Abstract
Mycobacteria secrete the siderophore exochelin when grown under iron-limiting conditions. In order to understand iron uptake mechanisms in mycobacteria, we have taken a genetic approach to identify those genes involved in exochelin biosynthesis and transport in Mycobacterium smegmatis. Of the 6,000 chemically mutagenized clones of M. smegmatis mc2155 screened on agar plates containing chrome azural S, 19 mutants that had lost the ability to produce or secrete exochelin were identified. Thirteen of these mutants were complemented by a single M. smegmatis cosmid. Sequence analysis of this cosmid revealed nine open reading frames, three of which are homologous to genes encoding transporter proteins, which are likely involved in exochelin transport. Complementation and Tn10 mutagenesis analysis identified two new genes, fxbB and fxbC, which are required for exochelin biosynthesis. The fxbB and fxbC genes encode large proteins of 257 and 497 kDa, respectively, which are highly homologous to peptide synthetases, indicating that exochelin biosynthesis occurs by a nonribosomal mechanism.
Iron is essential for microorganisms (31, 50), as it is needed for a variety of important cellular functions such as electron transport, the reduction of ribonucleotides and dinitrogen, and the decomposition of peroxide (24). Although there is abundant iron in the earth and in animals, it is not readily accessible to bacteria. When exposed to the earth’s atmosphere, iron is oxidized to the ferric state. Ferric iron easily forms an insoluble hydroxide in aqueous solution at neutral or alkaline pH. In animals, most iron is bound to proteins such as hemoglobin, myoglobin, and cytochromes or iron carriers such as transferrin, lactoferrin, and ferritin. The relative insolubility of iron in the environment and the iron-withholding mechanisms in animals limit the amount of iron available to bacteria. In order to survive and propagate, microorganisms have developed sophisticated mechanisms such as the siderophore system to scavenge iron from the environment.
Siderophores are low-molecular-weight iron chelators synthesized by most microorganisms to sequester and deliver iron (32). Organisms often have multiple types of siderophores and specific receptors for siderophores, presumably to ensure adequate acquisition of iron under many conditions. Siderophore synthesis is tightly regulated by the amount of iron in the environment (23). Since siderophores are involved in the acquisition of iron under iron starvation conditions (e.g., within a host environment), the production of siderophores has been shown to be associated with virulence as well as the survival of many microorganisms (24, 33, 51). The siderophore systems of Escherichia coli, Yersinia enterocolitica (24), Erwinia chrysanthemi (14), and Pseudomonas aeruginosa (29) are correlated with the virulence of these organisms. Siderophore pathways may also be involved in the virulence of pathogenic mycobacteria such as Mycobacterium tuberculosis. The identification of genes involved in iron sequestration pathways will facilitate our understanding of the mechanisms of iron uptake and the role of iron in the biology of mycobacteria.
We use Mycobacterium smegmatis as a model system to study siderophore-mediated iron uptake in mycobacteria because it is nonpathogenic, fast-growing, and more tractable genetically than M. tuberculosis. M. smegmatis is known to produce a cell envelope-associated siderophore called mycobactin (43) and two secreted siderophores—exochelin MS, a pentapeptide derivative (40), and carboxymycobactin, a modified form of mycobactin (22, 34). The gene fxbA, which is required for exochelin biosynthesis in M. smegmatis, was identified in a previous study (16). In this report, we describe the isolation of exochelin-deficient mutants of M. smegmatis generated by ethyl methanesulfonate (EMS) mutagenesis. Complementation studies and mutagenic analyses identified two genes, fxbB, and fxbC, which are essential for exochelin biosynthesis. The two biosynthesis enzymes, FxbB and FxbC, share significant homology with peptide synthetases (PPSs). Furthermore, we have identified three other open reading frames (ORFs) which may also have a role in export and uptake of exochelin.
(Data in this paper are from a thesis by Shengwei Yu to be submitted in partial fulfillment of the requirements for the Doctor of Philosophy degree from the Sue Golding Graduate Division of Medical Sciences, Albert Einstein College of Medicine, Yeshiva University, Bronx, N.Y.)
MATERIALS AND METHODS
Bacteria, media, and growth conditions.
The strains, cosmids, plasmids, and phage used in this study are listed in Table 1. E. coli cultures were routinely grown in Luria-Bertani (LB) medium (Difco Laboratories) at 37°C. Antibiotics, when required, were added at the following concentrations: ampicillin, 50 μg/ml; kanamycin, 50 μg/ml for E. coli and 20 μg/ml for M. smegmatis; and hygromycin B, 200 μg/ml for E. coli and 50 μg/ml for M. smegmatis. Hygromycin B was purchased from Boehringer Mannheim (50 mg/ml in phosphate-buffered saline); all other antibiotics were purchased from Sigma Chemical Co. (St. Louis, Mo.). Mycobacterial cultures were grown in Middlebrook 7H9 medium (Difco Laboratories) supplemented with glycerol (0.2%), glucose (0.2%), bovine serum albumin fraction V (0.5%), and NaCl (0.085%) at 37°C. Bovine serum albumin fraction V was purchased from Boehringer Mannheim; glycerol, glucose, and NaCl were purchased from Fisher Scientific. Minimal medium (MM) and the chrome azural S agar medium have been described previously (16). MM was used as the iron-limiting medium for mycobacteria (16).
TABLE 1.
Bacterial strains, cosmids, plasmids, and phage used in this study
| Strain, cosmid, plasmid, or phage | Relevant genotype or description | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK− mK+) phoA supE44 λ−thi-1 gyrA96 relA1 | GIBCOBRL |
| DH10B | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 deoR recA1 endA1 araD139 Δ(ara leu)7697 galU galK λ−rpsL nupG | GIBCOBRL |
| STBL2 | F−mcrA Δ(mcrBC-hsdRMS-mrr) recA1 endA1 gyrA96 thi supE44 relA1 λ− (lac-proAB) | GIBCOBRL |
| M. smegmatis | ||
| mc2155 | ept-1 | 42 |
| mc2848 | ept-1 fxbA1 | 16 |
| mc21287 | ept-1 fxbA2 | This study |
| mc21289 | ept-1 fxbB4 | This study |
| mc21608 | ept-1 fxbB21::Tn10 | This study |
| mc21609 | ept-1 fxbC22::Tn10 | This study |
| mc21610 | ept-1 fxbB21::Tn10/pYUB563 | This study |
| mc21611 | ept-1 fxbC22::Tn10/pYUB563 | This study |
| Cosmids or plasmids | ||
| pKSII+ | Ampr; high-copy-number cloning vector, ColE1, unable to replicate in mycobacteria | Stratagene |
| pMD31 | Kanr; E. coli-mycobacterium shuttle vector | 11 |
| pYUB412 | Ampr Hygr; E. coli-mycobacterium shuttle vector; ColE1 ori; int attP nonreplicative but integration proficient in mycobacteria | 6 |
| pYUB415 | Ampr Hygr; E. coli-mycobacterium shuttle vector; ColE1 ori; M. fortuitum pAL5000 ori; lambda cos | 6 |
| pYUB340 | pYUB18 with a 30-kb insert which can complement exochelin mutant mc2848 | 16 |
| pYUB347 | pMD31 with a 4.3-kb fragment which can complement exochelin mutant mc2848 | 16 |
| pYUB563 | pYUB415 with a 31-kb insert from mc2155 in the BamHI site | This study |
| pYUB908 | pYUB563 fxbB::Tn10 | This study |
| pYUB909 | pYUB563 fxbC::Tn10 | This study |
| Phage λNK1316 | Mini-Tn10 kan/Ptac-ATS transposase | 20 |
Generating exochelin-defective mutants of M. smegmatis.
