatility of the commonly occurring species (21). The group comprises various degraders, e.g., of chlorophenol (27), cyanide (11), or diphenyls (16) and amino acid producers and is therefore of environmental and biotechnological interest. The characterization and identification of coryneform bacteria are of particular interest because of their biotechnological potential and because conventional identification methods often fail for these organisms.
We have applied two chemotaxonomic methods, namely, gas chromatographic (GC) analysis of fatty acid methyl esters of glyco- and phospholipids and fast atom bombardment (FAB)-mass spectrometry (MS) of glyco- and phospholipids, to analyze strains belonging to the majority of the validly described species of the genera Agromyces, Aeromicrobium, Arthrobacter, Aureobacterium, Cellulomonas, Curtobacterium, Nocardioides, and Terrabacter. Analysis of the polar lipids of these strains by FAB-MS led to spectra which can serve as “fingerprints” of these lipids and allow rapid comparison between strains. These spectra revealed ions which are characteristic for taxa of the coryneform bacteria. Such compounds, which are characteristic for a group of organisms, are called biomarkers and are very valuable for the detection of these bacteria in bacterial communities. Especially in combination with tandem MS (MS/MS), these ions can be analyzed and the structures of the molecules from which they are derived can be elucidated. In this way, a number of rare or novel lipids were detected and their structures were elucidated (2, 32). The combination of nuclear magnetic resonance (NMR), leading to determination of the structure and configuration of the core of the polar lipid, with MS/MS, allowing determination of the structures and positions of the various fatty acids attached to this core, resulted in fast and sensitive elucidation of the structures of these biomarkers.
In the course of our study, we detected in one of the strains, LMG 3820, assigned to the species Arthrobacter globiformis, a composition of fatty acids quite different from that reported by other authors. Previously A. globiformis was reported to have mainly saturated iso- and anteiso-fatty acids and to produce mono-, digalacto-, and dimannopyranosyl lipids (34, 39), none of which are present in our strain. Consequently, we have studied and characterized strain LMG 3820 in depth. Here we report on the structures of the cardiolipins, phosphatidylglycerols, and phosphatidylinositol, together with those of the major unusual phospholipid, acyl phosphatidylglycerol (APG), found in this strain.
MATERIALS AND METHODS
Strains and culture medium.
Strain LMG 3820, assigned to A. globiformis, was obtained from the Laboratorium voor Microbiologie, Universiteit Gent, Ghent, Belgium; Corynebacterium variabilis DSM 20132T and C. variabilis DSM 20536 were obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ), Braunschweig, Germany. They were cultured at 37°C in 1-liter shake flasks in a medium containing 20 g of tryptone, 5 g of yeast extract, and 5 g of NaCl in 1 liter of deionized water. The biomass was harvested, in the late logarithmic phase of growth, after 72 h.
Polar lipid fatty acid analysis.
Lipids were extracted by a modified Bligh-Dyer procedure (3), and fatty acid methyl esters were generated and analyzed by GC as described previously (37).
Thin-layer chromatography.
Thin-layer chromatography was performed on 20- by 20-cm plates coated with 1-mm silica gel 60 with a fluorescence indicator. The solvent system was chloroform-methanol-ammonia (28%) (65/25/2.7, vol/vol/vol).
Quantitative determination of phospholipids.
Phospholipid content was determined by high-pressure liquid chromatography via a method recently described (17).
FAB-MS.
FAB-MS in the negative mode was performed on the first of two mass spectrometers of a tandem high-resolution instrument with an E1B1E2B2 configuration (JMS-HX/HX110A; JEOL, Tokyo, Japan) at a 10-kV accelerating voltage. Resolution was set to 1:1,500. The JEOL FAB gun was operated at 6 kV with xenon as the FAB gas. A mixture of triethanolamine and tetramethylurea (2:1, vol/vol) was used as the matrix.
MS/MS.
Negative daughter ion spectra were recorded by using all four sectors of the tandem mass spectrometer. High-energy collision-induced dissociation (CID) took place in the third field-free region. Helium served as the collision gas at a pressure sufficient to reduce the precursor ion signal to 30% of the original value. The collision cell was operated at ground potential. Resolution of MS2 was set to 1/1,000. FAB CID spectra (linked scans of MS2 at a constant magnetic-to-electric-field [B/E] ratio) were recorded with 300-Hz filtering and a JEOL DA 7000 data system.
One- and two-dimensional (1D and 2D) NMR spectra were recorded in 7:3 CDCl3-CD3OD at 300 K on Bruker DMX-600 NMR (1D, 1H; 2D, correlated spectroscopy [COSY] and total correlated spectroscopy [TOCSY] with a mixing time of 70 ms) and ARX-400 NMR (1D, 1H, 13C, and 31P; 2D, 1H detected one-bond and multiple-bond 13C multiple-quantum coherence spectra, HMQC and HMBC) spectrometers, respectively (35). 1H and 13C chemical shifts are given in parts/million relative to internal trimethylsilyl, 31P chemical shifts are relative to external H3PO4, and couplings are in hertz. Infrared (IR) spectra were measured on KBr, using the diffuse reflected IR Fourier transform (DRIFT) mode.
1,2-Diacylglycerylphospho-1′-monoacyl-glycerol.
Rf is 0.65 in chloroform-methanol-ammonia (28%) (65/25/2.7, vol/vol/vol). IR (KBr) spectral analysis yielded the following: 3,350 (br), 2,920, 2,850, 1,740, 1,600, 1,465, 1,385, 1,245, 1,175, 1,105, 1,075, 970, 820, and 720 cm−1.
16S rDNA sequencing.
