Abstract
We found that the previously reported SS-B drug-supersusceptible mutant of Salmonella typhimurium (S. Sukupolvi, M. Vaara, I. M. Helander, P. Viljanen, and P. H. Mäkelä, J. Bacteriol. 159:704–712, 1984) had a mutation in the acrAB operon. Comparison of this mutant with its parent strain and with an AcrAB-overproducing strain showed that the activity of the AcrAB efflux pump often produced significant resistance to β-lactam antibiotics in the complete absence of β-lactamase. The effect of AcrAB activity on resistance was more pronounced with agents containing more lipophilic side chains, suggesting that such compounds were better substrates for this pump. This correlation is consistent with the hypothesis that only those molecules that become at least partially partitioned into the lipid bilayer of the cytoplasmic membrane are captured by the AcrAB pump. According to this mechanism, the pump successfully excretes even those β-lactams that fail to traverse the cytoplasmic membrane, because these compounds are likely to become partitioned into the outer leaflet of the bilayer. Even the compounds with lipophilic side chains were shown to penetrate across the outer membrane relatively rapidly, if the pump was inactivated genetically or physiologically. The exclusion of such compounds, exemplified by nafcillin, from cells of the wild-type S. typhimurium was previously interpreted as the result of poor diffusion across the outer membrane (H. Nikaido, Biochim. Biophys. Acta 433:118–132, 1976), but it is now recognized as the consequence of efficient pumping out of entering antibiotics by the active efflux process.
During the last few years it has become increasingly clear that multidrug efflux pumps, especially those containing the resistance nodulation division (RND) family transporters (33), play a major role in producing both the intrinsic and the elevated levels of resistance to a very wide range of noxious compounds in gram-negative bacteria (21, 24, 25). Examples include the AcrAB system of Escherichia coli (19, 20, 22) and the MexAB-OprM system of Pseudomonas aeruginosa (17, 30), which increase in the wild-type cells of these organisms the MICs of agents such as fluoroquinolones, tetracycline, chloramphenicol, novobiocin, and fusidic acid. The former system also contributes significantly to resistance to erythromycin as well as to various dyes and detergents (21). Inactivation of genes coding for components of these systems was also shown to decrease the MICs of some β-lactams. For example, in P. aeruginosa inactivation of mexA decreases the MICs of ceftriaxone, cefoperazone, azlocillin, and carbenicillin by a factor of 8 to 128 (17), and in E. coli the deletion of acrAB decreases the MICs of ampicillin and benzylpenicillin by a factor of 2 to 4 (19).
Although these results suggested that such systems are capable of pumping out β-lactams and thereby making the cells more resistant to these antibiotics, convincing evidence has been difficult to obtain. The presence of chromosomally coded β-lactamases in these bacteria made interpreting data on benzylpenicillin accumulation and MICs quite difficult (16). Furthermore, the observation that some of the β-lactams do not diffuse into the cytoplasm during the time span of our experiments yet appear to be excluded from the cell (16) was rather unexpected for an efflux pump. This increased the skepticism of some workers in the field and led to attempts to determine whether the outer membrane channel alone could carry out the β-lactam efflux from the periplasm (42) or at least determine the specificity of the process (36).
In this study, we used a close relative of E. coli, Salmonella typhimurium, to study the efflux-based resistance to β-lactams. S. typhimurium does not contain the structural gene for β-lactamase (2) and is therefore a much simpler system to study. Earlier, a mutant of the S. typhimurium LT2 line, called SS-B, was isolated and shown to map in the region of the chromosome where acrAB is located (39). We show that this strain is indeed mutated in the salmonella homolog of acrAB and use it to demonstrate the function of the AcrAB system in pumping out various β-lactam compounds. Furthermore, we investigated the efflux of various β-lactam compounds to examine the structural requirements for substrates of AcrAB pump and propose a mechanism that would enable the cells to pump out various drugs from the cytoplasm and periplasm and would explain the surprisingly wide substrate specificity of the pump.
MATERIALS AND METHODS
Bacterial strains and growth media.
S. typhimurium SH5014 (LT2 rfaJ4041 ilv-1178 thr-914 his-6116 metA22 metE511 trpB2 xyl-404 H1-b H2-e,n,x flaA66 rpsL120 cured of Fels2) and its SS-B-type hypersusceptible mutant SH7616 have been described previously (39). A P1-sensitive derivative of SH7616, HN1003, was selected from SH7616 by first isolating a rare P1cml clr 100(ts) lysogen on a chloramphenicol-containing Luria-Bertani (LB) plate (see below) and then curing the phage by growth at 37°C, as described by Goldberg et al. (9). S. typhimurium TN716 (apt apeB galE) was obtained from the Salmonella genetics stock center, University of Calgary, Calgary, Canada. HN891 was a single-step multidrug-resistant mutant of SH5014, obtained as a colony growing on an LB plate containing 8 μg of chloramphenicol per ml inoculated with approximately 108 cells of unmutagenized SH5014.
LB broth (10 g of Bacto Tryptone, 10 g of Bacto Yeast Extract, and 5 g of NaCl per liter) and M63 minimal medium with 0.2% glucose as a carbon source (40) were used as growth media, with 1.5% Bacto Agar added when plates were made. Cultures were grown at 37°C [except in the transduction procedures with P1 clr 100(ts)], and liquid cultures were aerated by shaking.
Chemicals.
Most antibiotics and other inhibitors were obtained from Sigma. Latamoxef and penicillin N were gifts of Shionogi & Co. Ltd (Osaka, Japan) and Wyeth-Ayerst Laboratories (Philadelphia, Pa.).
