Abstract
Background
Breast cancer (BC) risk prediction models consider cancer family history (FH) and germline pathogenic variants (PVs) in risk genes. It remains elusive to what extent complementation with polygenic risk score (PRS) and non-genetic risk factor (NGRFs) data affects individual intensified breast surveillance (IBS) recommendations according to European guidelines.
Methods
For 425 cancer-free women with cancer FH (mean age 40·6 years, range 21–74), recruited in France, Germany and the Netherlands, germline PV status, NGRFs, and a 306 variant-based PRS (PRS306) were assessed to calculate estimated lifetime risks (eLTR) and estimated 10-year risks (e10YR) using CanRisk. The proportions of women changing country-specific European risk categories for IBS recommendations, i.e. ≥20 % and ≥30 % eLTR, or ≥5 % e10YR were determined.
Findings
Of the women with non-informative PV status, including PRS306 and NGRFs changed clinical recommendations for 31·0 %, (57/184, 20 % eLTR), 15·8 % (29/184, 30 % eLTR) and 22·4 % (41/183, 5 % e10YR), respectively whereas of the women tested negative for a PV observed in their family, clinical recommendations changed for 16·7 % (25/150), 1·3 % (2/150) and 9·5 % (14/147). No change was observed for 82 women with PVs in high-risk genes (BRCA1/2, PALB2). Combined consideration of eLTRs and e10YRs identified BRCA1/2 PV carriers benefitting from IBS <30 years, and women tested non-informative/negative for whom IBS may be postponed.
Interpretation
For women who tested non-informative/negative, PRS and NGRFs have a considerable impact on IBS recommendations. Combined consideration of eLTRs and e10YRs allows personalizing IBS starting age.
Funding
Horizon 2020, German Cancer Aid, Federal Ministry of Education and Research, Köln Fortune.
Keywords: Breast cancer, Genetic testing, Hereditary breast and ovarian cancer syndrome, Cancer prevention
Highlights
-
•
Breast cancer (BC) risk prediction considers cancer family history and germline pathogenic variants (PVs).
-
•
Polygenic risk score (PRS) and non-genetic risk factors (NGRFs) may affect intensified breast surveillance (IBS) recommendations.
-
•
For women without PVs, PRS and NGRFs considerably impact IBS recommendations.
-
•
Comprehensive risk calculation may personalize IBS starting ages.
1. Introduction
In the context of familial/hereditary breast and ovarian cancer (OC), breast cancer (BC) risk prediction models are widely used for assessing individual risks of developing BC. Established BC risk prediction models such as the Breast and Ovarian Analysis of Disease Incidence and Carrier Estimation Algorithm (BOADICEA) consider cancer family history (FH) and pathogenic variant (PV) status for BC risk genes. In addition, polygenic risk scores (PRSs) and non-genetic risk factors (NGRFs) have been integrated in BOADICEA version 5, implemented in the CE-marked CanRisk web interface [[1], [2], [3]]. Considering these additional risk factors further refines individual BC risk prediction and helps to identify women at moderate/high BC risk who may benefit from intensified breast surveillance (IBS) programmes while avoiding unnecessary preventive measures for women who are at low BC risk [4].
IBS programmes for individuals at elevated BC risk commonly include breast magnetic resonance imaging (MRI), ultrasound, and mammography; however, screening guidelines differ between European countries, especially for non-BRCA1/2 PV carriers [3]. In most European countries, the clinical recommendations for IBS are based on the individual estimated lifetime risk (eLTR). Most guidelines define eLTRs ≥30 % as high, generally justifying IBS, whereas eLTR thresholds of ≥20 % frequently define moderately increased BC risk, justifying less intense IBS modalities [3]. In Germany, however, clinical recommendations on IBS mainly depend on the estimated 10-year risk (e10YR) using a 5 % threshold.
CanRisk was shown to be well-calibrated for BC risk prediction in both the high-risk BC setting as well as the general population [5]. Yet, it remains unclear to what extent the complementation of CanRisk-based BC risk prediction with data on further genetic (PRS) [6] and non-genetic risk factors (NGRFs) affects clinical recommendations on an individual level according to country-specific guidelines. In this study, we assessed the clinical implications of a comprehensive CanRisk-based BC risk prediction in comparison to a risk prediction that only considers cancer FH and PV status. The study sample comprises 425 cancer-free women with a FH of BC and/or OC consecutively enrolled at three study sites in Germany, France and the Netherlands.
2. Material and methods
2.1. Study sample
As part of the BRIDGES project [7], 469 women with a FH of BC and/or OC were approached for participation in this study between November 2019 and June 2021. 452 study participants, all aged 18 years or above, were recruited at the Cologne Center for Hereditary Breast and Ovarian Cancer (CC-HBOC, Germany, n = 214 women), the Institut Curie, Paris (IC, France, n = 199 women), and the Leiden University Medical Center (LUMC, Netherlands, n = 39 women). Criteria for study inclusion differed between the three study sites (Table 1).
Table 1.
Criteria for study inclusion by study center. CC-HBOC, Cologne Center for Hereditary Breast and Ovarian Cancer; GC-HBOC, German Consortium for Hereditary Breast and Ovarian cancer; IC, Institut Curie; LUMC, Leiden University Medical Center; PV, pathogenic variant.
| Study center | Criteria for the study inclusion |
|---|---|
| CC-HBOC (Germany) | Cancer-free female relatives of index patients, irrespective of the germline PV status of the index patient. All index patients met the inclusion criteria of the German Consortium for Hereditary Breast and Ovarian cancer (GC-HBOC) for germline testing [8]. |
| IC (France) | Cancer-free first degree female relatives of index patients. Index patients carried pathogenic variants in BRCA1, BRCA2, or PALB2. |
| LUMC (Netherlands) | Cancer-free female relatives of index patients. Index patients were affected with BC and did not carry (likely) pathogenic variants in ATM, BRCA1, BRCA2, CHEK2, or PALB2. |
2.2. Ethics statement
Written informed consent was obtained from all study participants. Ethical approval was granted by the Ethics Committee of the University of Cologne, Germany (N°16–098), the Comité consultatif sur le traitement de l'information en matière de recherche dans le domaine de la santé (CCTIRS: Consultative committee for information management in health research – N°16.314) and the Comité de Protection des Personnes Ile-de-France V (CPP – N° 18.12.28.38743 CAT2), France, and the Medisch-Ethische Toetsingscommissie Leiden Den Haag Delft (NL68501.058.18), the Netherlands.