The protocol for EMS mutagenesis of M. smegmatis was adapted from that in Current Protocols in Molecular Biology (4). A 1-h 15-min EMS exposure yielded 25% survival of M. smegmatis. After mutagenesis, the mixture was sonicated and passed through a 5.0-μm-pore-size filter (Cameo 25NS; Micron Separations Inc.) to obtain suspension of single cells. The samples were diluted and transferred to plates containing Middlebrook 7H9 medium as the base, 0.2% glycerol, 10% albumin-dextrose-saline, 0.5% Casamino Acids, 0.1 mg of diaminopimelate per ml, and 0.02 mg of trytophan per ml, and the plates were incubated at 37°C (27). Individual colonies were then plated onto CAS medium agar plates and scored for the absence of a yellow halo, indicative of exochelin production (16).
Complementation analysis.
A cosmid library of M. smegmatis mc2155 genomic DNA in the replicating cosmid vector pYUB415 (6) was generated by Vaamonde and Jacobs (47) and electroporated into the M. smegmatis exochelin mutants. Transformants were grown on hygromycin-containing medium, selected, and transferred to CAS medium to screen for the complemented clones.
DNA manipulation.
Restriction enzymes, T4 DNA ligase, and DNA polymerase I large fragment (Klenow) were purchased from Promega and New England Biolabs. Shrimp alkaline phosphatase was obtained from USB/Amersham. Restriction fragments were isolated from agarose gels with the Qiaex II gel extraction kit (Qiagen Inc.). Plasmids were isolated by alkaline lysis (36) or with the Qiagen plasmid kit (Qiagen Inc.). All enzymes and kits were used in accordance with the manufacturer’s recommendations. Chromosomal DNA from M. smegmatis was purified as described previously (5).
Mini-Tn10 transposon mutagenesis of pYUB563 and allelic exchange.
In vivo mutagenesis of the complementing cosmid pYUB563 was performed in E. coli with a mini-Tn10 transposon delivered by phage lambda NK1316 (20). Briefly, DH10B cells transformed with pYUB563 were infected with phage lambda NK1316 (ATCC 77345) and plated on LB medium with kanamycin at 30 μg/ml. Cosmids were prepared from pools of the Kanr DH10B cells and used to transform E. coli DH5α cells. Transformants were selected on LB medium with kanamycin to isolate the Kanr cosmids which have mini-Tn10 insertions. Cosmid DNA was prepared from individual clones and analyzed by restriction endonuclease digestion.
The flanking regions of the Tn10 insertions in these cosmids were identified by first subcloning Sau3AI partial digestions of the cosmids, selecting for kanamycin-resistant subclones, followed by sequencing toward the mini-Tn10 insertion sites with the T3 and T7 universal primers. The DNA sequences of the flanking regions were compared to the cosmid sequence, and the mini-Tn10 insertion sites were identified.
The Tn10-mutagenized cosmids were utilized to construct site-directed insertion mutants in M. smegmatis. The cosmids were linearized by ScaI digestion and then electroporated into strain mc2155. Kanr colonies were selected, transferred to hygromycin plates to screen for Hygs colonies and then transferred to CAS plates to screen for halo production.
Southern blotting and DNA sequencing.
Southern blotting was done by the alkaline-denaturation procedure (26). DNA was transferred to Biotrans nylon membranes (ICN, Irvine, Calif.) by the capillary method. Hybridization and detection were performed as recommended by the manufacturer (ECL kit; Amersham), under high-stringency (0.1 M NaCl and 42°C) and low-stringency (0.5 M NaCl and 42°C) conditions for prehybridization and hybridization.
The sequencing strategy consisted of using the M13 universal primers with 100 subclones in pMD31, followed by designing primers using the newly acquired sequences to directly sequence the cosmid pYUB563. Nucleotide sequencing reactions were performed by using a Dye Terminator Cycle Sequencing Ready Reaction DNA sequencing kit, and the sequence runs were done by using an ABI PRISM 377 DNA sequencer (Perkin-Elmer Applied Biosystems). Sequence data were analyzed with the MacVector and AssemblyLIGN programs (Scientific Imaging Systems), and homology searches were performed by using the BLAST program of the National Center for Biotechnology Information (3).
Mycobactin and exochelin assays.
For the exochelin assay, mycobacteria were grown in liquid MM and MM containing 50 μM FeCl3. The cells were centrifuged at 5,000 × g for 15 min, and the supernatant was assayed for exochelin by the universal CAS method developed by Schwyn and Neilands (38). The mycobactin assay procedure of Hall and Ratledge (19) was used to isolate and determine the concentration of mycobactin. For the mycobactin assay, mycobacteria were spread onto MM, MM containing 50 μM 2,2′-dipyridyl (an iron chelator), and MM containing 50 μM FeCl3 to produce a lawn of growth. The cells were scraped from the plates and weighed. The mycobactin from the weighed cells was extracted overnight with ethanol.
Hydrogen peroxide killing.
Mycobacteria were grown (37°C, with shaking) in MM and MM plus 50 μM FeCl3 to an A600 of 0.35. Aliquots (0.9 ml) of the culture were added to 0.1 ml of hydrogen peroxide (obtained as a 30% solution from Sigma Chemical Co.) at concentrations from 0 to 35 mM. After 30 min of incubation at 37°C, the number of surviving cells was obtained by diluting the hydrogen peroxide-treated cells in phosphate-buffered saline with 0.05% Tween 80 and plating 20-μl aliquots onto Middlebrook 7H9 medium. The experiments were repeated twice.
Nucleotide sequence accession number.
The nucleotide sequence reported in this study (nucleotides 1 to 30683) has been submitted to the GenBank/EMBL data bank under accession no. AF027770.
RESULTS
Isolation of M. smegmatis exochelin biosynthesis mutants.
M. smegmatis exochelin is detectable on agar plates containing CAS (16). This medium turns blue when CAS binds to ferric iron; once the iron is deprived from the dye by a higher-affinity iron chelator such as exochelin, the blue medium will turn yellow. Wild-type M. smegmatis mc2155 produces and secretes exochelin, leading to the formation of yellow halos around the colonies on CAS plates; exochelin-defective mutants will fail to form yellow halos as previously shown (16). We screened 6,000 EMS-mutagenized M. smegmatis colonies to identify mutants lacking a yellow halo on CAS medium. Nineteen mutants were obtained and presumed to be defective in either exochelin synthesis or export and characterized further.
Complementation analysis of M. smegmatis exochelin mutants.