Individual colonies were picked from agar medium, suspended in 100 μl of Tris-EDTA buffer, and boiled for 15 min. The suspension was centrifuged briefly, and 1 μl of the supernatant was used for PCR (29), with forward primer 16F27 and reverse primer 16R1492 (Escherichia coli 16S rRNA gene position) (24). PCR was carried out with a GeneAmp 9600 thermocycler (Perkin-Elmer, Weiterstadt, Germany) and conditions described previously (23). Amplified DNA was purified with Microcon 100 microconcentrators (Amicon GmbH, Witten, Germany), and quality was controlled with gel electrophoresis on a 1% agarose gel with Tris-acetate-EDTA buffer and subsequent ethidium bromide staining. The sequence of the amplified 16S rDNA gene was determined directly, using an Applied Biosystems 373A DNA sequencer (Perkin-Elmer, Applied Biosystems GmbH, Weiterstadt, Germany), standard 16S rRNA sequencing primers (24), and the protocols recommended by the manufacturer for Taq polymerase-initiated cycle sequencing with fluorescent-dye-labeled dideoxynucleotides. Resulting sequences were aligned with reference 16S and 16S rRNA gene sequences (26, 38), using the evolutionarily conserved primary sequence and secondary structure (15, 31) as references. Evolutionary distances (22) were calculated from complete sequence pair dissimilarities, using only homologous, unambiguously determined nucleotide positions. Phylogenetic trees were constructed with the programs of the PHYLIP package (12).
DNA-DNA hybridization.
The analysis was done by DSMZ. The DNA was isolated by chromatography on hydroxyapatite (4), and DNA-DNA hybridizations were carried out by using a Gilford System 2600 spectrophotometer equipped with a Gilford 2527-R thermoprogrammer and plotter (10).
Nucleotide sequence accession number.
The 16S rRNA gene sequences determined have been deposited in the EMBL nucleotide sequence database under accession no. AJ222815 to AJ222817.
RESULTS
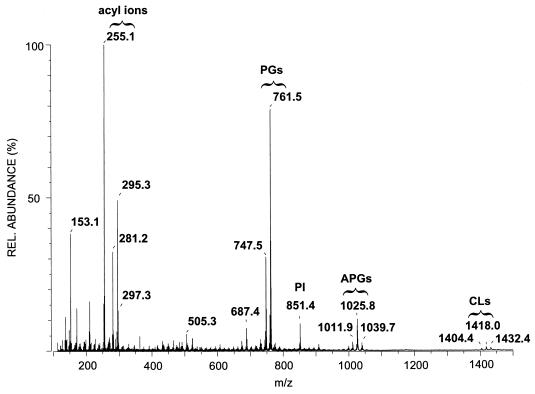
A combination of MS/MS and multidimensional NMR techniques provides a means of establishing the structure of the major component in the phospholipid fraction from the coryneform bacterium LMG 3820. Negative-mode FAB-MS of this fraction showed, in addition to phosphatidylglycerols and cardiolipins, deprotonated molecular ions at m/z 999, 1,011, 1,025, and 1,039 (Fig. 1). The differences in mass suggested the presence of a homologous series of molecules arising from differences in fatty acid composition. Indeed, major carboxylate anions for C15:0 (m/z 241), C16:0 (m/z 255), C18:1 (m/z 281), and C19:1 (m/z 295) were observed (Fig. 1). The C15:0, C16:0, and C18:1 fatty acids were identified from their fragmentation patterns in MS/MS experiments as n-pentadecanoic acid, n-hexadecanoic acid (palmitic acid), and 9-octadecenoic acid (oleic acid), respectively (data not shown) (18). The C19:1 fatty acid, however, displayed an unusual fragmentation pattern and needed further investigation.
FIG. 1.
Negative-mode FAB mass spectrum of the phospholipid fraction of strain LMG 3820, showing acyl ions between m/z 250 and 300, phosphatidylglycerol ions (PGs) around m/z 750, phosphatidylinositol ions (PI) at m/z 851, APGs between m/z 1,010 and 1,040, and cardiolipins (CLs) around m/z 1,420. Note that there is only one phosphatidylinositol, which is the only polar lipid to bear 10-methyl-octadecanoic acid. REL., relative.
Identification of this fatty acid by MS was performed with a chromatographically purified sample. Alkaline saponification yielded a solution containing the free carboxylic acids, which was then subjected to negative-mode FAB-MS. The signal intensity of the free carboxylic acids was much higher than those of the carboxylate anions generated by fragmentation of the intact molecule and resulted in CID spectra that were much more intense and more reliable to interpret.
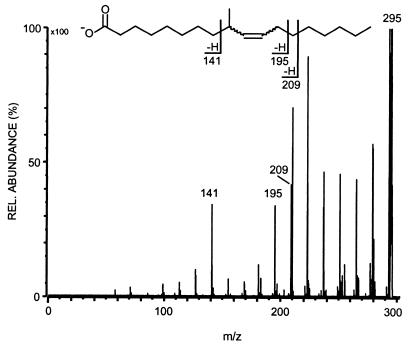
Branch points or double bonds in carboxylic acid residues are usually clearly indicated in the CID spectra. A saturated, methyl-branched carboxylic acid displays a dip of one signal in the evenly 14-amu (atomic mass unit)-spaced fragmentation pattern (18), while monounsaturated carboxylic acids show a gap of three signals due to allylic cleavage (19, 36). The carboxylate anion at m/z 295 exhibited a gap of four minor peaks surrounded by two larger ones, at m/z 210 and 141, resulting from allylic cleavage (Fig. 2). The observation of a fourth minor signal implies the presence of a methyl branch somewhere in the allylic unit. Whether this fragment, at m/z 195, results from vinylic cleavage at the double bond or from subsequent elimination of a methylene residue from the fragment at m/z 210 remains unclear. However, the allylic fragments indicate that the double bond is between carbons 10 and 11. A very similar fragmentation pattern has been described by Couderc and colleagues for 11-methyl-12-octadecenoic acid detected in mycobacteria (7, 8). To determine the position of the methyl branch a microhydrogenation (H2 over Pd-C) was performed with the saponified sample. The negative-mode FAB CID spectrum of the saturated product (m/z 297) clearly showed a single gap in the fragmentation pattern between m/z 169 and 141, establishing that the methyl group is located at carbon 9.
