Abstract
Burkholderia cepacia AC1100 metabolizes 2,4,5-trichlorophenoxyacetic acid (2,4,5-T) via formation of 5-chlorohydroxyquinol (5-CHQ), hydroxyquinol (HQ), maleylacetate, and β-oxoadipate. The step(s) leading to the dechlorination of 5-CHQ to HQ has remained unidentified. We demonstrate that a dechlorinating enzyme, TftG, catalyzes the conversion of 5-CHQ to hydroxybenzoquinone, which is then reduced to HQ by a hydroxybenzoquinone reductase (HBQ reductase). HQ is subsequently converted to maleylacetate by hydroxyquinol 1,2-dioxygenase (HQDO). All three enzymes were purified. We demonstrate specific product formation by colorimetric assay and mass spectrometry when 5-CHQ is treated successively with the three enzymes: TftG, TftG plus HBQ reductase, and TftG plus HBQ reductase plus HQDO. This study delineates the complete enzymatic pathway for the degradation of 5-CHQ to maleylacetate.
Microbial degradation of simple aromatic or chloroaromatic compounds involves catechol or chlorocatechol intermediates which are further metabolized via the ortho or modified ortho cleavage pathways (27). On the other hand, some polychlorinated compounds such as 2,4,5-trichlorophenoxyacetic acid (2,4,5-T) (4, 16, 23), 2,4,6-trichlorophenol (12, 19), γ-hexachlorocyclohexane (15, 21), and pentachlorophenol (1, 24) are converted to chlorohydroxyhydroquinone (chlorohydroxyquinol [CHQ]) intermediates. In these pathways, chlorinated phenols are subjected to two steps of hydroxylation of the benzene ring to yield CHQ intermediates (1, 13, 17, 18, 25, 28, 30).
Only a few reports have dealt with subsequent enzymatic steps of the CHQ pathways. For example, the enzyme 6-chlorohydroxyquinol 1,2-dioxygenase was purified from the 2,4,6-trichlorophenol-degrading bacteria Streptomyces rochei 303 and Azotobacter sp. strain GP1 (17, 30). These enzymes catalyze ortho cleavage of 6-chlorohydroxyquinol (6-CHQ) to yield chloromaleylacetate. In Rhodococcus chlorophenolicus PCP-1 and Mycobacterium fortuitum CG-2, CHQ as the intermediate of pentachlorophenol degradation is reductively dechlorinated to 1,2,4-trihydroxybenzene (hydroxyquinol, [HQ]), which is subsequently metabolized by a meta cleavage enzyme (1, 26). Recently, we purified the hydroxyquinol 1,2-dioxygenase (HQDO) enzyme encoded by the tftH gene of the 2,4,5-T-degrading bacterium Burkholderia cepacia AC1100 (8). Unlike the 6-chlorohydroxyquinol 1,2-dioxygenase, the B. cepacia AC1100 HQDO could use only HQ, not 6-CHQ or 5-CHQ, as a substrate for ortho cleavage (8). Since 5-CHQ is the central metabolite in the 2,4,5-T pathway (23), one or more enzymes in addition to HQDO (TftH) must be necessary to metabolize this intermediate.
We previously reported the isolation of a 2,4,5-T-negative mutant, PT88, that accumulates 5-CHQ when grown in the presence of glucose and 2,4,5-T (6, 23). Complementation of PT88 for growth on 2,4,5-T as a sole source of carbon identified a cluster of genes (tftEFGH) essential for the metabolism of the 5-CHQ intermediate (6, 7). The tftE gene encodes maleylacetate reductase, the tftF gene encodes glutathione reductase, and the tftH gene encodes HQDO (7, 8). The function of the tftG gene product was heretofore unknown. This study shows that tftG encodes a novel dechlorinase enzyme, which in concert with a novel reductase converts 5-CHQ to HQ, the substrate for HQDO (TftH). The purification of this novel reductase and its mode of action in concert with TftG are described in this report.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains, plasmids, and media used in this study have previously been described (6, 8).
Chemicals.
The compound HQ was purchased from Aldrich Chemical Co. (Milwaukee, Wis.). 5-CHQ was synthesized as described before (8).
Overproduction of TftG and TftH in Escherichia coli crude cell extracts.
Overproduction of the tftG and tftH gene products was accomplished by growing E. coli MV1184 containing plasmid pMMD3 or pMMD4 (6) at 37°C in 100 ml of Luria broth supplemented with 75 μg of ampicillin/ml. After 3 h of incubation, expression of the tftG and tftH genes from the tac promoter of the vector was induced by the addition of 1.0 mM isopropyl-β-d-thiogalactopyranoside (IPTG). The cells were incubated for an additional 4 h before being harvested by centrifugation. Cells were resuspended in 50 mM sodium phosphate buffer (pH 7.0) and lysed by sonication three times for 1 min each in a Branson Sonifier 450 (output level, 3.5 [25 Watts]; duty cycle, 50%). The cell debris was removed by centrifugation at 9,000 rpm in an SM-24 rotor for 20 min at 4°C. The control crude cell extract was prepared from E. coli MV1184 containing only the vector pMMB66HE (11).
Colorimetric assay for the conversion of 5-CHQ and HQ in E. coli crude cell extracts.
E. coli crude cell extracts (300 μg of protein) containing overproduced TftG or TftH, individually or together, were incubated at room temperature in 1.0 ml of 50 mM sodium phosphate buffer (pH 6.6). The substrate, 5-CHQ or HQ, was added at a final concentration of 0.5 to 1.0 mM. The cofactor NADH was added at a final concentration of 0.4 to 0.8 mM. Reaction mixtures were incubated at room temperature until the product of the reaction spontaneously oxidized from 5-CHQ to 5-chlorohydroxybenzoquinone (5-CHQox) or HQ to hydroxybenzoquinone (HQox). Control reactions were performed with crude cell extracts from cells containing only the vector. In addition, standard compounds alone (without cellular extracts) were incubated in 50 mM sodium phosphate buffer (pH 6.6). The spontaneously oxidized reaction products of the crude cell assay are colored quinones. The colored reaction tubes were photographed and compared to controls and standards.
B. cepacia PT88 crude cell assays.
Crude cell extracts of deletion mutant B. cepacia PT88 (ΔtftEFGH) and B. cepacia PT88 containing plasmid pRKD34 (tftGH under control of the lac promoter [6]) were prepared as E. coli cell extracts overproducing TftG and TftH. The crude cell extracts were tested for glutathione reductase activity as described by Davis et al. (9) and for HQDO activity as described previously (8). In addition, the crude cell extracts (150 μg of protein) were incubated with 5-CHQ as described for the conversion assays of E. coli.
Cloning of the tftG gene encoding TftG-His6.