The 19 mutants were transformed with an M. smegmatis cosmid library constructed in pYUB415. In addition, each mutant was transformed with pYUB347, a plasmid containing fxbA, a gene previously shown to be required for exochelin biosynthesis (16). We found that these mutants fell into at least two different complementation groups. The group I mutant, mc21287 (1 of 19), was complemented by pYUB347. Therefore, this mutant most likely has a mutation in the fxbA gene. Group II mutants (13 of 19) were not complemented by pYUB347 but were complemented by four independent but similar, overlapping cosmids. One of these cosmids, pYUB563, was chosen for further study. The remaining mutants (5 of 19) could not be complemented by pYUB347, pYUB563, or the cosmid library. These clones will not be discussed further.
DNA sequence analysis of the M. smegmatis exochelin biosynthesis gene cluster.
The cosmid pYUB563 was digested by BamHI, ClaI, KpnI, PstI, and SphI separately and subcloned into the corresponding sites in pMD31, a mycobacterial shuttle vector (11). After electroporating the group II mutants with the subcloned plasmids, we screened the transformed colonies on CAS plates for complementing clones. No complementation was observed with any of the subcloned constructs. In addition, a Sau3AI partial digestion of the cosmid pYUB563 was pooled for shotgun subcloning. DNA fragments of ∼10 kb were obtained and ligated to pMD31. DNA was prepared from a pool of 200 E. coli transformants and electroporated into the group II mutants. None of the transformants had the ability to form a halo when transferred to CAS plates. Restriction analysis showed that more than 80% of the colonies contained plasmids with random 10-kb inserts (data not shown). The failure to obtain a small complementing fragment suggests that either the complementing gene is very large or it is located in a large operon such that subclones would lack a promoter and the promoterless vector pMD31 could not express the gene.
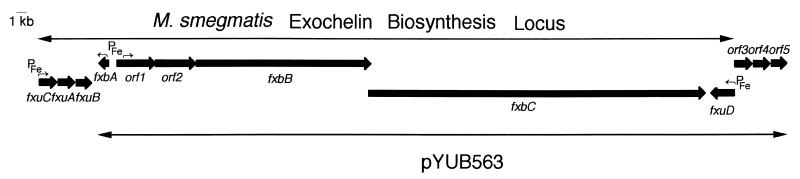
Since subcloning techniques had failed, the full-length cosmid pYUB563 was sequenced in order to identify the complementing gene(s). We sequenced 30,683 bp of DNA and identified nine ORFs (Fig. 1). The gene fxbA (for ferric exochelin biosynthesis) had been previously identified as an exochelin biosynthesis gene and was flanked at the 3′ end by genes fxuA (for ferric exochelin uptake), fxuB, and fxuC (16) (Fig. 1). The BLAST program (2) was used to search for homologous genes of the eight remaining ORFs in GenBank, EMBL, and Sanger Center databases. ORF1 and ORF2 have homology with the ABC transporters (Table 2). ABC transporters consist of four domains, two hydrophobic transmembrane domains which are associated with two hydrophilic domains containing ATP-binding cassettes. The ATP-binding domain and the membrane-spanning domain of an ABC transporter can be located on the same polypeptide or on separate polypeptides (15). The N-terminal halves of ORF1 (amino acids [aa] 1 to 302) and ORF2 (aa 1 to 285) contain five and four putative transmembrane domains respectively (30). Both of the C-terminal halves of ORF1 (aa 303 to 574) and ORF2 (aa 286 to 584) share homology with ATP-binding cassettes. FxuD was named according to the homology of this ORF to the genes encoding a family of periplasmic proteins involved in iron transport, including the ferrichrome receptor, FhuD, from Bacillus subtilis and the iron dicitrate transport system permease protein FepB of Synechocystis sp. (Table 2). These similarities suggest that FxuD might be a membrane-associated protein that may serve as a receptor for ferric exochelin. ORF3 has homology with ABC transporters found in M. tuberculosis and Mycobacterium leprae (Table 2). Unlike ORF1 and ORF2, ORF3 has only a hydrophilic ATP-binding domain and does not contain a hydrophobic transmembrane domain within the same peptide. ORF4 and ORF5 lack homology to any well-characterized proteins except to four putative M. tuberculosis and M. leprae membrane proteins (Table 2). ORF4 consists of six transmembrane domains, and ORF5 consists of five transmembrane domains (30), suggesting they are membrane proteins.
FIG. 1.
Exochelin biosynthesis locus. Nine ORFs were identified in cosmid pYUB563. Based on homologies to known biosynthesis and transport genes, the ORFs were named fxbA, orf1, orf2, fxbB, fxbC, fxuD, orf3, orf4, and orf5. Six of these ORFs and the previously identified putative iron uptake genes fxuC, fxuA, and fxuB which are located in the 3′ end of the fxbA gene are likely involved with exochelin biosynthesis and transport pathways. PFe→, the putative iron box with direction of transcription is indicated. The transcription of fxbA gene and the orf1, orf2, fxbB, and fxbC gene operon were under the control of divergent promoters and iron boxes.
TABLE 2.
Homologies of proteins in pYUB563 to selected sequences from other organismsa
| Protein in pYUB563 | Homologous protein | % Iden- tityc | % Simi- larityb,c | Accession no. or referenced |
|---|---|---|---|---|
| ORF1 | Streptomyces glaucescens StrV | 27 | 38 | 1361425 |
| ORF2 | S. glaucescens StrW | 31 | 46 | 1361426 |
| FxbB | Streptomyces roseosporus daptomycin biosynthesis protein subunit | 31 | 32 | AF021262 |
| Streptomyces pristinaespiralis SnbC | 38 | 50 | 9 | |
| FxbC | S. pristinaespiralis SnbED | 36 | 47 | 1906378 |
| FxuD | Corynebacterium diphtheriae ORF1 | 34 | 49 | 37 |
| Bacillus subtilis FhuD | 30 | 51 | 2226253 | |
| M. tuberculosis MTCY6A4.09C | 36 | 51 | 1850110 | |
| Synechocystis sp. strain FepB | 31 | 47 | 1651665 | |
| ORF3 | M. tuberculosis MTV007.05c ABC transporter ATP-binding protein | 86 | 94 | 2791392 |
| M. leprae ABC2 | 82 | 89 | 2145963 | |
| ORF4 | M. tuberculosis MTV007.04c | 56 | 61 | 2791391 |
| M. leprae B1496-F2-59 | 51 | 58 | 2145664 | |
| ORF5 | M. tuberculosis MTV007.03c | 44 | 52 | 2791390 |
| M. leprae B1496-F2-61 | 47 | 56 | 2145665 |
Homologies were calculated by using the gapped BLAST program (2) of the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov).
Includes conservative substitutions.
These homologies are over the entire length of these genes.
GenBank accession numbers or literature references.
The fxbB and fxbC gene products have homology to multidomain PPSs which use a nonribosomal enzyme thiotemplate mechanism for peptide synthesis (8–10, 21, 41, 46). The multiple domains in PPSs are homologous to each other and have been referred to as modules, each one catalyzing the activation and condensation of the amino or hydroxy acid in sequence (9). The FxbB sequence has two putative modules, while the FxbC sequence has four. These modules show a high degree of sequence similarity and identity, and the conserved amino acid sequences correspond to the ATP-binding site, the ATPase motif, and pantetheine attachment motif in the amino acid activation domains (Fig. 2). Besides the homology among these six modules, they also have a high degree of similarity to modules in several multifunctional peptide synthetases and adenylate-forming enzymes of diverse origin (Table 2).