Determination of MICs.
MICs were determined by twofold serial broth dilution in LB broth. The inoculum was 5 × 104 cells/ml, and the results were read after overnight incubation at 37°C.
Immunoblot analysis.
Whole-cell proteins were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and the protein bands were transferred to a nitrocellulose membrane which was stained with a mixture of nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate after the application of primary rabbit antibody and the alkaline-phosphatase-conjugated goat anti-rabbit-immunoglobulin G antibody (Bio-Rad) essentially as described earlier (5). The anti-AcrA antibody was generated in rabbits by using as an immunogen the AcrA tagged with hexahistidine at its C terminus, which had been purified by nickel column chromatography (44).
Assay of β-lactamase.
Cells were disintegrated in a Gallenkamp Soniprep 150 sonicator with four cycles of 30-s pulses each, and enzyme activity was assayed spectrophotometrically at 260 nm with 100 μM cephaloridine as the substrate (26).
Assay of outer membrane permeability.
This assay was carried out by one of two methods. By the first method, the influx of the β-lactams across the outer membrane caused by spontaneous diffusion was coupled with subsequent hydrolysis by periplasmic β-lactamase as described earlier (26). Plasmid pBR322, coding for type A β-lactamase, was introduced by transformation (34). Cells were grown in LB broth to mid-exponential phase, harvested, washed, and resuspended in an Mg-containing phosphate buffer as described earlier (26). The hydrolysis of cephalosporins was followed continuously at 260 nm with 1 mM substrate and a cell with a 1-mm light path as described previously (26). By the second method, the entry of the drugs across the outer membrane and then the cytoplasmic membrane was followed by measurement of the time-dependent decrease of drug concentration in the external medium in suspensions containing high concentrations of cells (23). Briefly, exponential-phase cultures in LB were centrifuged, and cells washed once with M63 containing 0.5% (wt/vol) glucose were suspended in the same medium at a concentration of about 0.47 g (wet weight)/ml. The suspensions were kept at room temperature (24°C) for about 5 min. Nafcillin (0.3 ml of a 25-mg/ml solution) was then added to 2.1 ml of cellular suspension at time 0, and portions of the suspensions were removed and placed at 1-min intervals into heavy-walled glass centrifuge tubes prechilled in an ice bath to stop further diffusion of nafcillin through the lipid bilayer regions of the membranes. After centrifugation, the supernatants were diluted 400-fold, and nafcillin concentration was determined from optical density at 227 nm (OD227). The data analysis was performed as described in reference 23.
RESULTS
Transductional mapping of SS-B mutation.
In the original description, conjugational mapping showed the SS-B mutation to be located in the region covering 8 to 12 min of the S. typhimurium chromosome (39). Although E. coli is known to contain three other homologs of acrAB (20), this region of the chromosome contains only acrAB among these homologs. Nevertheless, in order to further confirm the identity of the SS-B mutation, the cotransduction of the apt marker from the donor TN716 into a P1-sensitive derivative of SH7616, HN 1003, was tested. The apt gene is located only 0.14 min away from the acrAB cluster in E. coli (3). Since the arrangement of the mapped genes (such as lon, hupB, apt, and adk) is similar on the chromosomes of E. coli and S. typhimurium in the 9-to-12-min region (3, 35), we expected that acrAB of S. typhimurium would also be located close to the apt gene. When transductants containing the donor acr+ allele were selected on LB agar plates containing 100 μg of cloxacillin (an excellent selective agent for acr+, see below) per ml, 29 transductants out of 40 tested (73%) were shown to have coinherited the mutant apt allele from the donor, as they grew in M63-glucose medium containing 50 μg of an adenine analog, 2,6-diaminopurine (13), per ml (in addition to the required amino acids). Growth of the recipient strain, containing the wild-type apt+ allele, was inhibited even by 2 μg of 2,6-diaminopurine per ml under these conditions. This cotransduction frequency is similar to the predicted frequency (81%) for two markers that are 0.14 min away from each other (43) and confirms that SS-B corresponds to the acrAB homolog of S. typhimurium.
Isolation of a multidrug-resistant mutant, HN891.
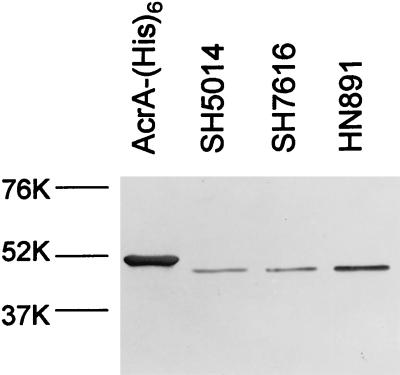
HN891 was isolated as a spontaneous chloramphenicol-resistant mutant of SH5014 (see Materials and Methods). This strain turned out to be more resistant to a number of antibiotics than the parent strain (Table 1). An examination of the level of AcrA protein by immunoblotting indicated that the strain produced significantly more AcrA than the parent (Fig. 1). (The acr mutant SH7616 produced an apparently normal amount of AcrA of normal size. The mutation therefore may be a missense mutation in acrA or may lie in the acrB gene.) It appears likely, therefore, that HN891 is multidrug resistant owing to the increased production of the AcrAB efflux pump, reminiscent of the situation already encountered in E. coli (28). Although the nature of the mutation is not known, it may be in marR, as efforts to isolate mutants of E. coli with a similar phenotype usually select for marR mutants (8).
TABLE 1.