PCR was used to generate the tftG gene containing six histidine codons at the 3′ end, just before the TGA stop codon (encoding TftG-His6), and the sequence of the PCR product was verified. Plasmid pMMD34 (6) was used as the template DNA, and primers for the PCR were synthesized by Gibco BRL (Grand Island, N.Y.). The 5′-end primer was 5′-GCA GAT CTA ATC GAA AGC TAC CGG GAA CG-3′, and the 3′-end primer was 5′-TCA ATG ATG ATG ATG ATG ATG TTT GGA GTC GTT GTG CTT GCC GTG C-3′. The PCR product was ligated into the pCRII expression vector from Invitrogen (San Diego, Calif.) Original TA Cloning kit. The new clone, designated pORF5, was transformed into E. coli TG1.
Purification of TftG (dechlorinase).
The one-step nickel affinity chromatography procedure was used for purification of TftG-His6 as described previously for purification of TftH-His6 (8).
Conversion of 5-CHQ by purified TftG for HPLC analysis.
Purified TftG at a final concentration of 20 μg/ml was incubated with 0.4 mM 5-CHQ in 10 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES; pH 7.0). The reaction mixture was incubated in an anaerobic glove box (5% H, 95% Ar) for 2 h. A final concentration of 1.0 mM titanium citrate was added to some reactions. Because sodium dithionite interfered with high-pressure liquid chromatography (HPLC) analysis, stock solutions of 5-CHQ and HQ were prepared without sodium dithionite under anaerobic conditions.
HPLC conditions.
Samples were analyzed with an HPLC apparatus equipped with a model 996 photodiode array detector (Waters, Milford, Mass.). Samples were directly transferred into minivials anaerobically and immediately injected onto a Nova-Pak C18 column (3.9 by 150 mm; pore size of 60 Å) equilibrated with 11 mM H3PO4. Samples were eluted at a flow rate of 1 ml/min with an 11 mM H3PO4–acetonitrile gradient (acetonitrile concentrations of 0% [isocratic, 5 min], 0 to 70% [linear, 5 min], 70% [isocratic, 5 min], and 100% [isocratic, 5 min]). The absorbance was monitored between 250 and 350 nm. Sample retention times and absorption maxima were compared to those of standard compounds. The HPLC standards HQ, 5-CHQ, and 5-CHQox were distinguished by their respective retention times of 3.1, 10.9, and 12.7 min and their absorption maxima of 290, 290, and 275 nm (data not shown). The compound 5-CHQox was prepared by allowing 5-CHQ to spontaneously oxidize in air. The compound HQox could not be easily detected by HPLC analysis because it is unstable and forms polymers.
Induction of hydroxybenzoquinone (HBQ) reductase activity in B. cepacia AC1100 cells.
Cells were grown for 5 h at 30°C in BSM medium [50 mM potassium phosphate buffer (pH 7.0), 15 mM (NH4)2SO4, 0.8 mM MgSO4, 2 μM FeSO4] containing 2 mg of succinate/ml. After inducer (2,4,5-T, benzoquinone, or methylbenzoquinone) was added, the cells were grown for an additional 12 h, after which the cells were collected, disrupted by sonication, and centrifuged. The reductase specific activity was determined in cell extracts.
Purification of HBQ reductase.
B. cepacia AC1100 cells were grown at 30°C in BSM medium containing 2,4,5-T (1 mg/ml) as the sole source of carbon and energy. After 2 days of incubation, cells were harvested by centrifugation, washed with 50 mM potassium phosphate buffer (pH 7.0), resuspended in 50 mM Tris-HCl (pH 7.5) supplemented with 5 μM flavin mononucleotide (FMN) (buffer F), and disrupted in a French press (12,000 lb/in2, three times). Cell debris was removed by centrifugation for 10 min at 12,000 rpm. The cell extract was saturated by ammonium sulfate up to 0.8 M and centrifuged to remove the pellet, and the supernatant was loaded on a Butyl-Sepharose column (26 by 100 cm) equilibrated with 0.8 M ammonium sulfate in buffer F. The reductase, which did not bind with Butyl-Sepharose, was collected from the first part of the flowthrough, dialyzed against buffer F, and applied to a Blue-Sepharose column (13 by 280 cm) on an FPLC (fast protein liquid chromatography) system equilibrated with the same buffer. A linear gradient (0 to 2.2 M KCl) was applied; the active fractions were collected at 2.2 M KCl and concentrated by ultrafiltration with Amicon YM10 and Centricon PM10 membranes to a final volume of 0.4 ml. The concentrated supernatant was further purified by FPLC using a Sephacryl S100 column (Hi Prep 16/60). The rate of elution was 0.5 ml/min, and the reductase activity was detected at an eluate volume of 44 to 46 ml.
Assay of HBQ reductase.
HBQ reductase activity was determined with a BioSpec-1601 UV/VIS spectrophotometer (Shimadzu). The reaction mixture contained 100 mM sodium phosphate buffer (pH 7.2), 0.2 mM NADH, 0.01 mM FMN, and 0.4 mM methylbenzoquinone as a substrate; the reaction was initiated with the addition of enzyme. One unit of activity was defined as the amount of enzyme that catalyzed the oxidation of 1 μmol of NADH per min. Absorption spectra of HBQ reductase were recorded on a Beckman DU-65 UV/VIS spectrophotometer at a protein concentration of 0.89 mg/ml.
NH2-terminal amino acid sequence.
HBQ reductase was electroblotted onto a polyvinylidene fluoride protein transfer and sequencing membrane (Schleicher & Schuell) and subjected to NH2-terminal amino acid sequencing. Automated Edman degradation in an ABI 477A protein sequencer (Applied Biosystems) was used for this purpose.
Purification of TftH protein (HQDO).
The tftH gene product (HQDO) was overproduced in E. coli BL21DE3/pLysS which had an IPTG-inducible T7 RNA polymerase and contained plasmid pDD2QΔ7 with ORF5 and ORF6 (6). The cells were grown to an optical density at 600 nm of 0.3 to 0.4 at 37°C in Luria broth supplemented with ampicillin (50 μg/ml) and chloramphenicol (34 μg/ml), and expression of the tftH gene was induced by the addition of 0.5 mM IPTG. The cells were incubated for an additional 2 to 2.5 h, harvested and washed with 50 mM potassium phosphate buffer by centrifugation, and lysed in 50 mM Tris-HCl buffer, pH 8.0 (buffer A). DNA was precipitated from the lysate by the addition of 1 ml of 10% streptomycin sulfate. After 15 min of incubation at room temperature, DNA precipitate and cell debris were removed by centrifugation (8,000 rpm, 20 min). As the first (fractionation) step of purification, 35 to 50% ammonium sulfate saturation was used. The pellet was resuspended in buffer A and applied to a Q-Sepharose column (0.5 by 8 cm) on an FPLC system equilibrated with the same buffer. A linear increasing gradient of 0 to 1.0 M KCl was used to elute the protein. Active fractions eluted at around 0.62 M KCl were collected, concentrated, mixed with 1.2 M ammonium sulfate in 50 mM Tris-HCl buffer, pH 8.0 (buffer B), and loaded on a Resource-Phenyl (HR 5/50) column on an FPLC system equilibrated with the same buffer. A linear decreasing gradient of 1.2 to 0 M (NH4)2SO4 was applied, and the active fractions were eluted at around 0.35 M (NH4)2SO4.