FIG. 2.
Alignment of the sequences of six modules of FxbB and FxbC. The number at the end of each line refers to the amino acid sequence position in FxbB and FxbC. Identical regions are boxed. The ATP-binding site, ATPase site, and pantetheine motif are underlined. Dots indicate variable amino acids, and dashes indicate gaps.
Previous studies showed that fxbA gene expression is iron regulated and that the fxbA promoter region has an “iron box” sequence recognized by the mycobacterial regulator protein IdeR (12, 13, 16). Sequence analysis showed that there are also iron boxes in the promoter and operator regions of the fxuC gene, orf1, and the fxuD gene (Table 3).
TABLE 3.
Putative iron binding sequences in the promoter regions of fxbA, orf1, fxuC, and fxuD genes
fxbA is flanked by orf1 and orf2 on one side and fxuA, fxuB and fxuC on the other side (16). fxuA, fxuB, and fxuC are highly homologous to genes encoding iron permeases belonging to a large family of ATP-dependent permeases in gram-negative organisms (16); therefore, the biosynthesis genes fxbA, fxbB, and fxbC are flanked by ABC transporter genes and a possible receptor gene, fxuD. All of these genes are strong candidates for genes involved in exochelin biosynthesis and transport pathways.
Mutational analysis of the fxbB and fxbC genes.
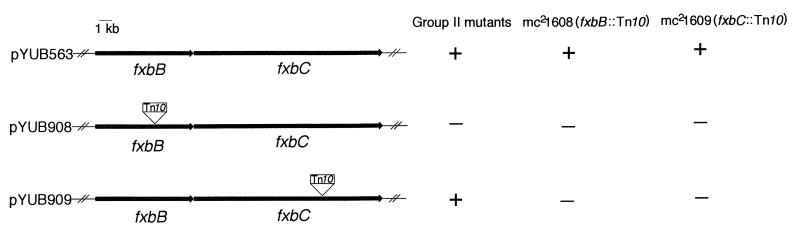
In order to identify the gene(s) that complement group II mutants, in vivo mutagenesis of the complementing cosmid was performed in E. coli with a Tn10 transposon (see Materials and Methods). Of the 50 arbitrarily picked Kanr cosmids, 30 had a transposon in the insert. All of the 30 Tn10-disrupted cosmids were electroporated into mc21289 (one of the group II mutants); eight failed to complement this mutant. These eight cosmids were electroporated into the rest of the mutants in group II and were also found to be unable to complement any of these mutants. By analyzing the DNA sequence flanking the region of the Tn10 insertion in these cosmids, we found that every noncomplementing cosmid had a Tn10 inserted in the fxbB gene.
Analysis of the sequence suggests that fxbB and fxbC are in the same transcription unit. Therefore, group II mutants could have mutations in the fxbB and/or fxbC genes. The Tn10 insertion in fxbB may exert a polar effect on expression of fxbC. To differentiate between the contributions of the fxbB and fxbC genes on the complementing phenotype, pYUB908, a cosmid containing a transposon insertion in fxbB, and pYUB909, a cosmid with an insertion in fxbC, were electroporated into group II mutants. pYUB908 failed to complement mutants in group II, whereas pYUB909 retained the ability to complement (Fig. 3). Therefore, all of the group II isolates have a mutation in the fxbB gene.
FIG. 3.
Complementation of exochelin biosynthesis mutants. The ability of the cosmids to complement exochelin biosynthesis gene mutations are indicated at the right. Symbols: +, complementation; −, no complementation.
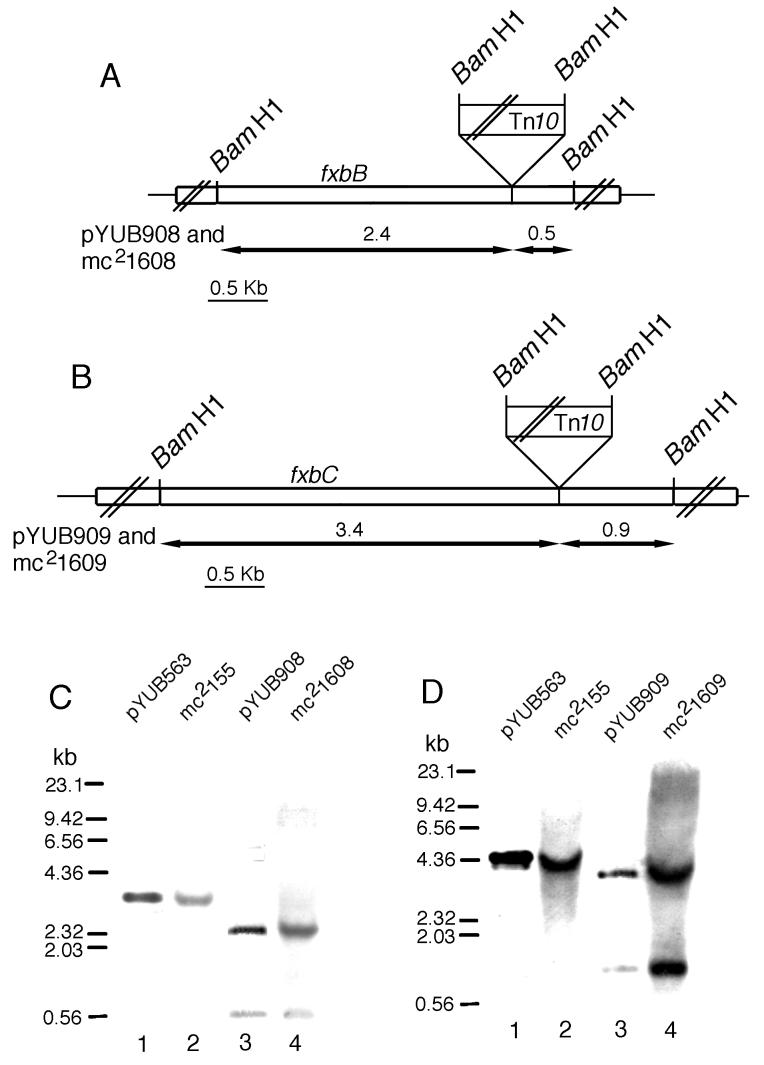
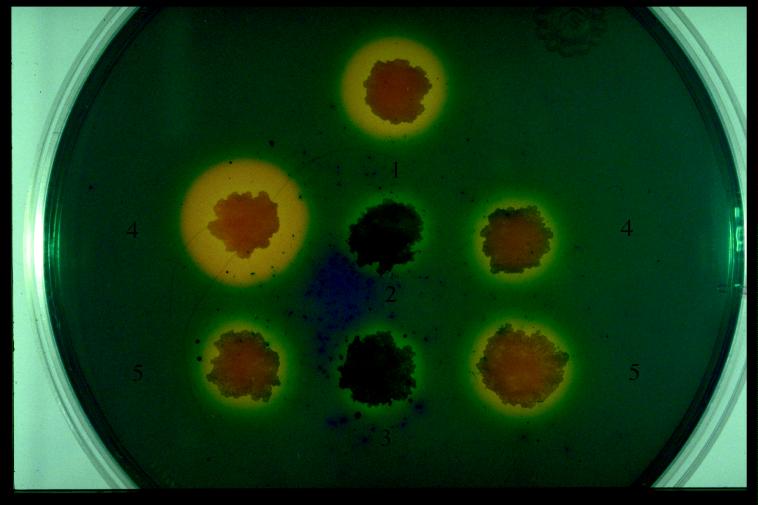
We used cosmids pYUB908 (fxbB::Tn10) and pYUB909 (fxbC::Tn10) to construct insertions in the genome of strain mc2155. Linearized pYUB908 and pYUB909 were electroporated into mc2155, and Kanr recombinants were selected. Two Kanr clones, mc21608 and mc21609, were analyzed by Southern hybridization (Fig. 4). Mutant mc21608 has a Tn10 insertion in the fxbB gene, and mutant mc21609 has a Tn10 insertion in the fxbC gene. Both mc21608 and mc21609 fail to form halos on CAS plates (Fig. 5). Thus, it appears that both FxbB and FxbC are essential for exochelin production in M. smegmatis.