FIG. 2.
Negative-mode FAB CID spectrum of the C19:1 fatty acid from the hydrolysate of APG. For a discussion of the observed daughter ions, see text.
Taking both results together, we conclude that the only possible structure for the unknown fatty acid is 9-methyl-10-octadecenoic acid. No differentiation between the cis and trans isomers of the underivatized carboxylic acid could be made from the CID spectra. However, the configuration was unambiguously established to be trans from the coupling constant data (J = 15.7 Hz) for this moiety in the 1H NMR spectrum. Such a fatty acid has not been reported so far in the literature.
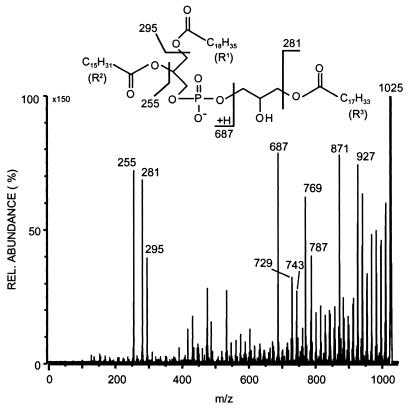
To elucidate the structure of the polar lipid bearing the novel fatty acid, detailed MS studies were performed. MS/MS studies of the major deprotonated molecular ion at m/z 1,025 revealed three different fatty acids, n-hexadecanoic acid, 9-octadecenoic acid, and 9-methyl-10-octadecenoic acid, as ions corresponding to the neutral loss of free fatty acids (m/z 769, 743, and 729) and the corresponding ketenes (m/z 787, 761, and 747) were observed (Fig. 3). Furthermore, there were a number of fragments in the upper mass region at intervals of about 14 amu. These correspond to fragmentations along the fatty acid acyl chains and represent charge remote fragmentation similar to that observed in other phospholipids (20). Included here are fragments due to allylic cleavages in the unsaturated acyl fatty acids (m/z 927 and 871).
FIG. 3.
Negative-mode FAB CID spectrum of the main APG at m/z 1,025. The insert shows the major fragmentation pathways of this phospholipid.
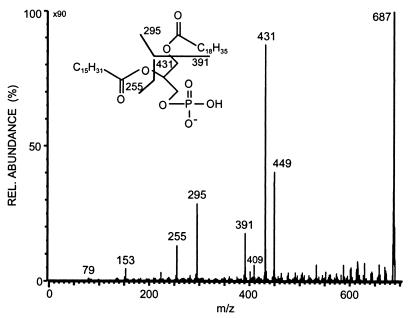
Diagnostically important was the daughter ion at m/z 687, which was therefore analyzed by additional CID experiments. The CID spectrum (Fig. 4) displayed the typical pattern of a glycerophosphatic acid (GPA) (30). CID of the (M-H)− ion from GPA yielded abundant carboxylate anions from both the sn-1 position and the sn-2 position (m/z 255 and 295), thus establishing that this fragment contained n-hexadecanoic acid and 9-methyl-10-octadecenoic acid. In addition, there were neutral losses of the sn-2 and sn-1 substituent as free carboxylic acid (m/z 431 and 391) as well as loss of each fatty acyl group as a substituted ketene (m/z 449 and 409). The intensity differences of these various ions indicated the positions of the different fatty acid moieties, as losses are most abundant for the substituent positioned at sn-2 (30). Therefore, palmitic acid must be at sn-2 and 9-methyl-10-octadecenoic acid must be at sn-1. The ions at m/z 153 and 79 are due to the phosphate unit. These data indicated that the fragment at m/z 687 is 1-(9-methyl-10-octadecenoyl)-2-hexadecanoyl-GPA.
FIG. 4.
Negative-mode FAB CID spectrum of the phosphatidyl acid at m/z 687 derived from the APG at m/z 1,025. The insert shows the major fragmentation pathways. Note the different intensities of the ions at m/z 449 and 391, which can be used to assign the two fatty acids to the different positions at the glycerol moiety.
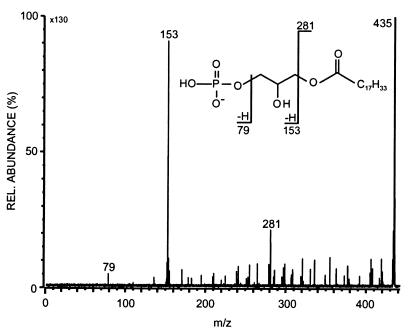
Subtracting the mass of this structural moiety from the molecular weight of 1,026 gives a difference of 339 amu. Taking into account that there must be an additional C18:1 fatty acid residue (281 amu), then there is a final remainder of 58 amu. This is in agreement with a double-esterified glycerol backbone, which carries one free hydroxyl group (C3H6O). This was shown to be the case, as acetylation caused an increase in the molecular weight to 1,068, corresponding to addition of one acetyl group. The other minor components in the phospholipid fraction behaved in a similar way. The phospholipid therefore seemed to be a phosphatidylglycerol which is additionally esterified at one position in the second glycerol unit. Under negative-mode FAB conditions, one would expect a fragment consisting of the phosphate unit, the glycerol residue, and the C18:1 acyl fatty acid. This would have a mass of 435 amu and was detected. The CID spectrum is shown in Fig. 5. The observed daughter ions are in full accord with the postulated partial structure as indicated in the fragmentation scheme.
