Abstract
Haemophilus influenzae is a ubiquitous colonizer of the human respiratory tract and causes diseases ranging from otitis media to meningitis. Many H. influenzae isolates express pili (fimbriae), which mediate adherence to epithelial cells and facilitate colonization. The pilus gene (hif) cluster of H. influenzae type b maps between purE and pepN and resembles a pathogenicity island: it is present in invasive strains, absent from the nonpathogenic Rd strain, and flanked by direct repeats of sequence at the insertion site. To investigate the evolution and role in pathogenesis of the hif cluster, we compared the purE-pepN regions of various H. influenzae laboratory strains and clinical isolates. Unlike Rd, most strains had an insert at this site, which usually was the only chromosomal locus of hif DNA. The inserts are diverse in length and organization: among 20 strains, nine different arrangements were found. Several nontypeable isolates lack hif genes but have two conserved open reading frames (hicA and hicB) upstream of purE; their inferred products are small proteins with no data bank homologs. Other isolates have hif genes but lack hic DNA or have combinations of hif and hic genes. By comparing these arrangements, we have reconstructed a hypothetical ancestral genotype, the extended hif cluster. The hif region of INT1, an invasive nontypeable isolate, resembles the hypothetical ancestor. We propose that a progenitor strain acquired the extended cluster by horizontal transfer and that other variants arose as deletions. The structure of the hif cluster may correlate with colonization site or pathogenicity.
Haemophilus influenzae is a commensal bacterium of humans and can be isolated from the respiratory mucosa of most healthy individuals (22). H. influenzae is responsible for a variety of localized respiratory infections (bronchitis, otitis media, and pneumonia) as well as invasive disease (meningitis, septicemia, and epiglottitis). Systemic disease is caused by a minority of the overall population, usually encapsulated strains of H. influenzae serotype b (Hib); isolates from mucosal infections of the respiratory tract and from healthy throats are generally nonencapsulated and hence nontypeable (42). Exceptions are strains such as INT1, a nonencapsulated but invasive strain isolated from a meningitis patient (31), and the agent of Brazilian purpuric fever (BPF), a member of the aegyptius subgroup of nontypeable H. influenzae (6, 33).
Virulent strains of H. influenzae differ genetically from nonpathogenic strains. Most isolates from invasive disease belong to a few, clonally related lineages (29). The genomes of certain pathogenic H. influenzae strains are larger than those of nonpathogenic strains: the genome of Hib Eagan, a virulent, meningitis-associated strain, is 270 kb larger than that of the nonpathogenic Rd strain (7). Genes found in Eagan and other Hib genomes but absent from Rd include the type b capsular polysaccharide biosynthesis locus, the pilus operon, and a tryptophanase gene cluster (23, 25, 44). The Rd genome has been fully sequenced, facilitating comparisons with pathogenic isolates and the identification of virulence-associated genes (12). Comparative studies of virulence-associated genes in different H. influenzae isolates should shed light on evolutionary adaptations underlying pathogenicity.
Virulence genes have been described as contingency loci, representing adaptations to unusual or swiftly changing host microenvironments (27). Contingency loci typically determine surface molecules which directly contact host cells and are subject to strong and varying selection for tissue tropism and immune evasion. Virulence determinants are often highly mutable compared to genes for housekeeping functions (27); their variability can provide clues to the nature of the selective forces driving pathogen evolution. We chose to examine diversity within a prototype virulence region, the hif (major pilus) cluster, which was known to vary among strains (12). Of particular interest was the hif region of nontypeable strains, that majority of H. influenzae strains whose evolutionary history, adherence structures, and pathogenicity mechanisms are just beginning to be explored (14, 15, 28, 35, 40).
Bacterial colonization of the respiratory tract or conjunctivae requires adherence to host cells. One structure mediating adherence is the pilus (also called fimbria). Various H. influenzae isolates express hemagglutinating pili similar to type I pili of Escherichia coli (reviewed in reference 16). Pili are required for H. influenzae adherence to respiratory epithelial cells (35) and facilitate colonization of the upper respiratory tract (2, 16, 17, 45). The pilus operon of Hib (hifABCDE) consists of five genes encoding proteins involved in cell adherence and pilus assembly (44). HifA is the major pilin subunit, HifB is a periplasmic chaperone, HifC is an outer membrane usher, and HifD and HifE are minor pilus subunits (44). HifE, located at the pilus tip, may be an adhesin (26). The hif operon has several characteristics of a pathogenicity island acquired by lateral transfer (9, 18, 19): it is present in virulent Hib strains but absent from the nonpathogenic Rd strain (12), it is flanked by 57-bp direct repeats of a sequence at its insertion site, and the insertion site includes inverted repeats (44).
Although hif pili are important for initial colonization of the human nasopharynx, their expression may hinder subsequent steps in infection, including persistence within the respiratory epithelium and systemic invasion. Piliated Hib strains become nonpiliated soon after nasal infection, and H. influenzae isolates from the blood and cerebrospinal fluid (CSF) are invariably found to be nonpiliated (34, 45). Expression of pili is reversibly regulated by phase variation, acting on a bidirectional promoter region between hifA and hifB (43). Variation in the number of TA repeat units in the hif promoter modulates transcription; 10 units confers maximal expression, 11 units confers reduced expression, and 9 units confers transcriptional silencing. Phase variation can occur during the course of an infection, converting a piliated variant adept in colonization to a nonpiliated variant capable of tissue invasion (44, 45).
In Salmonella enterica subspecies and in uropathogenic strains of E. coli, pilin genes are found in diverse combinations and arrangements, reflecting adaptation to different hosts or tissues (4, 21). Among H. influenzae strains, hifA is variable in sequence, perhaps facilitating immune evasion (10, 16). A recent survey of nontypeable H. influenzae found variability both (i) in the presence or absence of hif genes in the purE-pepN region and (ii) in the hifA-hifB intergenic region (14). To begin to trace the history of the pilin gene cluster, we examined the locations and arrangements of hif genes in 19 H. influenzae laboratory strains and isolates, including nontypeable as well as encapsulated variants. For a more detailed picture of relationships between the hif regions of different isolates, we sequenced either the entire purE-pepN region or purE-hifA junctions from a representative set of seven strains.
MATERIALS AND METHODS
Bacteria and plasmids.
Bacterial strains are described in Table 1. Growth of E. coli and H. influenzae and purification of E. coli plasmids were as previously described (25).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Sourcea | Relevant properties | Reference(s) |
|---|---|---|---|
| E. coli DH5α | GIBCO BRL | Host for E. coli plasmids | |
| E. coli plasmids | |||
| Bluescript pSK− | Stratagene | ||
| pHic | Cloned 468-bp fragment of R3001 purE-pepN region, obtained by PCR amplification with primers purE-F and pepN-R followed by subcloning of AluI partial digestion fragment into SmaI site of pSK− | This study | |
| pMCC1 | PstI (5.2-kb) fragment of the Hib Eagan hif operon in pUC19, subcloned from a cosmid library; includes the purE-hifA junction, hifA, hifB, and the 5′ end of hifC | This study; 13 | |
| H. influenzae strains | |||
| Eagan (E1A) | CSF from meningitis patient | Type b, streptomycin resistant, virulent in infant rat model | 38 |
| Rd (R906) = Goodgal | Nonencapsulated derivative of type d strain; avirulent in rat model | 8 | |
| INT1 | Blood from meningitis patient | Nonencapsulated; biotype V; virulent in rat model | 31 |
| R539 (ATCC 9006) | ATCC | Reference strain of serotype a | |
| R538 (ATCC 9795) | ATCC | Reference strain of serotype b | |
| R540 (ATCC 9007) | ATCC | Reference strain of serotype c | |
| R541 (ATCC 9008) | ATCC | Reference strain of serotype d | |
| R542 (ATCC 8142) | ATCC | Reference strain of serotype e | |
| R543 (ATCC 9796) | ATCC | Reference strain of serotype f | |
| U11 | CSF | Streptomycin-resistant derivative of U1 (Ramirez); nonencapsulated; avirulent in infant rat assay | 38 |
| R1965 (NCTC 8143) | NCTC | HK389; type strain of H. influenzae; nonencapsulated, biotype II | 22 |
| R1967 (ATCC 11116) | ATCC | Type strain of H. influenzae biotype aegyptius | |
| R2140 | Blood | H. influenzae biotype aegyptius isolate from BPF; hemagglutinating (CDC F3031) | 6 |
| R2141 | Blood | H. influenzae biotype aegyptius isolate from BPF; nonhemagglutinating (CDC F3035) | 6 |
| R2777 | CSF, from invasive disease | Nonencapsulated | This study |
| R3001 | Bronchial lavage of cystic fibrosis patient | Nonencapsulated | 25 |
| C2836 | Otitis media | Nonencapsulated | This study |
| C2840 | Tracheal infection in newborn | Nonencapsulated | This study |
| C2843 | Otitis media | Type b | This study |
| C2853 | Sputum from cystic fibrosis patient | Nonencapsulated | 25 |
| C2859 | CSF | Nonencapsulated | 25 |
| C2861 | CSF | Nonencapsulated | 25 |
ATCC, American Type Culture Collection; NCTC, National Collection of Type Cultures.