2.3. Targeted next generation sequencing
Genomic DNA was isolated from venous EDTA blood samples using standard techniques. For all 452 study participants, gene panel testing was performed centrally at the CC-HBOC in a routine diagnostic setting using the customized hybridization capture-based TruRisk® v3 gene panel for target enrichment (Agilent SureSelect, QXT protocol). The TruRisk® v3 gene panel covers the entire coding regions of 17 established cancer predisposition genes (ATM, NM_000051.3; BARD1, NM_000465.3; BRCA1, NM_007294.3; BRCA2, NM_00059.3; BRIP1, NM_032043.2; CDH1, NM_004360.4; CHEK2, NM_007194.3; MLH1, NM_000249.3; MSH2, NM_000251.2; MSH6, NM_000179.2; PALB2, NM_024675.3; PMS2, NM_000535.6; PTEN, NM_000314.6; RAD51C, NM_058216.2; RAD51D, NM_002878.3; STK11, NM_000455.4; TP53, NM_000546.5). In addition, the TruRisk® v3 gene panel covers 306 risk variants for the calculation of a BC PRS [6]. These 306 risk variants are implemented as 'BRIDGES_306_PRS' in the CanRisk web interface [1]. Next generation sequencing (NGS) was performed using a NextSeq 500 device (Illumina, San Diego, CA, U.S.).
2.4. Genotyping of common variants, quality control and test for ancestry
For genotyping, sequencing reads were mapped to the reference genome assembly GRCh37 including decoy sequences (hs37d5) using the BWA-MEM functionality of Burrows-Wheeler-Aligner v0.7.15 [9] and further processed according to the GATK Best Practices for germline variant calling using GATK v4.1 [10]. Genotyping was performed on a merged BAM file including all samples using FreeBayes v1.3.1 [11] under specification of common variants from dbSNP build 151 [12] as variant input. Quality control included checks for duplicated samples and was performed as described previously [13]. To identify study participants of non-European ancestry, minor allele frequencies of European (EUR), African (AFR), South-Asian (SAS) and East-Asian (EAS) samples in 1000 Genomes Project phase 3 [14] of 665 variants in linkage equilibrium served as input for principal component analysis (PCA) using R's prcomp () function. The resulting rotation matrix was applied to transform the samples' genotype data (with genotypes coded as 0, 0.5 and 1) using the predict () function, and samples that did not show the smallest Euclidean distance to EUR were excluded due to putative non-European ancestry.
2.5. Detection and classifications of rare variants
Detection of rare (likely) PVs was carried out using the SeqNext module of the SeqPilot Software Package (JSI medical systems GmbH, Ettenheim, Germany). The Alamut Visual version 2.13 analysis software tool (Interactive Biosoftware, Rouen, France) was applied for variant annotation and integration of up-to-date classifications from the ClinVar database (www.ncbi.nlm.nih.gov/clinvar/). Variant classification was performed using the criteria of the GC-HBOC for the classification of germline sequence variants in predisposition genes for hereditary BC and OC [15], which are based on the guidelines by the Evidence-Based Network for the Interpretation of Germline Mutant Alleles (ENIGMA, www.enigmaconsortium.org) and the American College of Medical Genetics and Genomics (ACMG) [16]. As proposed by the International Agency for Research on Cancer (IARC), a five-tier classification system was applied [17]. This classification system defines germline variants as pathogenic (class 5), likely pathogenic (class 4), variant of uncertain significance (VUS, class 3), likely benign (class 2), or benign (class 1). Protein-truncating variants (PTVs) were defined as nonsense, frameshift, or essential splice site variants affecting the invariant splice sites or the last nucleotide of an exon. PVs included (likely) pathogenic PTVs and (likely) pathogenic missense variants. All PVs were verified by Sanger sequencing. For the prediction of copy number variations (CNVs) in blood-derived DNA, we employed the CE-IVD-marked Sophia Genetics DDM pipeline v3.4.0–4.6.2 (Sophia Genetics, Saint-Sulpice, Switzerland), as described previously [18]. Predicted CNVs were verified by multiplex ligation-dependent probe amplification (MLPA) using SALSA® MLPA® kits (MRC Holland, Amsterdam, Netherlands).
2.6. Polygenic risk score (PRS) computation
The CanRisk Variant Call Format (VCF) upload utility was used for 'BRIDGES_306_PRS' computation. Genotyping was performed by running FreeBayes v1.3.1 [11] sample-wise under specification of the corresponding 306 variants as defined by CanRisk as variant input in PRS306 reference file BRIDGES 306 [6]. Mean read coverages over all PRS variant loci ranged from 35x to 396x. Variants which could not be genotyped due to technical reasons (indicated by GT = ./. in FreeBayes VCF output) were imputed by the corresponding doubled effect allele frequency as defined in the CanRisk specifications under usage of the dosage (DS) tag. This imputation approach was applied for 20 samples for a single variant each. For the remaining 432 samples, the entire BRIDGES 306 variant set could be genotyped.