FIG. 5.
Negative-mode FAB CID spectrum of the phosphatidyl acid at m/z 435 derived from the APG at m/z 1,025. The insert shows the major fragmentation pathways.
The nature and substitution patterns of the glycerol moieties were verified from a comprehensive set of NMR data. Initially homonuclear 2D COSY and TOCSY allowed unambiguous identification of two glycerol systems (Table 1). From the signal multiplicities, we could readily deduce which methylene groups were attached to the phosphate group. The difference in 1H shifts of the central methene groups of ca. 1.2 ppm was strong evidence that the moiety possessing the high-field one has a glycerol unit with carries a central free hydroxyl group. In addition the relative shifts of the non-phosphate-bound methylenes indicated that these must be acylated. The 13C shift differences between all the carbons in the two moieties, assigned via the heteronuclear correlations, are also compatible with these conclusions. Inspection of further groups of signals in the COSY spectrum and their correlations in the heteronuclear inverse 1H-detected one-bond correlation (HMQC data) suggested that a number of fatty acid systems were present, some of which contained signals characteristic of a central cis double bond flanked by a number of methylene groups and a further novel system incorporating a trans double bond in a -CH2-CH(CH3)-CH⩵CH-CH2- moiety. The intensity of the olefinic signals in the 1H spectrum suggested there were approximately one cis and one trans double-bond moiety per molecule. In conclusion, the NMR data allowed the position of the free hydroxyl group to be determined and unambiguously established the configuration and nature of the olefinic systems present. The exact positions of the different acyl groups in the molecule, however, could be deduced only from the detailed MS data reported above. MS and NMR data led to the identification of main component 4 as 1-(trans-9-methyl-10-octadecenoyl)-2-hexadecanoyl-glyceryl-phospho-1′-oleoyl-glycerol.
TABLE 1.
NMR data for the major component in the phospholipid fraction from Corynebacterium sp. strain LMG 3820
| Component | System | 1H shift | Multiplicity (couplings) | 13C shifta |
|---|---|---|---|---|
| Glycerol moieties | CH2Ob | 4.42 | dd (3.5, 12.0) | 60.9 |
| 4.19 | dd (6.6, 12.0) | |||
| CHO | 5.23 | m | 68.5 | |
| CH2O | 3.99 | m | 61.7 | |
| CH2Ob | 4.14 | d (5.8) | 63.1 | |
| CHO | 3.98 | m | 66.9 | |
| CH2Oc | 3.95 | ddd (3.5, 7.9, 11.1) | 65.1 | |
| 3.87 | ddd (5.6, 8.1, 10.7) | 65.1 | ||
| Fatty acids | CH3 | 0.89 | t (7.2) | 13.5 |
| CH2 | 1.27 | m | 22.3 | |
| ..(CH2)x.. | 1.27–1.35 | m | 28.8–29.3 | |
| CH2 | 1.63 | m | 24.6 | |
| CH2 | 2.34 | m (several close together) | 34.8, 34.9 | |
| CO | 173.1, 173.8 | |||
| ..CH2 | 1.31 | m | 28.8–29.3 | |
| CH2 | 2.02 | m | 26.8 | |
| CH⩵CH | 5.35 | m | 129.2 | |
| CH2 | 2.02 | m | 26.8 | |
| CH2.. | 1.31 | m | 28.8–29.3 | |
| ..CH (CH3) | 2.04 (CH) | m | 36.5 | |
| 0.94 (CH3) | d (6.8) | 20.3 | ||
| CH⩵CH | 5.22d | dd (6.3, 15.7) | 135.8 | |
| 5.33 | dt (6.3, 15.7) | 128.1 | ||
| CH2 | 1.96 | m | 31.9 | |
| CH2.. | 1.31 | m | 28.8–29.3 |
Taken from the one-bond (HMQC) and long-range (HMBC) 13C-1H correlations.
The protons of the methylene show no couplings to 31P.
The protons of the methylene group show couplings to 31P, which shows a single resonance at +2.2 ppm relative to external H3PO4.
Olefinic carbon adjacent to aliphatic methene carbon-carrying methyl group.
The remaining four minor phospholipids of this system (compounds 1 to 3 and 5) were investigated only by MS methods. The results are presented in Table 2. Dispositions of the substituents were determined by the masses of the GPA anions detected in the CID spectra. The negative CID spectrum of the ion at m/z 999 clearly indicated only palmitic acid and 9-methyl-10-octadecenoic acid and a GPA anion at m/z 687 (compound 1). The ion at m/z 1,011 consisted of two isobaric phospholipids. One contained oleic acid (C18:1) and palmitic acid (C16:0) (compound 2), and the other contained C18:1, pentadecanoic acid (C15:0), and 9-methyl-10-octadecenoic acid (3). For compound 5, only two fatty acid moieties were present in both parts of the molecule. The positions of the fatty acids in compounds 1 to 3 and 5 given in Table 2 were determined analogously to those in compound 4.
TABLE 2.