DNA isolation and PCR amplification.
H. influenzae DNA was isolated as described previously (5). DNA concentrations were determined by A260 or by fluorometry with a Hoefer TKO fluorometer and Hoechst 33258 (24). Long PCR was performed by using the Expand Long Template PCR kit (Boehringer Mannheim). The purE-pepN region was amplified with primers purE-F (5′-GACCCCATCACAACGCGAATTTGTG-3′), corresponding to nucleotides (nt) 447 to 472 in the Hib (AM30) hif region (GenBank accession no. Z33502), and pepN-R (5′-CTGTGACCGTAAAATCTGGTTGTTTGTAATC-3′), complementary to nt 7687 to 7717 in the same region (44). Amplification conditions were a 60-s hold at 94°C; 10 cycles of 94°C for 10 s, 55°C for 30 s, and 68°C for 6 min; 20 of the same cycles except with 20-s increments added to the extension phase; and a 12-min extension at 68°C.
Restriction fragment length polymorphism (RFLP) analysis.
Similarly sized PCR fragments from the purE-pepN region were compared by restriction digestion and gel electrophoresis. Fragments 0.9 to 1.1 kb in length were digested with AluI and compared after electrophoresis in Metaphor agarose (FMC). Fragments 7 to 8 kb in length were digested with PstI and electrophoresed in standard agarose.
DNA blotting.
Genomic DNA (0.5 to 1 μg) digests were electrophoresed and transferred to nylon membranes by standard methods (37). The hif probe was made by long PCR amplification of Hib Eagan DNA, using primers hifA-F (5′-GGGCGATAAAGTGGAGAGGAAGTTC-3′), corresponding to Hib AM30 nt 722 to 745 (hif cluster; accession no. Z33502 [44]), and hifE-R (5′-CCTTCGGTTAAAGCACCTCTTGTTGTG-3′), complementary to Hib AM30 nt 7376 to 7402. The resulting PCR fragment was gel purified with a Qiagen kit and digoxigenin labelled with random primers (Boehringer Mannheim). The hic probe was made by PCR labelling a cloned fragment of H. influenzae R3001 DNA in a Bluescript vector, using a Boehringer kit and phage promoter primers. Following hybridization, membranes were washed with a final stringency of 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% sodium dodecyl sulfate (SDS) at 56°C and developed for chemiluminescence detection. For reprobing, membranes were stripped by two 15-min washes with 0.2 N NaOH–0.1% SDS at 37°C and a 10-min wash with 2× SSC at 37°C, followed by 1 h of incubation in 2× SSC–0.1% SDS at 65°C and a rinse with 2× SSC at room temperature.
DNA sequencing.
PCR fragments were gel purified as described above and sequenced either directly or following subcloning of AluI fragments into a Bluescript vector; the sequence obtained by subcloning was confirmed by direct sequencing or repeated subcloning. Both strands were sequenced, using external primers and additional primers made to internal sequence. (The purE-hifA junction of reference type c was sequenced only partially.) Sequencing reactions were performed by using dye terminator chemistry (ABI dRhodamine and BigDyes) and run on an ABI Prism 377 sequencer. Sequences were analyzed by using BLASTX and BLASTN (1), the CLUSTAL program (20), and DNAStar software.
Hemagglutination assay.
Detection of H. influenzae variants capable of agglutinating human erythrocytes (RBC) was as described by Stull et al. (41). H. influenzae was grown to stationary phase in supplemented brain-heart infusion (sBHI), centrifuged, and resuspended in phosphate-buffered saline containing 0.1% (wt/vol) gelatin (PBSg) to an A600 of 0.6. One milliliter of bacterial suspension was added to 1 ml of a 3% (vol/vol) suspension of type O+ RBC in PBSg and 10 ml of sBHI. The mixture was allowed to settle, and the RBC pellet was plated on sBHI agar. After overnight incubation at 37°C, a loopful of bacteria was scraped off the plate and resuspended in 1 ml of PBSg. The enrichment procedure was repeated three to five times until individual colonies expressing pili were detectable in a control strain known to be pilus positive (Hib Eagan). Macro- and microhemagglutination were as previously defined (41): macrohemagglutination can be observed with the unaided eye, whereas microhemagglutination can be detected only with an inverted microscope.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the sequences presented here are as follows: R3001, AF071757; C2859, AF071758; C2861, AF071759; INT1, AF071760; ATCC 9796, AF071761; Eagan, AF071762.
RESULTS
The purE-pepN region of H. influenzae is genetically diverse.
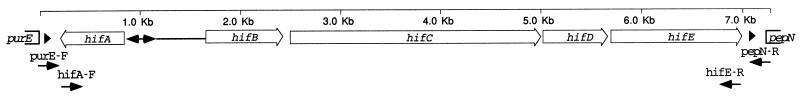
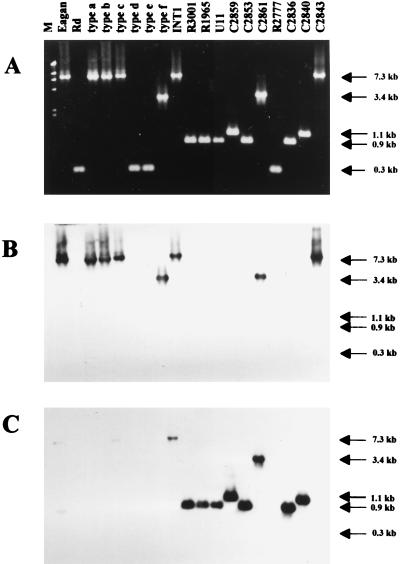
The hif operon of Hib AM30, shown in Fig. 1, is located between the divergently transcribed genes purE and pepN and flanked by direct repeats of a 57-bp sequence (44). In Rd, the 57-bp sequence (referred to here as the direct repeat unit) occurs once, at the position just upstream of purE as indicated in Fig. 1 (12). Long PCR with primers made to these neighboring genes was used to amplify the intervening region and yielded fragments of various lengths (Fig. 2A). Six strains had fragments equivalent in length to the entire hif operon. Four strains, including Rd, had 0.3-kb fragments, indicating that no insert was present. The remaining nine strains had fragments of intermediate length, ranging from 0.9 to 3.4 kb and insufficient to encode all five hif genes.
FIG. 1.
The hif operon of Hib AM30 (43), showing PCR primers used to amplify the region. Open arrows, genes; arrowheads, 57-bp direct repeats flanking the hif operon; double-headed arrow (not to scale), overlapping promoter region between hifA and hifB; closed arrows (not to scale), external primers purE-F and pepN-R, used to amplify the entire purE-pepN region, and primers hifA-F and hifE-R, used to make the hif-specific probe.
FIG. 2.