2.7. CanRisk-based breast cancer risk estimations
Carriers of PVs in genes not included in BOADICEA V5 were not considered for this analysis, as for these, no meaningful BC risk estimation can be provided (Supplementary Fig. 1, CONSORT diagram). CanRisk versions 1.0.4–1.2.2 were used for BC risk estimations. CanRisk updates did not affect the BOADICEA V5 algorithm. To allow for cross-country comparisons, cancer incidence rates and genetic test sensitivities were set to 0·9 (overall) and 1·0 (CHEK2). For each study participant, we carried out four different BC risk estimations (Table 2). The BASIC BC risk estimation considered FH and PV status from germline gene panel analyses in moderate- (ATM, CHEK2) and high-risk genes (BRCA1, BRCA2, PALB2). The PRS BC risk estimation considered FH, PV status and the BRIDGES PRS306. The NGRF risk estimation considered FH, PV status and NGRFs (i.e., age at menarche, parity, age at first live birth, use of oral contraception, use of menopause hormone therapy, body mass index, daily alcohol intake, height). The FULL BC risk estimation considered FH, PV status, the BRIDGES PRS306 and NGRFs. For NGRF and FULL risk calculation, mammographic density was not considered, as this information was not available for a sufficient number of study participants. Estimated BC risks were automatically extracted from the CanRisk reports using in-house scripts. For further analyses, we considered eLTRs, defined as the risk of developing BC between 20 and 80 years, and e10YRs for women aged <70 years, defined as the risk of developing BC in the next 10 years.
Table 2.
CanRisk-based BC risk estimation models used in this study. FH = family history of breast, ovarian, prostate and pancreatic cancer, age information on unaffected family members, information on birth cohort; PV status = genetic test result from gene panel analyses for pathogenic variants (PVs) in moderate- or high-risk predisposition genes (ATM, BRCA1, BRCA2, PALB2, CHEK2); NGRF = non-genetic risk factors (age of menarche, parity, age at first live birth, use of oral contraception, use of menopause hormone therapy, body mass index, daily alcohol intake, height). PRS306 = 306 variant-based BRIDGES PRS7.
| Risk estimation model | Risk factor category |
|||
|---|---|---|---|---|
| FH | PV status | NGRF | PRS306 | |
| BASIC | ● | ● | ||
| NGRF | ● | ● | ● | |
| PRS | ● | ● | ● | |
| FULL | ● | ● | ● | ● |
2.8. Statistical analyses
For each subgroup, we provide descriptive data on mean, standard deviation, and range of eLTRs and e10YRs. Two-sided Welch's two sample t-test was employed for comparison of age distributions, with p-values <0·05 considered statistically significant. Analyses were conducted using R statistical software (version 4.1.2).
3. Results
Of the 452 women with a FH of BC and/or OC enrolled at the three study sites, 27 were excluded (Supplementary Fig. 1, CONSORT diagram) and the remaining 425 (mean age 40·6 years, range 21–74 years) were assigned to subgroups defined by PV status. A majority of 184 women tested non-informative, i.e., no PVs were detected in established cancer predisposition risk genes covered by the TruRisk® v3.1 gene panel, and no PVs in these genes were reported in the respective index patients. Another 150 women tested negative for the PVs observed in the respective index patients, and 91 women tested positive for PVs. Of these 91 women, 82 carried PVs in any of the high-risk genes BRCA1, BRCA2, or PALB2, and nine in the moderate-risk genes ATM, or CHEK2.
The eLTRs varied broadly and differed between the subgroups (Fig. 1, Supplementary Table 1). Using the BASIC model, women who tested negative had the lowest mean eLTR of 13·74 % (range 7·2 % to 24·9 %), followed by women who tested non-informative (19·67 %, range 10·0 % to 37·0 %). For carriers of PVs in moderate-risk genes, the mean eLTR was 39·57 % (range 34·0 % to 46·1 %). For carriers of PVs in high-risk genes, the mean eLTR was 74·65 % (range 44·9 % to 94·1 %). The integration of additional risk factor categories (NGRF, PRS, FULL) resulted in larger standard deviations of eLTRs compared with the BASIC model (Fig. 1, Supplementary Table 1), suggesting a higher degree of risk discrimination.
Fig. 1.
Boxplots of estimated lifetime risks (eLTR) for breast cancer depending on the CanRisk estimation model and genetic test result. NGRF = non-genetic risk factors, PRS = polygenic risk score, FULL = comprehensive model including PRS306 and NGRFs.
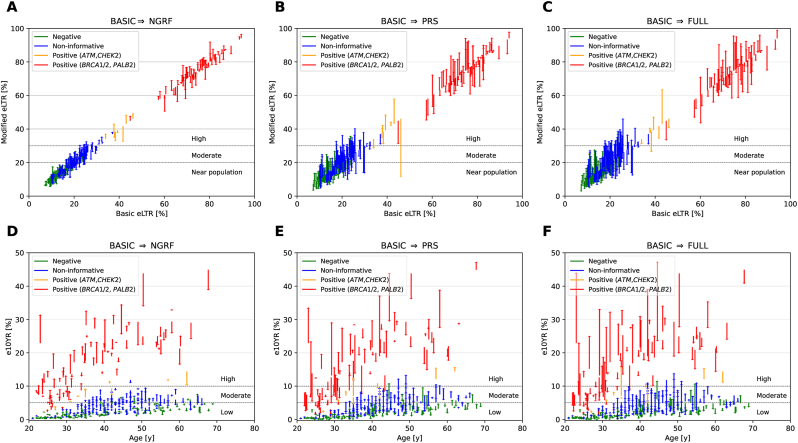
On the individual level, 88·7 % (133/150) of the women who tested negative were at eLTR near general population, 11·3 % (17/150) were at moderate, and none were at high eLTR when using the BASIC model (Table 3, Fig. 2 A–C). When using the FULL model, clinical recommendations changed for 16·7 % (25/150) when applying the 20 % eLTR threshold and for 1·3 % (2/150) when applying the 30 % eLTR threshold. More pronounced effects were observed for women who tested non-informative; 53·3 % (98/184) were at near general population eLTR, 44·0 % (81/184) were at moderate and 2·7 % (5/184) at high eLTR when using the BASIC model (Table 3, Fig. 2 A–C). When using the FULL model, clinical recommendations changed for 31·5 % (58/184) when applying the 20 % eLTR threshold and for 15·8 % (29/184) when applying the 30 % eLTR threshold. For women who tested either negative or non-informative, the inclusion of additional risk factors resulted in a decrease of patients with eLTRs near the general population in our study sample. The proportion of women with eLTRs near general population decreased by 10·0% points to 78·7 % (118/150) and by 6·6% points to 46·7 % (86/184), respectively, when applying the FULL model (Table 3). For nine women with PVs in moderate-risk genes, no category change was observed when applying the FULL model and the 20 % eLTR threshold. When using the 30 % eLTR threshold, clinical recommendations changed for one woman. The 82 women with PVs in high-risk genes all remained in the high eLTR category, irrespective of the underlying risk estimation model.