Phospholipids from Corynebacterium sp. strain LMG 3820
| Com- pound | Typea | Mass (amu) | Phospholipidb
|
|||
|---|---|---|---|---|---|---|
| R1COOH (sn-1) | R2COOH (sn-2) | R3COOH (sn-1′) | R4COOH (sn-2′) | |||
| 1 | APG | 1,000 | 19:1 | 16:0 | 16:0 | |
| 2 | APG | 1,012 | 18:1 | 16:0 | 18:1 | |
| 3 | APG | 1,012 | 18:1 | 15:0 | 19:1 | |
| 4 | APG | 1,026 | 19:1 | 16:0 | 18:1 | |
| 5 | APG | 1,040 | 19:1 | 16:0 | 19:1 | |
| 6 | PG | 748 | 18:1 | 16:0 | ||
| 7 | PG | 762 | 19:1 | 16:0 | ||
| 8 | PI | 852 | 19:0 | 16:0 | ||
| 9 | CL | 1,404 | 18:1 | 16:0 | 18:1 | 16:0 |
| 10 | CL | 1,418 | 18:1 | 16:0 | 19:1 | 16:0 |
| 11 | CL | 1,432 | 19:1 | 16:0 | 19:1 | 16:0 |
Abbreviations: APG, acyl phosphatidylglycerol; PG, phosphatidylglycerol; PI, phosphatidylinositol; CL, cardiolipin.
15:0, n-pentadecanoic acid; 16:0, palmitic acid, 18:1, 9-octadecenoic acid; 19:1, trans-9-methyl-10-octadecenoic acid; 19:0, 10-methyl-octadecanoic acid.
Three different cardiolipins were detected in the phospholipid fraction of strain LMG 3820. Their structures were elucidated from the negative CID spectra. The lightest of them, with a mass of 1,404 amu, displayed only one phosphatidyl ion at m/z 673 and two acyl ions at m/z 255 and 281 belonging to palmitate (C16:0) and oleate (C18:1). Because the fragment [M-C16:0]− is more intensive than [M-C18:1]− and [673-C16:0]− is more intensive than [673-C18:1]−, palmitic acid must be at sn-2 and oleic acid must be at sn-1 of the glycerol, resulting in structure 9 of this cardiolipin. Analogous reasoning with respect to the phosphatidyl ion at m/z 687 led to the structure of cardiolipin 11. The third cardiolipin, with [M-H]− at m/z 1,417, contains three acyl ions at m/z 255 (C16:0), 281 (C18:1), and 295 (C19:1). It is an asymmetric cardiolipin since phosphatidyl ions at m/z 673 and 687 are observed in the negative CID spectrum. The loss of palmitic acid from [M-H]− is again more intensive than the loss of oleic acid or 9-methyl-10-octadecenoic acid; thus, one oleate moiety of compound 9 must be replaced by 9-methyl-10-octadecenoate, and the structure is therefore that of compound 10. Although cardiolipin 9 was never found as a natural product and is reported here for the first time as such, it was synthesized more than 30 years ago (9), while cardiolipins 10 and 11 have not been described before.
In addition to APGs and cardiolipins, two other types of phospholipids were found: phosphatidylglycerol, where two different species were identified; and phosphatidylinositol, where only one compound could be detected. This lipid, 1-(10-methyl-octadecanoyl)-2-palmitoyl-phosphatidylinositol, was the only phospholipid where the characteristic 10-methyl-octadecanoic acid could be found.
Besides the very unusual C19:1 fatty acid, the unbranched even-numbered fatty acids found in strain LMG 3820 are not known to exist in Arthrobacter spp. in general. Also, the type of phospholipids found in this strain is quite unusual for Arthrobacter spp. Cardiolipins have been found in Arthrobacter spp., but the cardiolipins from strain LMG 3820 are much heavier than those from Arthrobacter spp. due to longer fatty acids attached to them. While the cardiolipins of Arthrobacter atrocyaneus and A. globiformis contain mainly C15:0 to C17:0 fatty acids, resulting in molecules of 1,300 to 1,350 amu (32), the cardiolipins of strain LMG 3820 have masses of 1,400 to 1,450 amu due to the presence of C16:0 to C19:1 in these phospholipids.
All of these differences from typical Arthrobacter strains raise the question of the correct identification of strain LMG 3820. To clarify the phylogenetic position of strain LMG 3820, we sequenced the 16S rDNA from this strain. Alignment of the sequence obtained revealed no close relationship to any sequence of Arthrobacter spp. but did show similarity to that of C. variabilis (5, 6) (Fig. 6). We sequenced the 16S rDNAs of strains DSM 20132T and DSM 20536 and found that sequences of 1,490 bp, that is, 95% of the presumed length of the whole 16S rRNA gene, were identical for all three strains. However, analysis of the phospholipids of these strains revealed clear differences between them. The ratios of the phospholipids APG, phosphatidylglycerol, phosphatidylinositol, and cardiolipin were 47:52:1:<1 for DSM 20132T, 63:32:5:<1 for DSM 20536, and 43:46:9:2 for LMG 3820. However, the differences in lipid species were much larger. We calculated ratios of 1:1.9 for phosphatidylglycerols 6 and 7 and 1:2.8:0.5 for APGs 2/3, 4, and 5 for DSM 20132T; the corresponding ratios were 1:0.1 and 1:0.2:0.1 for strain DSM 20536 and 1:2.9 and 1:4.4:1.6 for strain LMG 3820. These differences are found not only in the phospholipids but also in the glycolipids, as can be seen in the relative abundances of fatty acids in these lipids (Table 3). To prove that these DSM strains, although possessing different lipid compositions, belong to the same species DNA-DNA hybridizations performed, and a DNA-binding value of 90.5% was found.
FIG. 6.
Phylogenetic tree based on a comparison of the 16S rDNA sequences of strain LMG 3820, assigned to the species A. globiformis, and related organisms. The sequence data for all but the two strains of C. variabilis were obtained from the GenBank/EMBL and/or RDP databases. Scale bar represents one nucleotide substitution per 100 bases.