MICs (milligrams per liter) of various antimicrobial agents for HN891 (an overproducer of the Acr pump), SH5014 (parent strain), and SH7616 (an acr mutant)
| Antimicrobial agent | MIC for indicated strain
|
||
|---|---|---|---|
| HN891 | SH5014 | SH7616 | |
| Sodium dodecyl sulfate | >16,000 | >16,000 | 64 |
| Sodium deoxycholate | >16,000 | >16,000 | 1,024 |
| Sodium cholate | >64,000 | >64,000 | 1,024 |
| Triton X-100 | >64,000 | >64,000 | 256 |
| Crystal violet | >256 | >256 | 1 |
| Acriflavine | >128 | >128 | 32 |
| Fusidic acid | >512 | 512 | 4 |
| Novobiocin | >256 | 64 | 1 |
| Erythromycin A | 1,024 | 512 | 8 |
| Rifampin | 16 | 16 | 8 |
| Tetracycline | 2 | 1 | 0.25 |
| Chloramphenicol | 8 | 4 | 0.25 |
| Norfloxacin | 1 | 0.24 | 0.03 |
| Nalidixic acid | 8 | 4 | 0.5 |
| Penicillin G | 32 | 8 | 0.25 |
FIG. 1.
Immunoblot analysis of AcrA proteins. Purified AcrA (containing hexahistidine tag; see Materials and Methods) as well as identical amounts (1.5 μg of total protein each) of sonicates of bacterial cells were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and the proteins were detected by immunoblotting with anti-AcrA antibody as described in Materials and Methods. At left, positions of prestained molecular size standards (Bio-Rad) are shown. The purified AcrA migrates slightly more slowly because of the hexahistidine tag. Scanning showed that the level of AcrA in HN891 was more than twofold higher than that in SH5014 or SH7616.
Phenotypes of the acr mutant SH7616 and the Acr overproducer HN891.
As shown in Table 1, the acr mutant SH7616 was more susceptible to a wide variety of compounds, including detergents, dyes, and many antibiotics, confirming the earlier results obtained with a more limited range of agents (39). Among antibiotics other than β-lactams, SH7616 showed unaltered susceptibility only to aminoglycosides such as kanamycin and gentamicin and to bacitracin (data not shown). This hypersusceptibility phenotype is very similar to that of the E. coli acrAB mutants (19, 20).
The AcrAB overproducer HN891, in contrast, was more resistant to fusidic acid, novobiocin, tetracycline, norfloxacin, nalidixic acid, and penicillin G (Table 1) in addition to chloramphenicol, which was used for its selection. This multidrug-resistant phenotype is precisely what is expected of a strain producing higher levels of AcrAB-type, multidrug efflux pump(s) (1, 28).
S. typhimurium was reported to lack the structural gene coding for the class C β-lactamase, known to be present in most other species of the family Enterobacteriaceae (2). A β-lactamase assay with cephaloridine as the substrate showed that the activity was <60 pmol mg of protein−1 min−1 in both SH5014 and SH7616. This activity is less than 1% of the activity in the E. coli K-12 strain (27), which, in turn, is less than 1/1,000 of the activity seen in fully induced cells of Enterobacter cloacae (7, 41). We could thus confirm the absence of any detectable β-lactamase activity in SH5014 as well as SH7616 cells, yet the presence and the overproduction, respectively, of the AcrAB pump in SH5014 and HN891 significantly increased the MICs of penicillin (Table 1) and other β-lactams (Table 2) over that of SH7616, clearly indicating that the AcrAB system does pump out these compounds at significant rates.
TABLE 2.
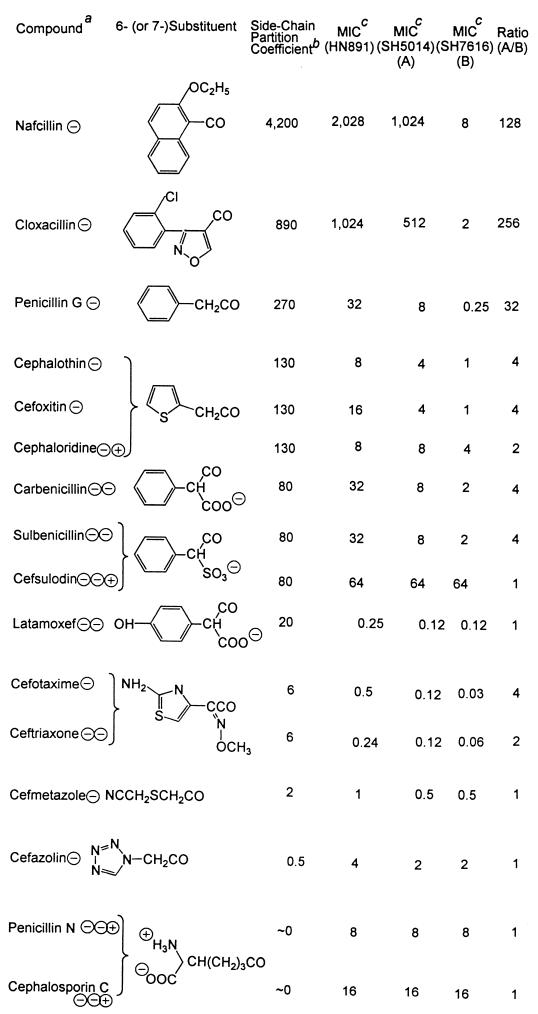
Lipophilicities of the side chains and the MICs of penicillins and cephalosporins
The electrical charges present on each compound at a neutral pH are shown after the name of the compound.