PCR amplification of the purE-pepN regions from various H. influenzae strains. (A) Genomic DNAs were amplified with primers purE-F and pepN-R (Fig. 1). Lane M, markers (λHindIII fragments). The sizes of PCR fragments, estimated from mobilities, are indicated at the right. The predicted size of the PCR fragment from Rd is 256 bp, and that from Hib AM30 is 7,270 bp. (B) The gel in panel A was blotted and probed with a hif-specific probe made by PCR with internal primers hifA-F and hifE-R. (C) The blot in panel B was stripped and reprobed with a hic probe, specific for the R3001 insert. The hic probe is a cloned, 468-bp AluI partial digestion fragment of the R3001 insert (Fig. 3).
Identification of novel DNA contiguous to the hif operon.
Hybridization with a hif-specific probe revealed that purE-pepN regions 0.9 and 1.1 kb in length contained no hif DNA (Fig. 2B). When novel DNA from this region of nontypeable strain R3001 was used as a probe (Fig. 2C), several purE-pepN regions were found to contain a combination of sequences homologous to the hif operon and to the novel DNA. Sequences identified by the R3001-specific probe are referred to as hic (for hif-contiguous) DNA.
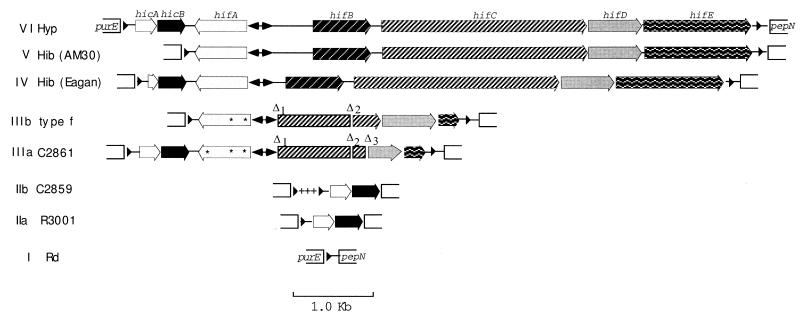
Classification of hif genotypes.
Genotypes of the purE-pepN region were grouped into classes by length, hybridization pattern, and RFLP analysis of purE-pepN PCR products. All genotypes grouped within a single class (IIa, IIb, or V) had identical or near-identical RFLPs. Nine distinct genotype classes were found (I, IIa, IIb, IIIa, IIIb, and IV to VII) (Table 2) (see Fig. 4 for a summary). Class I is typified by Rd (12), and class V is typified by Hib AM30. To examine the relationships between these and other classes, we sequenced the purE-pepN region or purE-hifA junctions from representative strains.
TABLE 2.
Strain classification by hif genotype and hemagglutination phenotype
| Straina | Sourceb | Classc | hifAd | hicABe | HAf |
|---|---|---|---|---|---|
| Hib Eagan | CSF, meningitis patient | IV | +, (AT)10 | − | + |
| Rd | L | I | − | − | − |
| Type a | L | V | + | − | + |
| Type b | L | V | + | − | + |
| Type c | L | VII | + | − | + |
| Type d | L | I | − | − | − |
| Type e | L | I | − | − | − |
| Type f | L | IIIb | −, (AT)5 | − | ± |
| INT1 | Blood, meningitis patient | VI | +, (AT)4 | + | − |
| R2777 | CSF | I | − | − | − |
| R3001 | Bronchial infection | IIa | − | + | − |
| R1965 | L | IIa | − | + | − |
| U11 | CSF | IIa | − | + | − |
| C2836 | Otitis media | IIa | − | + | − |
| C2840 | Tracheal infection | IIb | − | + | ± |
| Hib C2843 | Otitis media | V | + | − | − |
| C2853 | Sputum | IIa | − | + | − |
| C2859 | CSF | IIb | − | + | − |
| C2861 | CSF | IIIa | −, (AT)4 | + | ± |
| Hib AM30g | CSF, meningitis patient | V | +, (AT)10 | − | + |
Types a to f are the reference strains for each capsular serotype. All other isolates except those designated Hib are nontypeable.
L, laboratory reference strain.
hif genotype classification (Fig. 6).
Presence (+) or absence (−) of an intact hifA gene and number of AT repeats in the hifA promoter, if known. For strains showing phase variation, the number of repeats is that in a piliated variant; for INT1, C2861, and type f, the number of repeats is that in the original isolate.
Presence (+) or absence (−) of intact copies of both hicA and hicB within the region between purE and pepN.
HA, hemadsorption ability after enrichment for adherent variants (see Materials and Methods). +, macroagglutination; ±, microagglutination (see Materials and Methods).
FIG. 4.
Maps of purE-pepN genotypes. Hyp, hypothetical ancestor containing the extended hif cluster and consisting of the purE-hicB region of R3001 affixed to the hifA-pepN region of Hib AM30 by the hicB-hifA junction of INT1. Genes and ORFs are depicted as arrows; fill patterns distinguishing genes are as indicated for class VI. Double-headed arrows, position of the combined hifA-hifB promoter region (not to scale); arrowheads at left and right, direct repeat units; asterisks within hifA, single-base frameshift deletions. Relative to Hib AM30, Δ1 is a deletion removing 2,687 bp, including most of the hifA-hifB intergenic region on the hifB side of the promoter, hifB, and the first 1,226 bp of hifC (nt 1477 to 4164 in Hib AM30; accession no. Z33502); Δ2 removes 130 bp within hifC (nt 4973 to 5102); and Δ3 (534 bp) removes the 3′ terminus of hifC and 300 bp from the 5′ end of hifD (nt 5230 to 5763). In class III, another deletion (ΔhifE) removes 1,189 bp (nt 6327 to 7515) from the 3′ end of hifE and the hifE-pepN intragenic region, ending just before the right direct repeat unit. +++, a 189-bp sequence found in IIb but not IIa (part of a 246-bp addition); it is followed by a second copy of the direct repeat unit.
The hic region of R3001 contains two new ORFs.
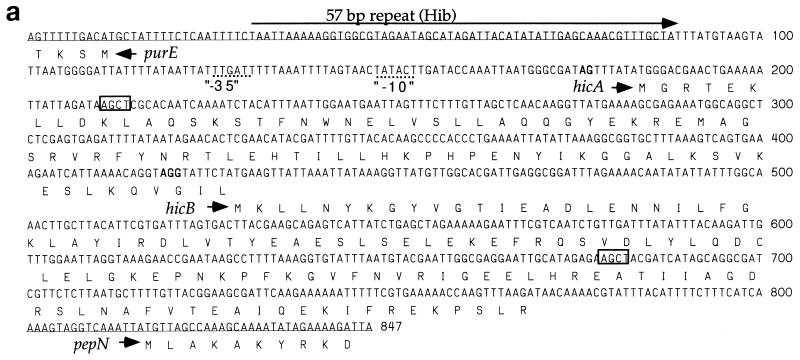
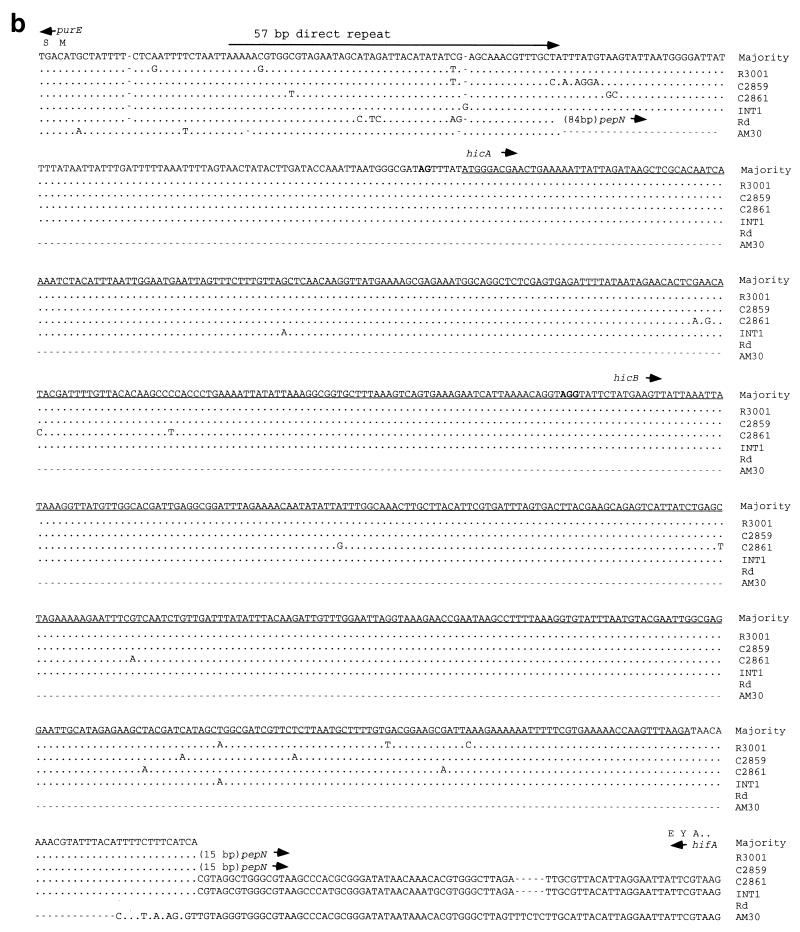
In R3001 (class IIa), the distance between initiating codons of purE and pepN (the “intergenic distance”) is 803 bp (Fig. 3a). A 76-bp sequence upstream of purE is similar to that of Hib AM30 (43) and Rd (12). Immediately past the direct repeat unit, however, is 722 bp of novel sequence. This includes two short open reading frames (ORFs), referred to as hicA and hicB. The ORFs are oriented oppositely to purE, and the stop codon of hicA overlaps the initiating codon of hicB. The novel sequence ends just 15 bp upstream of pepN.