FIG. 4.
Disruption of fxbB and fxbC. (A and B) Restriction maps of the fxbB and fxbC gene disruption in plasmids pYUB908 and pYUB909 and in strains mc21608 and mc21609. The double slashes indicate that the map is not to scale. (C and D) Southern blot analyses of the M. smegmatis mc2155 wild-type and fxbB- and fxbC-defective mutant strains mc21608 and mc21609. pYU563 was used as a control for the undisrupted gene, and pYUB908 and pYUB909 were used as controls for the disrupted genes. pYU563, pYUB908, pYUB909, and chromosomal DNAs from the three strains were digested with BamHI and analyzed by Southern blotting under high-stringency conditions with probes corresponding to the 2.89- and 4.30-kb BamHI fragments of the fxbB and fxbC genes, respectively.
FIG. 5.
CAS plate assay. The presence of a yellow halo indicates the production of the secreted exochelin. Colonies: 1, mc2155(pYUB415); 2, mc21608(pYUB415); 3, mc21609(pYUB415); 4, mc21610; 5, mc21611.
The cosmid pYUB908 (fxbB::Tn10) cannot complement mutant mc21608 (fxbB::Tn10), mutant mc21609 (fxbC::Tn10), and all of the group II mutants; pYUB909 (fxbC::Tn10) cannot complement mutant mc21609 (fxbC::Tn10) or mc21608 (fxbB::Tn10), but it can complement all of the group II mutants (Fig. 3). From these data, we conclude that Tn10 insertion in the fxbB gene in pYUB908 and mc21608 has a polar effect upon expression of the fxbC gene.
Characterization of fxbB and fxbC mutants.
Quantitative assays of mycobactin and exochelin from the different strains were performed to help us understand the regulation of the different siderophores under different iron conditions. Fiss had demonstrated that M. smegmatis with a mutation in the fxbA gene failed to produce exochelin but still produced mycobactin (16). Mutant strains mc21608 and mc21609 have defects in the fxbB and fxbC genes, respectively, and lack the ability to produce yellow halos on CAS medium (Fig. 5). The siderophores mycobactin and exochelin were partially purified from the parent strain mc2155 and mutant strains mc21608 and mc21609, mc21610, and mc21611 (Table 4). Exochelin production was diminished in mutant strains mc21608 and mc21609, but the iron binding activity of the supernatants was not totally abolished. The small amount of activity may be due to the other secreted siderophore, carboxymycobactin. Synthesis of exochelin was restored in the complementing strains mc21610 and mc21611. The mutant strains mc21609 and mc21608 synthesized mycobactin in amounts comparable to or greater than the wild-type strain mc2155.
TABLE 4.
Production of mycobactin grown under iron-deplete and -replete conditionsa
| Strain | Relevant genotype | Mycobactin production (mg of mycobactin/g [wet wt] of cells)
|
Exochelin productionb | ||
|---|---|---|---|---|---|
| MM plus dipyridyl | MM | MM plus iron | |||
| mc2155 | fxbA+fxbB+fxbC+ | 3.69 ± 0.57 | 1.63 ± 0.48 | 0.23 ± 0.21 | 1.11 ± 0.05 |
| mc21608 | fxbB21::Tn10 | 7.12 ± 1.05 | 4.61 ± 0.86 | 0.32 ± 0.20 | 0.30 ± 0.10 |
| mc21609 | fxbC22::Tn10 | 3.75 ± 0.68 | 2.53 ± 0.77 | 0.32 ± 0 | 0.18 ± 0.08 |
| mc21610 | fxbB21::Tn10/pYUB563 | 3.44 ± 0.44 | 2.81 ± 0.37 | 0.30 ± 0.27 | 1.16 ± 0.11 |
| mc21611 | fxbC22::Tn10/pYUB563 | 2.49 ± 0.07 | 1.68 ± 0.11 | 0.27 ± 0.32 | 1.13 ± 0.10 |
The bacteria were grown for 3 days in MM alone or MM supplemented with 50 μM 2,2′-dipyridyl (iron deplete) or with 50 μM iron (iron replete). The data are from a single experiment performed in triplicate and are representative of three separate experiments.
Exochelin production (measured by the change in optical density at 630 nm per milliliter) for cells grown in MM.
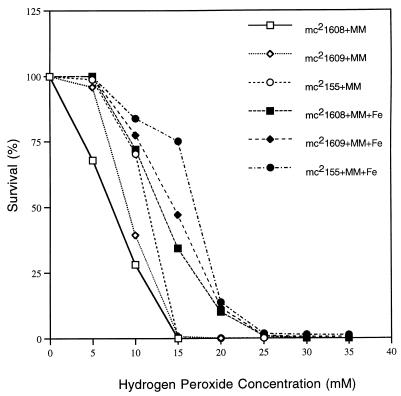
A previous study had shown that iron-starved M. smegmatis cells are more susceptible to hydrogen peroxide killing than iron-loaded cells are (25). The susceptibility of the iron-deficient cells was due to their inability to upregulate two of the major oxidative stress proteins, DnaK and GroEL, when stressed with hydrogen peroxide. Based on this phenomenon, we hypothesized that the exochelin deficient mutants are more sensitive to hydrogen peroxide killing than wild-type M. smegmatis. The fxbB and fxbC mutants and wild-type mc2155 were tested for hydrogen peroxide sensitivity under high- and low-iron conditions. The mutant strains mc21608 and mc21609 were more sensitive to hydrogen peroxide killing than wild-type mc2155 under both high- and low-iron conditions (Fig. 6). The wild-type mc2155 had a comparable magnitude of killing in this study under both high- and low-iron conditions, unlike results of the previous study (25). The sensitivities toward hydrogen peroxide of the complemented strains mc21610 and mc21611 were similar to that of the wild-type mc2155 (data not shown).