b This column shows the calculated octanol-water partition coefficients of the 6-substituents (in penicillins) and the 7-substituents (in cephalosporins). The terminal carbonyl group was excluded from calculations of partition coefficients. When charged groups were present close to the carbonyl group, as in carbenicillin, sulbenicillin, and cefsulodin, partition coefficients were calculated only on the part of the substituent that was farther away from the charged group; that is, calculation included only the phenyl group. Calculation was performed as follows. First, the logarithm of the partition coefficient (logP) was calculated by the fragment approach (31), based on the measured octanol-water partition coefficient of various compounds (10). For example, the calculation of the logP value of the benzyl group in penicillin G is 2.13 (logP of benzene) − 0.225 (fragmental constant for H) + 0.528 (fragmental constant for −CH2−) = 2.43. The logP value of the side chain for cefotaxime and ceftriaxone was calculated by starting from the measured logP value for O-methylbenzaldoxime (2.53) and comparing the logP value of benzene (2.13) to that of 2-aminothiazole (0.38). Partition coefficients were then calculated from logP values.
MICs are in milligrams per liter.
Entry of nafcillin.
To demonstrate more directly that the intracellular accumulation of β-lactams is affected by AcrAB activity, we first attempted to examine the entry of [3H]benzylpenicillin by silicone oil centrifugation assay (16). However, the R-type lipopolysaccharide produced by these strains (note the rfaJ4041 mutation present in SH5014; Materials and Methods) caused strong aggregation of cells, which resulted in a poor and nonreproducible recovery of the cells at the bottom of the tubes.
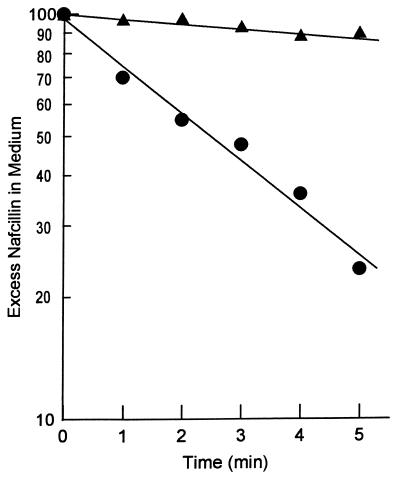
We therefore used an alternative assay and incubated a dense suspension of intact cells (about 0.5 g [wet weight]/ml [see reference 23]) with nafcillin, a strongly hydrophobic penicillin. Under these conditions, we can observe the time-dependent decrease in the external concentration of nafcillin as it diffuses into first the periplasm and then the cytoplasm of the cells (23). As shown in Fig. 2, nafcillin entered the acr mutant SH7616 relatively rapidly but was not accumulated by the cells of wild-type parent strain SH5014. Since the only difference between the two strains was presumably the activity of the AcrAB pump, this result suggests that the nonaccumulation of nafcillin in wild-type S. typhimurium was due to the active efflux of the drug and that nafcillin is a good substrate of AcrAB. It should also be noted that Fig. 2 plots the excess nafcillin concentration, i.e., the drug concentration in the medium minus the expected equilibrium drug concentration when the intracellular concentration becomes equal to the external concentration. The exponential decrease of this excess nafcillin concentration in the SH7616 experiment suggests that the intracellular concentration is indeed approaching the expected equilibrium concentration and that there is no active accumulation or active extrusion of the drug by SH7616.
FIG. 2.
Entry of nafcillin into whole cells of S. typhimurium. Time course of entry of nafcillin at room temperature (24°C) was followed by the use of thick suspensions of the exponential-phase cells of the wild-type strain (SH5014) (triangles) and its acr mutant (SH7616) (circles) as described in Materials and Methods. Excess nafcillin in medium was calculated by subtracting the calculated nafcillin concentration at complete equilibrium (C∞ in reference 23) from the measured nafcillin concentration in the external medium. The result was expressed as a percentage of the initial nafcillin concentration in the medium.
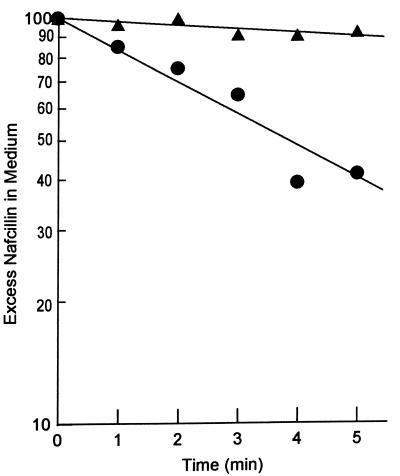
To confirm that the lack of nafcillin accumulation in the wild-type strain SH5014 was due to active efflux, the SH5014 cells were incubated in the presence and absence of a proton conductor, carbonylcyanide m-chlorophenylhydrazone (CCCP) (Fig. 3). When the cytoplasmic membrane was deenergized with CCCP and the proton-motive-force-dependent AcrAB pump ceased to function, a rapid entry of nafcillin occurred. Since CCCP is unlikely to affect the permeability of the outer membrane, these results show that the absence of net entry of nafcillin into wild-type S. typhimurium cells (Fig. 2 and reference 23) resulted from the activity of the AcrAB efflux pump working synergistically (25) with the retardation of drug entry by the outer membrane.
FIG. 3.
Entry of nafcillin into energized and deenergized cells of SH5014. Exponential-phase cells of the wild-type strain SH5014 were used in an experiment similar to that shown in Fig. 1 except that the cell suspension was divided into two equal portions, and CCCP (final concentration, 100 μM) was added to one portion. Triangles, cells without CCCP; circles, cells treated with CCCP.