Table 3.
Changes in individual-level breast cancer (BC) risk categories according to CanRisk estimations. ↑(↓) indicating switches to higher (lower) risk categories compared to the BASIC model. a = three women aged ≥70 years excluded; b = one woman aged ≥70 years excluded. # = Only in this subgroup, four women switched two categories from <20 % eLTR (BASIC) to ≥30 % eLTR (FULL). A switch from ≥30 % eLTR to <20 % eLTR was not observed in any subgroup when comparing FULL vs BASIC model. ## In this subgroup, one woman switched two categories from <5 % e10YR to ≥10 % e10YR. ### In this subgroup, one woman switched two categories from ≥10 % e10YR to <5 % e10YR. Abbreviations: BC, breast cancer; eLTR, estimated BC lifetime risk; e10YR, estimated 10-year BC risk; NGRF, non-genetic risk factors; PRS, polygenic risk score.
| eLTR near general population (<20 %) | eLTR moderate (≥20 % and <30 %) | eLTR high (≥30 %) | category changes (<20 % vs ≥ 20 %) | category changes (<30 % vs ≥ 30 %) | ||
|---|---|---|---|---|---|---|
| negative, n = 150 | ||||||
| BASIC | 88.7 % (133) | 11.3 % (17) | 0.0 % (0) | – | – | |
| NGRF | 84.7 % (127) | 15.3 % (23) | 0.0 % (0) | 5.3 % (8: 7 ↑ 1 ↓) | 0.0 % (0: 0 ↑ 0 ↓) | |
| PRS | 84.7 % (127) | 14.0 % (21) | 1.3 % (2) | 13.3 % (20: 13 ↑ 7 ↓) | 1.3 % (2: 2 ↑ 0 ↓) | |
| FULL | 78.7 % (118) | 20.0 % (30) | 1.3 % (2) | 16.7 % (25: 20 ↑ 5 ↓) | 1.3 % (2: 2 ↑ 0 ↓) | |
| non-informative, n = 184 | ||||||
| BASIC | 53.3 % (98) | 44.0 % (81) | 2.7 % (5) | – | – | |
| NGRF | 47.3 % (87) | 46.2 % (85) | 6.5 % (12) | 22.3 % (41: 26 ↑ 15 ↓) | 3.8 % (7: 7 ↑ 0 ↓) | |
| PRS | 50.0 % (92) | 37.5 % (69) | 12.5 % (23) | 26.1 % (48: 27 ↑ 21 ↓) | 12.0 % (22: 20 ↑ 2 ↓) | |
| FULL | 46.7 % (86) | 37.0 % (68) | 16.3 % (30) | 31.5 % (58: 35 ↑ 23 ↓) | 15.8 % (29: 27 ↑ 2 ↓)# | |
| positive (ATM, CHEK2), n = 9 | ||||||
| BASIC | 0.0 % (0) | 0.0 % (0) | 100 % (9) | – | – | |
| NGRF | 0.0 % (0) | 0.0 % (0) | 100 % (9) | 0.0 % (0: 0 ↑ 0 ↓) | 0.0 % (0: 0 ↑ 0 ↓) | |
| PRS | 11.1 % (1) | 11.1 % (1) | 77.8 % (7) | 11.1 % (1: 0 ↑ 1 ↓) | 22.2 % (2: 0 ↑ 2 ↓) | |
| FULL | 0.0 % (0) | 11.1 % (1) | 88.9 % (8) | 0.0 % (0: 0 ↑ 0 ↓) | 11.1 % (1: 0 ↑ 1 ↓) | |
| positive (BRCA1/2, PALB2), n = 82 | ||||||
| BASIC | 0.0 % (0) | 0.0 % (0) | 100 % (82) | – | – | |
| NGRF | 0.0 % (0) | 0.0 % (0) | 100 % (82) | 0.0 % (0: 0 ↑ 0 ↓) | 0.0 % (0: 0 ↑ 0 ↓) | |
| PRS | 0.0 % (0) | 0.0 % (0) | 100 % (82) | 0.0 % (0: 0 ↑ 0 ↓) | 0.0 % (0: 0 ↑ 0 ↓) | |
| FULL |
0.0 % (0) |
0.0 % (0) |
100 % (82) |
0.0 % (0: 0 ↑ 0 ↓) |
0.0 % (0: 0 ↑ 0 ↓) |
|
|
e10YR low (<5 %) |
e10YR moderate (≥5 % and <10 %) |
e10YR high (≥10 %) |
category changes (<5 % vs ≥ 5 %) |
category changes (<10 % vs ≥ 10 %) |
||
| negative, n = 147a | ||||||
| BASIC | 94.6 % (139) | 5.4 % (8) | 0.0 % (0) | – | – | |