FIG. 3.
The region upstream from purE in H. influenzae. (a) Sequence of the R3001 insert and inferred amino acid sequences of hicA and hicB. AluI sites flanking the hic probe are shown (boxes). (b) Alignment of the purE-hifA region in various strains. Dots signify identity, letters indicate sequence differences, and dashes are gaps. Position 1 is the initiating A of the purE coding sequence, and the carboxy-terminal amino acids of HifA (E Y A) are indicated. C2859 has a 246-bp insertion immediately following the 57-bp repeat.
Potential ribosomal binding sites are found next to hicA and hicB, although the hicA site is weak. The inferred translational product of hicA is an 82-amino-acid, basic (pI = 9.9) polypeptide. The inferred translation product of hicB is a 114-amino-acid, acidic (pI = 5.0) polypeptide. Both putative proteins can form amphipathic α-helices. BLAST searches did not reveal significant homologies within the hic region in GenBank. The G+C content (34%) is similar to the species average of 38%, and the codon usages of hicA and hicB are typical of H. influenzae.
Sequence comparisons of the hic regions in H. influenzae strains.
The region upstream from purE from several nontypeable isolates, as well as from Hib Eagan, was sequenced. C2859 (IIb), C2861 (IIIa), and INT1 (VI) sequences can be aligned with the R3001 segment containing hicA and hicB (Fig. 3b). Among these four strains, the hic region is highly conserved; average pairwise DNA sequence differences are 1.6% (range, 0.4 to 2.7%). Hib Eagan has hicB but only the 3′ end of hicA (data not shown). The hicB gene of Hib Eagan is identical to that of C2861.
Conservation of inferred amino acid sequence in the hic region is also high. Among the strains having hicA, there are five DNA polymorphisms (Fig. 4), only two of which change the amino acid sequence from the consensus: Ala28→Thr in INT1 and Glu50→Lys in C2861. Similarly, there are 10 DNA polymorphisms among the strains having hicB, all but 2 of which are silent: Arg48→His in C2861 and Hib Eagan and Lys103→His in R3001.
The extended hif cluster.
Sequence comparisons among genotype classes allowed reconstruction of a hypothetical ancestral cluster, termed the extended hif cluster (Fig. 4). The extended hif cluster contains both hic and hif genes, between flanking copies of the direct repeat unit. Although the hypothetical ancestral cluster is envisaged as a composite of Hib AM30 and R3001, it closely resembles the structure of INT1 between purE and hifB and between pepN and hifE. INT1 has tentatively been assigned to the same class (VI) as the hypothetical ancestor.
Class II genotypes have lost hif DNA.
R3001 (IIa) is related to the extended hif cluster by a 7.1-kb deletion, corresponding to Hib AM30 nt 561 to 7644 (44) (Fig. 4). This deletion removes the second copy of the direct repeat sequence and ends just upstream of pepN.
C2859 (IIb) can be aligned with IIa at the pepN end and has the same deletion. However, IIb has an additional 246 bp immediately distal to the direct repeat unit. The additional sequence includes a second copy of the direct repeat unit (Fig. 4).
Class III genotypes have partial deletions of the hif cluster.
C2861 (IIIa) and reference type f (IIIb) have related hif genotypes (Fig. 4). Each has multiple mutations and deletions within a severely deleted hif region. They each have a hifA pseudogene, with two shared frameshift mutations (IIIa has a third). The hifA-B promoter region has a reduced number of TA repeats (4 in C2861 and 5 in type f) relative to that of Hib (9 to 11 repeats). A 2,687-bp deletion (Δ1 in Fig. 4) has excised the region from upstream of hifB through approximately half of hifC; the coordinates of Δ1 are identical in the two class III isolates. The strains share a 130-bp deletion within hifC (Δ2) and a third (ΔhifE) removing all but the 5′-proximal 197 bp of hifE. Their genotypes must have diverged from a common class III ancestor with shared mutations. Each strain has additional, nonshared mutations: IIIb has lost most of the hic region, which is retained in IIIa, and IIIa has a deletion (Δ3) which excises the 3′ end of hifC and 300 bp from the 5′ end of hifD. In contrast, IIIb has an intact hifD gene.
Class IV has a partial deletion of hic DNA.
Hib Eagan has a complete copy of the hif operon (14). The sequence of the purE-hifA junction revealed that it has a complete copy of hicB and a 5′ deleted copy of hicA (Fig. 4). The deletion begins just after the left direct repeat unit. The hifA-hifB intergenic region of Hib Eagan is much shorter than that of Hib AM30, lacking all but one of the 10 REP sequences found in this region of Hib AM30 (14).
Class V has a complete hic deletion.
The published sequence of the purE-pepN region from Hib AM30, compared to those of the IIIb, IV, and VI junctions, indicates that it has arisen by deletion of the hic region (44) (Fig. 4).
Class VI contains both hic and hif regions.
The virulent nontypeable strain INT1 has both hic genes, an intact copy of hifA, the hifA-hifB promoter region, and at least the 5′ end of hifB and the 3′ end of hifE (Fig. 4). Although the interior of the hif cluster has not been sequenced, its length is sufficient to include hifA to hifE. The hifA-hifB promoter region has only four TA repeats, and the distance from the promoter to hifB is shorter than that in Hib AM30, having a single full REP sequence. Although Hib Eagan and INT1 each have a short (approximately 300-bp) hifA-hifB intergenic region, relative to that of Hib AM30, their sequences are different, suggesting that different REP units have been lost.
The structures of IIb and VII are composites of other genotype classes.
