Abstract
A gene encoding β-galactosidase, designated mbgA, was isolated from Bacillus megaterium ATCC 14581. Chromosomal β-galactosidase production could be dramatically induced by lactose but not by isopropyl-β-d-thiogalactopyranoside (IPTG) and was subject to catabolite repression by glucose. Disruption of mbgA in the B. megaterium chromosome resulted in loss of lactose-inducible β-galactosidase production. A 27-bp inverted repeat was found to overlap the mbgA promoter sequence. Two partially overlapping catabolite-responsive elements (CREs) were identified within the inverted repeat. Base substitutions within CRE-I and/or CRE-II caused partial relief from catabolite repression. The results suggest that the 27-bp inverted repeat may serve as a target for a catabolite repressor(s).
Many catabolic enzymes in Bacillus spp. are subjected to catabolite repression by glucose or other rapidly metabolizable carbon sources (4, 11, 27, 33). The mechanism underlying catabolite repression in Bacillus spp. is quite different from the cyclic AMP (cAMP)-dependent positive regulatory mechanism operative in Escherichia coli (28). The latter involves a positive regulatory protein, cAMP receptor protein (19). In contrast, Bacillus does not contain detectable cAMP or cAMP receptor protein-like proteins (2, 13), and addition of exogeneous cAMP does not affect catabolite repression in B. subtilis (24). A negative transcription regulator, CcpA (7, 9, 10), which is a member of the LacI/GalR family (36), is believed to mediate catabolite repression by interacting with a cis-acting catabolite-responsive element (CRE) located in the regulatory region or coding region of catabolic-enzyme-encoding genes subjected to carbon catabolite repression (12, 35). The consensus sequences 5′-TGWNANCGNTNWCA-3′ (35), where W stands for adenine or thymine, and 5′-(T/A)GNAA(C/G)CGN(T/A)(T/A)NCA-3′ (12) have been proposed for the CRE. In this report, we describe the cloning, sequencing, expression, and regulation of the B. megaterium β-galactosidase gene (mbgA). A 27-bp inverted repeat was identified in the mbgA promoter region. Two CRE-like sequences within the 27-bp inverted repeat were found to be able to exert catabolite repression on β-galactosidase production in B. megaterium.
Cloning of the β-galactosidase gene.
To clone the β-galactosidase gene, chromosomal DNA of B. megaterium ATCC 14581 was isolated as described previously (37) and was partially digested with the restriction enzyme Sau3AI. DNA fragments ranging in size from 6 to 9 kb were gel purified and ligated into BamHI-cut and dephosphorylated vector pQE30 (Qiagen, Inc.). E. coli lac mutant JM109 (29) was transformed with the ligated DNA to generate a genomic library. The β-galactosidase gene was isolated by direct selection for enzyme activity on 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) plates. One plasmid isolated from a blue colony was found to contain an insert of about 9 kb and was subjected to further analysis. A 5-kb PstI fragment derived from the 9-kb insert was further subcloned into PstI-cut pQE30 in both orientations. Both of the resulting plasmids, when introduced into E. coli JM109, still gave blue colonies on Luria-Bertani (LB) medium plates (29) containing X-Gal. These results suggest that the 5-kb PstI fragment contains the cloned β-galactosidase gene and the promoter-like sequences from which transcription can start in E. coli.