| NGRF | 94.6 % (139) | 5.4 % (8) | 0.0 % (0) | 5.4 % (8: 4 ↑ 4 ↓) | 0.0 % (0: 0 ↑ 0 ↓) | |
| PRS | 92.5 % (136) | 6.8 % (10) | 0.7 % (1) | 4.8 % (7: 5 ↑ 2 ↓) | 0.7 % (1: 1 ↑ 0 ↓) | |
| FULL | overall | 90.5 % (133) | 8.2 % (12) | 1.4 % (2) | 9.5 % (14: 10 ↑ 4 ↓) | 1.4 % (2: 2 ↑ 0 ↓)## |
| <40 yrs (n = 81) | 97.5 % (79) | 2.5 % (2) | 0.0 % (0) | 2.5 % (2: 2 ↑ 0 ↓) | 0.0 % (0: 0 ↑ 0 ↓) | |
| 40–49 yrs (n = 35) | 82.9 % (29) | 14.3 % (5) | 2.8 % (1) | 14.3 % (5: 4 ↑ 1 ↓) | 2.9 % (1: 1 ↑ 0 ↓) | |
| ≥50 yrs (n = 31) | 80.6 % (25) | 16.1 % (5) | 3.2 % (1) | 22.6 % (7: 4 ↑ 3 ↓) | 3.2 % (1: 1 ↑ 0 ↓)## | |
| non-informative, n = 183b | ||||||
| BASIC | 63.4 % (116) | 36.1 % (66) | 0.5 % (1) | – | – | |
| NGRF | 59.6 % (109) | 39.9 % (73) | 0.5 % (1) | 11.5 % (21: 14 ↑ 7 ↓) | 0.0 % (0: 0 ↑ 0 ↓) | |
| PRS | 61.2 % (112) | 36.1 % (66) | 2.7 % (5) | 17.5 % (32: 18 ↑ 14 ↓) | 3.3 % (6: 5 ↑ 1 ↓) | |
| FULL | overall | 58.5 % (107) | 37.7 % (69) | 3.8 % (7) | 22.4 % (41: 25 ↑ 16 ↓) | 4.4 % (8: 7 ↑ 1 ↓) |
| <40 yrs (n = 68) | 77.9 % (53) | 22.1 % (15) | 0.0 % (0) | 19.1 % (13: 11 ↑ 2 ↓) | 0.0 % (0: 0 ↑ 0 ↓) | |
| 40–49 yrs (n = 77) | 48.1 % (37) | 46.8 % (36) | 5.2 % (4) | 24.7 % (19: 13 ↑ 6 ↓) | 6.5 % (5: 4 ↑ 1 ↓) | |
| ≥50 yrs (n = 38) | 44.7 % (17) | 47.4 % (18) | 7.9 % (3) | 23.7 % (9: 1 ↑ 8 ↓) | 7.9 % (3: 3 ↑ 0 ↓) | |
| positive (ATM, CHEK2), n = 9 | ||||||
| BASIC | 11.1 % (1) | 55.6 % (5) | 33.3 % (3) | – | – | |
| NGRF | 11.1 % (1) | 55.6 % (5) | 33.3 % (3) | 0.0 % (0: 0 ↑ 0 ↓) | 0.0 % (0: 0 ↑ 0 ↓) | |
| PRS | 22.2 % (2) | 44.4 % (4) | 33.3 % (3) | 11.1 % (1: 0 ↑ 1 ↓) | 22.2 % (2: 1 ↑ 1 ↓) | |
| FULL | overall | 11.1 % (1) | 55.6 % (5) | 33.3 % (3) | 0.0 % (0: 0 ↑ 0 ↓) | 22.2 % (2: 1 ↑ 1 ↓) |
| <40 yrs (n = 5) | 20.0 % (1) | 80.0 % (4) | 0.0 % (0) | 0.0 % (0: 0 ↑ 0 ↓) | 20.0 % (1: 1 ↑ 0 ↓) | |
| 40–49 yrs (n = 2) | 0.0 % (0) | 100.0 % (2) | 0.0 % (0) | 0.0 % (0: 0 ↑ 0 ↓) | 50.0 % (1: 0 ↑ 1 ↓) | |
| ≥50 yrs (n = 2) | 0.0 % (0) | 0.0 % (0) | 100.0 % (2) | 0.0 % (0: 0 ↑ 0 ↓) | 0.0 % (0: 0 ↑ 0 ↓) | |
| positive (BRCA1/2, PALB2), n = 82 | ||||||
| BASIC | 6.1 % (5) | 29.3 % (24) | 64.6 % (53) | – | – | |
| NGRF | 6.1 % (5) | 23.2 % (19) | 70.7 % (58) | 2.4 % (2: 1 ↑ 1 ↓) | 6.1 % (5: 5 ↑ 0 ↓) | |
| PRS | 15.9 % (13) | 19.5 % (16) | 64.6 % (53) | 9.8 % (8: 0 ↑ 8 ↓) | 14.6 % (12: 6 ↑ 6 ↓) | |
| FULL | overall | 13.4 % (11) | 20.7 % (17) | 65.9 % (54) | 9.8 % (8: 1 ↑ 7 ↓) | 13.4 % (11: 6 ↑ 5 ↓)### |
| <40 yrs (n = 50) | 22.0 % (11) | 34.0 % (17) | 44.0 % (22) | 16.0 % (8: 1 ↑ 7 ↓) | 22.0 % (11: 6 ↑ 5 ↓)### | |
| 40–49 yrs (n = 17) | 0.0 % (0) | 0.0 % (0) | 100.0 % (17) | 0.0 % (0: 0 ↑ 0 ↓) | 0.0 % (0: 0 ↑ 0 ↓) | |
| ≥50 yrs (n = 15) | 0.0 % (0) | 0.0 % (0) | 100.0 % (15) | 0.0 % (0: 0 ↑ 0 ↓) | 0.0 % (0: 0 ↑ 0 ↓) | |
Fig. 2.