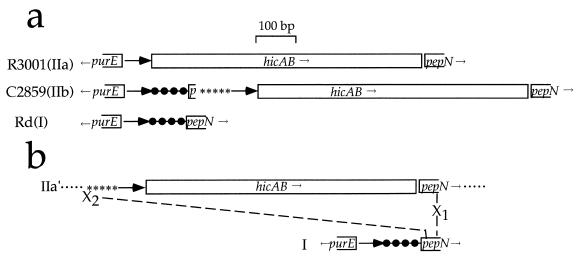
Although at first sight IIb seems to have a simple insertion of 246 bp within IIa, sequence details suggest a more complex relationship (Fig. 5a). The addition consists of three segments: (i) 91 bp identical to the corresponding Rd (class I) intergenic region, including 7 bp from the 5′ end of the pepN coding sequence; (ii) 97 bp of repetitive DNA, including two complete copies and one partial copy of a palindromic repeat (REP) found in multiple copies in the hif operon and elsewhere in the H. influenzae genome; and (iii) a second copy of the direct repeat unit. Sequence distal to the second direct repeat unit is nearly identical to that in IIa. Unlike class IIa, IIb has an uninterrupted class I purE-pepN region, with an insertion after the start of pepN. Thus, IIb appears to have arisen by uptake of REP sequences and hic genes within the coding sequence of pepN. This might have happened by lateral transfer of a fragment from a class IIa-bearing strain to a class I strain, followed by partially homologous recombination within pepN (Fig. 5b). According to this idea, a class II fragment bordered by REP sequences was incorporated into pepN by a single crossover within homologous sequence (X1); a nonhomologous exchange (X2), between REP sequence and the 5′ end of pepN restored the chromosome.
FIG. 5.
The purE-pepN region of class IIb as a composite of I and IIa. (a) Comparison of classes I, IIa, and IIb. Heavy arrows, direct repeat units; filled circles, 91 bp identical in Rd and C2859, including p, the first 7 bp of pepN; ∗∗∗∗∗, a 97-bp region containing REP sequences; and hicAB, the hic region. (b) Hypothesized origin of class IIb. A horizontally transferred fragment (IIa′) from a class IIa genome is incorporated into a class I genome, facilitated by homology within pepN. X1, homologous exchange between pepN genes of donor and recipient, occurring downstream from X2, a nonhomologous exchange between the REP region of the donor fragment and pepN nt 7 and 8 of the recipient genome.
The left junction of class VII (corresponding to reference type c strain) is similarly complex (not shown). Sequence to the right of the leftmost direct repeat unit matches the class I intergenic region until just 5′ of pepN; this is followed by REP sequences, a second copy of the direct repeat unit, and a partially deleted version of the hic region, followed by hifA. The length of the class VII purE-pepN region is sufficient to include hifA to E. Class VII may also have arisen by lateral transfer and partially homologous recombination near pepN.
Deletions of hif genes have occurred between short regions of homology.
Class III genotypes have multiple deletions relative to that of Hib AM30. Considering Hib AM30 as the ancestral genotype, analysis of sequence bordering the four deletions in C2861 (Table 3) suggests that these deletions have occurred within short interspersed regions of homology, ranging from 6 to 10 bp. The right junction of ΔhifE, but not the left, is part of a REP sequence; the other repeats are not REP related.
TABLE 3.
Junctions of C2861 deletions
| Deletiona | Deleted sequence (Hib AM30 nt) | Sequence at bordersb |
|---|---|---|
| Δ1 | 1477–4164 | ATTTTGAATTAAT…2687 bp…GTTTTGGATTAAA |
| AttttgGATTAAA | ||
| Δ2 | 4973–5102 | CACGCTAAAG…130 bp…AACGCCAAAT |
| CacgcCAAAT | ||
| Δ3 | 5230–5763 | GCCAGTGCCA…534 bp…CCCCGTGCCT |
| GccCGTGCCT | ||
| ΔhifE | 6327–7515 | GCTACCACA…1189 bp…CCTACAACT |
| GcgactacT |
Deletions are shown in Fig. 4.
For each deletion, the AM30 sequence is shown above the C2861 sequence. Matching Hib AM30 sequences at the right and left junctions are underlined, and the sequence within which an exchange is thought to have occurred is indicated in lowercase. The C2861 ΔhifE sequence differs from Hib AM30 junctions at two positions (the third and sixth from its 5′ end). Hib AM30 sequences are from reference 44 (accession no. Z33502). Hib AM30 coordinates are as follows: Δ1, nt 1474 to 1486 and 4162 to 4174; Δ2, nt 4968 to 4977 and 5098 to 5107; Δ3, nt 5228 to 5237 and 5762 to 5771; ΔhifE, nt 6324 to 6332 and 7510 to 7518.
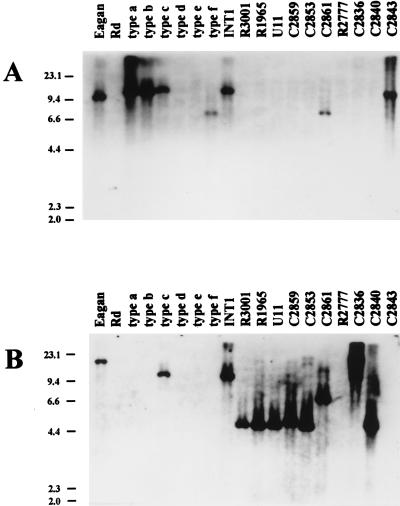
Genomic DNA blots with hif and hic probes.
To identify hif-related DNA in H. influenzae isolates, genomic DNA was digested with BglII, which does not cut within the Hib AM30 hif region, and probed with hif (Fig. 6A) and hic (Fig. 6B) DNAs. For each genotype class, genomic blotting confirmed the organization and location of genes predicted by sequence analysis. Additionally, Southern blotting showed that hif sequences are located uniquely at the purE-pepN site (Fig. 6A). In hic-positive strains, hic DNA was found at the same site. Hib Eagan (class IV) has a BglII site near the 3′ end of hifA (13) (accession no. M64334), between regions specified by hic and hif probes; as expected, hif and hic probes bound different fragments of Hib Eagan DNA (Fig. 6). Isolate C2840 (genotype class IIb) had two slightly resolved ∼4.5-kb BglII fragments bound by the hic probe, both similar in size to class IIb fragments; assuming that BglII sites are conserved, this may signify a duplication of hic sequences in C2840.
FIG. 6.
Genomic blots with hif- and hic-specific probes. Genomic DNAs were digested with BglII, blotted, and probed as indicated. Assuming conservation of flanking sequence, RFLPs can be predicted from Rd and the purE-pepN sequence by adding 3,700 bp to the distance between purE and pepN translational start sites. Predicted RFLPs are 3,860 bp for Rd (class I), 10,875 for Hib AM30 (class V), 4,503 bp for R3001 (class IIa), 4,749 bp for C2859 (class IIb), and 6,984 bp for C2861 (class IIIa). The positions and lengths (in kilobases) of λHindIII marker fragments are indicated on the left. (A) Blot with the hif probe. Fragment lengths, estimated from mobilities, are 10 to 11 kb for Hib Eagan, Hib C2843, INT1, and reference strains of type a, type b, and type c and 7.5 kb for C2861 and the type f reference strain. (B) The same blot stripped and reprobed with the hic probe. Fragment lengths, estimated from mobilities, are >22 kb for Hib Eagan, >22 kb for C2836, 10 to 11 kb for reference type c and INT1, 7.5 kb for C2861, 5 kb for C2859 and C2840, and 4.4 kb for R3001, R1965, U11, and C2853. C2840 had two slightly resolved fragments of similar size, suggesting a duplication of hic DNA, and C2836 and Hib Eagan have RFLPs for BglII.
H. influenzae strains associated with BPF have undergone a duplication of the hif operon (36). Genomic DNAs from two BPF isolates (R2140 and R2141) and from the reference H. influenzae biotype aegyptius strain were probed with hif- and hic-specific DNAs. As expected, the hif probe identified two BglII fragments of different lengths in BPF-associated H. influenzae strains; a single, low-mobility fragment was identified in the H. influenzae biotype aegyptius reference strain (data not shown). H. influenzae biotype aegyptius genomic DNA did not hybridize with the hic probe.
Hemadsorption phenotypes.