TABLE 3.
Fatty acid content of the glycolipid and phospholipid fractions of Corynebacterium sp. strain LMG 3820, C. variabilis DSM 20132T, and C. variabilis DSM 20536
| Fatty acid | Equivalent chain length | Fatty acid content (mean % of total)
|
|||||
|---|---|---|---|---|---|---|---|
| Glycolipid
|
Phospholipid
|
||||||
| LMG 3820 | DSM 20132 | DSM 20536 | LMG 3820 | DSM 20132 | DSM 20536 | ||
| C12:0 | 12.000 | tra | tr | ||||
| C11 3-OH | 12.355 | tr | tr | ||||
| C14:1ω11 | 13.764 | ||||||
| C14:0 | 14.000 | tr | 1.6 | tr | tr | tr | |
| C15:0i | 14.624 | tr | |||||
| C15:0a | 14.707 | tr | tr | tr | tr | tr | |
| C15:1ω6 | 14.850 | 1.2 | |||||
| C15:0 | 15.000 | tr | tr | tr | tr | tr | |
| C16:1ω6i | 15.464 | 1.8 | 5.5 | ||||
| C16:0i | 15.628 | 1.1 | 3.7 | tr | |||
| C16:1ω7 | 15.793 | 1.6 | 1.1 | 2.4 | tr | ||
| C16:0 | 16.000 | 16.2 | 20.7 | 16.8 | 42.5 | 39.8 | 35.3 |
| C17:1ω7i | 16.416 | tr | tr | tr | |||
| C17:0i | 16.613 | 1.2 | 2.4 | ||||
| C17:0a | 16.740 | 3.5 | 4.6 | tr | tr | ||
| C17:0 | 17.000 | tr | tr | tr | tr | tr | |
| C18:1ω9 | 17.742 | 41.0 | 31.1 | 23.1 | 17.7 | 25.4 | 48.9 |
| C18:1ω7 | 17.798 | 1.2 | tr | 4.5 | tr | tr | 1.8 |
| C18:1ω8 9-Me | 18.000 | 21.9 | 19.3 | 20.3 | 28.4 | 28.5 | 4.5 |
| C18:0 10-Me | 18.376 | 9.1 | 10.1 | 5.8 | 7.8 | 5 | 7.6 |
| C19:0 d8,9 | 18.871 | tr | tr | ||||
| C20:1ω9 | 19.743 | tr | tr | 1.5 | tr | tr | |
| C22:1ω9 | 21.750 | tr | tr | ||||
tr, trace (less than 1% of total).
DISCUSSION
APGs have been reported to exist in Corynebacterium bovis, Nocardioides albus, Micromonospora coerulea, Actinomadura dassonvillei, and Promicromonospora spp. (25), and that from Corynebacterium amycolatum has only recently been further characterized (40). Yagüe and coworkers could not determine the position of the fatty acid at the polar head group, but we have unambiguously determined it by using NMR techniques (Table 1). The APGs from C. amycolatum displayed only oleic acid as the fatty acid on the polar head, while we found oleic acid as the most abundant fatty acid in this position but detected also some others as minor species. We also found a reversed order of the fatty acids in the diacylglycerol moiety of APG 2 (the only APG found in both C. amycolatum and LMG 3820) in strain LMG 3820 compared to that described for C. amycolatum. The distribution of fatty acids within the APGs of strain LMG 3820 shows some regularity. Only 9-octadecenoic acid and trans-9-methyl-10-octadecenoic acid were found at sn-1 of the nonpolar glycerol moiety whereas only saturated fatty acids are attached to sn-2, a characteristic also found in the phosphatidylglycerols and cardiolipins of this strain. The sn-1 position of the polar glycerol showed a range of different fatty acids much broader than that reported for C. amycolatum.
The results from the 16S rRNA sequences corroborated completely the findings from the lipid analysis. Lipid analysis of C. variabilis DSM 20132T also revealed APG but in lower amounts than in strain LMG 3820. C. variabilis DSM 20536, however, displayed a phospholipid composition different from that observed in strain LMG 3820. As in C. amycolatum, the main APG of C. variabilis DSM 20536 has a mass of 1,012 amu, in contrast to the type strain and to strain LMG 3820, with the main APG of 1,026 amu. All three strains possess the novel fatty acid trans-9-methyl-11-octadecanoic acid. This fatty acid has different abundances in the glycolipids and phospholipids. Again, this ratio is similar for LMG 3820 and the type strain but different from that found in DSM 20536. We have here one of the rare cases where different fatty acids and lipid compositions are found in strains with identical 16S rDNA sequences.
As these phospholipids are among the major phospholipids detected in this strain, however, an important function in the cell wall of Corynebacterium sp. strain LMG 3820 can be assumed. We could not deduce other functions of the APGs within the cell. 1,2-Diacyl-glyceryl-phospho-1′-acyl-glycerols 1 to 5 displayed no antimicrobial, antifungal, or cytotoxic activities (1).
The fatty acid trans-9-methyl-10-octadecenoic acid is reported here for the first time; hence its distribution within the genus Corynebacterium is unknown. Its routine detection is hampered by the fact that the equivalent chain length number, usually used for the automated identification of fatty acid methyl esters, is the same as that for stearic acid (C18:0). Because of this difficulty, one can speculate that trans-9-methyl-10-octadecenoic acid has sometimes been misidentified as stearic acid. Such a misidentification can be avoided by the use of GC-MS.
APGs are widespread in the genus Corynebacterium (40) and are furthermore among the major phospholipids in C. amycolatum. This species, however, is not very closely related to Corynebacterium sp. strain LMG 3820 (13), and so the amount of APG cannot be used as a biomarker for these species as proposed by Yagüe and coworkers (40). Further studies are required to determine the distribution of the different APGs and their substitution patterns of fatty acids within the genus Corynebacterium.