(A–C) Individual-level estimated modified lifetime risks (eLTR) for breast cancer (BC) depending on CanRisk estimation model and genetic test result, using the BASIC model as baseline. (D–F) Individual-level estimated 10-year risks (e10YR) for BC, which consider the age at the time of consultation, depending on CanRisk estimation model and genetic test result. (A, D) BASIC vs NGRF (B, E) BASIC vs PRS (C, F) BASIC vs FULL. NGRF = non-genetic risk factors, PRS = polygenic risk score, FULL = comprehensive model including PRS306 and NGRFs.
Following the German guidelines, clinical recommendations on IBS are guided by e10YR that takes the age at the time of consultation into account. An e10YR <5 % is defined as low, e10YRs between 5 % and 10 % as moderate, and an e10YR ≥ 10 % as high. The mean e10YR was lowest in women who tested negative, followed by women who tested non-informative and women who carried PVs in moderate- or high-risk genes (Fig. 2 D–F).
On the individual level, 94·6 % (139/147) of the women who tested negative were at low e10YR, 5·4 % (8/147) were at moderate, and none were at high e10YR when using the BASIC model (Table 3). The FULL model changed clinical recommendations for 9·5 % (14/147) of women when applying the 5 % e10YR threshold. More pronounced effects were observed for women who tested non-informative. The FULL model changed clinical recommendations for 22·4 % (41/183) of the women using the 5 % e10YR threshold, compared to the BASIC model. In the 9 women with PVs in moderate-risk genes, no changes in risk category occurred when applying the 5 % e10YR-threshold. Of the women with PVs in high-risk genes, 6·10 % (5/82) were at low e10YRs, 29·3 % (24/82) were at moderate, and 64·6 % (53/82) were at high e10YRs when using the BASIC model. As expected, the 29 women with low/moderate e10YRs were statistically significantly younger (mean age 27·0 years, range 22·0 to 33·9 years) than the 52 women who were at high e10YRs (mean age 42·5 years, range 23·0 to 67·7 years, p < 10−13). When applying the FULL versus the BASIC model, risk category changed for 9·8 % (8/82) of women with PVs in high-risk genes. IBS for women with PVs in BRCA1/2 usually starts at 25 or 30 years of age, respectively, with country-specific differences [3]. Of the 78 BRCA1/2 PV carriers, 19 of the 28 women younger than 30 years (mean age 26·5 years at consultation, range 23–29 years) had an e10YR ≥ 5 % (FULL model) and IBS may start before 30 (n = 19) or 25 years of age (n = 3). In contrast, only one of the 50 BRCA1/2 PV carriers aged ≥30 years (BRCA2 PV carrier, aged 31 years) had an e10YR <5 % (FULL model) and IBS may be postponed.
4. Discussion
For cancer-free women who tested non-informative in our study, the incorporation of PRS306 and NGRF data into CanRisk-based risk prediction substantially affected clinical recommendations, regardless of country-specific guidelines. Clinical recommendations changed for 31·5 % (58/184) of these women when applying the 20 % eLTR threshold, 15·8 % (29/184) when applying the 30 % eLTR threshold, and 22·4 % (41/183) when applying the 5 % e10YR threshold. Lakeman et al. recently described that integrating PRS313 data in CanRisk-based risk prediction affects clinical recommendations for 26·8 % (635/2369) of women tested non-informative when applying the 20 % eLTR threshold, and 7·1 % (167/2369) when applying the 30 % eLTR threshold [19]. This is similar to the effects observed in our investigation using the PRS risk estimation model (20 % eLTR: 26·1 %, 48/184; 30 % eLTR: 12·0 %, 22/184, Table 3). The addition of NGRF without PRS data affected clinical recommendations especially for women tested non-informative (20 % eLTR, 22·3 %), indicating the clinical relevance of NGRFs (Table 3).
The addition of both PRS306 and NGRF data, however, lead to the highest number of risk category changes compared to the BASIC risk estimation model in our study (Fig. 2 A-C, Table 3). Similar effects, but less pronounced, were observed for women who tested negative. Of note, 21·3 % with a negative PV status had an eLTR ≥20 %, compared to 11·3 % using the BASIC model.
In the overall study sample, the 20 % eLTR and 5 % e10YR thresholds resulted in concordant risk categories for 84·8 % (357/421) when applying the FULL model.
For most women with discrepant risk categories, namely 90·6 % (58/64), the FULL model resulted in an eLTR ≥20 % but an e10YR <5 %. On average, these 58 women were statistically significantly younger than the 357 women with concordant results (29·7 versus 41·7 years, p < 10−15).
Women with PVs in BRCA1/2 were generally at high eLTR and incorporating PRS306 and NGRF did not alter individual risk category, irrespective of the applied eLTR threshold. Nonetheless, when considering the BASIC vs. FULL model, the strongest upward risk change was +21·8 % in a BRCA2 PV carrier (68·3 % to 90·1 % eLTR) and the strongest downward change was -19·2 % in a BRCA1 PV carrier (81·1 % to 61·9 %); this might provide additional arguments for decision-making regarding prophylactic bilateral mastectomy. No participant aged <30 years with a non-informative or negative PV status with an eLTR ≥20 % had an e10YR ≥5 % using the FULL model, giving no indication for recommendation of an earlier start of IBS (i.e., before 30 years of age). Conversely, postponing screening onset for non-informatively tested women aged ≥30 years (34·4 years, range 30–41 years) may be considered, as 21·7 % (20/92) had an e10YR <5 % while being identified at elevated BC risk through eLTR ≥20 %. For women who tested negative with an age at consultation of ≥30 years and an eLTR ≥20 %, 33·3 % (6/18, mean age at consultation 37·9 years, range 30–48 years) had an e10YR <5 %, and IBS may be postponed. Considering the eLTR along with e10YR using thresholds of 20 % and 5 %, respectively, identifies those women for whom the age of onset of IBS may deviate from the age specified by national guidelines.