MICs of various β-lactam agents in strains with different levels of AcrAB pump activity.
In an effort to gain knowledge about the substrate specificity of the AcrAB pump, we determined the MICs of various β-lactam compounds in the acr mutant SH7616, the parent SH5014, and the AcrAB overexpressor HN891 (Table 2). For compounds containing lipophilic side chains, such as nafcillin and cloxacillin, the acr mutation in SH7616 lowered the MIC by a factor of more than 100, suggesting that they are good substrates of the AcrAB pump. Furthermore, MICs of these more lipophilic compounds became even higher in HN891 than in SH5014. On the other hand, MICs of compounds containing hydrophilic side chains, such as penicillin N and cefazolin, were affected little by the presence or absence or the overproduction of the functional AcrAB system. We will evaluate in the Discussion section the correlation between the lipophilicity of the side chains and the magnitude of the change in MIC upon the introduction of the acr mutation.
Entry rates of several cephalosporins.
MICs will be affected not only by the rate of excretion of various compounds but also by their rate of entry through the outer membrane. We therefore determined the rate of diffusion of three cephalosporins covering a wide range of lipophilicity, cephalothin, cefazolin, and cephalosporin C, by measuring the rate of hydrolysis of these compounds with intact cells of SH7616 and SH5014 containing pBR322, using a method established earlier (26). The outer membrane of SH7616 had permeability coefficients of 1.3 × 10−5, 5.0 × 10−5, and 1.5 × 10−5 cm s−1, respectively, for the three compounds mentioned above. With SH5014, the permeability coefficients obtained were slightly lower, by about 30% on average, than the values with SH7616; this result may be due to some efflux of these compounds from the periplasm through the AcrAB system.
DISCUSSION
In this study, we first showed that an SS-B mutant of S. typhimurium, SH7616, had a mutation in the homolog of acrAB of E. coli, based on its phenotype as well as the high frequency of cotransduction observed with the apt marker. This conclusion is consistent with results from other laboratories that were reported during the course of this study. Thus, the S. typhimurium acrB gene was identified by partial sequencing of DNA (15), and insertional inactivation of this gene produced a drug sensitivity phenotype very similar to that of SH7616 (reference 14 and Table 1). Mapping by another laboratory also confirmed the location of acrB on the Salmonella chromosome at a position corresponding to that of the E. coli gene. Thus, a TnphoA insertion into a locus within a few minutes of purE (mutant SM7) was shown to make Salmonella enteritidis noninvasive in tissue culture cells (37), presumably because of its hypersensitivity to detergents (see reference 38). Sequencing of DNA from the insertion site showed that the sequence was similar to that of the E. coli acrB gene (12).
Once nafcillin reaches the periplasm after the rate-limiting diffusion across the outer membrane, it crosses the cytoplasmic membrane rapidly, owing to its high lipophilicity (23, 29); this feature makes the nafcillin accumulation assay much easier to perform than assays with some β-lactam compounds that enter only into the periplasm. The nafcillin assay showed that the drug entered into the mutant SH7616 cells much more rapidly than into the wild-type parent cells, suggesting that the hypersusceptibility to β-lactams in SH7616 is caused by either a higher permeability of its outer membrane or the lack of active efflux of these compounds. The former possibility can be ruled out because (a) the inhibition of efflux by deenergization with CCCP also allowed the rapid influx of nafcillin into SH5014 (Fig. 3) and (b) the direct assay of outer membrane permeability with both moderately lipophilic (cephalothin) and hydrophilic (cefazolin and cephalosporin C) cephalosporins showed that there is only a small apparent difference between the two strains (see Results).
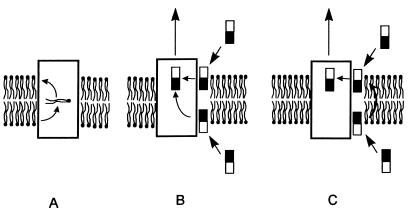
Some of the β-lactam compounds did not enter the cytoplasm during our experimental time period (16), and it was unexpected that active efflux mechanism would be able to capture its substrates without their entry into the cytoplasm. However, similar observations have been made repeatedly with the MDR pump of mammalian cells. Thus, Homolya et al. (11) showed that 3T3 cells expressing the MDR protein capture and exclude the neutral pentaester of a dye, FURA-2, from within the membrane bilayer. Furthermore, Ruetz and Gros (32) showed that mouse MDR2, a homolog of MDR, is a phosphatidylcholine flippase that translocates the lipid molecule from the inner leaflet to the outer leaflet (Fig. 4A). Thus, both MDR and MDR2 capture substrates from within the bilayer.
FIG. 4.
Hypothetical mechanisms of the efflux pumps. (A) The mouse MDR2 protein flips over the phosphatidylcholine from the inner leaflet to the outer leaflet, as shown by Ruetz and Gros (32). (B) The AcrB pump captures the substrates from both the inner and outer leaflets of the membrane bilayer. In this manner, any lipophilic or amphiphilic nonphospholipid molecule that becomes inserted spontaneously into the bilayer can be captured and pumped out. In this mechanism, the AcrB protein is assumed to have a flippase-like activity so that it can excrete drugs from the cytosol (below the bilayer in the figure) as well as from the periplasm (above the bilayer). (C) The AcrB protein is seen to be capable of capturing substrates only from the outer leaflet. In this case, the substrate in the inner leaflet, originally coming from the cytosol, must be flipped over (possibly spontaneously) to the outer leaflet before being captured by the pump protein.