Activity of HQDO.
HQDO activity was determined as described before (8).
Protein concentrations.
Protein concentrations were determined by the Bradford method (3). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was used with Coomassie blue staining to visualize purified protein.
Color reactions of 5-CHQ conversion by purified TftG, TftH, and HBQ reductase.
5-CHQ was added at a final concentration of 1 mM to 0.1 ml of 0.1 M potassium phosphate buffer (pH 6.6) containing 2 mM NADH, 0.05 mM FMN, and TftG (1.5 μg) either alone or with HBQ reductase (1.6 μg) or HBQ reductase (1.6 μg) and TftH (4 μg). As a control, either 5-CHQ or HQ was added at the same concentration to 0.1 M potassium phosphate buffer (pH 6.6) containing 2 mM NADH and 0.05 mM FMN. Reaction mixtures were incubated for 2 h at room temperature.
Detection of products of 5-CHQ conversion by purified TftG, TftH, and HBQ reductase by mass spectrometric analysis.
Electrospray mass spectrometry was used to detect the products of 5-CHQ conversion. To prepare the samples, 5-CHQ as substrate was added at a final concentration of 0.1 mM to 1 ml of 0.1 M ammonium formate buffer (pH 6.6) containing 0.2 mM NADH, 0.01 mM FMN, and TftG (1.5 μg) either alone or with HBQ reductase (1.6 μg) or HBQ reductase (1.6 μg) and TftH (4 μg). For standards, either 5-CHQ, HQ, or maleylacetate (MA) was added at final concentration of 0.1 mM to 0.1 M ammonium formate buffer (pH 6.6) containing 0.2 mM NADH and 0.01 mM FMN. The reaction mixture was incubated for 5 min at room temperature, diluted with isopropanol, and immediately injected.
Electrospray mass spectrometry conditions.
Electrospray mass spectrometry was done on a Micromass (Manchester, England) Quattro II tandem mass spectrometer operated in negative-ion mode. Samples were introduced by flow injection using a mobile phase consisting of isopropanol-water (1:2) at a flow rate of 12 μl/min. Typical electrospray source parameters were as follows: capillary, 2.45 kV; counter electrode, 0.64 kV; and cone voltage, 25 V. For MA, cone voltage was kept at 15 V because higher voltages caused extensive in source fragmentation. A range 100 to 180 was scanned at 3 s/scan rate at unit resolution. During tandem mass spectrometry, collision-induced dissociation was carried out using a collision energy of 25 eV and argon gas pressure of 3.0 × 10−3 mbar.
RESULTS
Conversion of 5-CHQ or HQ in the presence or absence of TftG and/or TftH.
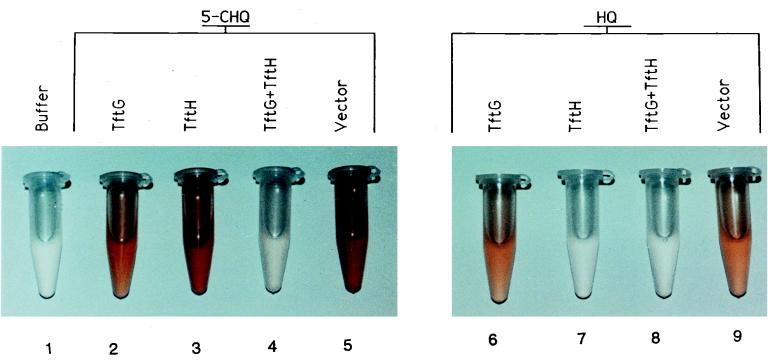
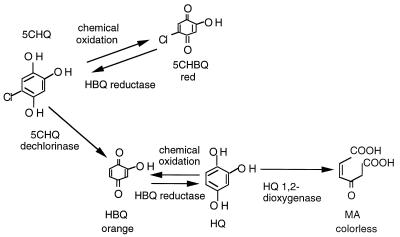
To determine the enzymes and cofactors required for the metabolism of 5-CHQ, an assay using E. coli crude cell extracts overproducing the B. cepacia AC1100 enzymes TftG and TftH was developed. Spontaneous oxidation of HQ and 5-CHQ is delayed by E. coli crude cell extracts in the presence of NADH, presumably due to enzymes which reduce quinones or scavenge oxygen radicals (2). When NADH was depleted, the reaction products spontaneously oxidized to the corresponding colored quinones; this was the initial basis of product detection and analysis (Fig. 1). Thus, in absence of TftG, 5-CHQ was spontaneously oxidized to the corresponding chlorohydroxybenzoquinone, a red-colored product (Fig. 1, tubes 3 and 5). The presence of TftG allowed formation of an orange-colored product (tube 2) corresponding to HQox (tubes 6 and 9). Addition of both TftG and TftH produced a colorless solution (Fig. 1, tube 4), suggesting that the HQ was converted to a colorless product by TftH. 5-CHQ was unaffected by TftH (Fig. 1, tube 3), while HQ was unaffected by TftG (tube 6), suggesting that 5-CHQ is a substrate for TftG but not for TftH, while HQ is a substrate for TftH but not for TftG.
FIG. 1.
Oxidized, colored reaction products of 5-CHQ and HQ from crude cell conversion assays. The reactions used crude cell extracts overproducing TftG or TftH, individually or together. Extracts were incubated with either 5-CHQ or HQ. Buffer is 50 mM sodium phosphate buffer (pH 6.6) and TftG crude cell extract without a chemical substrate; “Vector” denotes assays using crude cell extracts from cells containing only the vector plasmid.
The reaction using HQ as a substrate remained colorless as long as TftH crude cell extracts were present because TftH catalyzes ortho cleavage of HQ to yield MA, a colorless product, as previously described (8) (Fig. 1, tubes 7 and 8).
Conversion of 5-CHQ by purified TftG and TftH.
Purified TftH was able to catalyze the conversion of HQ to MA, but not 5-CHQ, as previously determined (8). In crude cell extracts, TftG and TftH together catalyze the conversion of 5-CHQ to MA (based on data presented in Fig. 1). However, in reactions using purified TftG and purified TftH, 5-CHQ immediately turned reddish-orange and no MA could be detected (data not shown). This result suggests that TftG and TftH are not solely responsible for converting 5-CHQ to MA in crude cell extracts, and one or more enzymes present in the cell extract of E. coli were also needed.