The BOADICEA algorithm embedded in the CanRisk interface has been the subject of numerous validation studies considering different risk populations, model components and time scales, where the model has generally been shown to be well-calibrated with good performance in comparison to other risk models [5, [20], [21], [22], [23], [24], [25]].
Improved risk prediction and discrimination, e.g. by the additional consideration of BC risk factors such as PRS and NGRFs, is anticipated to lead to a more favorable ratio between risks and benefits of IBS [26]. However, the differing screening modalities and risk thresholds across European countries [3,27] lead to differing clinical recommendations for IBS for the same woman. A harmonization of European guidelines on indication for IBS for women at increased risk of developing BC is warranted.
4.1. Limitations of the study
The NGRFs considered in this investigation excluded mammographic density, which has a substantial effect on individual BC risk estimates using the BOADICEA model [1]. Social desirability for patient-reported information on alcohol consumption, height, and body weight may affect NGRF data [28,29]. The PRS306 is based on European-descent data, and we therefore excluded women of presumed non-European ancestry. To ensure health equity, the performance of the PRSs should be evaluated and clinically implemented for non-European ancestries. For women with PVs in moderate-risk genes ATM and CHEK2 (n = 9), and PALB2, defined as high-risk gene in our investigation (n = 4), numbers were too low for statistically meaningful conclusions.
5. Conclusion
This clinical investigation provides a rationale for considering PRS306 and NGRFs for individualized BC risk prediction in routine clinical care, and highlights the dependence of clinical recommendations on the factors included in BC risk prediction and IBS thresholds used. Differing risk threshold definitions and IBS modalities across European countries potentially result in different clinical recommendations for the same woman.
Funding
This work was supported by the European Union's Horizon 2020 research and innovation programme [grant agreement number 634935 (BRIDGES)]. The CC-HBOC (Germany) was supported by the German Cancer Aid (grant no 110837 and grant no 70114178, coordinator: Rita K. Schmutzler, Cologne) and the Federal Ministry of Education and Research (BMBF), Germany (grant no 01GY1901). Genetic analyses were supported by the Köln Fortune Program, Faculty of Medicine, University of Cologne, Germany. The funding sources had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; nor in the decision to submit the manuscript for publication.
Credit author statement
Julia Dick: Investigation, Resources, Writing – review & editing. Nienke van der Stoep: Investigation, Resources, Writing – review & editing. Christi J. van Asperen: Investigation, Resources, Writing – review & editing. Monika Maringa: Investigation, Resources, Writing – review & editing. Natalie Herold: Investigation, Resources, Writing – review & editing. Britta Blümcke: Investigation, Resources, Writing – review & editing. Robert Remy: Investigation, Resources, Software, Writing – review & editing. Dominique Stoppa-Lyonnet: Conceptualization, Funding acquisition, Investigation, Resources, Supervision, Writing – review & editing. Rita K. Schmutzler: Conceptualization, Funding acquisition, Investigation, Resources, Supervision, Writing – review & editing. Corinna Ernst: Data curation, Formal analysis, Investigation, Methodology, Resources, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. Eric Hahnen: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing. Amélie Anota: Data curation, Formal analysis, Investigation, Methodology, Resources, Validation, Writing – original draft, Writing – review & editing. Inge M. M. Lakeman: Investigation, Resources, Writing – review & editing. Lisa Golmard: Investigation, Resources, Writing – review & editing. Nadine Kütting: Investigation, Resources, Writing – review & editing. Chrystelle Colas: Investigation, Resources, Writing – review & editing. Barbara Wappenschmidt: Investigation, Resources, Writing – review & editing. Kerstin Rhiem: Investigation, Resources, Writing – review & editing. Peter Devilee: Funding acquisition, Investigation, Resources, Supervision, Writing – review & editing. Anja Tüchler: Data curation, Formal analysis, Investigation, Methodology, Resources, Validation, Writing – original draft, Writing – review & editing. Antoine De Pauw: Data curation, Formal analysis, Investigation, Methodology, Resources, Validation, Writing – original draft, Writing – review & editing. Anke Westerhoff: Investigation, Resources, Writing – review & editing. Denise J. Stommel-Jenner: Investigation, Resources, Writing – review & editing. Eléonore Frouin: Investigation, Resources, Writing – review & editing. Lisa Richters: Investigation, Resources, Writing – review & editing.
Declaration of competing interest
The authors declare no conflicts of interest.