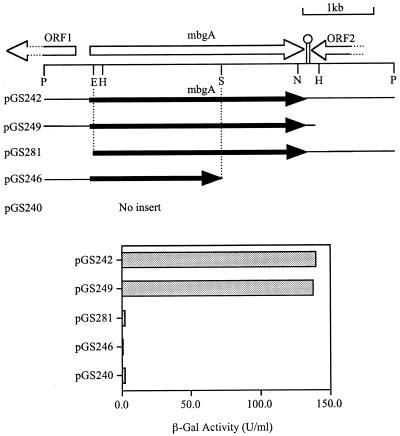
The 5-kb PstI fragment was further cloned into PstI-cut pBluescript KS(+) (Stratagene, Inc.) in both orientations for DNA sequencing by the dideoxy-chain termination method (30). The restriction map is shown in Fig. 1. One complete open reading frame (ORF) and two incomplete ORFs were identified. Based on its sequence homology to several published β-galactosidase genes, this complete ORF is assumed to be the structural gene for the β-galactosidase of B. megaterium and is designated here mbgA (megaterium β-galactosidase gene). This position of mbgA in the 5-kb PstI fragment is reinforced by assaying for changes in the β-galactosidase activity of various deletion derivatives of the PstI fragment shown in Fig. 1. pGS240, a modified form of pBluescript KS(+) which loses α-complementation ability (29), was constructed by inversion of the 0.45-kb PvuII fragment containing part of the coding sequence of the lacZ α fragment of pBluescript KS(+). Insertion of the 5-kb PstI fragment which contains the cloned mbgA gene into the PstI site of pGS240 yields plasmid pGS242. pGS246 was constructed by digestion of plasmid pGS242 with SalI to delete a 2.3-kb fragment, followed by religation. pGS281 was constructed by digestion of plasmid pGS242 with EcoRV to delete a 0.8-kb fragment, followed by religation. pGS249 was constructed by unidirectional deletion (6) with exonuclease III of a 1.2-kb fragment downstream of the cloned β-galactosidase gene.
FIG. 1.
Restriction map and genetic organization of the B. megaterium β-galactosidase gene (mbgA) and β-galactosidase activities of deletion derivatives of the 5-kb PstI fragment in E. coli. E, EcoRV; H, HindIII; N, NaeI; P, PstI; S, SalI.
Sequence analysis revealed that mbgA consists of 3,102 bp, which corresponds to a protein monomer of 1,034 amino acids with a calculated molecular mass of 118,088 Da (data not shown). Comparison of the deduced amino acid sequence encoded by the mbgA gene with those of the β-galactosidase of E. coli (15), Klebsiella pneumoniae (3), and Streptococcus thermophilus (31) revealed 43.7, 45.4, and 49.5% overall similarity, respectively. Immediately downstream of mbgA is an inverted repeat with the sequence AAGAAGGACTTTCATTTGAAAGTCCTTTTT, which may serve as a putative ρ-independent transcription terminator. Further downstream is an incomplete ORF transcribed in the opposite direction (Fig. 1). Further upstream is an incomplete ORF transcribed in the direction opposite to that of the mbgA gene (Fig. 1). The genetic organization of these ORFs suggests that B. megaterium contains a β-galactosidase gene that exists in a monocistronic operon. This is unlike the case in many other bacteria, where the β-galactosidase gene and the lactose permease gene usually coexist in an operon (15, 18, 26, 38). The nucleotide sequence of the mbgA regulatory region is shown in Fig. 2.
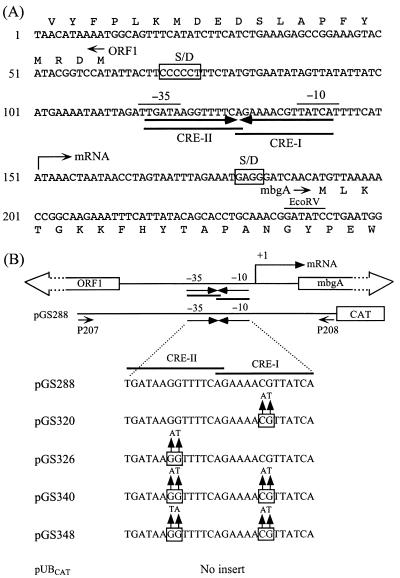
FIG. 2.