FIG. 6.
Hydrogen peroxide killing of M. smegmatis mc2155, mc21608, and mc21609 grown in liquid MM alone and liquid MM plus 50 μM FeCl3 (MM+Fe). Exponential-growth-phase cultures were exposed to the indicated concentrations of H2O2, and the surviving fraction of the population was determined by counting CFU. 100% survival corresponds to ≈108 CFU per ml.
DISCUSSION
We report results from the sequencing of a cosmid capable of complementing mutants defective in exochelin biosynthesis. This endeavor revealed the presence not only of the previously described fxbA gene but also of five ORFs which are likely involved with exochelin synthesis and export. Two genes, fxbB and fxbC, have striking homology with the PPSs and as was the case with fxbA, are required for the synthesis of exochelin MS.
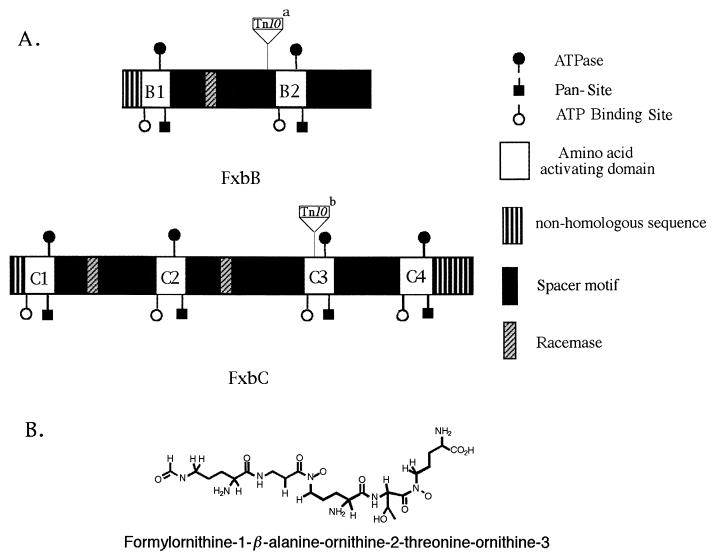
PPSs are large multifunctional enzyme complexes made of multiple homologous domains which catalyze the synthesis of linear and cyclic peptides including antibiotics and siderophores by a nonribosomal peptide synthesis mechanism (1, 7–10, 21, 28, 35, 41, 45, 46, 48, 49). PPSs form a complex spatial organization to direct activation and racemization reactions and sequential polymerization of the component amino acids. The homology between FxbB and FxbC to PPSs indicates that M. smegmatis exochelin is synthesized by a nonribosomal peptide biosynthesis pathway. M. smegmatis exochelin has the structure N-(δ-N-formyl–(R)-δ-N-hydroxyornithine-1)–β-alanine–(R)-δ-N-hydroxyornithine-2–(R)-allo- threonine–(S)-δ-N-hydroxyornithine-3 (Fig. 7B). The structure of this is similar to those of pseudobactin, pyoverdin, and azotobactin of Pseudomonas species but unique in that all of the conventional peptide linkages involve R-amino acids. The number of activation modules in PPSs usually corresponds to the number of amino acids in the peptide produced (8–10, 21, 41, 46, 49). By analyzing the amino acid sequences encoded by the fxbB and fxbC genes, we identified six amino acid activation modules: FxbB1, FxbB2, FxbC1, FxbC2, FxbC3, and FxbC4 encoded by these two genes. Each module contains a ATP-binding motif, an ATPase motif, and a pantetheine attachment motif. In addition, there are three epimerase domains within FxbB and FxbC to epimerize the S-amino acids to R-amino acids (Fig. 7A). These three epimerase domains are located in the C termini of FxbB1, FxbC1, and FxbC2 (17). M. smegmatis exochelin is a linear, formylated pentapeptide containing one β-alanine, one (R)-allo-threonine, two (R)-δ-N-hydroxyornithines, one (S)-δ-N-hydroxyornithine, and a formyl group. The FxbB1 and its C-terminal epimerase domain correspond to the first R-amino acid in the exochelin structure, (R)-δ-N-hydroxyornitheine-1; the FxbC1 and FxbC2 and their C-terminal epimerase domains correspond to the third and fourth R-amino acid (R)-δ-N-hydroxyornitheine-2 and (R)-allo-threonine. The number and position of R-amino acids in exochelin correspond with the presence of the three epimerase domains found in FxbB and FxbC. The FxbB2, FxbC3, and FxbC4 modules do not have any epimerase domain in their C termini. We believe that FxbB2 corresponds to the second amino acid β-alanine, while one of the FxbC3 or FxbC4 domains corresponds to the S-δ-N-hydroxyornithine-3. The fxbA gene product is likely to transfer a formyl group to the (R)-δ-N-hydroxyornithine-1 at the N terminus (16). Sequence analysis of the deduced amino acid of the fxbB and fxbC genes shows that there are a total of six amino acid activation domains. However, the final secreted exochelin is a pentapeptide. It is not clear how the final exochelin (pentapeptide) is formed. One possibility is that the hexapeptide is formed, but it is an unstable addition and only the pentapeptide is detected. Alternatively, the sixth module could be inactive. Unfortunately, we were unable to find precedence for either possibility in the PPS literature. This six-domain/pentapeptide discrepancy will require additional experiments to explain.
FIG. 7.
(A) Diagram of the domain organization of the fxbB and fxbC products. White boxes indicate the amino acid activating domains. Each activating domain has an ATP-binding site, an ATPase motif, and a pantetheine motif (Pan- Site) and is separated by space motifs. B1 and B2 are the two amino acid activating domains in FxbB; C1, C2, C3, and C4 are the four amino acid activating domains in FxbC. There is one epimerase domain in FxbB and two epimerase domains in FxbC. The Tn10 insertion site in pYUB908 and mc21608 and the Tn10 insertion site in pYUB909 and mc21609 are indicated by superscript letters a and b, respectively. (B) Covalent structure of M. smegmatis exochelin (40).
There are two putative ABC transporters, possibly in the same operon as the exochelin biosynthesis genes fxbB and fxbC (orf1 and orf2 in Fig. 1). We propose that these genes, due to their homology and location in the exochelin biosynthesis operon, would likely encode proteins involved in transporting the exochelin MS outside of the M. smegmatis cell, and experiments are under way to test this hypothesis.
In this study, we also found that exochelin-deficient mutants had an increased sensitivity to hydrogen peroxide under both high- and low-iron conditions. These results suggest a role for exochelin in the maintenance of internal iron concentrations within the cell and in the regulation of genes turned on during oxidative stress. While the mechanism of increased sensitivity in the exochelin-deficient mutants to H2O2 is unclear, the isolation of suppressor mutants resistant to H2O2 killing might provide a means to elucidate this phenomenon.