Acknowledgements
We thank all BRIDGES study participants.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.breast.2023.103615.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
References
- 1.Lee A., Mavaddat N., Wilcox A.N., et al. BOADICEA: a comprehensive breast cancer risk prediction model incorporating genetic and nongenetic risk factors. Genet Med. 2019;21:1708–1718. doi: 10.1038/s41436-018-0406-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carver T., Hartley S., Lee A., et al. CanRisk tool-A web interface for the prediction of breast and ovarian cancer risk and the likelihood of carrying genetic pathogenic variants. Cancer Epidemiol Biomarkers Prev. 2021;30:469–473. doi: 10.1158/1055-9965.EPI-20-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marmolejo D.H., Wong M.Y.Z., Bajalica-Lagercrantz S., et al. Overview of hereditary breast and ovarian cancer (HBOC) guidelines across Europe. Eur J Med Genet. 2021;64 doi: 10.1016/j.ejmg.2021.104350. [DOI] [PubMed] [Google Scholar]
- 4.Wolfson M., Gribble S., Pashayan N., et al. Potential of polygenic risk scores for improving population estimates of women's breast cancer genetic risks. Genet Med. 2021;23:2114–2121. doi: 10.1038/s41436-021-01258-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang X., Eriksson M., Czene K., et al. Prospective validation of the BOADICEA multifactorial breast cancer risk prediction model in a large prospective cohort study. J Med Genet. 2022;59:1196–1205. doi: 10.1136/jmg-2022-108806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mavaddat N., Ficorella L., Carver T., et al. Incorporating alternative polygenic risk scores into the BOADICEA breast cancer risk prediction model. Cancer Epidemiol Biomarkers Prev. 2023;32:422–427. doi: 10.1158/1055-9965.EPI-22-0756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breast Cancer Association C., Dorling L., Carvalho S., et al. Breast cancer risk genes - association analysis in more than 113,000 women. N Engl J Med. 2021;384:428–439. doi: 10.1056/NEJMoa1913948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kast K., Rhiem K., Wappenschmidt B., et al. Prevalence of BRCA1/2 germline mutations in 21 401 families with breast and ovarian cancer. J Med Genet. 2016;53:465–471. doi: 10.1136/jmedgenet-2015-103672. [DOI] [PubMed] [Google Scholar]
- 9.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:1303.3997. 2013 [Google Scholar]
- 10.Van der Auwera G.A., Carneiro M.O., Hartl C., et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11. doi: 10.1002/0471250953.bi1110s43. 10.11-11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garrison E., Marth G. Haplotype-based variant detection from short-read sequencing. arXiv preprint arXiv:1207.3907. 2012 [Google Scholar]
- 12.Sherry S.T., Ward M.-H., Kholodov M., et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borde J., Ernst C., Wappenschmidt B., et al. Performance of breast cancer polygenic risk scores in 760 female CHEK2 germline mutation carriers. J Natl Cancer Inst. 2020 doi: 10.1093/jnci/djaa203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Genomes Project C., Auton A., Brooks L.D., et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wappenschmidt B., Hauke J., Faust U., et al. Criteria of the German Consortium for hereditary breast and ovarian cancer for the classification of germline sequence variants in risk genes for hereditary breast and ovarian cancer. Geburtshilfe Frauenheilkd. 2020;80:410–429. doi: 10.1055/a-1110-0909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richards S., Aziz N., Bale S., et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of medical genetics and Genomics and the association for molecular pathology. Genet Med. 2015;17:405–423. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Plon S.E., Eccles D.M., Easton D., et al. Sequence variant classification and reporting: recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum Mutat. 2008;29:1282–1291. doi: 10.1002/humu.20880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lepkes L., Kayali M., Blümcke B., et al. Performance of in silico prediction tools for the detection of germline copy number variations in cancer predisposition genes in 4208 female index patients with familial breast and ovarian cancer. Cancers. 2021;13:118. doi: 10.3390/cancers13010118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lakeman I.M.M., Rodríguez-Girondo M.D.M., Lee A., et al. Clinical applicability of the Polygenic Risk Score for breast cancer risk prediction in familial cases. J Med Genet. 2022 doi: 10.1136/jmg-2022-108502. [DOI] [PubMed] [Google Scholar]
- 20.Pal Choudhury P., Brook M.N., Hurson A.N., et al. Comparative validation of the BOADICEA and Tyrer-Cuzick breast cancer risk models incorporating classical risk factors and polygenic risk in a population-based prospective cohort of women of European ancestry. Breast Cancer Res. 2021;23:22. doi: 10.1186/s13058-021-01399-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li S.X., Milne R.L., Nguyen-Dumont T., et al. Prospective evaluation over 15 Years of six breast cancer risk models. Cancers. 2021;13 doi: 10.3390/cancers13205194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Danladi C., Serakinci N. Performance of risk prediction models in breast cancer screening among women in Cyprus. East Mediterr Health J. 2022;28:888–895. doi: 10.26719/emhj.22.089. [DOI] [PubMed] [Google Scholar]
- 23.Li S.X., Milne R.L., Nguyen-Dumont T., et al. Prospective evaluation of the addition of polygenic risk scores to breast cancer risk models. JNCI Cancer Spectr. 2021:5. doi: 10.1093/jncics/pkab021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lakeman I.M.M., Rodríguez-Girondo M., Lee A., et al. Validation of the BOADICEA model and a 313-variant polygenic risk score for breast cancer risk prediction in a Dutch prospective cohort. Genet Med. 2020;22:1803–1811. doi: 10.1038/s41436-020-0884-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacInnis R.J., Knight J.A., Chung W.K., et al. Comparing 5-year and lifetime risks of breast cancer using the prospective family study cohort. JNCI: Journal of the National Cancer Institute. 2020;113:785–791. doi: 10.1093/jnci/djaa178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brooks J.D., Nabi H.H., Andrulis I.L., et al. Personalized risk assessment for prevention and early detection of breast cancer: integration and implementation (PERSPECTIVE I&I) J Personalized Med. 2021:11. doi: 10.3390/jpm11060511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pashayan N., Antoniou A.C., Lee A., et al. Should age-dependent absolute risk thresholds Be used for risk stratification in risk-stratified breast cancer screening? J Personalized Med. 2021:11. doi: 10.3390/jpm11090916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burke M.A., Carman K.G. You can be too thin (but not too tall): social desirability bias in self-reports of weight and height. Econ Hum Biol. 2017;27:198–222. doi: 10.1016/j.ehb.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 29.Davis C.G., Thake J., Vilhena N. Social desirability biases in self-reported alcohol consumption and harms. Addict Behav. 2010;35:302–311. doi: 10.1016/j.addbeh.2009.11.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.