Nucleotide sequence of the regulatory region of the B. megaterium mbgA gene and base substitutions in CREs. (A) The ςA-like promoter sequence is overlined. The 27-bp inverted repeat that overlaps the promoter sequence is indicated by a pair of solid inverted arrows. Two partially overlapping CREs which are contained in the 27-bp inverted repeat are designated CRE-I and CRE-II. The N-terminal amino acids of ORF1 and mbgA are also shown. The putative ribosome-binding sites (S/D) are boxed. (B) pUBCAT-based plasmids pGS288, pGS320, pGS326, pGS340, and pGS348 contain a cat reporter gene transcriptionally fused to the mbgA promoter. pGS288 carries the wild-type mbgA promoter, whereas pGS320, pGS326, pGS340, and pGS348 carry various base substitutions in CREs. The positions of the two flanking primers, P207 (5′-GCGGGTCGACAAGCTCGCCTCTTCCTT-3′) and P208 (5′-GCGGAAGCTTTGGTAGTAAACAGA-3′), are also indicated.
Identification of the transcription initiation site of mbgA.
To determine the precise transcription initiation site of mbgA, RNA was isolated from log-phase B. megaterium cells harboring plasmid pGS288 (Fig. 2). A 16-mer oligonucleotide (5′-CGTTTGCAGGTGCTGT-3′) complementary to a region extending from position +71 to position +86 relative to the transcription start site of the mbgA gene was used as the primer. Primer extension was carried out as described previously (29), and the result is shown in Fig. 3. Only one major extension product was detected, indicating that the 5′ end of the mRNA for mbgA is located 40 bp upstream of its translation start site (Fig. 2). High-resolution S1 nuclease mapping (1) was also performed to further confirm this result. Two major protected fragments with only a one-base difference in length were observed (Fig. 3). One of them corresponds to a transcription initiation site which is identical to that identified by primer extension analysis. This transcription initiation site is at an appropriate distance from a putative promoter sequence (TTGATA for the −35 box and TATCAT for the −10 box) that may be recognized by the B. megaterium equivalent of B. subtilis ςA (23). A 27-bp inverted repeat overlaps the −35 and −10 regions of this putative promoter (Fig. 2). Within the 27-bp inverted repeat are two partially overlapping sequences (CRE-I and CRE-II) that exhibit considerable sequence similarity to the CRE consensus sequence (12).
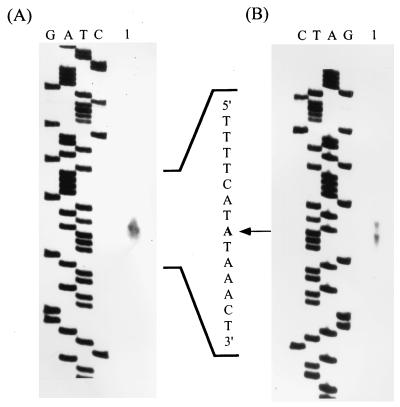
FIG. 3.
Determination of the transcription initiation site for mbgA by primer extension analysis and S1 nuclease mapping. (A) Primer extension analysis of transcripts was performed with RNA from B. megaterium cells harboring pGS288. Lane 1 shows the result of this analysis. (B) S1 nuclease mapping was carried out with RNA isolated from the same source as that used for panel A. Lane 1 shows the protected products. For both panels, dideoxy sequencing ladders obtained with the same primer used to make the probes for S1 nuclease mapping and primer extension experiments are shown. The sequence indicated is complementary to that read from the ladder.
Effects of lactose, glucose, and IPTG on β-galactosidase production in wild-type B. megaterium and the mbgA mutant.