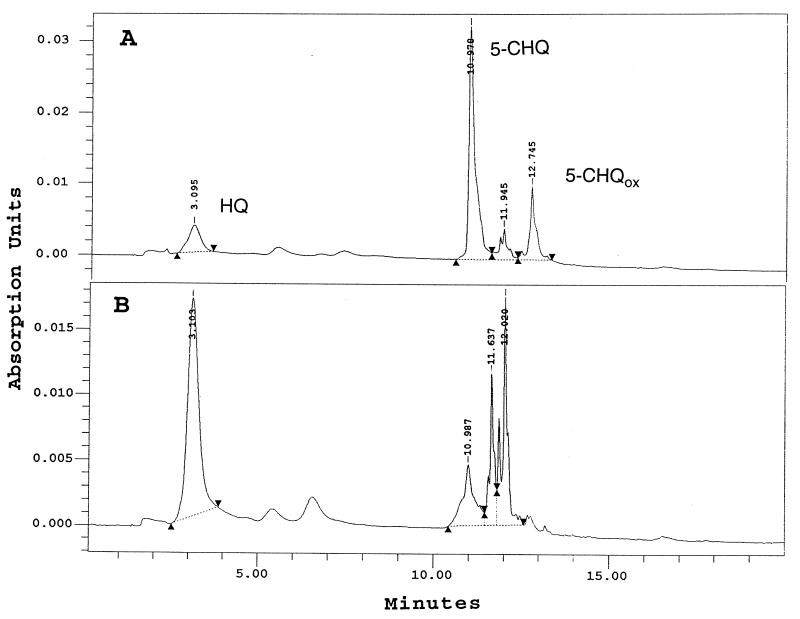
HPLC analysis of 5-CHQ conversion by purified TftG.
To determine the reaction product when purified TftG is incubated with 5-CHQ, reaction mixtures were incubated under anaerobic conditions and then analyzed by HPLC. As soon as purified TftG was added to the reaction, a red color developed. After 2 h of incubation, three compounds were identified: 52 μM HQ (3.1 min), 263 μM CHQ (10.9 min), and 19 μM 5-CHQ-ox (12.7 min) (Fig. 2A). 5-CHQ incubated alone under anaerobic conditions was not oxidized to 5-CHQox. Only when purified TftG was added to the reaction mixture was 5-CHQox identified together with HQ. HQox could not be detected by HPLC, probably due to its polymerization. Our hypothesis was that 5-CHQ is dechlorinated and converted to HQox by TftG, which then chemically reacts with 5-CHQ to produce small amounts of HQ and 5-CHQox. To test this hypothesis, 1.0 mM titanium citrate was added to the reaction mixture to reduce the compounds which were becoming oxidized. When 1.0 mM titanium citrate was added, 227 μM HQ and 38 μM 5-CHQ were detected (Fig. 2B), but even after 2 h of incubation, 5-CHQox was not present (Fig. 2B).
FIG. 2.
HPLC chromatograms of the conversion of 5-CHQ by purified TftG. Reactions were performed anaerobically with purified TftG protein. The standard compounds HQ, 5-CHQ, and 5-CHQox have retention times of 3.1, 10.9, and 12.7 min, respectively. The reaction mixture analyzed in panel B contained 1.0 mM titanium citrate.
B. cepacia PT88 crude cell assays.
Based on the purified enzyme assays, we hypothesized that a common cellular reductase was involved in the conversion of 5-CHQ to MA. Previously, the tftF gene of AC1100 had been shown to encode a glutathione reductase (6). We speculated that the enzyme encoded by TftF may be the reductase component. Crude cell extracts of mutant B. cepacia PT88 (ΔtftEFGH) and of B. cepacia PT88 complemented with the tftG and tftH genes did not have glutathione reductase activity because of lack of TftF. The crude cell extracts of B. cepacia PT88 complemented with tftG and tftH were able to catalyze the conversion of 5-CHQ to MA, based on a clear reaction tube after incubation with 5-CHQ (data not shown). This result shows that TftF (glutathione reductase) is not essential for this reaction. Indeed, purified glutathione reductase encoded by the tftF gene had no HBQ reductase activity in vitro (data not shown).
Induction of HBQ reductase activity in B. cepacia AC1100 cells.
To purify the presumptive reductase that keeps 5-CHQ reduced and converts the dechlorinated, oxidized form of 5-CHQ to HQ, we attempted to detect this activity in B. cepacia AC1100. Quinone reductase activity was detected at a low level (around 3 U/mg of protein) in succinate-grown cells. This reductase activity was twofold higher when 0.05 mM benzoquinone or 0.1 mM methylbenzoquinone was added in the growth medium. Addition of 0.1 and 0.5 mM 2,4,5-T in the growth medium increased the reductase activity 3- and 10-fold, respectively. During growth on 2,4,5-T (1 mg/ml) as the sole source of carbon, the activity of the reductase reached its maximal level after 2 days, at the end of the log phase of growth.
Purification and properties of HBQ reductase.
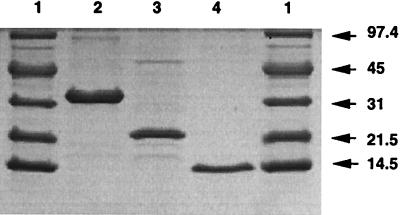
While the nature of the primary substrate of the reductase is unknown, we infer from its ability to reduce HQox to HQ as well as its inducibility in the presence of benzoquinone or methylbenzoquinone that it likely is an HBQ reductase. The substrate specificities of this reductase are similar for HBQ, 5-CHQox, and methylbenzoquinone. Since the induction experiment suggested that the HBQ reductase is induced during growth in the presence of 2,4,5-T, we attempted to purify this enzyme from a 2,4,5-T-grown AC1100 culture. We obtained a relatively pure (Fig. 3) preparation of HBQ reductase in 1.9% yield with a specific activity of 1,831 U/mg of protein, representing a 27-fold enrichment over the fully induced level in crude extract (Table 1). During hydrophobic chromatography, the major activity detected was the HBQ reductase shown in Fig. 3, lane 3; however, two other small peaks of activity (varying from 5 to 10%) were seen during elution with decreasing ammonium sulfate concentration. These smaller peaks have not been characterized; however, to indicate the presence of possible isoenzymes, we have designated the purified enzyme HBQ reductase I.
FIG. 3.
SDS-PAGE analysis of the enzymes involved in the transformation of 5-CHQ to MA. Lanes: 1, low-molecular-mass protein standards (sizes are indicated in kilodaltons); 2, HQDO (4 μg); 3, HBQ reductase I (2.6 μg); 4, 5-CHQ dechlorinase (2.5 μg).
TABLE 1.