Most clinical isolates of H. influenzae do not express pili, even if they are genetically capable of doing so, presumably because persistence within the host selects against piliation and for pilus-negative phase variants (34, 44). However, in strains having an intact hif operon, a small minority of the population consists of phase variants expressing hif pili; these are capable of agglutinating O+ human RBC and can be selected by repeated cycles of enrichment for adherent phase variants (41). As expected, only strains with an intact hif operon were positive in the hemadsorption enrichment assay (Table 2); these included representatives of classes IV, V, and VII. The presence of hif genes was not sufficient for pilus expression, as INT1 (class VI) and Hib C2843 (class V) failed to hemagglutinate, even after repeated cycles of enrichment. Three nontypeable strains (in classes IIb, IIIa, and IIIb) displayed microhemagglutination, despite lacking hifA.
DISCUSSION
As humans are the exclusive host of H. influenzae (22), all contemporary strains are likely to have had a common ancestor within the past 5 million years of hominid evolution. Strain evolution can be viewed as adaptative radiation to different niches within the human body and to different modes of transmission. Assuming this relatively recent divergence time, limited DNA sequence diversity is expected among strains, except where selection is strong and disruptive (25, 32). The region of the H. influenzae genome containing the hif operon is unusually diverse: nine different arrangements were noted among 20 strains. This diversity contrasts with the apparent conservation of neighboring chromosomal DNA sequence, suggesting that the hif cluster has evolved rapidly under selective pressure within different host microenvironments. Rapid evolutionary change is typical of pathogenicity-related gene clusters (19).
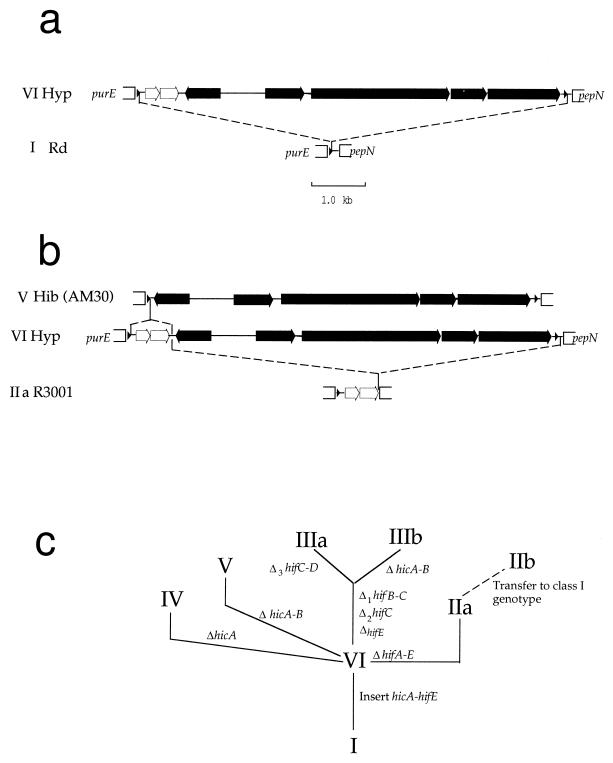
A hypothetical evolutionary tree for hif is shown in Fig. 7. H. influenzae is likely initially to have acquired the extended hif cluster by horizontal transfer (44). Inverted repeats within the target site (44) and duplication of that site suggest transduction by a phage whose integration site differs from those of known H. influenzae phages (9). Integration within the direct repeat unit of a class I genome generated class VI (Fig. 7a). Most other genotypes arose by subsequent deletions (Fig. 7b); deletion of the hif operon produced class IIa, and deletion of hic genes produced class V. Following initial radiation of hif genotypes (Fig. 7c), two genotypes (classes IIb and VII) probably arose by lateral transfer and integration of extended hif cluster DNA into a class I strain, facilitated by homology in or near pepN. The potential for horizontal transfer across lineages means that strains with similar hif arrangements need not be clonally related. A similar combination of lateral transfer and deletions has shaped the pilus gene clusters of enteric bacteria (4).
FIG. 7.
Proposed model of hif evolution. (a) Insertion (dashed lines) of the extended hif cluster within the purE-pepN intergenic region of a class I strain generates genotype VI (hypothetical [Hyp]). The hic ORFs are indicated with unfilled arrows, and hif operon genes are indicated with filled arrows. (b) Deletions (dashed lines) within the extended hif cluster give rise to genotypes IIa and V. (c) Proposed phylogeny of hif genotypes, based on shared rearrangements only. The relationships shown refer only to genotypes of the purE-pepN region and not to strains or groups of organisms. Branch lengths and the order of mutations between nodes are arbitrary. Δ1, Δ2, and Δ3 are as in Fig. 4.
Recently, Geluk et al. (14) have examined the purE-pepN region in a larger collection of nontypeable H. influenzae isolates by hybridization analysis and partial sequencing. In eight nontypeable strains having the hif gene cluster, novel DNA (570 to 720 bp) was found between purE and hifA; this DNA, described as noncoding, was homologous among the strains and probably corresponds to partial or complete hic regions. Four of nine nonencapsulated strains which lacked hif DNA had a short intergenic region like that of Rd. The remaining five had insert sequences ranging up to 1.1 kb; these had homology to the novel DNA downstream of hifA in hif-containing strains. These five variants may correspond to our class IIa and IIb genotypes, as 1.1 kb is the length of the intergenic region in class IIb. Taken together, our data and those of Geluk et al. (14) indicate a high prevalence for hic sequences among nontypeable H. influenzae and a general tendency among respiratory isolates to lose hif genes. The strains described by Geluk et al. either had all of the hif genes or none of them; they did not observe any partial deletions of the hif operon such as in classes IIIa and IIIb. They offer a contrasting model of hif evolution, in which the direct repeat unit has been the target of multiple insertion events. However, the relatively simple relationships between genotype classes, including seeming evolutionary intermediates, make it unnecessary to invoke multiple insertions.
When was the extended hif cluster acquired by H. influenzae? The hic region, including noncoding DNA, is similar among strains in different lineages, differing by less than 3% in the most disparate genotypes. This degree of similarity suggests that it was acquired recently, probably after H. influenzae first colonized human ancestors (25, 32). Certain hif genes, especially hifC, are also highly conserved. hifA is more variable; however, it is likely to have evolved under strong selection for immune evasion (10).
Do hicA and hicB encode proteins? Their high DNA sequence similarity among strains might simply reflect recent acquisition. However, hicB in particular shows a bias toward silent substitutions that is consistent with coding sequence. We are attempting to resolve this question by raising antipeptide antibodies to identify putative hic products.
Deletions within the hif cluster are not merely private mutations localized to individual clinical isolates. Some mutations and deletion genotypes are shared by apparently unrelated strains from different locales, time periods, and host sites. For instance, genotype class IIa is found in a commensal organism from sputum, in isolates from upper and lower respiratory tract infections, and in CSF isolates from meningitis patients. The widespread distribution of such variants suggests a selective advantage to certain hif genotypes.
The hif region contains unusual repetitive sequences, including multiple palindromic REP sequences (14, 44). It has been suggested that REP sequences could facilitate rearrangement within the hif cluster (44). This may have happened in the hifA-hifB promoter region, where a total of 10 repeats in Hib AM30 have been reduced to one in Hib Eagan and INT1. In contrast, deletions in class III genomes have occurred between short interspersed regions of homology and, with one exception, do not involve REP sequences.
With few exceptions, the hif variants described here fall into two groups: those which have disabled their hif genes but retain the hic region and those which have shed hic DNA but retain hif genes. In this small sample, expression of pili and retention of hic genes were mutually exclusive: of nine hic-positive strains, none were hemagglutination positive, and of five hemagglutination-positive strains, none had intact hic genes. A similarly mutually exclusive relationship has been noted for expression of two high-molecular-weight adherence proteins in H. influenzae (40). These opposing trends suggest divergent ecological adaptations in the human host, at least for pathogenic isolates.