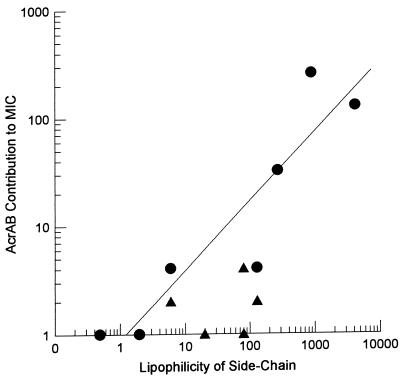
Although mammalian MDR and bacterial RND pumps have no sequence homology and are energized in different ways, they may have a similar mechanism for substrate capture. We hypothesize that the RND pump binds substrates that have already been partitioned into the outer leaflet of the membrane bilayer and that even β-lactam compounds that do not penetrate into the cytoplasm can be pumped out efficiently in this manner (Fig. 4B and C). (A similar mechanism involving the capture of substrates from within the bilayer was independently proposed for a multidrug efflux pump of a totally different type [4]; in this case, the capture appears to occur exclusively from the inner leaflet of the bilayer.) This model also explains the unusually wide specificity of the AcrAB system. The major requirement for any drug to become a substrate of this pump is an efficient, at least partial, insertion into the membrane bilayer (25), and this is precisely what was shown by the correlation between the side-chain lipophilicity of the β-lactams and their ability to become substrates of the AcrAB efflux pump. Thus, those compounds with strongly lipophilic side chains that are likely to partition spontaneously into the bilayer, such as nafcillin and cloxacillin, appear to be good substrates of the pump because the parent strain producing the functional AcrAB system was far more resistant to them than the acr mutant SH7616 (Table 2 and Fig. 5). In contrast, compounds with hydrophilic side chains, such as penicillin N, cefazolin, or cefmetazole, were apparently not pumped out efficiently, as judged by the similar MICs shown by the AcrAB overproducer, the parent strain, and the acr mutant (Table 2 and Fig. 5). Thus, our hypothesis predicts that the outer membrane channel alone would obviously be incapable of pumping out drug molecules from the periplasm, a prediction confirmed by recent studies with P. aeruginosa (36, 42). It must be pointed out, however, that the observed correlation (Fig. 5) can also be explained by assuming a highly lipophilic substrate-binding site within the transporter protein but not that the substrates first enter the lipid bilayer. This hypothesis is not attractive because it forces the assumption of two substrate-binding sites, one facing the periplasmic space and the other facing the cytosol.
FIG. 5.
Correlation between the lipophilicity of the 6- (or 7-) substituent group and the MIC increase caused by the AcrAB pump. The former parameter is expressed as the calculated octanol-water partition coefficient of the side-chain group as detailed in the footnotes to Table 2. The latter was calculated as the MIC for the wild type (SH5014) divided by the MIC for the acr mutant (SH7616) and is presented as a ratio (A/B) in Table 2. Circles, data on monoanionic compounds; triangles, data on compounds carrying multiple charges.
An examination of Table 2 shows that the correlation between lipophilicity and efflux is not perfect. The discrepancy is most prominent with cefsulodin, which showed equal but low activity on the three strains used. Possibly the three charges on this compound hinder even a partial partitioning of the molecule into bilayer. Indeed, the presence of the AcrAB pump has a much weaker effect on the efficacy of compounds with more than one charged group (Fig. 5, triangles). However, this does not alter our conclusion on the correlation of side-chain hydrophobicity and the effectiveness of the efflux process, as monoanionic compounds containing hydrophilic side chains, such as cefazolin and cefmetazole, were apparently poor substrates of the AcrAB pump (Table 2).
The final proof, however, of the correlation between side-chain lipophilicity and susceptibility to efflux requires the elimination of another interpretation of the data of Table 2, that the more hydrophilic compounds may not be affected significantly by the presence or absence of the AcrAB pump simply because they can cross the outer membrane much more rapidly through porin channels than the hydrophobic compounds (see reference 26). However, one of the most lipophilic compounds tested, nafcillin, appears to penetrate the outer membrane with a permeability coefficient in the range of 2 × 10−5 cm/s at 37°C. (This result was estimated from the measured half-equilibration time of 2 to 3 min [Fig. 2 and 3] at 24 to 26°C and from the high-temperature coefficient of the nafcillin penetration rate [23].) This is comparable to the coefficients for hydrophilic β-lactam molecule permeability through E. coli porin channels (26) and to those for the permeability of moderately lipophilic and hydrophilic cephalosporins determined in the S. typhimurium strains in this study, that is, between 1.3 and 5 × 10−5 cm/s (see Results). It is thus clear that overall permeation rates across the outer membrane differed relatively little (less than fourfold) among various β-lactam compounds tested (although the more lipophilic compounds must have used the lipid bilayer pathway more than the porin channels for diffusion). This observation, then, rules out the alternative interpretation described above, which is also contradicted by the finding that the presence of AcrAB had no effect on the MIC of cephalosporin C (Table 2), whose permeability coefficient was one of the lowest among those examined (see Results).