Purification of B. cepacia HBQ reductase
| Fraction | Vol (ml) | Act (U/ml) | Total activity (U) | Protein (mg/ml) | Sp act (U/mg) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|---|---|
| Crude extract | 44 | 449.2 | 19,769 | 6.6 | 68 | 100 | 1.0 |
| Butyl-Sepharose | 50 | 52 | 2,600 | 0.7 | 74.3 | 13.1 | 1.1 |
| Blue-Sepharose (concentrated) | 0.8 | 1,750 | 1,400 | 3.76 | 466 | 7.1 | 6.8 |
| Sephacryl S100 (concentrated) | 0.4 | 952.5 | 381 | 0.52 | 1,831 | 1.9 | 26.9 |
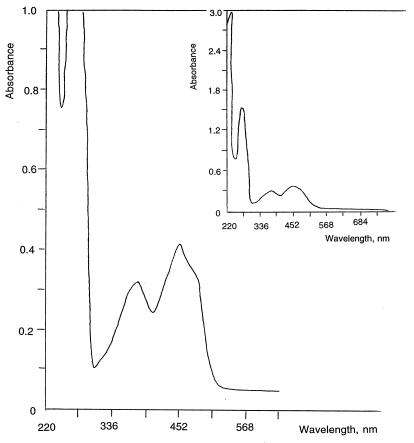
The molecular weight of the HBQ reductase I on SDS-PAGE was shown to be about 22,000, while the molecular weight of the native enzyme during gel filtration was observed to be around 50,000. Thus, HBQ reductase I appears to be a homodimer. The enzyme has a light green color at a protein concentration of 0.89 mg/ml. The UV/VIS spectrum of the reductase contains peaks which are characteristic for flavoproteins (22) (Fig. 4). FMN was essential for reductase activity and could not be replaced by flavin adenine dinucleotide. The activity was about four times lower when exogenous FMN was not added to the reaction mixture. Moreover, the enzyme was unstable during the purification procedure unless FMN was added to all buffers. We conclude that HBQ reductase I is an FMN-specific protein. Analysis of the NH2-terminal amino acid sequence (MLTTKRIATLVG) did not reveal significant similarity with any known quinone reductases.
FIG. 4.
UV-VIS spectrum of HBQ reductase I at a protein concentration of 0.89 mg/ml. Buffer F, containing 5 μM FMN, was used as a reference control.
Purification of the TftH protein.
The procedure for purifying HQDO is presented in Table 2. Pure enzyme had a red color at a protein concentration of 4.9 mg/ml. A gel filtration step was eliminated from the purification procedure due to a drastic loss of activity, presumably because of the removal of iron from the active center.
TABLE 2.
Purification of B. cepacia HQDO
| Fraction | Vol (ml) | Activity (U/ml) | Total activity (U) | Protein (mg/ml) | Sp act (U/mg) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|---|---|
| Crude extract | 10 | 3.52 | 35.2 | 8.6 | 0.4 | 1.0 | |
| 35–50% (NH4)2SO4 | 5 | 7.9 | 39.5 | 7.2 | 1.09 | 100 | 4.75 |
| Q-Sepharose (concentrated) | 1 | 18.8 | 18.8 | 9.5 | 1.99 | 53.4 | 4.97 |
| Resource-Phenyl (concentrated) | 0.08 | 180 | 14.4 | 4.9 | 36.7 | 40.9 | 91.7 |
It should be noted that the present scheme of purification results in an enzyme with specific activity 10 times higher than that of TftH-His6 enzyme purified by nickel affinity chromatography (8). It is likely that the active center iron is loosely bound to the enzyme and readily replaced by nickel during the nickel affinity chromatography step, leading to a sharp decrease in the specific activity of the His-tagged enzyme. This may be a limitation of the His-tag approach for enzymes with metal centers or metal requirements. Comparison of substrate specificity shows that both TftH and TftH-His6 are able to use HQ but not 5-CHQ as a substrate.
Color reactions of enzymatic and chemical conversion of 5-CHQ and HQ.
The color reactions shown in Fig. 1 suggested that the autooxidation product of 5-CHQ was a red-colored chlorobenzoquinone (Fig. 1, tube 5), while that of HQ was an orange-colored benzoquinone product (Fig. 1, tube 9). The availability of purified dechlorinase (TftG), HBQ reductase, and HQDO (TftH) allowed us to sequentially examine the nature of each product formed through colorimetric assay as well as chemical characterization via mass spectrometry. The colors of the substrates, presumptive spontaneous chemical oxidation products, and enzymatic reaction products are shown in Fig. 5. To eliminate any effect due to the presence of reducing agents, NADH and FMN were included in all reactions. Chemical oxidation of 5-CHQ, in the absence of the dechlorinase, always led to the formation of the red-colored product. In the presence of dechlorinase but absence of HBQ reductase, a mixture of red and orange (reddish-orange)-colored products was formed. When both dechlorinase and reductase were present, the color turned to orange because of the predominant formation of HQox. When 5-CHQ was exposed to all three enzymes in the presence of NADH and FMN, the reaction mixture was light yellow (due to the presence of FMN) but otherwise clear because of the sequential conversion of 5-CHQ to HQ and further to MA (Fig. 5).
FIG. 5.
Color reactions of enzymatic and spontaneous chemical conversion of 5-CHQ and HQ.
Mass spectrometric analysis of 5-CHQ conversion by purified TftG, TftH, and HBQ reductase.