The effects of lactose, glucose, and IPTG on β-galactosidase production were examined by using B. megaterium cells grown in LB medium. β-Galactosidase activities were measured at 37°C with o-nitrophenyl-β-d-galactopyranoside (ONPG) as the substrate by the method of Miller (20). It was found that 2% lactose could increase β-galactosidase production about 18-fold (Table 1). In the presence of 2% lactose plus 2% glucose, about 25-fold repression exerted by glucose was observed. Treatment with either 1 or 6 mM IPTG caused only a slight increase in β-galactosidase production compared to 2% lactose treatment. These results suggest that β-galactosidase production in B. megaterium is lactose inducible and is subject to catabolite repression by glucose. To examine the contribution of mbgA to lactose-mediated induction of β-galactosidase production in B. megaterium, we disrupted the mbgA gene in the B. megaterium chromosome by using integrative plasmid pGS278, which was constructed as follows. A 600-bp EcoRI-HindIII fragment carrying an internal sequence of the mbgA gene was generated by PCR using synthetic oligonucleotides P194 (5′-GCCAAAGCTTAAATGGAAGC-3′) and P195 (5′-GCGGAATTCAGCTGAAACGCTAAGTT-3′). This fragment was cloned into EcoRI- and HindIII-cut plasmid pDG1515 (5) to get plasmid pGS278. Disruption of the mbgA gene by a Campbell-like single-crossover recombination event was confirmed by Southern blot analysis (29) (data not shown). The mbgA mutant was then grown in LB medium in the absence or presence of 2% lactose to an optical density at 600 nm of 0.6 for β-galactosidase activity assay. As shown in Table 1, disruption of the mbgA gene in the B. megaterium chromosome abolished the lactose-mediated induction of β-galactosidase production. This suggests that the mbgA gene is the only lactose-inducible β-galactosidase gene in B. megaterium.
TABLE 1.
Effects of lactose, glucose, and IPTG on β-galactosidase production in wild-type B. megaterium and an mbgA mutant
| B. megaterium strain | Additiona | Mean β-galactosidase activityb (U/ml) ± SD |
|---|---|---|
| Wild type | None | 0.31 ± 0.03 |
| Lac | 5.54 ± 0.39 | |
| Lac + Glu | 0.22 ± 0.02 | |
| 1 mM IPTG | 0.39 ± 0.04 | |
| 6 mM IPTG | 0.35 ± 0.05 | |
| mbgA mutant | None | 0.15 ± 0.02 |
| Lac | 0.21 ± 0.02 |
Abbreviations: Lac, lactose; Glu, glucose.
B. megaterium cells were grown in LB medium to an optical density at 600 nm of 0.6 in the absence or presence of 2% sugar or IPTG, as specified. β-Galactosidase activities of the chloroform-treated B. megaterium cells were determined as described previously (20). Each value is the mean of four experiments.
Effects of mutations in CREs on catabolite repression of the mbgA promoter-cat transcriptional fusions.
To assess the contributions of the two CRE-like sequences to catabolite repression of β-galactosidase production, we firstly introduced a DNA fragment carrying the mbgA promoter and CRE-like sequences into promoter probe vector pUBCAT (37). The resulting plasmid, pGS288, bears an mbgA promoter-cat transcriptional fusion (Fig. 2). B. megaterium cells transformed with plasmid pGS288 were grown in LB medium in the absence or presence of lactose or lactose plus glucose. Chloramphenicol acetyltransferase (CAT) activities were measured at 37°C by the spectrophotometric method of Shaw (32). It was found that 2% glucose could still exert about ninefold catabolite repression (Table 2).
TABLE 2.
Effects of base substitutions in CREs on catabolite repression of mbgA promoter-cat transcriptional fusions
| Plasmid in B. megateriuma | Additionb | Mean CAT sp actc (mU/mg of protein) ± SD | Fold repressiond |
|---|---|---|---|
| pGS288 | None | 1,310 ± 67 | |
| Lac | 1,300 ± 82 | ||
| Lac + Glu | 140 ± 21 | 9.3 | |
| pGS320 | None | 1,290 ± 59 | |
| Lac | 1,310 ± 73 | ||
| Lac + Glu | 730 ± 34 | 1.8 | |
| pGS326 | None | 2,210 ± 117 | |
| Lac | 1,880 ± 110 | ||
| Lac + Glu | 1,320 ± 61 | 1.4 | |
| pGS340 | None | 400 ± 28 | |
| Lac | 380 ± 24 | ||
| Lac + Glu | 180 ± 17 | 2.1 | |
| pGS348 | None | 1,340 ± 69 | |
| Lac | 1,420 ± 75 | ||
| Lac + Glu | 1,090 ± 58 | 1.3 | |
| pUBCAT | None | 90 ± 18 | |
| Lac | 85 ± 14 | ||
| Lac + Glu | 89 ± 12 |
The plasmids listed are shown in Fig. 2.