What might such divergent host adaptations be? Commensal strains of H. influenzae are isolated from the nasopharynx, whereas disease isolates are isolated from the lower respiratory tract, middle ear, blood, or CSF. Each environment may select for different adherence adaptations. Hib pili attach to squamous cells of the nasopharynx, where they presumably assist in providing a large starting inoculum for efficient colonization (45). Infecting bacteria soon lose their pili, however, perhaps to allow movement from initially seeded cells to the respiratory mucosa, where other adhesins are required. In long-term airway infections, pili might be a liability in exposing bacteria to immune clearance. Hib strains persist within the host by reversibly shutting off hif expression by phase variation. Irreversible loss of hif genes, as has occurred in most of the nontypeable isolates described here, may be a convergent adaptation.
Forms of H. influenzae which lack long pili attach to certain human cell types by other mechanisms, including nonhemagglutinating pili, outer membrane adhesins, and nonprotein hemagglutinins (3, 11, 15, 39, 46). Three strains analyzed in this study lacked pilin genes but had weak hemagglutinating activity, indicating some means of adhering to RBC. Microhemagglutination activity does not require hic DNA, because it was observed for the type f reference strain, which lacks these genes. The function, if any, of hic genes is unknown; an attractive and testable hypothesis is that they are implicated in a different nonpilus adherence mechanism.
An apparent exception to the mutual exclusion of hif and hic genes is INT1, an isolate which is also exceptional in being both nonencapsulated and virulent. At the junctions of the hif cluster, the INT1 genome resembles the reconstructed ancestral genome, having both hic genes, hifA, and at least the 5′ terminus of hifB and the 3′ terminus of hifE. Although the INT1 sequence between hifB and hifE has not been determined, its length is consistent with a full-length hif operon. Phenotypically, however, INT1 conforms to the mutual exclusion of hif and hic. Despite its intact hifA gene, INT1 did not express pili, as judged by the hemadsorption enrichment assay. A likely explanation is that its hif promoter has only 4 TA repeats, compared to 10 required for maximal expression (43); 4 repeats provide inadequate spacing between −35 and −10 promoter regions. Unlike other strains which are unable to express pili, INT1 has not suffered major hif deletions or hifA frameshift mutations. Perhaps its promoter mutation is recent, the result of mispaired strand slippage in a few replicative cycles. Presumably, loss of most TA repeats would irreversibly inactivate the operon. As with reversible phase variation, loss of pili might confer a short-term advantage on a potentially invasive clone. If so, wild-type relatives of INT1 which contain an active, extended hif operon may exist.
The population genetics of H. influenzae has been investigated extensively, and much information on relationships among encapsulated and nontypeable strains is available (28–30, 40), including recent information on hif genes (14). This study has not taken advantage of population genetics, focusing instead on more intensive sequencing of a limited set of isolates. The two approaches are complementary. Without detailed sequence information, it is difficult to recognize evolutionary relationships among genotype classes. For instance, IIIa and IIIb are closely related but have dissimilar RFLPs and hybridization patterns, and IIa and IIb are superficially similar but IIb probably arose by uptake of a IIa-like region within a different lineage. However, many questions raised here can be framed only within population genetics. Does the distribution of widespread genotypes represent clonal relationships among strains? How common is horizontal transmission of hif genes, and does it correlate with site or pathogenicity? Are hif genotypes shared among encapsulated and nontypeable strains? Answering these questions requires placing hif evolution in the context of H. influenzae population genetics.
ACKNOWLEDGMENTS
This work was supported by separate University of Missouri Research Board grants to M.G. and A.L.S. and by NIH grant ESO4889 to A.E.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Anderson P W, Pichichero M E, Connor E M. Enhanced nasopharyngeal colonization of rats by piliated Haemophilus influenzae type b. Infect Immun. 1986;48:565–568. doi: 10.1128/iai.48.2.565-568.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barenkamp S J, St. Geme J W., III Identification of a second family of high-molecular weight adhesion protein expressed by non-typeable Haemophilus influenzae. Mol Microbiol. 1996;19:1215–1223. doi: 10.1111/j.1365-2958.1996.tb02467.x. [DOI] [PubMed] [Google Scholar]
- 4.Baumler A J, Gilde A I, Tsolis R M, van der Velden A W M, Ahmer B M M, Heffron F. Contribution of horizontal gene transfer and deletion events to development of distinctive patterns of fimbrial operons during evolution of Salmonella serotypes. J Bacteriol. 1997;179:317–322. doi: 10.1128/jb.179.2.317-322.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berns K I, Thomas C A. Isolation of high molecular weight DNA from Haemophilus influenzae. J Mol Biol. 1965;11:476–490. doi: 10.1016/s0022-2836(65)80004-3. [DOI] [PubMed] [Google Scholar]
- 6.Brenner D J, Mayer L W, Carlone G M, Harrison L H, Bibb W F, Brandileone M C, Sottnek F O, Iriono K, Reeves M W, Swenson J M, Birkness K A, Weyant R S, Berkley S F, Woods T C, Steigerwalt A G, Grimont P A D, McKinney R M, Fleming D W, Gheesling L L, Cooksey R C, Arko R J, Broome C V The Brazilian Purpuric Fever Study Group. Biochemical, genetic, and epidemiological characterization of Haemophilus influenzae biogroup aegyptius (Haemophilus aegyptius) strains associated with Brazilian purpuric fever. J Clin Microbiol. 1988;26:1524–1534. doi: 10.1128/jcm.26.8.1524-1534.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butler P D, Moxon E R. A physical map of the genome of Haemophilus influenzae type b. J Gen Microbiol. 1990;136:2333–2342. doi: 10.1099/00221287-136-12-2333. [DOI] [PubMed] [Google Scholar]
- 8.Catlin B W, Bendler J W, Goodgal S H. The type b capsulation locus of Haemophilus influenzae: map location and size. J Gen Microbiol. 1972;70:411–422. doi: 10.1099/00221287-70-3-411. [DOI] [PubMed] [Google Scholar]
- 9.Cheetham B F, Katz M E. A role for bacteriophages in the evolution and transfer of bacterial virulence determinants. Mol Microbiol. 1995;18:201–208. doi: 10.1111/j.1365-2958.1995.mmi_18020201.x. [DOI] [PubMed] [Google Scholar]
- 10.Clemans D L, Marrs C F, Patel M, Duncan M, Gilsdorf J R. Comparative analysis of Haemophilus influenzae pilin genes. Infect Immun. 1998;66:656–663. doi: 10.1128/iai.66.2.656-663.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farley M M, Stephens S S, Kaplan S L, Mason E O J. Pilus- and non-pilus mediated interactions of Haemophilus influenzae type b with human erythrocytes and human nasopharyngeal mucosa. J Infect Dis. 1990;161:274–280. doi: 10.1093/infdis/161.2.274. [DOI] [PubMed] [Google Scholar]