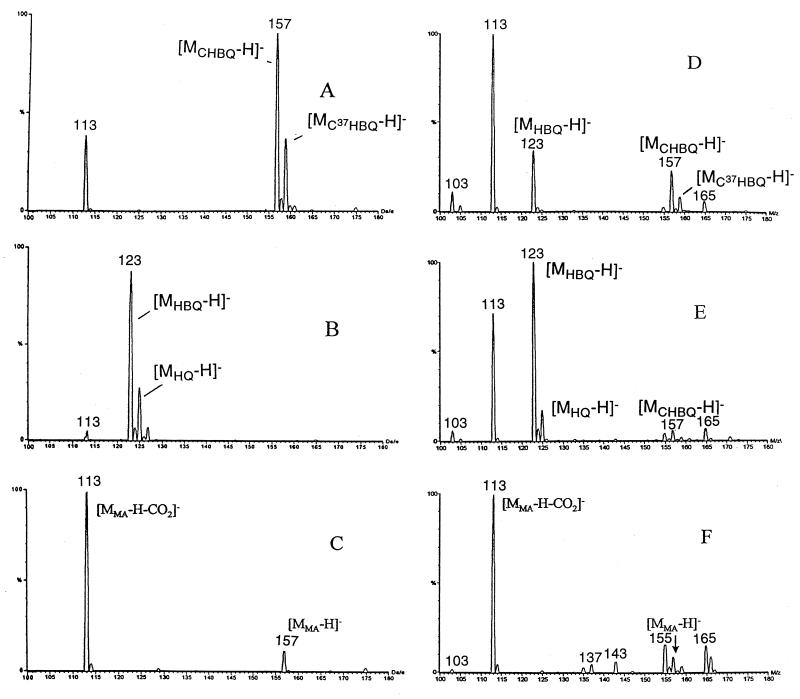
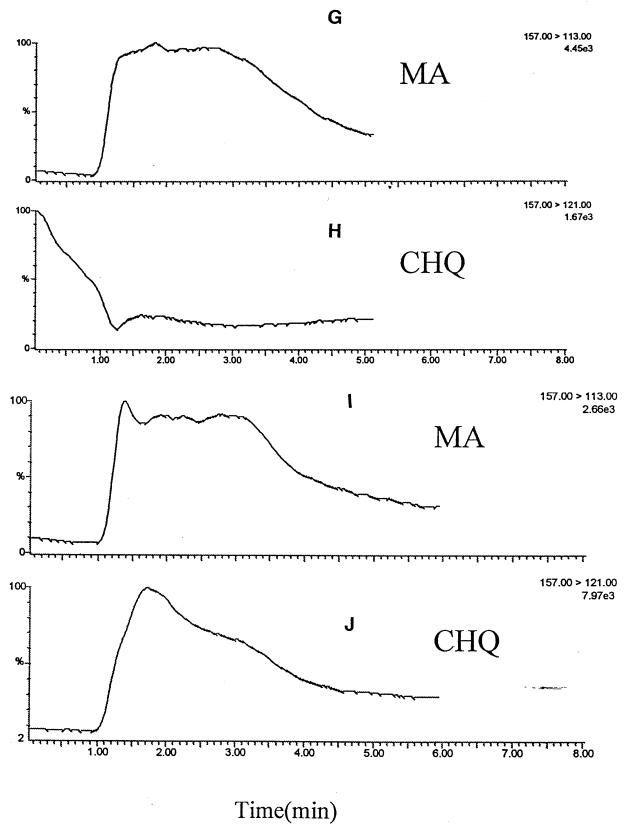
Mass spectra of 5-CHQ (Fig. 6A), HQ (Fig. 6B), and MA (Fig. 6C) were compared to those of intermediates generated in various reaction mixtures. Since 5-CHQ is oxidized very rapidly to 5-CHQox, it is this oxidation product that is primarily detected in the mass spectrum (Fig. 6A). This analysis showed that when 5-CHQ is added to the reaction mixture containing the dechlorinating enzyme (Fig. 6D), the intensity of peak [MCHQox-H]− decreased, while a new peak, [MHQox-H]−, appeared. When 5-CHQ was added to the reaction mixture containing the dechlorinase plus HBQ reductase (Fig. 6E), the intensity of peak corresponding to 5-CHQox decreased significantly, while an additional new peak, [MHQ-H]−, corresponding to HQ appeared. When 5-CHQ was added to the reaction mixture containing the dechlorinating enzyme plus HBQ reductase plus HQDO (Fig. 6F), only peaks [MMA-H]− and [MMA-H-CO2]−, corresponding to MA and its first fragment under ionization conditions, were detected. To distinguish [MMA-H]− and [MCHQox-H], which are similar in molecular mass, tandem mass spectrometric experiments were performed (Fig. 6G to J). The major fragmentation pathway for MA was loss of CO2, giving rise to ion 113, while CHQ showed m/z 121 as the major fragment ion. This ion corresponds to loss of HCl from the oxidized form. Single reaction monitoring experiments using transitions from 157 to 113 and 157 to 121 for detection of MA and CHQ, respectively, were then performed. The results for an incubation mixture consisting of CHQ and all three enzymes (Fig. G and H) clearly show the presence of MA and the corresponding absence of 5-CHQox. To show that the ionization of MA does not suppress the ionization of 5-CHQ, a mixture of standards 5-CHQ and MA at concentrations 0.02 and 0.2 mM, respectively, was subjected to tandem mass spectrometric analysis. As Fig. 6I and J demonstrate, 5-CHQ, despite being present at a 10-times-lower concentration, gave a signal comparable to that of MA, proving that lack of signal of 5-CHQ in the incubation mixture was not due to signal suppression. It is therefore clear that MA, but not 5-CHQox, is detected in Fig. 6F, and MA is really the product of 5-CHQ conversion by the mixture of dechlorinase plus HBQ reductase plus HQDO.
FIG. 6.
Mass spectra of enzymatic and chemical oxidation of 5-CHQ and HQ. (A) 5-CHQ, 0.1 mM; (B) HQ, 0.1 mM; (C) MA, 0.1 mM; (D) 5-CHQ (0.1 mM) + TftG (5-CHQ dechlorinase; 1.5 μg); (E) 5-CHQ (0.1 mM) + TftG (1.5 μg) + HBQ reductase I (1.6 μg); (F) 5-CHQ (0.1 mM) + TftG (1.5 μg) + HBQ reductase I (1.6 μg) + TftH (HQDO; 4 μg); (G) 5-CHQ (0.1 mM) + TftG (1.5 μg) + HBQ reductase I (1.6 μg) + TftH (4 μg), tandem mass spectrometric experiment to follow the fragmentation of MA, [MMA-H-CO2]− (M113); (H) the same as panel G to follow the fragmentation of 5-CHQox, [MCHQox-H-HCl]− (M121); (I) 5-CHQ (0.02 mM) + MA (0.2 mM), tandem mass spectrometric experiment to follow the fragmentation of MA, [MMA-H-CO2]−(M113); (J) 5-CHQ (0.02 mM) + MA (0.2 mM), tandem mass spectrometric experiment to follow the fragmentation of 5-CHQox, [MCHQox-H-HCl]− (M121).
DISCUSSION
The tftG gene product, with 31% sequence identity to E. coli Ycil, a protein of unknown function (20), had previously been shown to be essential in 2,4,5-T metabolism; however no obvious function could be discerned (6). E. coli crude cell extracts overproducing TftG converted 5-CHQ to an oxidized orange-colored reaction product (Fig. 1) that was shown to be HQox. In reactions where 5-CHQ was incubated with both TftG- and TftH-hyperproducing cell extracts, 5-CHQox and HQox disappeared. This observation was strictly dependent on both gene products; TftH could not catalyze this reaction without TftG, nor could TftG catalyze it without TftH. TftH was able to catalyze the conversion of HQ to MA (clear product) but not of 5-CHQ. The TftG-containing crude cell extracts alone catalyzed changes in the 5-CHQ substrate but not in the HQ substrate (Fig. 1). These data indicated that in crude cell extracts, TftG catalyzed the dechlorination of 5-CHQ to produce HQ, the substrate for TftH.
To determine the reaction product when purified TftG is incubated with 5-CHQ, HPLC analysis was performed. The chromatogram in Fig. 2B shows that under reducing conditions, a significant amount of 5-CHQ was converted to HQ and no 5-CHQox was detected. This result suggested that 5-CHQ was dechlorinated by TftG and oxidized to HQox, but that titanium citrate reduced HQox to HQ before it could chemically react with 5-CHQ. The mechanism of dechlorination by TftG may be similar to the elimination reaction catalyzed by LinA during lindane degradation (15). The tftG gene product shows 19.1% amino acid sequence identity and 45.7% similarity with LinA and may therefore act as a 5-CHQ dehydrochlorinase. The 17-kDa LinA polypeptide is slightly larger than the 12-kDa TftG polypeptide, and the similarity is found primarily between TftG and the carboxy terminus of LinA. The conserved amino acid residues may provide information on the reaction mechanism of this type of dechlorination reaction.