Abbreviations: Lac, lactose; Glu, glucose.
At least two independent clones for each construct were chosen for CAT activity assays. B. megaterium cells were grown in LB medium to an optical density at 600 nm of 1.0 in the absence or presence of 2% sugar, as specified. After sonication and centrifugation, crude extracts were subjected to CAT assays (32). Each value is the mean of at least six determinations in separate experiments.
Lac/(Lac + Glu).
We also made some mbgA promoter-cat transcriptional fusion constructs in which CRE-I, CRE-II, or both were mutated by the two-step PCR method (8) (Fig. 2). We then examined the responses of these fusions to lactose and glucose. For each construct, at least two independent clones were chosen for CAT activity assays in order to obtain consistent results. The results showed that mutations in either of the two CREs caused partial relief of catabolite repression (Table 2). Mutations in CRE-I of pGS320 and CRE-II of pGS326 decreased catabolite repression from about 9-fold in the wild type to about 1.8-fold and 1.4-fold, respectively. These results suggest that both CRE-I and CRE-II can function as an active CRE for catabolite repression of the β-galactosidase gene of B. megaterium. When CRE-I and CRE-II were mutated together (pGS340 and pGS348 in Fig. 2), catabolite repression was still not completely abolished (Table 2). These results suggest that either another mechanism(s) is involved in catabolite repression of mbgA expression or another CRE-like sequence(s) is located in the coding region of the mbgA gene. Previous studies indicated that, in addition to a promoter-proximal CRE, a cis-acting site required for catabolite repression was also identified in the coding regions of the hutP (25, 39), gntR (21, 22), and xylA (14) genes of B. subtilis. We cannot exclude the possibility that base substitutions in the central CG or GG residues of the two CREs could only partially reduce the affinity of the catabolite repressor for these two sites. However, previous in vitro studies indicated that alterations of the central residues could prevent binding of the catabolite repressor to CREs identified in some Bacillus genes (17, 22). It remains to be experimentally determined whether two catabolite repressor molecules can bind to these two CREs simultaneously. Investigation by in vitro gel mobility shift assays and in vivo studies of the possible role of CcpA in catabolite repression of mbgA expression via the two CREs should help to clarify these issues.
On the other hand, mutations of CRE-I, CRE-II, or both had differential effects on basal expression from the cat reporter gene. Mutations in CRE-I or pGS320 had little or no effect on basal expression, whereas mutations in CRE-II of pGS326 caused about a 1.7-fold increase in the basal expression (Table 2). The estimated ΔG values for the potential hairpin structure of the 27-bp inverted repeat (an 11-bp stem and a 5-base loop) in pGS288, pGS320, pGS326, pGS340, and pGS348 are −5.8, −3.6, −3.6, −9.8, and −3.6 kcal/mol, respectively, according to the method of Tinoco et al. (34). Site-directed mutagenesis in pGS348 reduced the dyad symmetry (ΔG = −3.6 kcal/mol) of the 27-bp inverted repeat, whereas mutagenesis in pGS340 enhanced the dyad symmetry (ΔG = −9.8 kcal/mol) in such a way that a perfect 27-bp inverted repeat was obtained (Fig. 2). Mutations in both CRE-I and CRE-II of pGS348 had little or no effect on the basal expression, whereas mutations in pGS340, which strengthen the hairpin, caused about a 3.3-fold reduction in the basal expression. Base substitutions in either half of the 27-bp inverted repeat did not directly change the sequences in the −35 and −10 boxes of the mbgA promoter. However, it was still possible that alterations of their flanking sequences might affect RNA polymerase recognition and binding, thus altering the promoter strength.