- 12.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J-F, Dougherty B A, Merrick J M, McKenney K, Sutton G, FitzHugh W, Fields C, Gocayne J D, Scott J, Shirley R, Liu L-I, Glodek A, Kelley J M, Weidmann J F, Phillips C A, Spriggs T, Hedblom E, Cotton M D, Utterback T R, Hanna M C, Nguyen L D, Saudek D M, Brandon R C, Fine L D, Fritchman J L, Geoghagen N S M, Gnehm C L, McDonald L A, Small K V, Fraser C M, Smith H O, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 13.Forney L J, Marrs C F, Bektesh S L, Gilsdorf J R. Comparison and analysis of nucleotide sequence of pilin genes from Haemophilus influenzae type b strains Eagan and M43. Infect Immun. 1991;59:1991–1996. doi: 10.1128/iai.59.6.1991-1996.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geluk F, Eijk P P, van Ham M, Jansen H M, van Alphen L. The fimbrial gene cluster of nontypeable Haemophilus influenzae. Infect Immun. 1998;66:406–417. doi: 10.1128/iai.66.2.406-417.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilsdorf J R, Chang H Y, McCrea K W, Bakaletz L O. Comparison of hemagglutinating pili of Haemophilus influenzae type b with similar structures of nontypeable H. influenzae. Infect Immun. 1992;60:374–379. doi: 10.1128/iai.60.2.374-379.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilsdorf J R, McCrea K W, Marrs C F. Role of pili in Haemophilus influenzae adherence and colonization. Infect Immun. 1997;65:2997–3002. doi: 10.1128/iai.65.8.2997-3002.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilsdorf J R, Tucci M, Marrs C F. Role of pili in Haemophilus influenzae adherence to, and internalization by, respiratory cells. Pediatr Res. 1996;39:343–348. doi: 10.1203/00006450-199602000-00025. [DOI] [PubMed] [Google Scholar]
- 18.Groisman E A, Ochman H. Pathogenicity islands: bacterial evolution in quantum leaps. Cell. 1996;87:791–794. doi: 10.1016/s0092-8674(00)81985-6. [DOI] [PubMed] [Google Scholar]
- 19.Hacker J, Blum-Oehler G, Muhldorfer I, Tschape H. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 20.Higgins D G, Sharp P M. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 21.Kao J-S, Stucker D, Warren J W, Mobley H L T. Pathogenicity island sequences of pyelonephritogenic Escherichia coli CFT073 are associated with virulent uropathogenic strains. Infect Immun. 1997;65:2812–2820. doi: 10.1128/iai.65.7.2812-2820.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kilian M. A taxonomic study of the genus Haemophilus, with the proposal of a new species. J Gen Microbiol. 1976;93:9–62. doi: 10.1099/00221287-93-1-9. [DOI] [PubMed] [Google Scholar]
- 23.Kroll J S, Loynds B M, Moxon E R. The Haemophilus influenzae capsulation gene cluster: a compound transposon. Mol Microbiol. 1991;5:1549–1560. doi: 10.1111/j.1365-2958.1991.tb00802.x. [DOI] [PubMed] [Google Scholar]
- 24.Labarca C, Paigen K. A simple, rapid, and sensitive DNA assay procedure. Anal Biochem. 1980;102:344–352. doi: 10.1016/0003-2697(80)90165-7. [DOI] [PubMed] [Google Scholar]
- 25.Martin K, Morlin G, Smith A, Nordyke A, Eisenstark A, Golomb M. The tryptophanase gene cluster of Haemophilus influenzae type b: evidence for horizontal gene transfer. J Bacteriol. 1998;180:107–118. doi: 10.1128/jb.180.1.107-118.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCrea K W, Watson W J, Gilsdorf J R, Marrs C F. Identification of two minor subunits in the pilus of Haemophilus influenzae. J Bacteriol. 1997;179:4227–4231. doi: 10.1128/jb.179.13.4227-4231.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moxon E R, Rainey P B, Nowak M A, Lenski R E. Adaptive evolution of highly mutable loci in pathogenic bacteria. Curr Biol. 1994;4:24–33. doi: 10.1016/s0960-9822(00)00005-1. [DOI] [PubMed] [Google Scholar]
- 28.Musser J M, Barenkamp S J, Granoff D M, Selander R K. Genetic relationships of serologically nontypable and serotype b strains of Haemophilus influenzae. Infect Immun. 1986;52:183–191. doi: 10.1128/iai.52.1.183-191.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Musser J M, Kroll J S, Moxon E R, Selander R K. Clonal population structure of encapsulated Haemophilus influenzae. Infect Immun. 1988;56:1837–1845. doi: 10.1128/iai.56.8.1837-1845.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Musser J M, Kroll J S, Selander R K. Evolutionary genetics of the encapsulated strains of Haemophilus influenzae. Proc Natl Acad Sci USA. 1988;85:7758–7762. doi: 10.1073/pnas.85.20.7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nizet V, Colina K, Rubens A J, C E, Smith A L. A virulent nonencapsulated Haemophilus influenzae. J Infect Dis. 1996;173:180–186. doi: 10.1093/infdis/173.1.180. [DOI] [PubMed] [Google Scholar]
- 32.Ochman H, Wilson A C. Evolution in bacteria: evidence for a universal substitution rate in cellular genomes. J Mol Evol. 1987;26:74–86. doi: 10.1007/BF02111283. [DOI] [PubMed] [Google Scholar]
- 33.Perskins B A Brazilian Purpuric Fever Study Group. Brazilian purpuric fever identified in a new region of Brazil. J Infect Dis. 1992;165:S16–S19. doi: 10.1093/infdis/165-supplement_1-s16. [DOI] [PubMed] [Google Scholar]
- 34.Pichichero M E, Anderson P, Loeb M, Smith D H. Do pili play a role in pathogenicity of Haemophilus influenzae type b? Lancet. 1982;ii:960–962. doi: 10.1016/s0140-6736(82)90161-1. [DOI] [PubMed] [Google Scholar]
- 35.Read R C, Wilson R, Rutman A, Lund V, Todd C H, Brain A P R, Jeffery P K, Cole P J. Interaction of nontypable Haemophilus influenzae with human respiratory mucosa in vitro. J Infect Dis. 1991;163:549–558. doi: 10.1093/infdis/163.3.549. [DOI] [PubMed] [Google Scholar]
- 36.Read T D, Dowdell M, Satola S W, Farley M M. Duplication of pilus gene complexes of Haemophilus influenzae biogroup aegyptius. J Bacteriol. 1996;178:6564–6570. doi: 10.1128/jb.178.22.6564-6570.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 38.Smith A L, Smith D H, Averill D R, Marino J, Moxon E R. Production of Haemophilus influenzae b meningitis in infant rats by intraperitoneal inoculation. Infect Immun. 1973;8:278–290. doi: 10.1128/iai.8.2.278-290.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.St. Geme J W, III, Gilsdorf J R, Falkow S. Surface structures and adherence properties of diverse strains of Haemophilus influenzae biogroup aegyptius. Infect Immun. 1991;59:3366–3371. doi: 10.1128/iai.59.10.3366-3371.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.St. Geme J W, III, Kumar V V, Cutter D, Barenkamp S J. Prevalence and distribution of the hmw and hia genes and the HMW and Hia adhesins among genetically diverse strains of nontypeable Haemophilus influenzae. Infect Immun. 1998;66:364–368. doi: 10.1128/iai.66.1.364-368.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stull T, Mendelman P M, Haas J E, Schoenborn M A, Mack K D, Smith A L. Characterization of Haemophilus influenzae type b fimbriae. Infect Immun. 1984;46:787–796. doi: 10.1128/iai.46.3.787-796.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Turk D C. Clinical importance of Haemophilus influenzae—1981. In: Sell S H, Wright P F, editors. Haemophilus influenzae epidemiology, immunology, and prevention of disease. New York, N.Y: Elsevier/North Holland; 1982. pp. 3–9. [Google Scholar]
- 43.van Ham S M, van Alphen L, Mooi F, van Putten J P M. Phase variation of H. influenzae fimbriae: transcriptional control of two divergent genes through a variable combined promoter region. Cell. 1993;73:1187–1196. doi: 10.1016/0092-8674(93)90647-9. [DOI] [PubMed] [Google Scholar]
- 44.van Ham S M, van Alphen L, Mooi F R, van Putten J P M. The fimbrial gene cluster of Haemophilus influenzae type b. Mol Microbiol. 1994;6:277–282. doi: 10.1111/j.1365-2958.1994.tb00461.x. [DOI] [PubMed] [Google Scholar]
- 45.Weber A, Harris K, Lohrke S, Forney L, Smith A L. Inability to express fimbriae results in impaired ability of Haemophilus influenzae type b to colonize the nasopharynx. Infect Immun. 1991;59:4724–4728. doi: 10.1128/iai.59.12.4724-4728.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weiser J N, Chong S T H, Greenberg D, Fong W. Identification and characterization of a cell envelope protein of Haemophilus influenzae contributing to phase variation in colony opacity and nasopharyngeal colonization. Mol Microbiol. 1995;17:555–564. doi: 10.1111/j.1365-2958.1995.mmi_17030555.x. [DOI] [PubMed] [Google Scholar]