In crude cell extracts, TftG catalyzed the dechlorination of 5-CHQ to produce HQox, which was immediately reduced by cellular enzymes using NADH to yield HQ. The cellular reductase was not specific to B. cepacia AC1100, because the reactions were performed in E. coli crude cell extracts which have a number of reductase enzymes. One such enzyme is glutathione reductase, which is also encoded by the B. cepacia AC1100 tftF gene. B. cepacia PT88 crude cell assays in which the tftF gene is deleted and no glutathione reductase activity could be detected showed that the tftF-encoded glutathione reductase was not the enzyme responsible for the reduction of HQox to HQ. Purified glutathione reductase (TftF) could not catalyze the reduction of HQox to HQ, suggesting that HBQ reductase and not glutathione reductase is involved in this reductive step. Many flavin-containing reductases such as cytochrome P-450 monooxygenase reductase and glutathione reductase can reduce quinones via one-electron transfer to produce semiquinones. The semiquinones spontaneously react with oxygen to regenerate quinone and produce hydrogen peroxide. This leads to the continuous depletion of NADH and the production of activated oxygen species. In mammalian systems, NAD(P)H:quinone reductase reduces quinones directly to hydroquinones by a two-electron transfer process (10). This enzyme protects organisms from the toxic effects of quinones.
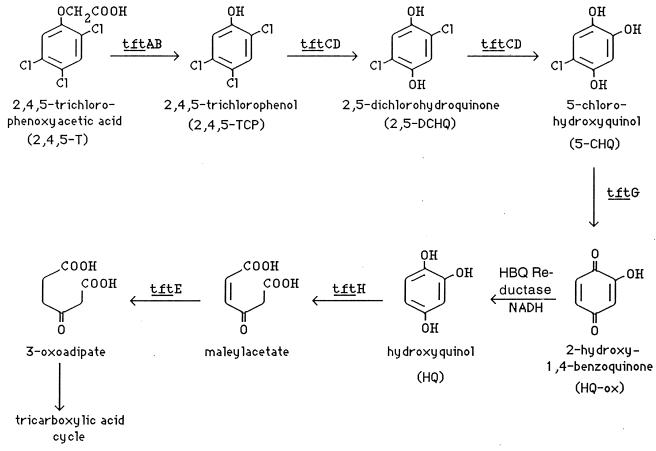
To determine what kind of a reductase might be involved in 5-CHQox or HQox reduction, we purified a major HBQ reductase (Fig. 3) which appeared to be induced during growth with 2,4,5-T. Both colored-product formation from (chloro)hydroxybenzoquinone intermediates (Fig. 1 and 5), as well as mass spectrometric analysis (Fig. 6) revealed that TftG (the dechlorinase) and the HBQ reductase were essential for the conversion of 5-CHQ to HQ. The 2,4,5-T-inducible HBQ reductase appears to differ in substrate specificity from other quinone reductases. Further characterization of this enzyme, as well as its gene, is under way. The product of the tftG gene, the dechlorinase, also appears to be novel since no known enzyme with sequence similarity has been submitted to the database. It has recently been reported that in Sphingomonas paucimobilis, the enzyme LinD catalyzes the conversion of 2,5-dichlorohydroquinone to chlorohydroquinone and slowly to hydroquinone. The low rate of in vitro conversion of chlorohydroquinone to hydroquinone by LinD, compared to rapid conversion of chlorohydroquinone to hydroquinone by resting cells, suggests that an alternative pathway for the degradation of chlorohydroquinone in S. paucimobilis may exist (21). It would be interesting to investigate whether this alternative pathway involves a reductase as reported herein. Based on the data presented in this report, a pathway of 2,4,5-T degradation that shows the individual enzymatic steps is given in Fig. 7.
FIG. 7.
Pathway of 2,4,5-T degradation. The tftA and tftB genes encode two subunits of the 2,4,5-T oxygenase responsible for the conversion of 2,4,5-T to 2,4,5-TCP (5, 29). A two-component flavin-containing monooxygenase encoded by the tftC and tftD genes (14) catalyzes the para-hydroxylation of 2,4,5-TCP to yield 2,5-DCHQ (25, 28). A second hydroxylation step by the same enzyme converts 2,5-DCHQ to 5-CHQ (28). The tftG gene product catalyzes dechlorination of 5-CHQ to yield HQox (this work). An HBQ reductase reduces HQox to HQ before HQDO, encoded by the tftH gene, can catalyze ring cleavage to yield MA (this work and reference 8). Maleylacetate reductase, encoded by the tftE gene, catalyzes the reduction of MA to 3-oxoadipate (8), which ultimately is converted to tricarboxylic acid cycle intermediates.
ACKNOWLEDGMENTS
Olga Zaborina and Dayna Daubaras contributed equally to this work.
This work was supported by Public Health Service grant ES 04050-12 from the National Institute of Environmental Health Sciences. Luying Xun is supported by NSF grant MCB-9218783.