In contrast to the dramatic induction of chromosomal β-galactosidase production by lactose, cat expressions from the various mbgA promoter-cat transcriptional fusions on the pUB110-derived high-copy plasmids (16) (ca. 50 copies per chromosome) were not lactose inducible (Table 2). One possible reason is that the concentration of the putative lactose-responsive repressor (not the catabolite repressor) in B. megaterium is relatively very low. A limited amount of the repressor might efficiently repress expression of the single-copy mbgA gene in the chromosome (lactose inducible) but might not be enough to repress expression of multiple copies of the mbgA gene on a high-copy plasmid (not lactose inducible). Another possibility is that the lactose-responsive cis-acting element is not contained in the mbgA promoter region mentioned above.
In conclusion, we have identified two CRE-like sequences within the 27-bp inverted repeat which can exert catabolite repression on β-galactosidase production in B. megaterium. To our knowledge, this is the first example of two functional overlapping CREs that exist in the promoter region of a Bacillus gene. In CRE-II, the central two residues are GG instead of the highly conserved CG residues. The sequencing markers shown in Fig. 3 clearly indicated that this was not a sequencing error. Moreover, this is not unprecedented; it has been reported that a functional CRE identified in the promoter region of gntR contains central TG residues (22). Although we have demonstrated that the β-galactosidase of B. megaterium is capable of hydrolyzing ONPG and X-Gal, the physiological substrates for the β-galactosidase of B. megaterium remain undetermined. Work using the purified β-galactosidase of B. megaterium is needed to determine if it can use lactose as a substrate.
Nucleotide sequence accession number.
The nucleotide sequence reported here has been assigned GenBank accession no. AF047824.
Acknowledgments
We are grateful to Douglas J. Cork, a Visiting Professor from the Department of Biological, Chemical and Physical Sciences, Illinois Institute of Technology, Chicago, for reviewing the manuscript.
This research was supported by grant NSC 86-2316-B-010-013-B19 from the National Science Council of the Republic of China.
REFERENCES
- 1.Berk A J, Sharp P A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977;12:721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- 2.Bernlohr R W, Maddock M K, Goldberg N D. Cyclic guanosine 3′:5′ monophosphate in Escherichia coli and Bacillus licheniformis. J Biol Chem. 1974;249:4329–4331. [PubMed] [Google Scholar]
- 3.Buvinger W E, Riley M. Nucleotide sequence of Klebsiella pneumoniae lac genes. J Bacteriol. 1985;163:850–857. doi: 10.1128/jb.163.3.850-857.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chambliss G H. Carbon source-mediated catabolite repression. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus and other gram-positive bacteria—1993. Washington, D.C: American Society for Microbiology; 1993. pp. 213–219. [Google Scholar]
- 5.Guerout-Fleury A-M, Shazand K, Frandsen N, Stragier P. Antibiotic-resistance cassettes for Bacillus subtilis. Gene. 1995;167:335–336. doi: 10.1016/0378-1119(95)00652-4. [DOI] [PubMed] [Google Scholar]
- 6.Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984;28:351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- 7.Henkin T M, Grundy F J, Nicholson W L, Chambliss G H. Catabolite repression of α-amylase gene expression in Bacillus subtilis involves a trans-acting gene product homologous to the Escherichia coli LacI and GalR repressors. Mol Microbiol. 1991;5:575–584. doi: 10.1111/j.1365-2958.1991.tb00728.x. [DOI] [PubMed] [Google Scholar]