The configuration of the double bond gives some clues to the biosynthesis of this acid. It is probably formed from a cis-9-octadecenoic acid precursor by methylation at C-9, giving way to the rearrangement of the double bond to a trans-configuration one at C-10 (14). The branching of fatty acids leads to a decrease in their melting temperature, making cell membranes more flexible (33). The acylation of phosphatidylglycerol to APG also results via reduced hydrogen bond interactions to more flexible lipids. If such a transition from unbranched to branched fatty acids is observed within a species, the species should show a variety of morphological forms due to the different transition temperatures of its cell membrane lipids. This is exactly what is found in C. variabilis, which is known for its variable cell morphology and its Gram strain reaction variability, hence its name (28). The genome sequences of the strains are very similar, as shown by DNA-DNA hybridization, which suggests either very recently acquired differences in lipid synthesis or a diversity in lipid compositions at the strain level due to different ecological niches requiring mainly flexibility in cell wall adaptation. Because intraspecific lipid variations seem to be rare and fatty acid compositions are usually very similar at the species level (some identification systems rely on this fact), the interpretation that strains of C. variabilis are adapted by the formation of different lipids to their particular environment is more probable. Obviously, the differences in the genotypes of C. variabilis strains are only minor ones whereas the polar lipids, influencing the phenotypes, vary over a rather broad range which favors survival of each strain in a multitude of ecological niches.
ACKNOWLEDGMENTS
We thank P. Wolff for excellent technical assistance, C. Kakoschke and B. Jaschok-Kentner for recording NMR spectra, and F. Sasse for performing the bioassays.
This work was partly supported by funds from the German Federal Ministry for Science, Education and Research (projects 0319433B and 0319433C) and the European Union within the T project “High Resolution Automated Identification and Application to Biotechnologically Relevant Ecosystems.”
REFERENCES
- 1.Abate D, Abraham W-R. Antimicrobial metabolites from Lentinus crinitus. J Antibiot. 1994;47:1348–1350. doi: 10.7164/antibiotics.47.1348. [DOI] [PubMed] [Google Scholar]
- 2.Abraham W-R, Meyer H, Lindholst S, Vancanneyt M, Smit J. Phospho- and sulfolipids as biomarkers of Caulobacter, Brevundimonas and Hyphomonas. Syst Appl Microbiol. 1997;20:522–539. [Google Scholar]
- 3.Bligh E G, Dyer W J. A rapid method for total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 4.Cashion P, Holder-Franklin M A, McCully J, Franklin M. A rapid method for the base ratio determination of bacterial DNA. Anal Biochem. 1977;81:461–466. doi: 10.1016/0003-2697(77)90720-5. [DOI] [PubMed] [Google Scholar]
- 5.Collins M D. Transfer of Arthrobacter variabilis (Müller) to the genus Corynebacterium, as Corynebacterium variabilis comb. nov. Int J Syst Bacteriol. 1987;37:287–288. [Google Scholar]
- 6.Collins M D, Smida J, Stackebrandt E. Phylogenetic evidence for the transfer of Caseobacter polymorphus (Crombach) to the genus Corynebacterium. Int J Syst Bacteriol. 1989;39:7–9. [Google Scholar]
- 7.Couderc F. Gas chromatography/tandem mass spectrometry as an analytical tool for the identification of fatty acids. Lipids. 1995;30:691–699. doi: 10.1007/BF02537794. [DOI] [PubMed] [Google Scholar]
- 8.Couderc F, Roche P, Raynaud C, Pougny J, Promé J C. Characterization of fatty acids with a methyl branch near to a double bond by MS/MS analysis of their carboxylate anions. Adv Mass Spectrom. 1989;11B:1496–1497. [Google Scholar]
- 9.de Haas G H, van Deenen L L M. Cardiolipin and derivatives. I. Synthesis of an acyl derivative of diphosphatidylglycerol. Rev Trav Chim. 1963;82:1163–1172. [Google Scholar]
- 10.Deley J, Cattoir H, Reynaerts A. The quantitative measurement of DNA hybridization from renaturation rates. Eur J Biochem. 1979;12:133–142. doi: 10.1111/j.1432-1033.1970.tb00830.x. [DOI] [PubMed] [Google Scholar]
- 11.Dubey S K, Holmes D S. Biological cyanide destruction mediated by microorganisms. World J Microbiol Biotechnol. 1995;11:257–265. doi: 10.1007/BF00367095. [DOI] [PubMed] [Google Scholar]
- 12.Felsenstein J. PHYLIP—phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 13.Funke G, Lawson P A, Collins M D. Corynebacterium mucifaciens sp. nov., an unusual species from human clinical material. Int J Syst Bacteriol. 1997;47:952–957. doi: 10.1099/00207713-47-4-952. [DOI] [PubMed] [Google Scholar]
- 14.Garton G A. Aspects of the chemistry and biochemistry of branched-chain fatty acids. Chem Ind (London) 1985;1985:295–300. [Google Scholar]
- 15.Gutell R R, Weiser B, Woese C R, Noller H F. Comparative anatomy of 16S-like ribosomal RNA. Prog Nucleic Acid Res Mol Biol. 1985;32:155–216. doi: 10.1016/s0079-6603(08)60348-7. [DOI] [PubMed] [Google Scholar]