The results presented show that active efflux has a strong influence on the efficacy and MICs of more lipophilic β-lactams. Earlier, one of the present authors found that considerations of only the permeability barrier and the kinetic parameters of periplasmic β-lactamase allowed the prediction of the efficacy of various β-lactam compounds in E. coli (27), and this was confirmed by another laboratory (6). In retrospect, these predictions were successful probably because most of the compounds used were those with hydrophilic side chains, which were reasonably effective against the wild-type E. coli. These compounds are now known to be poor substrates of the pump. We now realize that a more complete and accurate prediction of MICs, especially those of the more lipophilic compounds, will require consideration of the active efflux. Furthermore, if an organism has an outer membrane of unusually low permeability or expresses efflux pump(s) more strongly, models that do not include efflux processes will be less useful. As shown by Livermore and Davy (18), this is precisely what happens with P. aeruginosa, whose outer membrane has low permeability.
ACKNOWLEDGMENTS
This study was supported in part by a research grant from the U.S. Public Health Service, AI-09644.
We thank M. Vaara, K. E. Sanderson, and V. L. Miller for bacterial strains and for sharing unpublished information. We also thank W. Pan for his contribution in the early stage of this work.
REFERENCES
- 1.Alekshun M N, Levy S B. Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob Agents Chemother. 1997;41:2067–2075. doi: 10.1128/aac.41.10.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergström S, Lindberg F P, Olsson O, Normark S. Comparison of the overlapping frd and ampC operons of Escherichia coli with the corresponding DNA sequences in other gram-negative bacteria. J Bacteriol. 1983;155:1297–1305. doi: 10.1128/jb.155.3.1297-1305.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berlyn M K B, Low K B, Rudd K E. Linkage map of Escherichia coli K-12, edition 9. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1715–1902. [Google Scholar]
- 4.Bolhuis H, van Veen H W, Molenaar D, Poolman B, Driessen A J M, Konings W N. Multidrug resistance in Lactococcus lacticus: evidence for ATP-dependent drug extrusion from the inner leaflet of the cytoplasmic membrane. EMBO J. 1996;15:4239–4245. [PMC free article] [PubMed] [Google Scholar]
- 5.Davidson A L, Nikaido H. Purification and characterization of the membrane-associated components of the maltose transport system from Escherichia coli. J Biol Chem. 1991;266:8946–8951. [PubMed] [Google Scholar]
- 6.Frère J-M. Quantitative relationship between sensitivity to β-lactam antibiotics and β-lactamase production in Gram-negative bacteria. I. Steady-state treatment. Biochem Pharmacol. 1989;38:1415–1426. doi: 10.1016/0006-2952(89)90180-9. [DOI] [PubMed] [Google Scholar]
- 7.Galleni M, Amicosante G, Frère J-M. Survey of kinetic parameters of class C β-lactamases. Cephalosporins and other β-lactam compounds. Biochem J. 1988;255:123–129. doi: 10.1042/bj2550123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.George A M, Levy S B. Genes in major cotransductional gap of Escherichia coli linkage map required for the expression of chromosomal resistance to tetracycline and other antibiotics. J Bacteriol. 1983;155:541–548. doi: 10.1128/jb.155.2.541-548.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldberg R B, Bender R A, Streicher S L. Direct selection for P1-sensitive mutants of enteric bacteria. J Bacteriol. 1974;118:810–814. doi: 10.1128/jb.118.3.810-814.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hansch C, Leo A, Hoekman D. Exploring QSAR: hydrophobic, electronic, and steric constants. Washington, D.C: American Chemical Society; 1995. [Google Scholar]
- 11.Homolya L, Hollo Z, Germann U A, Pastan I, Gottesman M M, Sarkadi B. Fluorescent cellular indicators are extruded by the multidrug resistance protein. J Biol Chem. 1993;268:21493–21496. [PubMed] [Google Scholar]
- 12.Hong, K. H., and V. L. Miller. Personal communication.
- 13.Kalle G P, Gots J S. Alterations in purine nucleotide pyrophosphorylases and resistance to purine analogs. Biochim Biophys Acta. 1961;53:166–173. doi: 10.1016/0006-3002(61)90803-4. [DOI] [PubMed] [Google Scholar]
- 14.Lacroix F J C, Avoyne C, Pinault C, Popoff M Y, Pardon P. Salmonella typhimurium TnphoA mutants with increased sensitivity to biological and chemical detergents. Res Microbiol. 1995;146:659–670. doi: 10.1016/0923-2508(96)81063-1. [DOI] [PubMed] [Google Scholar]
- 15.Lacroix F J C, Cloeckaert A, Grepinet O, Pinault C, Popoff M Y, Waxin H, Pardon P. Salmonella typhimurium acrB-like gene: identification and role in resistance to biliary salts and detergents and in murine infection. FEMS Microbiol Lett. 1996;135:161–167. doi: 10.1111/j.1574-6968.1996.tb07983.x. [DOI] [PubMed] [Google Scholar]
- 16.Li X-Z, Ma D, Livermore D M, Nikaido H. Role of efflux pump(s) in intrinsic resistance of Pseudomonas aeruginosa: active efflux as a contributing factor to β-lactam resistance. Antimicrob Agents Chemother. 1994;38:1742–1752. doi: 10.1128/aac.38.8.1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X-Z, Nikaido H, Poole K. Role of MexA-MexB-OprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:1948–1953. doi: 10.1128/aac.39.9.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livermore D M, Davy K W M. Invalidity for Pseudomonas aeruginosa of an accepted model of bacterial permeability to antibiotics. Antimicrob Agents Chemother. 1991;35:916–921. doi: 10.1128/aac.35.5.916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma D, Cook D N, Alberti M, Pon N G, Nikaido H, Hearst J E. Molecular cloning and characterization of acrA and acrE genes of Escherichia coli. J Bacteriol. 1993;175:6299–6313. doi: 10.1128/jb.175.19.6299-6313.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma D, Cook D N, Alberti M, Pon N G, Nikaido H, Hearst J E. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol Microbiol. 1995;16:45–55. doi: 10.1111/j.1365-2958.1995.tb02390.x. [DOI] [PubMed] [Google Scholar]