- 8.Higuchi R, Krummel B, Saiki R K. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hueck C J, Kraus A, Hillen W. Sequences of ccpA and two downstream Bacillus megaterium genes with homology to the motAB operon from Bacillus subtilis. Gene. 1994;143:147–148. doi: 10.1016/0378-1119(94)90621-1. [DOI] [PubMed] [Google Scholar]
- 10.Hueck C J, Kraus A, Schmiedel D, Hillen W. Cloning, expression, and functional analysis of the catabolite control protein CcpA from Bacillus megaterium. Mol Microbiol. 1995;16:855–864. doi: 10.1111/j.1365-2958.1995.tb02313.x. [DOI] [PubMed] [Google Scholar]
- 11.Hueck C J, Hillen W. Catabolite repression in Bacillus subtilis: a global regulatory mechanism for the Gram-positive bacteria? Mol Microbiol. 1995;15:395–401. doi: 10.1111/j.1365-2958.1995.tb02252.x. [DOI] [PubMed] [Google Scholar]
- 12.Hueck C J, Hillen W, Saier M H., Jr Analysis of a cis-active sequence mediating catabolite repression in Gram-positive bacteria. Res Microbiol. 1994;145:503–518. doi: 10.1016/0923-2508(94)90028-0. [DOI] [PubMed] [Google Scholar]
- 13.Ide M. Adenyl cyclase of bacteria. Arch Biochem Biophys. 1971;144:262–268. [Google Scholar]
- 14.Jacob S, Allmansberger R, Gartner D, Hillen W. Catabolite repression of the operon for xylose utilization from Bacillus subtilis W23 is mediated at the level of transcription and depends on a cis site in the xylA reading frame. Mol Gen Genet. 1991;229:189–196. doi: 10.1007/BF00272155. [DOI] [PubMed] [Google Scholar]
- 15.Kalnins A, Otto K, Ruther U, Muller-Hill B. Sequence of the lacZ gene of Escherichia coli. EMBO J. 1983;2:593–597. doi: 10.1002/j.1460-2075.1983.tb01468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keggins K M, Lovett P S, Duvall E J. Molecular cloning of genetically active fragments of Bacillus DNA in Bacillus subtilis and properties of the vector plasmid pUB110. Proc Natl Acad Sci USA. 1978;75:1423–1427. doi: 10.1073/pnas.75.3.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim J H, Guvener Z T, Cho J Y, Chung K-C, Chambliss G H. Specificity of DNA binding activity of the Bacillus subtilis catabolite control protein CcpA. J Bacteriol. 1995;177:5129–5134. doi: 10.1128/jb.177.17.5129-5134.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leong-Morgenthaler P M, Zwahlen M C, Hottinger H. Lactose metabolism in Lactobacillus bulgaricus: analysis of the primary structure and expression of the genes involved. J Bacteriol. 1991;173:1951–1957. doi: 10.1128/jb.173.6.1951-1957.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Magasanik B, Neidhardt F C. Regulation of carbon and nitrogen utilization. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1987. pp. 1318–1325. [Google Scholar]
- 20.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- 21.Miwa Y, Fujita Y. Promoter-independent catabolite repression of the Bacillus subtilis gnt operon. J Biochem. 1993;113:665–671. doi: 10.1093/oxfordjournals.jbchem.a124100. [DOI] [PubMed] [Google Scholar]
- 22.Miwa Y, Nagura K, Eguchi S, Fukuda H, Deutscher J, Fujita Y. Catabolite repression of the Bacillus subtilis gnt operon exerted by two catabolite-responsive elements. Mol Microbiol. 1997;23:1203–1213. doi: 10.1046/j.1365-2958.1997.2921662.x. [DOI] [PubMed] [Google Scholar]
- 23.Moran C P, Jr, Lang N, LeGrice S F J, Lee G, Stephens M, Sonenshein A L, Pero J, Losick R. Nucleotide sequences that signal the initiation of transcription and translation in Bacillus subtilis. Mol Gen Genet. 1982;186:339–346. doi: 10.1007/BF00729452. [DOI] [PubMed] [Google Scholar]
- 24.Nihashi J, Fujita Y. Catabolite repression of inositol dehydrogenase and gluconate kinase syntheses in Bacillus subtilis. Biochim Biophys Acta. 1984;798:88–95. doi: 10.1016/0304-4165(84)90014-x. [DOI] [PubMed] [Google Scholar]