Peptide exochelins have so far been identified only in M. smegmatis (40) and Mycobacterium neoaurum (39). It appears unlikely that M. tuberculosis produces peptide exochelins. We have been unable to complement the M. smegmatis exochelin mutants with cosmid genomic libraries of M. tuberculosis or observe hybridization of the fxbA, fxbB, and fxbC genes to M. tuberculosis chromosomal DNA by Southern analysis. Comparison of the sequences of genes in pYUB563 to genes in the databases of the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov) and Sanger Sequence Center (Sanger Center, Cambridge, United Kingdom) (www.sanger.ac.uk) revealed strong homologies of four ORFs in the M. tuberculosis genome to the M. smegmatis fxuD gene, orf3, orf4, and orf5 (Table 2). In contrast, no strong homologies in the M. tuberculosis genome were observed to exochelin biosynthesis genes fxbA, fxbB, and fxbC, the putative ABC transporter genes orf1 and orf2, or the putative ATP-dependent iron permease genes fxuA, fxuB, and fxuC. Taken together, our data and data of a previous study (18) strongly suggest that M. tuberculosis does not produce compounds similar to peptide exochelins of M. smegmatis. However, since M. smegmatis and M. tuberculosis both produce carboxymycobactin and mycobactin (18, 34), our M. smegmatis exochelin mutants represent tractable surrogate organisms for genetic and biochemical analyses of the genes required for carboxymycobactin and mycobactin biosyntheses.
ACKNOWLEDGMENTS
We thank John McKinney for kindly providing his library of 6,000 EMS-mutagenized M. smegmatis clones, Deepti Thomas for helping us to screen the library on CAS plates, Carlos Vaamonde for generously providing the M. smegmatis mc2155/pYUB415 library, and Martin Pavelka, Miriam Braunstein, John McKinney, and Lynn Miesel for helpful discussions. We are indebted to Martin Pavelka, Miriam Braunstein, Benson Yeh, and Sing Sing Way for critical reading of the manuscript.
This work was supported in part by National Institutes of Health grant AI26170.
REFERENCES
- 1.Adams C, Dowling D N, O’Sullivan D J, O’Gara F. Isolation of a gene (pbsC) required for siderophore biosynthesis in fluorescent Pseudomonas sp. strain M114. Mol Gen Genet. 1994;243:515–524. doi: 10.1007/BF00284199. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul S M, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1994. pp. 13.3.1–13.3.4. [Google Scholar]
- 5.Balasubramanian V, Pavelka M S, Jr, Bardarow S S, Martin J, Weisbrod T W, McAdam R A, Bloom B R, Jacobs W R., Jr Allelic exchange in Mycobacterium tuberculosis with long linear recombination substrates. J Bacteriol. 1996;178:273–279. doi: 10.1128/jb.178.1.273-279.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bardarov, S., and W. R. Jacobs, Jr. Unpublished data.
- 7.Blanc V, Gil P, Bamas-Jacques N, Lorenzon S, Zagorec M, Schleuniger J, Bisch D, Blanche F, Debussche L, Crouzet J, Thibaut D. Identification and analysis of genes from Streptomyces pristinaespiralis encoding enzymes involved in the biosynthesis of the 4-dimethylamino-L-phenylalanine precursor of pristinamycin I. Mol Microbiol. 1997;23:191–202. doi: 10.1046/j.1365-2958.1997.2031574.x. [DOI] [PubMed] [Google Scholar]
- 8.Coque J J, Martin J F, Calzada J G, Liras P. The cephamycin biosynthetic genes pcbAB, encoding a large multidomain peptide synthetase, and pcbC of Nocardia lactamdurans are clustered together in an organization different from the same genes in Acremonium chrysogenum and Penicillium chrysogenum. Mol Microbiol. 1991;5:1125–1133. doi: 10.1111/j.1365-2958.1991.tb01885.x. [DOI] [PubMed] [Google Scholar]
- 9.de Crecy-Lagard V, Blanc V, Gil P, Naudin L, Lorenzon S, Famechon A, Bamas-Jacques N, Crouzet J, Thibaut D. Pristinamycin I biosynthesis in Streptomyces pristinaespiralis: molecular characterization of the first two structural peptide synthetase genes. J Bacteriol. 1997;179:705–713. doi: 10.1128/jb.179.3.705-713.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diez B, Gutierrez S, Barredo J L, van Solingen P, van der Voort L H, Martin J F. The cluster of penicillin biosynthetic genes. Identification and characterization of the pcbAB gene encoding the alpha-aminoadipyl-cysteinyl-valine synthetase and linkage to the pcbC and penDE genes. J Biol Chem. 1990;265:16358–16365. [PubMed] [Google Scholar]
- 11.Donnely-Wu M, Jacobs W R, Jr, Hatfull G F. Superinfection immunity of mycobacteriophage L5: applications for genetic transformation of mycobacteria. Mol Microbiol. 1993;7:407–417. doi: 10.1111/j.1365-2958.1993.tb01132.x. [DOI] [PubMed] [Google Scholar]
- 12.Dussurget, O., J. Timm, M. Gomez, S. Yu, S. Z. Sabol, R. K. Holmes, W. R. Jacobs, Jr., and I. Smith. Transcriptional control of the iron-responsive fxbA gene by the mycobacterial regulator IdeR. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 13.Dussurget O, Rodriguez M, Smith I. An ideR mutant of Mycobacterium smegmatis has derepressed siderophore production and an altered oxidative-stress response. Mol Microbiol. 1996;22:535–544. doi: 10.1046/j.1365-2958.1996.1461511.x. [DOI] [PubMed] [Google Scholar]
- 14.Enard C, Diolez A, Expert D. Systemic virulence of Erwinia chrysanthemi 3937 requires a functional iron assimilation system. J Bacteriol. 1988;170:2419–2426. doi: 10.1128/jb.170.6.2419-2426.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fath M J, Kolter R. ABC transporters: bacterial exporters. Microbiol Rev. 1993;57:995–1017. doi: 10.1128/mr.57.4.995-1017.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fiss E H, Yu S, Jacobs W R., Jr Identification of genes involved in the sequestration of iron in mycobacteria: the ferric exochelin biosynthetic and uptake pathways. Mol Microbiol. 1994;14:557–569. doi: 10.1111/j.1365-2958.1994.tb02189.x. [DOI] [PubMed] [Google Scholar]
- 17.Fuma S, Fujishima Y, Corbell N, D’Souza C, Nakano M M, Zuber P, Yamane K. Nucleotide sequence of 5′ portion of srfA that contains the region required for competence establishment in Bacillus subtilus. Nucleic Acids Res. 1993;21:93–97. doi: 10.1093/nar/21.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gobin J, Moore C H, Reeve J R, Wong D K, Gibson B W, Horwitz M A. Iron acquisition by Mycobacterium tuberculosis: isolation and characterization of a family of iron-binding exochelins. Proc Natl Acad Sci USA. 1995;92:5189–5193. doi: 10.1073/pnas.92.11.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall R M, Ratledge C. A simple method for the production of mycobactin, the lipid-soluble siderophore, from mycobacteria. FEMS Microbiol Lett. 1982;15:133–136. [Google Scholar]
- 20.Kleckner N, Bender J, Gottesman S. Uses of transposons with emphasis on Tn10. Methods Enzymol. 1991;204:139–180. doi: 10.1016/0076-6879(91)04009-d. [DOI] [PubMed] [Google Scholar]
- 21.Krätzschmar J, Krause M, Marahiel M A. Gramicidin S biosynthesis operon containing the structural genes grsA and grsB has an open reading frame encoding a protein homologous to fatty acid thioesterases. J Bacteriol. 1989;171:5422–5429. doi: 10.1128/jb.171.10.5422-5429.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lane S J, Marshall P S, Upton R J, Ratledge C, Ewing M. Novel extracellular mycobactins, the carboxymycobactins from Mycobacterium avium. Tetrahedron Lett. 1995;36:4129–4132. [Google Scholar]
- 23.Lewin R. How microorganisms transport iron. Science. 1984;225:401–402. doi: 10.1126/science.225.4660.401. [DOI] [PubMed] [Google Scholar]
- 24.Litwin C M, Calderwood S B. Role of iron in regulation of virulence genes. Clin Microbiol Rev. 1993;6:137–147. doi: 10.1128/cmr.6.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lundrigan M D, Arceneaux J E L, Zhu W, Byers B R. Enhanced hydrogen peroxide sensitivity and altered stress protein expression in iron-starved Mycobacterium smegmatis. Biometals. 1997;10:215–225. doi: 10.1023/a:1018355928990. [DOI] [PubMed] [Google Scholar]
- 26.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 27.McKinney, J. D. Personal communication.