- 16.Higson F K. Microbial degradation of biphenyl and its derivatives. Adv Appl Microbiol. 1992;37:135–164. doi: 10.1016/s0065-2164(08)70254-5. [DOI] [PubMed] [Google Scholar]
- 17.Hoischen C, Gura K, Luge C, Gumpert J. Lipid and fatty acid composition of cytoplasmic membranes from Streptomyces hygroscopicus and its stable protoplast-type L form. J Bacteriol. 1997;179:3430–3436. doi: 10.1128/jb.179.11.3430-3436.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jensen N J, Gross M L. Fast atom bombardment and tandem mass spectrometry for determining iso- and anteiso-fatty acids. Lipids. 1986;5:362–365. doi: 10.1007/BF02534056. [DOI] [PubMed] [Google Scholar]
- 19.Jensen N J, Tomer K B, Gross M L. Collisional activation decomposition mass spectra for locating double bonds in polyunsaturated fatty acids. Anal Chem. 1985;57:2018–2021. [Google Scholar]
- 20.Jensen N J, Tomer K B, Gross M L. FAB MSMS for phosphatidylinositol, -glycerol, -ethanolamine and other complex phospholipids. Lipids. 1987;7:480–489. doi: 10.1007/BF02540363. [DOI] [PubMed] [Google Scholar]
- 21.Jones D, Keddie R M. The genus Arthrobacter. In: Starr M P, Stolp H, Trüper H G, Balows A, Schlegel H G, editors. The procaryotes. A handbook on habitats, isolation and identification of bacteria. Berlin, Germany: Springer-Verlag; 1981. pp. 1281–1299. [Google Scholar]
- 22.Jukes T H, Cantor C R. Evolution of protein molecules. In: Munro H H, editor. Mammalian protein metabolism. New York, N.Y: Academic Press; 1969. pp. 21–132. [Google Scholar]
- 23.Karlson U, Dwyer D F, Hooper S W, Moore E R B, Timmis K N, Eltis L D. Two independently regulated cytochromes P-450 in a Rhodococcus rhodochrous strain that degrades 2-ethoxyphenol and 4-methoxybenzoate. J Bacteriol. 1993;175:1467–1474. doi: 10.1128/jb.175.5.1467-1474.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lane D J. 16S/23S sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester, England: John Wiley; 1991. pp. 115–175. [Google Scholar]
- 25.Lechevalier M P, De Bievre C, Lechevalier H. Chemotaxonomy of aerobic Actinomycetes: phospholipid composition. Biochem Syst Ecol. 1977;5:249–60. [Google Scholar]
- 26.Maidak B J, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The ribosomal RNA database project (RDP) Nucleic Acids Res. 1996;24:82–85. doi: 10.1093/nar/24.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McAllister K A, Lee H, Trevors J T. Microbial degradation of pentachlorophenol. Biodegradation. 1996;7:1–40. [Google Scholar]
- 28.Müller G. Mikrobiologische Untersuchungen über die “Futterverpilzung durch Selbsterhitzung.” 3. Mitteilung: Ausführliche Beschreibung neuer Bakterien-Species. Zentralbl Bakteriol Parasitenkd Infektionskr Hyg Abt 2 Orig Reihe A. 1961;114:520–537. [Google Scholar]
- 29.Mullis K B, Faloona F. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 1987;155:335–350. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- 30.Murphy R C, Harrison K A. Fast atom bombardment mass spectrometry of phospholipids. Mass Spectrom Rev. 1994;13:57–75. [Google Scholar]
- 31.Neefs, J.-M., Y. V. Van der Peer, P. De Rijk, A. Gloris, and R. De Wachter. 1991. Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res. 19(Suppl.):1987–2015. [DOI] [PMC free article] [PubMed]
- 32.Niepel T, Meyer H, Wray V, Abraham W-R. A new type of glycolipid, 1-[α-mannopyranosyl-(1α-3)-(6-O-acyl-α-mannopyranosyl)-3-O-acyl-glycerol, from Arthrobacter atrocyaneus. Tetrahedron. 1997;53:3593–3602. [Google Scholar]
- 33.Nuhn P, Gutheil M, Dobner B. Occurrence, biosynthesis and biological meaning of branched fatty acids. Fette Seifen Anstrichm. 1985;87:135–140. [Google Scholar]
- 34.Shaw N, Stead D. Lipid composition of some species of Arthrobacter. J Bacteriol. 1971;107:130–133. doi: 10.1128/jb.107.1.130-133.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Summers M F, Marzilii L G, Bax A. Complete 1H and 13C assignments of coenzyme B12 through the use of new two-dimensional NMR experiments. J Am Chem Soc. 1986;108:4285–4294. [Google Scholar]
- 36.Tomer K B, Crow F W, Gross M L. Location of double bonds position in unsaturated fatty acids by negative ion MS/MS. J Am Chem Soc. 1983;105:5487–5488. [Google Scholar]
- 37.Vancanneyt M, Witt S, Abraham W-R, Kersters K, Fredrickson H L. Fatty acid content in whole-cell hydrolysates and phospholipid fractions of pseudomonads: a taxonomic evaluation. Syst Appl Microbiol. 1996;19:528–540. [Google Scholar]
- 38.Van De Peer Y, Nicolai S, De Rijk P, De Wachter R. Database on the structure of small ribosomal subunit RNA. Nucleic Acids Res. 1996;24:86–91. doi: 10.1093/nar/24.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walker W, Bastl C P. The glycolipids from Arthrobacter globiformis. Carbohydr Res. 1967;4:49–54. [Google Scholar]
- 40.Yagüe G, Segovia M, Valero-Guillen P L. Acyl phosphatidylglycerol: a major phospholipid of Corynebacterium amycolatum. FEMS Microbiol Lett. 1997;151:125–130. doi: 10.1111/j.1574-6968.1997.tb12559.x. [DOI] [PubMed] [Google Scholar]