- 25.Oda M, Katagai T, Tomura D, Shoun H, Hoshino T, Furukawa K. Analysis of the transcriptional activity of the hut promoter in Bacillus subtilis and identification of a cis-acting regulatory region associated with catabolite repression downstream from the site of transcription. Mol Microbiol. 1992;6:2573–2582. doi: 10.1111/j.1365-2958.1992.tb01434.x. [DOI] [PubMed] [Google Scholar]
- 26.Poolman B, Royer T J, Mainzer S E, Schmidt B F. Lactose transport system of Streptococcus thermophilus: a hybrid protein with homology to the melibiose carrier and enzyme III of phosphoenolpyruvate-dependent phosphotransferase systems. J Bacteriol. 1989;171:244–253. doi: 10.1128/jb.171.1.244-253.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saier M H, Jr, Chauvaux S, Cook G M, Deutscher J, Paulsen I T, Reizer J, Ye J-J. Catabolite repression and inducer control in Gram-positive bacteria. Microbiology. 1996;142:217–230. doi: 10.1099/13500872-142-2-217. [DOI] [PubMed] [Google Scholar]
- 28.Saier M H, Jr, Chauvaux S, Deutscher J, Reizer J, Ye J-J. Protein phosphorylation and regulation of carbon metabolism in Gram-negative versus Gram-positive bacteria. Trends Biochem Sci. 1995;20:267–271. doi: 10.1016/s0968-0004(00)89041-6. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schroeder C J, Robert C, Lenzen G, McKay L L, Mercenier A. Analysis of the lacZ sequences from two Streptococcus thermophilus strains: comparison with the Escherichia coli and Lactobacillus bulgaricus β-galactosidase sequences. J Gen Microbiol. 1991;137:369–380. doi: 10.1099/00221287-137-2-369. [DOI] [PubMed] [Google Scholar]
- 32.Shaw W V. Chloramphenicol acetyltransferase from chloramphenicol-resistant bacteria. Methods Enzymol. 1975;43:737–755. doi: 10.1016/0076-6879(75)43141-x. [DOI] [PubMed] [Google Scholar]
- 33.Stewart G C. Catabolite repression in the Gram-positive bacteria: generation of negative regulators of transcription. J Cell Biochem. 1993;51:25–28. doi: 10.1002/jcb.240510106. [DOI] [PubMed] [Google Scholar]
- 34.Tinoco I, Borer P N, Dengler B, Levine M D, Uhlenbeck O C, Crothers D M, Gralla J. Improved estimation of secondary structure in ribonucleic acids. Nature (London) New Biol. 1973;246:40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- 35.Weickert M J, Chambliss G H. Site-directed mutagenesis of a catabolite repression operator sequence in Bacillus subtilis. Proc Natl Acad Sci USA. 1990;87:6238–6242. doi: 10.1073/pnas.87.16.6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weickert M J, Adhya S. A family of bacterial regulators homologous to Gal and Lac repressors. J Biol Chem. 1992;267:15869–15874. [PubMed] [Google Scholar]
- 37.Wen L P, Fulco A J. Cloning of the gene encoding a catalytically self-sufficient cytochrome P450 fatty acid monooxygenase induced by barbiturates in Bacillus megaterium and its functional expression and regulation in heterologous (Escherichia coli) and homologous (Bacillus megaterium) hosts. J Biol Chem. 1987;262:6676–6682. [PubMed] [Google Scholar]
- 38.Williams S G, Greenwood J A, Jones C W. Molecular analysis of the lac operon encoding the binding-protein-dependent lactose transport system and β-galactosidase in Agrobacterium radiobacter. Mol Microbiol. 1992;6:1755–1768. doi: 10.1111/j.1365-2958.1992.tb01348.x. [DOI] [PubMed] [Google Scholar]
- 39.Wray L V, Jr, Pettengill F K, Fisher S H. Catabolite repression of the Bacillus subtilis hut operon requires a cis-acting site located downstream of the transcription initiation site. J Bacteriol. 1994;176:1894–1902. doi: 10.1128/jb.176.7.1894-1902.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]