- 28.Merriman T R, Merriman M E, Lamont I L. Nucleotide sequence of pvdD, a pyoverdine biosynthetic gene from Pseudomonas aeruginosa: PvdD has similarity to peptide synthetases. J Bacteriol. 1995;177:252–258. doi: 10.1128/jb.177.1.252-258.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyer J M, Neely A, Stintzi A, Georges C, Holder I A. Pyoverdin is essential for virulence of Pseudomonas aeruginosa. Infect Immun. 1996;64:518–523. doi: 10.1128/iai.64.2.518-523.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakai K, Kanehisa M. Expert system for predicting protein localization sites in Gram-negative bacteria. Proteins Struct Funct Genet. 1991;11:95–110. doi: 10.1002/prot.340110203. [DOI] [PubMed] [Google Scholar]
- 31.Neilands J B. Microbial iron compounds. Annu Rev Biochem. 1981;50:715–731. doi: 10.1146/annurev.bi.50.070181.003435. [DOI] [PubMed] [Google Scholar]
- 32.Neilands J B. Siderophores: structure and function of microbial iron transport compounds. J Biol Chem. 1995;270:26723–26726. doi: 10.1074/jbc.270.45.26723. [DOI] [PubMed] [Google Scholar]
- 33.Payne S M. Iron and virulence in the family Enterobacteriaceae. Crit Rev Microbiol. 1980;16:81–111. doi: 10.3109/10408418809104468. [DOI] [PubMed] [Google Scholar]
- 34.Ratledge C, Ewing M. The occurrence of carboxymycobactin, the siderophore of pathogenic mycobacteria, as a second extracellular siderophore in Mycobacterium smegmatis. Microbiology. 1996;142:2207–2212. doi: 10.1099/13500872-142-8-2207. [DOI] [PubMed] [Google Scholar]
- 35.Rusnak F, Sakaitani M, Drueckhammer D, Reichert J, Walsh C T. Biosynthesis of the Escherichia coli siderophore enterobactin: sequence of the entF gene, expression and purification of EntF, and analysis of covalent phosphopantetheine. Biochemistry. 1991;30:2916–2927. doi: 10.1021/bi00225a027. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 37.Schmitt M P, Holmes R K. Cloning, sequence, and footprint analysis of two promoter/operators from Corynebacterium diphtheriae that are regulated by the Diphtheria toxin repressor (DtxR) and iron. J Bacteriol. 1994;176:1141–1149. doi: 10.1128/jb.176.4.1141-1149.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwyn B, Neilands J B. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 39.Sharman G J, Williams D H, Ewing D F, Ratledge C. Determination of the structure of exochelin MN, the extracellular siderophore from Mycobacterium neoaurum. Chem Biol. 1995;2:553–561. doi: 10.1016/1074-5521(95)90189-2. [DOI] [PubMed] [Google Scholar]
- 40.Sharman G J, Williams D H, Ewing D F, Ratledge C. Isolation, purification and structure of exochelin MS, the extracellular siderophore from Mycobacterium smegmatis. Biochem J. 1995;305:187–196. doi: 10.1042/bj3050187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith D J, Earl A J, Turner G. The multifunctional peptide synthetase performing the first step of penicillin biosynthesis in Penicillium chrysogenum is a 421,073 dalton protein similar to Bacillus brevis peptide antibiotic synthetases. EMBO J. 1990;9:2743–2750. doi: 10.1002/j.1460-2075.1990.tb07461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Snapper S B, Melton R E, Mustafa S, Kieser T, Jacobs W R., Jr Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol. 1990;4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 43.Snow G A. Mycobactins: iron-chelating growth factors from mycobacteria. Bacteriol Rev. 1970;34:99–125. doi: 10.1128/br.34.2.99-125.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tao X, Schiering N, Zeng H-Y, Ringe D, Murphy J R. Iron, DtxR, and the regulation of diphtheria toxin expression. Mol Microbiol. 1994;14:191–197. doi: 10.1111/j.1365-2958.1994.tb01280.x. [DOI] [PubMed] [Google Scholar]
- 45.Tognoni A, Franchi E, Magistrelli C, Colombo E, Cosmina P, Grandi G. A putative new peptide synthase operon in Bacillus subtilis: partial characterization. Microbiology. 1995;141:645–648. doi: 10.1099/13500872-141-3-645. [DOI] [PubMed] [Google Scholar]
- 46.Turgay K, Krause M, Marahiel M A. Four homologous domains in the primary structure of GrsB are related to domains in a superfamily of adenylate-forming enzymes. Mol Microbiol. 1992;6:529–546. doi: 10.1111/j.1365-2958.1992.tb01498.x. [DOI] [PubMed] [Google Scholar]
- 47.Vaamonde, C., and W. R. Jacobs, Jr. Unpublished data.
- 48.van Sinderen D, Galli G, Cosmina P, de Ferra F, Withoff S, Venema G, Grandi G. Characterization of the srfA locus of Bacillus subtilis: only the valine-activating domain of srfA is involved in the establishment of genetic competence. Mol Microbiol. 1993;8:833–841. doi: 10.1111/j.1365-2958.1993.tb01630.x. [DOI] [PubMed] [Google Scholar]
- 49.Weckermann R, Furbab R, Marahiel M A. Complete nucleotide sequence of tycA gene coding the tyrocidine synthetase 1 from Bacillus brevis. Nucleic Acids Res. 1988;16:11841. doi: 10.1093/nar/16.24.11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weinberg E D. Iron and infection. Microbiol Rev. 1978;42:45–66. doi: 10.1128/mr.42.1.45-66.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weinberg E D. The development of awareness of iron withholding defense. Perspect Biol Med. 1993;36:215–221. doi: 10.1353/pbm.1993.0063. [DOI] [PubMed] [Google Scholar]