- 21.Ma D, Cook D N, Hearst J E, Nikaido H. Efflux pumps and drug resistance in Gram-negative bacteria. Trends Microbiol. 1994;2:489–493. doi: 10.1016/0966-842x(94)90654-8. [DOI] [PubMed] [Google Scholar]
- 22.Nakamura H. Acriflavine-binding capacity of Escherichia coli in relation to acriflavine sensitivity and metabolic activity. J Bacteriol. 1966;92:1447–1452. doi: 10.1128/jb.92.5.1447-1452.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nikaido H. Outer membrane of Salmonella typhimurium. Transmembrane diffusion of some hydrophobic substances. Biochim Biophys Acta. 1976;433:118–132. doi: 10.1016/0005-2736(76)90182-6. [DOI] [PubMed] [Google Scholar]
- 24.Nikaido H. Prevention of drug access to bacterial targets: role of permeability barriers and active efflux. Science. 1994;264:382–388. doi: 10.1126/science.8153625. [DOI] [PubMed] [Google Scholar]
- 25.Nikaido H. Multidrug efflux pumps of gram-negative bacteria. J Bacteriol. 1996;178:5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nikaido H, Foulds J, Rosenberg E Y. Porin channels in Escherichia coli. Studies with β-lactams in intact cells. J Bacteriol. 1983;153:232–240. doi: 10.1128/jb.153.1.232-240.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nikaido H, Normark S. Sensitivity of Escherichia coli to various β-lactams is determined by the interplay of outer membrane permeability and degradation by periplasmic β-lactamases: a quantitative predictive treatment. Mol Microbiol. 1987;1:29–36. doi: 10.1111/j.1365-2958.1987.tb00523.x. [DOI] [PubMed] [Google Scholar]
- 28.Okusu H, Ma D, Nikaido H. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J Bacteriol. 1996;178:306–308. doi: 10.1128/jb.178.1.306-308.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Plésiat P, Nikaido H. Outer membranes of Gram-negative bacteria are permeable to steroid probes. Mol Microbiol. 1992;6:1323–1333. doi: 10.1111/j.1365-2958.1992.tb00853.x. [DOI] [PubMed] [Google Scholar]
- 30.Poole K, Krebes K, McNally C, Neshat S. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J Bacteriol. 1993;175:7363–7372. doi: 10.1128/jb.175.22.7363-7372.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rekker R F. The hydrophobic fragmental constant. Its derivation and application, a means of characterizing membrane systems. Amsterdam, The Netherlands: Elsevier Scientific Publishing Co.; 1977. [Google Scholar]
- 32.Ruetz S, Gros P. A phospholipid translocase: a physiological role for the mdr2 gene. Cell. 1994;77:1071–1081. doi: 10.1016/0092-8674(94)90446-4. [DOI] [PubMed] [Google Scholar]
- 33.Saier M H, Jr, Tam R, Reizer A, Reizer J. Two novel families of bacterial membrane proteins concerned with nodulation, cell division and transport. Mol Microbiol. 1994;11:841–847. doi: 10.1111/j.1365-2958.1994.tb00362.x. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Sanderson K E, Hessel A, Liu S-L, Rudd K E. The genetic map of Salmonella typhimurium, edition VIII. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1903–1999. [Google Scholar]
- 36.Srikumar R, Li X-Z, Poole K. Inner membrane efflux components are responsible for β-lactam specificity of multidrug efflux pumps in Pseudomonas aeruginosa. J Bacteriol. 1997;179:7875–7881. doi: 10.1128/jb.179.24.7875-7881.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stone B J, Garcia C M, Badger J L, Hassett T, Smith R I F, Miller V L. Identification of novel loci affecting entry of Salmonella enteritidis into eukaryotic cells. J Bacteriol. 1992;174:3945–3952. doi: 10.1128/jb.174.12.3945-3952.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stone B J, Miller V L. Salmonella enteritidis has a homologue of tolC that is required for virulence in BALB/c mice. Mol Microbiol. 1995;17:701–712. doi: 10.1111/j.1365-2958.1995.mmi_17040701.x. [DOI] [PubMed] [Google Scholar]
- 39.Sukupolvi S, Vaara M, Helander I M, Viljanen P, Mäkelä P H. New Salmonella typhimurium mutants with altered outer membrane permeability. J Bacteriol. 1984;159:704–712. doi: 10.1128/jb.159.2.704-712.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thanassi D G, Cheng L W, Nikaido H. Active efflux of bile salts by Escherichia coli. J Bacteriol. 1997;179:2512–2518. doi: 10.1128/jb.179.8.2512-2518.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vu H, Nikaido H. Role of β-lactam hydrolysis in the mechanism of resistance of a β-lactamase-constitutive Enterobacter cloacae strain to expanded spectrum β-lactams. Antimicrob Agents Chemother. 1985;27:393–398. doi: 10.1128/aac.27.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wong K K Y, Poole K, Gotoh N, Hancock R E W. Influence of OprM expression on multiple antibiotic resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1997;41:2009–2012. doi: 10.1128/aac.41.9.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu T T. A model for three-point analysis of random general transduction. Genetics. 1966;54:405–410. doi: 10.1093/genetics/54.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zgurskaya, H., E. R. Rosenberg, and H. Nikaido. Unpublished results.