REFERENCES
- 1.Apajalahti J H A, Salkinoja-Salonen M S. Complete dechlorination of tetrachlorohydroquinone by cell extracts of pentachlorophenol-induced Rhodococcus chlorophenolicus. J Bacteriol. 1987;169:5125–5130. doi: 10.1128/jb.169.11.5125-5130.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong S, Patel T R, Whalen M. Detoxification mechanisms for 1,2,4-benzenetriol employed by a Rhodococcus sp. BPG-8. Arch Microbiol. 1993;159:136–140. [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Chapman P J, Sangodkar U M X, Chakrabarty A M. Abstracts of the Eighth Annual Meeting of the Society for Environmental Toxicology and Chemistry, Pensacola, Fla. 1987. 2,4,5-T degradation pathway in Pseudomonas cepacia AC1100; p. 127. [Google Scholar]
- 5.Danganan C E, Ye R W, Daubaras D L, Xun L, Chakrabarty A M. Nucleotide sequence and functional analysis of the genes encoding 2,4,5-trichlorophenoxyacetic acid oxygenase in Pseudomonas cepacia AC1100. Appl Environ Microbiol. 1994;60:4100–4106. doi: 10.1128/aem.60.11.4100-4106.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daubaras D L, Hershberger C D, Kitano K, Chakrabarty A M. Sequence analysis of a gene cluster involved in the metabolism of 2,4,5-trichlorophenoxyacetic acid by Burkholderia cepacia AC1100. Appl Environ Microbiol. 1995;61:1279–1289. doi: 10.1128/aem.61.4.1279-1289.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daubaras D L, Danganan C E, Ye R W, Hubner A, Hendrickson W, Chakrabarty A M. Biodegradation of 2,4,5-trichlorophenoxyacetic acid by Burkholderia cepacia strain AC1100: evolutionary insight. Gene. 1996;179:1–8. doi: 10.1016/s0378-1119(96)00326-5. [DOI] [PubMed] [Google Scholar]
- 8.Daubaras D L, Saido K, Chakrabarty A M. Purification of hydroxyquinol 1,2-dioxygenase and maleylacetate reductase: the lower pathway of 2,4,5-trichlorophenoxyacetic acid metabolism by Burkholderia cepacia AC1100. Appl Environ Microbiol. 1996;62:4276–4279. doi: 10.1128/aem.62.11.4276-4279.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis N K, Greer S, Jones-Mortimer M C, Perham R N. Isolation and mapping of glutathione reductase-negative of Escherichia coli K12. J Gen Microbiol. 1982;128:1631–1634. doi: 10.1099/00221287-128-7-1631. [DOI] [PubMed] [Google Scholar]
- 10.Ernster L. DT diaphorase: a historical review. Chem Sci. 1987;27A:1–13. [Google Scholar]
- 11.Furste J P, Pansegrau W, Frank R, Blocker H, Scholz P, Bagdasarian M, Lanka E. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene. 1986;48:119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- 12.Golovleva L A, Zaborina O, Pertsova R, Baskunov B, Schurukhin J, Kuzmin S. Degradation of polychlorinated phenols by Streptomyces rochei 303. Biodegradation. 1992;2:201–208. doi: 10.1007/BF00124494. [DOI] [PubMed] [Google Scholar]
- 13.Haggblom M M, Janke D, Salkinoja-Salonen M S. Hydroxylation and dechlorination of tetrachlorohydroquinone by Rhodococcus sp. strain CP-2 cell extracts. Appl Environ Microbiol. 1989;55:516–519. doi: 10.1128/aem.55.2.516-519.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hübner A, Danganan C E, Xun L, Chakrabarty A M, Hendrickson W. Genes for 2,4,5-trichlorophenoxyacetic acid metabolism in Burkholderia cepacia strain AC1100: characterization of the tftCD genes and location of the tft operons on multiple replicons. Appl Environ Microbiol. 1998;64:2086–2093. doi: 10.1128/aem.64.6.2086-2093.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imai R, Nagata Y, Fukuda M, Takagi M, Yano K. Molecular cloning of a Pseudomonas paucimobilis gene encoding a 17-kilodalton polypeptide that eliminates HCl molecules from γ-hexachlorocyclohexane. J Bacteriol. 1991;173:6811–6819. doi: 10.1128/jb.173.21.6811-6819.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozyreva L P, Shurukhin Y U, Finkel’shtein Z I, Baskunov B P, Golovleva L A. Metabolism of the herbicide 2,4-D by a Nocardioides simplex strain. Mikrobiologiya. 1993;62:78–85. [Google Scholar]
- 17.Latus M, Seitz H-J, Eberspacher J, Lingens F. Purification and characterization of hydroxyquinol 1,2-dioxygenase from Azotobacter sp. strain GP1. Appl Environ Microbiol. 1995;61:2453–2460. doi: 10.1128/aem.61.7.2453-2460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee J-Y, Xun L. Purification and characterization of 2,6-dichloro-p-hydroquinone chlorohydrolase from Flavobacterium sp. strain ATCC 39723. J Bacteriol. 1997;179:1521–1524. doi: 10.1128/jb.179.5.1521-1524.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li D-Y, Eberspacher J, Wagner B, Kuntzer J, Lingens F. Degradation of 2,4,6-trichlorophenol by Azotobacter sp. strain GP1. Appl Environ Microbiol. 1991;57:1920–1928. doi: 10.1128/aem.57.7.1920-1928.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Milkman R. An Escherichia coli homologue of eukaryotic potassium channel proteins. Proc Natl Acad Sci USA. 1994;91:3510–3514. doi: 10.1073/pnas.91.9.3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyauchi K, Suh S-K, Nagata Y, Takagi M. Cloning and sequencing of a 2,5-dichlorohydroquinone reductive dehalogenase gene whose product is involved in degradation of γ-hexachlorocyclohexane by Sphingomonas paucimobilis. J Bacteriol. 1998;180:1354–1359. doi: 10.1128/jb.180.6.1354-1359.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muller F, Mayhew S. Temperature-difference spectra of flavins and flavoproteins. Methods Enzymol. 1980;66:350–360. doi: 10.1016/0076-6879(80)66480-5. [DOI] [PubMed] [Google Scholar]
- 23.Sangodkar U M X, Chapman P J, Chakrabarty A M. Cloning, physical mapping and expression of chromosomal genes specifying degradation of the herbicide 2,4,5-T by Pseudomonas cepacia AC1100. Gene. 1988;71:267–277. doi: 10.1016/0378-1119(88)90043-1. [DOI] [PubMed] [Google Scholar]
- 24.Steiert J G, Pignatello J J, Crawford R L. Degradation of chlorinated phenols by a pentachlorophenol-degrading bacterium. Appl Environ Microbiol. 1987;53:907–910. doi: 10.1128/aem.53.5.907-910.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tomasi I, Artaud I, Bertheau Y, Mansuy D. Metabolism of polychlorinated phenols by Pseudomonas cepacia AC1100: determination of the first two steps and specific inhibitory effect of methimazole. J Bacteriol. 1995;177:307–311. doi: 10.1128/jb.177.2.307-311.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uotila J S. Dehalogenases for polychlorinated aromatic compounds in Rhodococcus chlorophenolicus PCP-1 and Mycobacterium fortuitum CG-2. Ph.D. thesis. Helsinki, Finland: University of Helsinki; 1993. [Google Scholar]
- 27.van der Meer J R, de Vos W M, Harayama S, Zehnder A J B. Molecular mechanisms of genetic adaptation to xenobiotic compounds. Microbiol Rev. 1992;56:677–694. doi: 10.1128/mr.56.4.677-694.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xun L. Purification and characterization of chlorophenol 4-monooxygenase from Burkholderia cepacia AC1100. J Bacteriol. 1996;178:2645–2649. doi: 10.1128/jb.178.9.2645-2649.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xun L, Wagnon K. Purification and properties of component B of 2,4,5-trichlorophenoxyacetate oxygenase from Pseudomonas cepacia AC1100. Appl Environ Microbiol. 1995;61:3499–3502. doi: 10.1128/aem.61.9.3499-3502.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zaborina O, Latus M, Eberspacher J, Golovleva L A, Lingens F. Purification and characterization of 6-chlorohydroxyquinol 1,2-dioxygenase from Streptomyces rochei 303: comparison with an analogous enzyme from Azotobacter sp. strain GP1. J Bacteriol. 1995;177:229–234. doi: 10.1128/jb.177.1.229-234.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]